Abstract
We report the discovery of HD5-CD, an unprecedented C2-symmetric β-barrel-like covalent dimer of the cysteine-rich host-defense peptide human defensin 5 (HD5). Dimerization results from intermonomer disulfide exchange between the canonical α-defensin CysII–CysIV (Cys5–Cys20) bonds located at the hydrophobic interface. This disulfide-locked dimeric assembly provides a new element of structural diversity for cysteine-rich peptides as well as increased protease resistance, broad-spectrum antimicrobial activity, and enhanced potency against the opportunistic human pathogen Acinetobacter baumannii.
Disulfide bonds provide structures and enable the biological functions of various naturally occurring and synthetic peptides.1 Defensins are prominent examples of disulfide-linked peptides found in Nature.2 These cysteine-rich host-defense peptides (2–5 kDa) are ribosomally synthesized by lower and higher eukaryotes, and participate in the innate immune response.2 In humans, oxidized defensins exhibit three regiospecific disulfide bonds with the patterns of CysI–CysVI/CysII–CysIV/CysIII–CysV (α-defensins) or CysI–CysV/CysII–CysIV/CysIII–CysVI (β-defensins) that maintain a three-stranded β-sheet fold. Defensins self-associate to afford noncovalent oligomers.3 Despite many structure/activity relationship studies,2b,3 our understanding of what defensin structures (i.e., oxidized or reduced, various oligomeric states) are physiologically relevant in various biological microenvironments, and precisely how these structures contribute to innate immunity or other biological phenomena, remains unclear. To address such questions in the context of the human gut, our laboratory is investigating human α-defensin 5 (HD5), which is abundantly produced by small intestinal Paneth cells and is an important contributor to mucosal immunity.2b,4 This peptide exhibits broad-spectrum antimicrobial activity in vitro and may help control the composition of the resident gut microbiota.5,6 Here, we describe an unappreciated facet of the molecular gymnastics displayed by defensins and report that HD5 has the propensity to assemble into a novel disulfide-locked C2-symmetric covalent dimer.
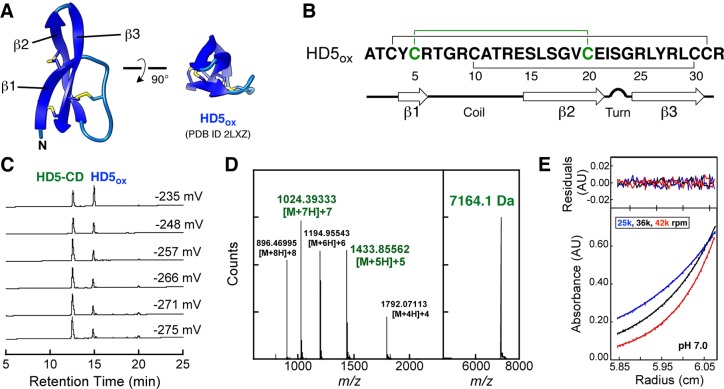
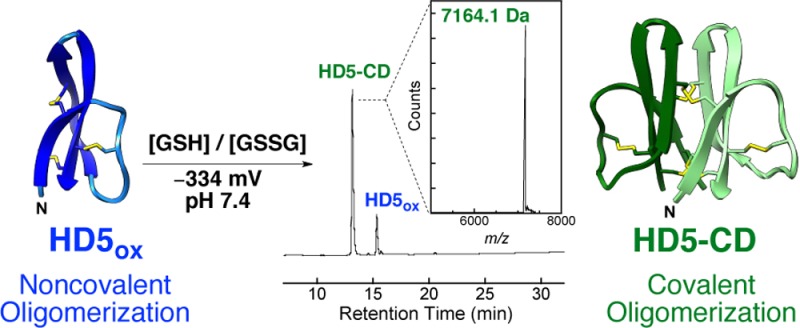
During an exploratory set of oxidative folding assays, 32-residue HD5 (Figure 1A,B) in its reduced (HD5red) or oxidized (HD5ox) form was incubated at pH 7.0 (75 mM HEPES) under anaerobic conditions in buffers of varying redox potentials (−275 to −235 mV) defined by the GSH/GSSG redox couple. The presence of a new peak with a HPLC retention time of 13.1 min (HD5red, 20.0 min; HD5ox, 15.0 min) was observed in the chromatogram of every sample (Figure 1C). This species constituted ca. 45% to 70% of the total HPLC peak area and was most abundantly formed from HD5ox at −275 mV. LC-MS of the new peak (denaturing conditions) provided a deconvoluted m/z value of 7164.1, which is twice the m/z value of HD5ox (m/z calc 3582.2) (Figure 1D), and thiol quantification supported no (0.55 ± 0.39) free Cys residues. Moreover, two ions corresponding to [M + 5H]+5 and [M + 7H]+7 identified in the mass spectrum of the new species were not observed for HD5ox (Supporting Information).
Figure 1.
(A) Previously determined NMR structure of the HD5ox monomer (PDB 2LXZ)3e with disulfide bonds shown. (B) Primary sequence of HD5. The solid lines indicate the regiospecific disulfide bonds of HD5ox. The Cys5–Cys20 disulfide bond is highlighted in green. The secondary structure elements are taken from the HD5ox NMR structure (PDB 2LXZ).3e (C) HPLC traces showing HD5-CD formation from 25 μM HD5ox in redox buffers containing GSH/GSSG (75 mM HEPES, pH 7.0). The minor peak at 20.0 min is HD5red. (D) ESI-TOF ionization pattern and deconvolution for HD5-CD. (E) Example sedimentation equilibrium data, fits, and residuals for 62 μM HD5-CD (Supporting Information).
The paucity of other new HPLC peaks observed with peptide folding, which would be suggestive of other non-native disulfide-linked species or glutathione adducts, was striking. In total, this work suggested that anaerobic buffers containing the GSH/GSSG redox couple enable an unexpected HD5 congener to predominate. We reasoned that the new species is a disulfide-linked covalent dimer (CD) of HD5 monomers with six disulfide bonds. We named this peptide HD5-CD.
Under the anaerobic redox buffer conditions, HD5-CD formed irrespective of initial peptide concentration (5–100 μM) and in the presence of millimolar concentrations of NaCl (100 mM), MgCl2 (10 mM), or CaCl2 (10 mM). Using the same GSH/GSSG-containing buffer and aerobic conditions, HD5-CD also formed, albeit to a lesser degree. In total, the conditions and redox potentials that afford HD5-CD suggest that this species has the potential to exist in vivo. HD5-CD formation was attenuated when GuHCl (≥250 mM) was added to the buffer, which increased the amount of HD5red in the mixture. A standard folding protocol performed aerobically that affords HD5ox in high yield employs a 10:1 GSH/GSSG ratio as well as ∼2 M GuHCl.7 Evaluation of HPLC traces from prior HD5ox preparations revealed that a peak with the HD5-CD retention time is often observed as a minor (<10%) product. Moreover, when studying oxidative folding of HD5red in the absence of GuHCl, a minor side product with a retention time of ∼13.0 min was observed,8 which we now assign to HD5-CD. To further characterize HD5-CD, we established preparative-scale folding conditions that afford milligram quantities of the CD in high purity (Supporting Information).
First, we conducted analytical ultracentrifugation (AUC) to ascertain the molecular weight of HD5-CD and probe its self-association properties. Sedimentation velocity experiments revealed that HD5-CD sedimented as a single species with a sedimentation coefficient of ca. 1.2 S and a diffusion coefficient of 13.6 F over a range of peptide concentrations (40–120 μM) and pH (2.0–8.0) (Supporting Information). Hydrodynamic modeling of HD5ox afforded a S20,w value of 0.69 S for the monomer determined by solution NMR (PDB 2LXZ)3e and 1.16 S for the noncovalent dimer determined by X-ray crystallography (PDB 1ZMP).9 The molecular weight of HD5-CD was determined to be 6985 Da with a 95% confidence interval of 6598–7330 Da from global fitting of sedimentation equilibrium data. In total, AUC confirmed the HD5-CD molecular weight and demonstrated that it does not form higher-order oligomers in aqueous solution (Figure 1E, Supporting Information), which contrasts the behavior of canonical α-defensins.3
Next, we prepared 15N- and 15N,13C-labeled HD5-CD for NMR spectroscopic investigations.3e The 2D 1H-15N HSQC spectrum of 15N-HD5-CD contained only 24 peaks, which contrasted the 62 amide protons expected for HD5-CD. Greater than 90% of these peaks were visually assigned upon comparison with the 1H-15N HSQC spectrum of HD5ox.3e This analysis suggested that HD5-CD adopts a symmetric structure where each protomer is similar in fold to the HD5ox monomer. 15N-R1, 15N-R2, and {1H}-15N heteronuclear NOE experiments confirmed that HD5-CD is a rigid, dimeric molecule under the NMR sample conditions (Supporting Information).
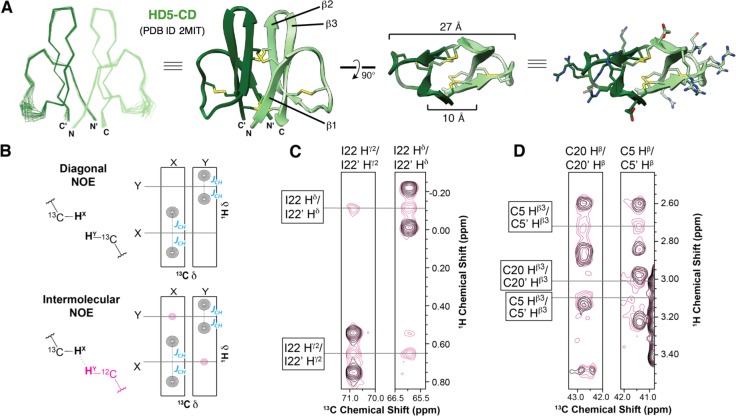
To solve the HD5-CD solution structure (Figure 2A), the standard suite of 3D triple-resonance experiments were performed on uniformly labeled 15N,13C-HD5-CD using a nonuniform sampling protocol.10 The peptide was assigned to 93.5% completeness with the exception of the amide resonances of residues 11–16, the Hα of Thr,12 and the Hα and Hβ of Glu14 (Supporting Information).11
Figure 2.
NMR solution structure of HD5-CD. (A) Overlay of the 20 lowest energy structures, ribbon diagrams, and structures showing disulfides and the hydrophilic residues. (B–D) 2D 13C-edited/15N,13C-filtered HSQC-NOESY indicating Ile22 packing and the intermolecular disulfide bonds (see text and Supporting Information).
To unambiguously identify NOEs at the dimer interface, we prepared a mixed-labeled sample by oxidative folding of a 1:1 molar ratio of 15N,13C-HD5red and unlabeled HD5red. This procedure afforded a statistical mixture of uniformly labeled, mix-labeled, and unlabeled HD5-CD, which was employed in a 2D 13C-edited/15N,13C-filtered HSQC-NOESY (Figure 2B–D). In this experiment, diagonal NOE peaks, or NOEs originating from protons directly bonded to 13C-resonances, are coupled yielding two peaks offset from the chemical shift by the respective JCH constant (Figure 2B). In contrast, intermolecular NOE crosspeaks, in which the proton is bonded to a 12C-resonance, exhibit only a single crosspeak at the chemical shift. Numerous intermolecular NOEs confirmed that HD5-CD possesses 2-fold rotational symmetry with Ile22 and Leu29 each packing against its symmetry mate in the other protomer, a feature that differs from the crystal structures of dimeric HD5ox (Supporting Information).3b,9
The HD5-CD solution structure reveals an unprecedented disulfide-locked C2-symmetric β-sheet cylinder (Figure 2A). Each protomer within HD5-CD exhibits the canonical α-defensin fold composed of three antiparallel β-strands with a type 1 β-hairpin connecting β2 and β3 and a flexible loop between β1 and β2. The final collection of 20 lowest energy structures (Figure 2A) was calculated with explicit water refinement to obtain a heavy atom backbone RMSD of 0.46 Å. Each HD5-CD protomer aligns to the NMR solution structure of HD5ox (PDB 2LXZ)3e with a Cα RMSD of 1.36 Å. The HD5-CD disulfide topology is like that of α-defensins except that the Cys5–Cys20 (CysII–CysIV, Figure 1B) bond is intermolecular. The intramolecular Arg6–Glu14 salt bridge of HD5ox is not observed in the HD5-CD solution structure. The NOE data indicate that the guanidino group of Arg6 is pointed toward, rather than away from, the β3 strand. Nevertheless, residues 11–16 appear to be in intermediate exchange on the NMR time scale (Supporting Information) and possess few inter-residue NOE restraints. Thus, it is possible that Glu14 can adopt a position consistent with formation of the canonical α-defensin salt bridge.
HD5-CD exhibits a hydrophobic interior and a hydrophilic exterior. The intermolecular disulfide bonds constrain the majority of hydrophobic residues within a 10-Å diameter β-barrel-like core. Polar moieties, including all 12 Arg residues, decorate the surface (Figure 2A). The proximity of the interprotomer β1 and β2 strands in HD5-CD provides a cylindrical array that is found to be unique among β-sheet structural families and most closely matches β-sandwich folds.12
Canonical defensin scaffolds confer protease resistance and antimicrobial activity. Remarkably, HD5-CD exhibits greater stability than HD5ox (Supporting Information). HD5-CD is significantly more resistant to proteolysis, and no hydrolysis of HD5-CD was observed under the harsh double-digest conditions required for HD5ox degradation (≥10 μg/mL trypsin and chymotrypsin, 37 °C).13 We also found that HD5-CD is more difficult to chemically reduce than HD5ox.
The distribution of Arg residues gives HD5-CD an extensive cationic surface (Figure 2A) and we hypothesized that this feature would afford bactericidal activity.14 We compared the antimicrobial activity (AMA) of HD5-CD and HD5ox against 10 strains. Both peptides exhibit broad-spectrum AMA;5 however, some strains exhibit variable sensitivities to these peptides (Figure 3). The enhanced killing (ca. 104-fold reduction in cfu) of the opportunistic human pathogen Acinetobacter baumannii by HD5-CD relative to HD5ox is noteworthy. Multidrug resistant A. baumannii is a serious public health threat and cause of nosocomial infections with high mortality rates,15 and new therapeutic strategies to combat infections caused by this pathogen are needed.
Figure 3.
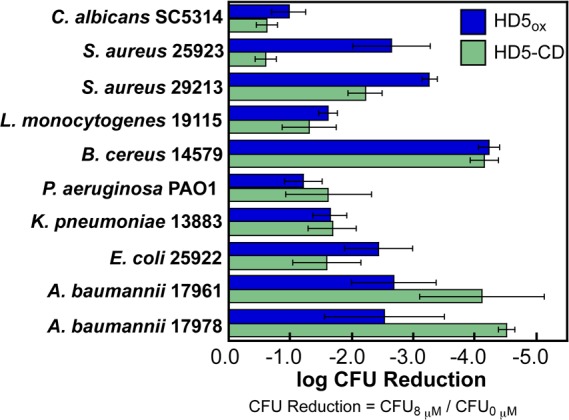
Antimicrobial activity of HD5-CD and HD5ox (mean ± SDM, n = 3).
In closing, HD5-CD provides, to the best of our knowledge, the first example of a disulfide-locked C2-symmetric defensin scaffold and expands the structural diversity of this peptide family. It will be interesting to ascertain whether other defensins form such covalent oligomers. The physiological relevance and practical utility of HD5-CD, as well as the design and characterization of related peptide scaffolds, also merit further investigation.
Acknowledgments
We thank the NIH (DP2OD007045 to E.M.N., PO1 GM047467 to G.W., and F32GM103005 to J.J.Z.) for financial support; Dr. C. Turner and Dr. S. Robson for assistance with the NMR experiments; I.-L. Chiang for technical assistance; Dr. P. Kaczmarek for preliminary synthetic work on HD5; Prof. S. Lindquist and Prof. K. Ribbeck for providing strains. The MIT Francis Bitter Magnet Laboratory is supported by the NIH (EB-002026). The MIT Biophysical Instrumentation Facility is supported by Grant NSF-007031.
Supporting Information Available
Experimental methods and additional data. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Góngora-Benítez M.; Tulla-Puche J.; Albericio F. Chem. Rev. 2014, 114, 901–926. [DOI] [PubMed] [Google Scholar]
- a Wilmes M.; Sahl H.-G. Int. J. Med. Microbiol. 2014, 304, 93–99. [DOI] [PubMed] [Google Scholar]; b Lehrer R. I.; Lu W. Immunol. Rev. 2012, 245, 84–112and references therein. [DOI] [PubMed] [Google Scholar]; c Ganz T. Nat. Rev. Immunol. 2003, 3, 710–720. [DOI] [PubMed] [Google Scholar]; d Selsted M. E.; Ouellette A. J. Nat. Immunol. 2005, 6, 551–557. [DOI] [PubMed] [Google Scholar]
- a Hill C. P.; Yee J.; Selsted M. E.; Eisenberg D. Science 1991, 251, 1481–1485. [DOI] [PubMed] [Google Scholar]; b Rajabi M.; Ericksen B.; Wu X.; de Leeuw E.; Zhao L.; Pazgier M.; Lu W. J. Biol. Chem. 2012, 287, 21615–21627. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Zhao L.; Ericksen B.; Wu X.; Zhang C.; Yuan W.; Li X.; Pazgier M.; Lu W. J. Biol. Chem. 2012, 287, 18900–18912. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Pazgier M.; Wei G.; Ericksen B.; Jung G.; Wu Z.; de Leeuw E.; Yuan W.; Szmacinski H.; Lu W.-Y.; Lubkowski J.; Lehrer R. I.; Lu W. J. Biol. Chem. 2012, 287, 8944–8953. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Wommack A. J.; Robson S. A.; Wanniarachchi Y. A.; Wan A.; Turner C. J.; Wagner G.; Nolan E. M. Biochemistry 2012, 51, 9624–9637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. C.; Bevins C. L. Annu. Rev. Physiol. 2013, 75, 289–311. [DOI] [PubMed] [Google Scholar]
- Porter E. M.; van Dam E.; Valore E. V.; Ganz T. Infect. Immun. 1997, 65, 2396–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman N. H.; Hung K.; Haribhai D.; Chu H.; Karlsson-Sjöberg J.; Amir E.; Teggatz P.; Barman M.; Hayward M.; Eastwood D.; Stoel M.; Zhou Y.; Sodergren E.; Weinstock G. M.; Bevins C. L.; Williams C. B.; Bos N. A. Nat. Immunol. 2010, 11, 76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Wu Z.; Ericksen B.; Tucker K.; Lubkowski J.; Lu W. J. Peptide Res. 2004, 64, 118–125. [DOI] [PubMed] [Google Scholar]; b Wanniarachchi Y. A.; Kaczmarek P.; Wan A.; Nolan E. M. Biochemistry 2011, 50, 8005–8017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Cougnon F. B. L.; Wanniarachchi Y. A.; Hayden J. A.; Nolan E. M. ACS Chem. Biol. 2013, 8, 1907–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyk A.; Wu Z.; Tucker K.; Yang D.; Lu W.; Lubkowski J. Protein Sci. 2006, 15, 2749–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyberts S. G.; Milbradt A. G.; Wagner A. B.; Arthanari H.; Wagner G. J. Biomol. NMR 2012, 52, 315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- These residues are located in the flexible loop and are in intermediate exchange on the NMR time-scale.
- Krissinel E.; Henrick K. Acta Crystallogr. 2004, D60, 2256–2268. [DOI] [PubMed] [Google Scholar]
- Kubo S.; Tanimura K.; Nishio H.; Chino N.; Teshima T.; Kimura T.; Nishiuchi Y. Int. J. Pept. Res. Ther. 2008, 14, 341–349. [Google Scholar]
- a Peschel A.; Sahl H.-G. Nat. Rev. Microbiol. 2006, 4, 529–536. [DOI] [PubMed] [Google Scholar]; b Zasloff M. Nature 2002, 415, 389–395. [DOI] [PubMed] [Google Scholar]
- Antunes L. C. S.; Visca P.; Towner K. J. Pathog. Dis. 2014, 71, 292–301. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.