Abstract

The capture of biotin by streptavidin is an inspiration for supramolecular chemistry and a central tool for biological chemistry and nanotechnology, because of the rapid and exceptionally stable interaction. However, there is no robust orthogonal interaction to this hub, limiting the size and complexity of molecular assemblies that can be created. Here we combined traptavidin (a streptavidin variant maximizing biotin binding strength) with an orthogonal irreversible interaction. SpyTag is a peptide engineered to form a spontaneous isopeptide bond to its protein partner SpyCatcher. SpyTag or SpyCatcher was successfully fused to the C-terminus of Dead streptavidin subunits. We were able to generate chimeric tetramers with n (0 ≤ n ≤ 4) biotin binding sites and 4-n SpyTag or SpyCatcher binding sites. Chimeric SpyAvidin tetramers bound precise numbers of ligands fused to biotin or SpyTag/SpyCatcher. Mixing chimeric tetramers enabled assembly of SpyAvidin octamers (8 subunits) or eicosamers (20 subunits). We validated assemblies using electrophoresis and native mass spectrometry. Eicosameric SpyAvidin was used to cluster trimeric major histocompatibility complex (MHC) class I:β2-microglobulin:peptide complexes, generating an assembly with up to 56 components. MHC eicosamers surpassed the conventional MHC tetramers in acting as a powerful stimulus to T cell signaling. Combining ultrastable noncovalent with irreversible covalent interaction, SpyAvidins enable a simple route to create robust nanoarchitectures.
Introduction
To advance bottom-up nanoassembly, it is important to develop building blocks for modular and robust construction, able to withstand real-world challenges.1 The rapid and exceptionally tight binding of biotin conjugates to avidin or streptavidin provides a ubiquitous tool for nanoassembly.2 As well as (strept)avidin’s ability to sensitively detect and immobilize biotinylated ligands (which have included drugs, nonbiological building blocks, and all classes of biomolecule), (strept)avidin clusters ligands into tetramers.2 This tetravalent assembly gives an avidity enhancement which is valuable in diverse contexts, notably for monitoring the immune response to cancer, autoimmune disease, and infection.3−5
The possibilities for nanoassembly would be greatly extended by a second robust interaction to (strept)avidin. There are a range of other ligands for (strept)avidin including biotin analogs (e.g., iminobiotin, desthiobiotin), small molecule ligands dissimilar to biotin (notably 4′-hydroxyazobenzene-2-carboxylic acid, HABA), peptides, and nucleic acid aptamers; however, their binding strengths are all far from the femtomolar stability of biotin binding and these ligands are generally released by competition with biotin.2,6−11 A unique cysteine can be introduced into streptavidin for chemical labeling,12,13 but it is limiting to have to work under precise oxidation states for enabling reaction, while new cysteines often impair folding of potential partner proteins.
To address this challenge we made use of spontaneous amide bond formation, creating a streptavidin tetramer able to form two orthogonal ultrastable interactions. SpyTag is a peptide we developed which forms a spontaneous isopeptide bond to its protein partner SpyCatcher simply upon mixing.14 Both SpyTag and SpyCatcher components are genetically encodable, can be positioned at various locations in the protein chain, are reactive under a wide range of conditions, and do not possess cysteines.14,15 Here we explore how SpyTag and SpyCatcher can be linked to streptavidin components, how these chimeric assemblies enable assembly of complex protein architectures, and how such nanoassemblies can powerfully stimulate cellular signaling.
Experimental Section
Plasmid Constructs
pET21-core traptavidin (Tr),16 pET21-core streptavidin (SA),17 pET21-Dead (D),17 and pET21-Dead-d-loop (Dd)18 have been described. PCR was performed with KOD Hot Start polymerase (EMD Millipore). For the addition of a C-terminal hexaglutamate tag to core traptavidin to give pET21-Tre (GenBank KJ906519, Addgene ID 59549), 5′-ATACATATGGCTGAAGCTGGTATCACCGGCACCTGG and 5′-CGCAAGCTTTTATTACTCTTCCTCTTCCTCTTCGGAAGCAGCGGACGGTTTAACTTTGG were used for PCR, digested with NdeI and HindIII, and subcloned into pET21. pET21-DTag (GenBank KJ906520, Addgene ID 59548) was generated by insertion of SpyTag (lacking the terminal Lys)15 at the C-terminus via a Gly/Ser spacer into pET21-Dead: site-directed ligase-independent mutagenesis (SLIM)19 was performed with 5′-GTGATGGTGGATGCCTACAAACCTACGTAATAAAAGCTTGCGGCCGCACTCG, 5′-TAATAAAAGCTTGCGGCCGCACTCG, 5′-GATGTGGGCGCCGCCTGATCCTGATCCGGAAGCAGCGGACGGTTTAACTTTG, and 5′-GGAAGCAGCGGACGGTTTAACTTTG. To make pET21-DCatch (GenBank KJ906521, Addgene ID 59547), for the initial insertion of SpyCatcher at the C-terminus with a Gly/Ser spacer on pET21-Dead d-loop, overlap extension PCR was performed. The forward primer 5′-ATCTCATATGGCTGAAGCTGGTATCACC and the reverse primer 5′-CCTGATCCTGAAGCAGCGGACGGTTTAAC were used to amplify the Dd portion, and the forward primer 5′-CCGCTGCTTCCGGATCAGGATCAGGAGATTACGACATCCCAACGACC and the reverse primer 5′-TACTAAGCTTTTAAATATGAGCGTCACCTTTAGTTG were used to amplify the SpyCatcher portion from pDEST14-SpyCatcher.14 The resulting two PCR products were mixed and reamplified using the Dd portion forward primer and the SpyCatcher portion reverse primer, digested with NdeI and HindIII, and subcloned into pET21. To shorten the N-terminus of the SpyCatcher region,15 to create pET21-DCatch, inverse PCR was performed with 5′-GATAGTGCTACCCATATTAAATTCTC and 5′-TCCTGATCCTGATCCTGAAGCAG.
pET21-SAe and pET21-AP-AffiIGF1R with an acceptor peptide (AP tag)20 for BirA-mediated biotinylation have been described previously.18 pET28-KTag-AffiHER2-SpyTag (here termed AffiHER2-SpyTag) has been described.21 All constructs were verified by Sanger sequencing (Source Bioscience).
Protein Expression and Purification
SA, SAe, Tr, Tre, DTag, and DCatch were expressed in E. coli BL21 [DE3] RIPL (Stratagene) and refolded from inclusion bodies by dilution into PBS in a similar method to that previously described.22 After IPTG induction, cells were harvested by centrifugation at 4000 g. The pellet was resuspended in 10 mL of PBS and stored at −80 °C. To isolate the inclusion bodies, 10 mL of PBS (10.1 mM Na2HPO4.2H2O, 2.7 mM KH2PO4, 2.7 mM KCl, 137 mM NaCl pH 7.4) containing EDTA-free mixed protease inhibitors (Roche), 1 mM phenylmethylsulfonyl fluoride (PMSF), 1% Triton X-100, 10 mM EDTA, and 0.1 mg/mL hen egg lysozyme (Sigma) were added to the cell suspension. Once thawed, this mixture was kept on ice for 1 h and then sonicated on ice using 4× 30 s pulses, with 1 min of rest between each pulse. Lysed cells were then centrifuged at 10 000g for 30 min at 4 °C, and the supernatant was discarded. The pellet was thoroughly resuspended in PBS containing 1% Triton X-100 and 10 mM EDTA and again centrifuged at 10 000g for 30 min at 4 °C. Inclusion bodies were then washed in PBS containing 10 mM EDTA and again centrifuged at 10 000g for 30 min at 4 °C. The washed inclusion body pellet was then dissolved in 5 mL of 6 M guanidinium hydrochloride pH 1.0 on ice, and any insoluble matter was removed by centrifugation at 10 000g for 30 min at 4 °C. The A280 of the dissolved inclusion bodies was then measured (typically 30–40), and an equivalent number of absorbance units of the appropriate variant was mixed together; e.g., 5 mL of Tre dissolved inclusion bodies with an A280 of 30 were mixed with 3.75 mL of DTag dissolved inclusion bodies with an A280 of 40. Homotetramers SA4, SAe4, Tre4, and DTag4 were generated by refolding without mixing with any other streptavidin variant.
For refolding, the inclusion bodies were diluted dropwise (by gravity through a 10 μL pipet tip secured with Parafilm to a 20 mL syringe barrel) into 200 mL of stirred precooled PBS plus 10 mM EDTA at 4 °C and left stirring for 16 h. To the refolded inclusion bodies we then added 60 g of (NH4)2SO4 (99%, Acros Organics), and the mixture was stirred for 1 h at 4 °C. This solution was then crudely filtered through several paper towels, and a further 60 g (NH4)2SO4 were added to the clarified solution. After a further 1 h of stirring at 4 °C, the protein was collected by centrifugation at 15 000g for 30 min. The pelleted protein was then dissolved in 50 mL of 50 mM boric acid with 300 mM NaCl (pH adjusted to 11.0 with 4 M NaOH) and again centrifuged at 10 000g for 30 min at 4 °C. The clarified mixture of either Tre/DTag or Tr/DCatch was first purified on a 5 mL iminobiotin-sepharose affinity column (Affiland, S.A.) using 50 mM sodium borate, 300 mM NaCl pH 11.0 as the binding and wash buffer and 20 mM KH2PO4 pH 2.2 as the elution buffer.
The eluate was then exchanged into 20 mM Tris·HCl pH 8.0 by dialysis and loaded onto a 5 mL Q-HP column (GE Healthcare). The different forms were isolated by using a 50 column volumes (i.e., 250 mL) linear gradient of 0.15–0.4 M NaCl and collecting 5 mL fractions with a 4 mL/min flow rate using 20 mM Tris·HCl pH 8.0 as the running buffer. For closely eluting peaks, SDS-PAGE sometimes showed that separation was incomplete, in which case a second round of ion-exchange chromatography was performed by loading samples onto a 1 mL Mono-P 5/50 GL column (GE Healthcare) with 1 mL/min flow rate. Separation was performed by applying a 60 column volumes (i.e., 60 mL) linear gradient of 0.25–0.4 M NaCl and collecting 2 mL fractions. The eluted fractions were concentrated to 5–10 mg/mL using a Vivaspin centrifugal concentrator 30 kDa cutoff (GE Healthcare), dialyzed into PBS, and stored at −80 °C. All purification steps were performed using an ÄKTA purifier 10 (GE Healthcare).
Glutathione-S-Transferase-BirA (plasmid a kind gift from Chris O’Callaghan, University of Oxford) was expressed in E. coli and purified using glutathione-sepharose as described.23 AP-AffiIGF1R and AffiHER2-SpyTag were expressed in E. coli and purified using Ni-NTA (Qiagen). BirA biotinylation was performed as described previously22 to generate bio-AffiIGF1R.
Protein concentrations were determined from A280 using the extinction coefficient calculated from the amino acid sequence using the ProtParam tool. Concentrations of all streptavidin forms refer to the concentration of monomer. The pI of each monomer was estimated from the sequence using the ProtParam tool. The tetramer pI was estimated from the weighted mean of the pI of each monomer.
Octamer and Eicosamer Formation and Isolation
To generate an octamer, 40 μM Tr3DCatch1 and 40 μM Tre3DTag1 were mixed together and incubated for 1 h at room temperature. The sample was then concentrated 2-fold using a centrifugal concentrator 6 kDa cutoff and incubated for 16 h at room temperature. Octamer was then isolated by size exclusion chromatography on a Superdex 200 GL 10/300 column (GE Healthcare) using PBS pH 7.4 as a running buffer and a flow rate of 0.5 mL/min.
To generate an eicosamer, 400 μM Tr3DCatch1 was mixed with 100 μM DTag4 to give a final concentration of 250 μM Tr3DCatch1 and 40 μM DTag4. Samples were incubated for 1 h at room temperature and then concentrated 2-fold using a centrifugal concentrator 6 kDa cutoff and further incubated for 16 h at room temperature. The eicosamer was then isolated using size exclusion chromatography on a Superdex 200 GL 10/300 column (GE Healthcare) using PBS pH 7.4 as a running buffer and a flow rate of 0.5 mL/min.
SDS-PAGE
SDS-PAGE was performed on 10%, 14%, and 16% Tris-glycine gels using an XCell SureLock system (Life Technologies). Protein samples (10 μM) were mixed with an equal volume of 2 × SDS loading buffer (20% glycerol, 100 mM Tris·HCl, 4% SDS, 0.2% bromophenol blue, pH 6.8). Unboiled samples were loaded on the gel without further treatment. Boiled samples were heated for 5 min at 95 °C. Variants of streptavidin and traptavidin remain folded, assembled in a tetramer, and bound to biotinylated ligands on SDS-PAGE without boiling; however, since they are not linear their mobility does not match the molecular weight16 and binding of biotinylated ligands can sometimes increase mobility. Also, the relative mobility of a folded tetramer depends on the polyacrylamide percentage, such as for Tr4 and Tr3DCatch1 in Figure 3C versus Figure S2B. All samples were unboiled, unless otherwise marked. Gels were typically run for 1 h at room temperature at 200 V in Tris-glycine running buffer (25 mM Tris·HCl, 192 mM glycine, and 0.1% SDS, pH 8.2). SDS-PAGE was also performed on 4% and 7% Tris-acetate polyacrylamide gels. Nondenatured samples were loaded without boiling in 5 × loading buffer (150 mM Tris-Acetate pH 7.0, 10% SDS, 25% sucrose, 0.2% bromophenol blue). Gels were typically run for 90 min on ice at 150 V in an XCell SureLock in running buffer (50 mM Tricine, 50 mM Tris·HCl, and 0.1% SDS, pH 8.2). Gels were stained with InstantBlue (Expedeon) and imaged using a ChemiDoc XRS imager and QuantityOne (version 4.6) software (Bio-Rad).
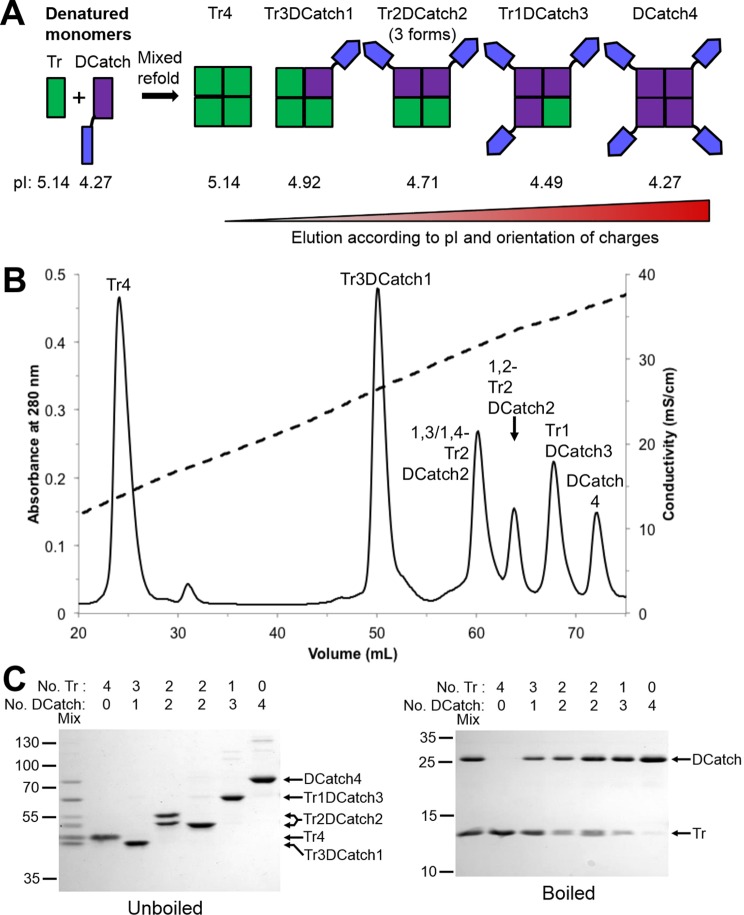
Figure 3.
Generation of traptavidin tetramers linked to SpyCatcher. (A) Formation of chimeric tetramer forms of traptavidin (Tr) and Dead streptavidin-SpyCatcher (DCatch) by mixed refolding of inclusion bodies and separation by ion-exchange chromatography. Predicted pI values are shown for the constituent monomers and the assembled tetramers. (B) Ion-exchange chromatogram showing separation of the mixed refold into individual species bearing different numbers of functional biotin binding sites and SpyCatchers. A280 is plotted with a solid line, and conductivity, with a dotted line. (C) 14% SDS-PAGE with Coomassie staining of Tr/DCatch tetramers showing the tetramer mobilities in unboiled samples (left) and the subunit compositions in boiled samples (right). Mix refers to the input sample.
Binding and Stability Analysis of SpyAvidin Tetramers
Tre3DTag1 was incubated with the indicated concentrations of bio-AffiIGF1R and SpyCatcher for 1 h at room temperature in PBS. Tr3DCatch1 was incubated with the indicated concentration of AffiHER2-SpyTag and bio-AffiIGF1R for 1 h at room temperature in PBS. Samples were then analyzed by SDS-PAGE with Coomassie staining.
For testing binding by MHC, we used biotinylated HLA-A2 bound to NY-ESO-1157–165 9V (see below). Proteins were incubated together overnight at room temperature in PBS and then analyzed by SDS-PAGE with Coomassie staining (for SA4 and Tr4 a 10% polyacrylamide Tris-glycine gel; for the octamer a 7% polyacrylamide Tris-acetate gel; for the eicosamer a 4% polyacrylamide Tris-acetate gel).
For stability tests, Tre3DTag1 or Tr3DCatch1 at 20 μM were incubated in PBS, 0.1% sodium azide, 1 mM PMSF, 1 mM EDTA, and EDTA-free mixed protease inhibitors (Roche) for 1–8 d at 25 or 37 °C before SDS-PAGE.
Mass Spectrometry
Samples were concentrated to ∼15 μM and buffer-exchanged into 200 mM ammonium acetate using Amicon Ultra 0.5 mL centrifugal filters with a 10 kDa cutoff (Millipore). Measurements were carried out on a modified Synapt G1 High Definition Mass Spectrometry (HDMS) Quadrupole Time of Flight (Q-ToF) mass spectrometer (Waters),24 calibrated using 10 mg/mL cesium iodide in 250 mM ammonium acetate. 2.5 μL aliquots of sample were delivered by nanoelectrospray ionization via gold-coated capillaries, prepared in house.25 Instrumental parameters for tetramers and octamers were as follows: source pressure 6.0 mbar, capillary voltage 1.20 kV, cone voltage 50 V, trap energy 10 V, bias voltage 5 V, and trap pressure 0.0163 mbar. For eicosamers, the cone voltage was 100 V and the trap energy was 25 V. Mass spectra were smoothed and peak-centered, and masses were assigned using MassLynx v4.1 (Waters).
Thermal Stability of the Eicosamer and Octamer
The eicosamer, octamer, and SAe4 in PBS at 10 μM in a final volume of 50 μL were incubated for 3 min at 25, 37, 50, 60, 70, 80, or 90 °C and then cooled to 10 °C in a Bio-Rad C1000 Thermal Cycler at 3 °C/s. SDS loading buffer was added to each sample directly before loading onto SDS-PAGE. As a control, the eicosamer, octamer, and SAe4 were incubated for 3 min at 95 °C in the presence of SDS loading buffer. SDS-PAGE was performed for the eicosamer on a 4% Tris-acetate gel, octamer on a 7% Tris-acetate gel, and SAe4 on an 18% Tris-glycine gel.
Peptide–MHC Complex Generation
Peptide–MHC complexes were generated by expressing the HLA-A2 MHC heavy chain and human β2-microglobulin as inclusion bodies in E. coli and then refolding with solid-phase synthesized peptide (Sigma-Genosys).26 Cognate peptide was NY-ESO-1157–165 9V, with sequence SLLMWITQV (a variant of the original tumor-derived peptide SLLMWITQC); the control peptide (sequence GLGGGGGGV) still had the P2 and P9 anchor residues for efficient binding to HLA-A2.26 The heavy chain contained an AP-tag at its C-terminus and was biotinylated in vitro with BirA. The biotinylated complexes were purified by FPLC on a Superdex-75 column (GE Healthcare) using 50 mM Tris·HCl pH 8.0 with 150 mM NaCl as the running buffer.
1 μM cognate or control peptide–MHC complex was incubated with a concentration of the SAe4, Tre4, octamer, or eicosamer such that there was 1.4 MHC to 1 biotin binding site. Incubation was for 1 h at room temperature in PBS, and then samples were stored at 4 °C before T cell assay.
T Cell Activation Assay
The human leukemia Jurkat T cell line J.RT3-T3.5 was stably transduced with the human 1G4 T cell receptor and the human CD8α coreceptor.26 The 1G4 T cell receptor has a binding affinity for the cognate peptide–MHC complex of 7.2 μM as measured by surface plasmon resonance.26 Cells were cultured in DMEM with 10% fetal calf serum and 1% penicillin/streptomycin and incubated at 37 °C and 10% CO2.
T cells at 107 cells/mL in DMEM were incubated with an equal volume of DMEM (unstimulated samples) or the streptavidin/traptavidin constructs in DMEM to give a 25 or 100 nM final concentration at 37 °C for 3 min. Cells were then fixed in 4% paraformaldehyde (Thermo Fisher) in PBS for 15 min at 37 °C. Cells were washed twice with PBS + 1% BSA and permeabilized on ice for 1 h in PBS-saponin [PBS with 1% BSA, 0.1% saponin (Sigma-Aldrich), and 0.05% sodium azide]. Staining for phospho-ERK was performed using 1:100 dilution phycoerythrin-labeled anti-phosphoERK antibody [Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (D13.14.4E) XP Rabbit monoclonal antibody, catalog number #5682 from Cell Signaling Technology, Inc.] in PBS-saponin for 1 h on ice in the dark. Cells were washed thrice in PBS. Cells were then analyzed by flow cytometry using FACSCalibur and CellQuest Pro software (Becton-Dickinson). 10 000 cells were analyzed per sample, and live cells were gated based on forward and side scatter. Each experiment was conducted with duplicate samples and repeated on a separate day.
Results
Chimeric SpyAvidin Tetramer Isolation
For maximal stability of the core assembly, we used traptavidin rather than streptavidin. Traptavidin is an S52G R53D mutant of streptavidin we developed with an ∼10-fold lower off-rate for biotin conjugates, as well as greater thermostability and mechanostability, and is the most stable binder of biotin conjugates.2,16,27 We aimed to combine the ultrastable biotin binding of traptavidin (Figure 1A) with the covalent binding of SpyTag/SpyCatcher to create SpyAvidins (Figure 1B). Dead streptavidin (D) subunits contain the mutations N23A, S27D, and S45A, which leads to negligible biotin binding but unchanged tetramer stability.17,27 By mixed refolding from inclusion bodies of traptavidin subunits and Dead subunits linked to SpyTag (DTag) or SpyCatcher (DCatch), we aimed to create chimeric tetramers where each subunit could bind biotin or SpyTag/SpyCatcher (Figure 1C).
Figure 1.

Nanohubs with biotin binding and SpyTag/SpyCatcher ligation. (A) Multiple noncovalent interactions in biotin binding. The tight binding of biotin (carbons in yellow) by traptavidin from the formation of eight hydrogen bonds as well as a hydrophobic cage of tryptophans, with residues mutated in traptavidin in cyan (from PDB 2Y3F). (B) Spontaneous isopeptide bond formation leads to a covalent linkage between a SpyTag-labeled protein target (cyan) and SpyCatcher (dark blue) (from PDB 4MLS). (C) Combining SpyTag/SpyCatcher with traptavidin/biotin allows the formation of tetramers with subunits able to bind biotin (Tr or Tre), bind SpyTag (DCatch), or bind SpyCatcher (DTag).
We previously showed that streptavidin tetramers containing a precise number of biotin binding or Dead subunits could be purified by ion-exchange chromatography, if there was sufficient difference in pI of the subunits.18 We linked SpyTag to the C-terminus of D, to create DTag subunits, and a tag of 6 glutamic acids to the C-terminus of traptavidin, to create Tre subunits (Figures 2A and S1A). With increasing numbers of hexaglutamate tags, the tetramers required increasing amounts of NaCl for elution from the ion-exchange resin. Six main peaks were eluted by ion exchange (Figure 2B). As previously, we saw two distinct peaks from the different arrangement of divalent tetramers (1,2 or 1,3 or 1,4 orientation) (Figures 2B and S1).18 We verified the ion-exchange separation by SDS-PAGE, with the negatively charged tags increasing tetramer mobility (Figure 2C). Traptavidin and the streptavidin variants in this study remain as a tetramer on SDS-PAGE unless boiled. The subunit composition of the resultant tetramer is then visible by SDS-PAGE of boiled samples (Figure 2C).
Figure 2.
Generation of traptavidin tetramers linked to SpyTag. (A) Formation of chimeric tetramers of traptavidin-glutamate tag (Tre) and Dead streptavidin-SpyTag (DTag) by mixed refolding of inclusion bodies and separation by ion-exchange chromatography. Predicted pI values are shown for the constituent monomers and the assembled tetramers. (B) Ion-exchange chromatogram showing separation of the mixed refold into individual species bearing different numbers of functional biotin binding sites and SpyTags. A280 is plotted with a solid line, and conductivity, with a dotted line. (C) SDS-PAGE with Coomassie staining of Tre/DTag tetramers showing the tetramer mobilities in unboiled samples (left) and the subunit compositions in boiled samples (right). Mix refers to the input sample.
To generate hybrid tetramers bearing SpyCatcher, Dead-d-loop subunits were connected at the C-terminus through a glycine/serine linker to SpyCatcher. Given the pI of SpyCatcher, we introduced the negative charge onto the Dead subunit by inserting multiple Asp residues into its 3,4 loop (Figure S1B).18 These DCatch subunits were refolded with core traptavidin (Tr) subunits (Figure S1B and 3A). Again, ion-exchange chromatography enabled resolution of 6 peaks (Figure 3B). The tetramer forms separated by ion exchange were confirmed by SDS-PAGE with or without boiling (Figure 3C). Tetramers containing SpyCatcher showed reduced gel mobility although, since streptavidin variants remain globular in the unboiled gel, the change in mobility was not according to molecular weight. We found that Tr2DCatch2 gave two distinct gel mobilities, since in the 1,2 tetramer arrangement the SpyCatcher chains will be close together, but in the 1,3 and 1,4 arrangement the two chains will be on opposite sides of the tetramer (Figure S1C).
Orthogonal Reactivity of SpyAvidin Tetramers
Clearly, many different SpyAvidin forms can be generated from these mixed tetramers bearing DCatch or DTag. We focused our attention on Tre3DTag1, which is monovalent for SpyCatcher binding, and Tr3DCatch1, which is monovalent for SpyTag binding.
Affibodies are nonimmunoglobulin scaffolds, selected by phage display for high affinity and selective binding.28 Incubating Tre3DTag1 with a site-specifically biotinylated affibody against Type I insulin-like growth factor receptor (IGF1R),29 we observed three major shifts in mobility when adding substoichiometric affibody concentrations (lane 5, Figure 4A) and complete occupancy of the three biotin-binding sites with a small excess of affibody (lane 6, Figure 4A). An extra shift upward was seen when SpyCatcher was also added (lane 7, Figure 4A), validating the bifunctional reactivity of Tre3DTag1.
Figure 4.
Validation of binding capacity of Tre3DTag1 and Tr3DCatch1. (A) SDS-PAGE with Coomassie staining of Tre3DTag1, showing the reactivity of DTag with free SpyCatcher and the ability of the Tre subunits to bind up to three biotinylated affibodies (bio-AffiIGF1R). (B) SDS-PAGE with Coomassie staining of Tr3DCatch1, showing the reactivity of DCatch with a SpyTag-bearing affibody (AffiHER2-SpyTag) and the ability of the Tr subunits to bind up to three biotinylated affibodies (bio-AffiIGF1R).
Similarly, we saw three major bands when adding substoichiometric levels of biotinylated anti-IGF1R affibodies to Tr3DCatch1 (Figure 4B). We further found a single shift upward, corresponding to binding of one SpyTag-linked affibody of a separate specificity, against the cancer-associated tyrosine kinase HER230 (Figure 4B), so validating the reactivity of Tr3DCatch1. Minor bands are also seen in Figure 4A and 4B that are likely to correspond to the altered mobility depending on whether the biotinylated ligand binds a traptavidin subunit proximal or distal (Figure S1C) to the SpyTag- or SpyCatcher-linked subunit.
We also confirmed that the SpyAvidin tetramers did not rearrange subunit composition over time, consistent with previous traptavidin tetramers:27 no new band appeared after 8 days of incubation at 37 °C for Tre3DTag1 (Figure S2A) or Tr3DCatch1 (Figure S2B).
Generation of Higher-Order SpyAvidin Multimers
(Strept)avidin is an obligate tetramer, and the vast majority of its applications have had a maximal valency of four, but higher order valency may enable new possibilities. Our generation of Tr3DCatch1 and Tre3DTag1 provided a facile route to a streptavidin octamer (8 subunits), by mixing these two forms (Figure 5A). Octamer formation was validated by SDS-PAGE (Figure 5B). To illustrate the progress of reaction, we show incubation at an early time point (lane 3, Figure 5B), but the final preparation, obtained after overnight reaction, was efficiently purified from any remaining tetramer by gel filtration chromatography (lane 4, Figure 5B). Native mass spectrometry, using a quadrupole time-of-flight mass spectrometer,24 enabled us to analyze the octameric assembly: octamer expected mass = 121 116 Da (with one isopeptide bond); octamer observed mass = 121 223 ± 16 Da (107 Da or 0.088% discrepancy to the expected mass) (Figure 5C).
Figure 5.
Generation of a SpyAvidin octamer and eicosamer. (A) Cartoon of tetramer, octamer, and eicosamer construction. (B) Reaction of Tre3DTag1 with Tr3DCatch1 to generate an octamer (lane 3), which can subsequently be purified via gel filtration (lane 4), analyzed by SDS-PAGE with Coomassie staining. (C) Mass spectrometry of the octamer. Peaks corresponding to the octamer are marked with circles, along with the charge state of the highest peak. (D) Reaction of DTag4 and Tr3DCatch1 to generate an eicosamer, which can subsequently be purified via gel filtration (far right lane), analyzed by SDS-PAGE with Coomassie staining. (E) Mass spectrometry of the eicosamer. Peaks corresponding to an eicosamer and a hexadecamer impurity are marked with circles, along with the charge state of the highest peak for each.
Building further, with DTag4 at the heart, mixing with Tr3DCatch1 should lead to an eicosamer (20 streptavidin subunits) (Figure 5A). Eicosamer formation was analyzed by SDS-PAGE. Tris-acetate gels enable analysis in this high molecular weight range, although mobility deviates from molecular weight because streptavidin-derived subunits remain folded (Figure 5D). Again proximal or distal orientations of (DTag4)1(Tr3DCatch1)2 are likely to generate distinct bands on SDS-PAGE (Figure 5D). Mass spectrometry measurement was consistent with formation of the desired eicosamer: eicosamer expected mass = 316 767 Da (with four isopeptide bonds); eicosamer observed mass = 318 523 ± 51 Da (1756 Da or 0.55% discrepancy to the expected mass) (Figure 5E). From analysis of these large noncovalent assemblies by mass spectrometry, where complete removal of water and buffer ions is challenging, these values represent a good match between observed and expected.17,31 Both SDS-PAGE and mass spectrometry detected an impurity of some hexadecamer (16 subunits) (Figure 5D,E).
The robustness of the eicosamer and octamer assemblies was tested by a thermostability assay, which revealed stability comparable to the wild-type streptavidin tetramer.16 The eicosamer and octamer remained intact up to 70 °C but started to rearrange at 80 °C (Figure S3).
SpyAvidin Octamers and Eicosamers Drove Enhanced T Cell Signaling
MHC class I on the cell surface enables cytotoxic T cells to monitor the events taking place inside the cell, to detect viral infection or tumorigenesis. MHC class I is a trimeric complex of a heavy chain bound to β2-microglobulin and loaded with an 8–10 amino acid peptide (Figure 6A). Binding of peptide–MHC complexes to the T cell receptor, for T cell activation, typically has a half-life of seconds. Therefore, tetramerization of peptide–MHC by streptavidin conferred stable binding to T cells and led to a revolution in the ability to monitor specific immune responses in infectious disease and cancer.4,5
Figure 6.

Eicosamers and octamers strongly stimulated T cell signaling. (A) Cartoon of how an eicosamer may bind multiple biotinylated peptide–MHC complexes. (B) T cell activation determined by flow cytometry of ERK phosphorylation, unstimulated or after stimulation with 25 nM (left panel) or 100 nM (right panel) cognate peptide–MHC incubated with the streptavidin tetramer (SAe4), traptavidin tetramer (Tre4), octamer, or eicosamer. Representative results from experiments run in duplicate on two separate days. The percentage of cells with staining above the threshold (dotted line) is marked. (C) T cell stimulation as in (B) but with control peptide–MHC.
We initially validated binding of peptide–MHC complexes to different biotin-binding assemblies, using SDS-PAGE. The steric clash from binding of the peptide–MHC complex to adjacent biotin binding sites makes it hard to achieve complete occupancy of 4 MHC around one streptavidin (Figure S4A). However, the extra stability of traptavidin’s biotin binding led to more uniform and high occupancy complexes with MHC (Figure S4B). We also analyzed binding of MHC to the octamer and eicosamer by SDS-PAGE, although with so many intermediate stoichiometries and spatial orientations it is not possible to confirm the identity of each band (Figure S4C and S4D).
To see how higher order clustering might change signal transduction, we determined the activation of T cell signaling caused by a given concentration of the biotinylated peptide–MHC complex bound to the streptavidin, traptavidin, octamer, or eicosamer. Figure 6A illustrates the potential clustering from binding of MHC class I to the eicosamer. T cell activation was measured from the phosphorylation of Extracellular Signal-related Kinase (ERK), an early downstream event following T cell receptor activation, as assessed by intracellular flow cytometry.32 As a negative control, we used a glycine-rich peptide with efficient binding to MHC but minimal binding to the T cell receptor. With 25 nM cognate peptide–MHC, the streptavidin tetramer and traptavidin tetramer gave a similar slight degree of activation (Figure 6B). However, activation was stronger with the octamer, while the eicosamer managed to activate a major fraction of the T cells (Figure 6B). With 100 nM cognate peptide–MHC, the streptavidin tetramer and traptavidin tetramer were able to activate a higher proportion of T cells than at 25 nM, but the activation with the octamer was superior to tetramers and was greater still with the eicosamer (Figure 6B). Importantly, none of the assemblies activated the cells when bound to the control peptide, even at 100 nM (Figure 6C). Therefore, activation depended on T cell receptor recognition and not any other feature of the assembly, such as binding to the coreceptor CD8 in a T cell receptor-independent fashion.4
Discussion
Here we have generated SpyAvidin hubs, tetramers with a defined number of orthogonal binding sites for biotin or SpyTag/SpyCatcher. These hubs can then be linked to create large modular protein architectures, including the octamers or eicosamers which were able to achieve highly sensitive and specific T cell activation.
The extra stability of biotin–conjugate binding by traptavidin compared to streptavidin16 is highlighted by the difficulty in assembling four biotinylated peptide–MHC complexes around streptavidin. Streptavidin binding is exceptionally strong for biotin itself and for small biotin conjugates, but when linking large ligands, especially at adjacent binding sites in the tetramer,18 binding efficiency declines33,34 and the extra stability of traptavidin is shown here to be valuable. Similarly SpyTag/SpyCatcher interaction is slower and requires more ligand excess when bringing together large ligands around a single hub (e.g., in generating eicosamers). This reflects a general challenge in nanoassembly and points to the importance of starting with reactions that are highly efficient at a small size range to be able to tolerate the challenges of building on the larger scale.
Sophisticated assemblies can be made with DNA nanotechnology, although stability is often limited if integrity depends solely on short regions base pairing35 and synthesis is generally on the microgram scale.36 Oligonucleotides are frequently biotinylated to enable (strept)avidin linkage, but it has not been straightforward to bridge precisely from this (strep)avidin to a subsequent protein. Coiled-coils or modular repeat proteins enable impressive polypeptide nanostructures,37−39 but without disulfides the binding affinity is often moderate and these structural elements do not share the extensive infrastructure of biotinylated ligands accessible to SpyAvidin. SpyLigase peptide–peptide ligation is an alternative route to connect irreversibly proteins into polymers, via isopeptide bonds, but it is not yet possible to control polymer size.21 We anticipate that SpyAvidins hubs will be valuable in interfacing with a range of different nanoassembly scaffolds. We have demonstrated spontaneous isopeptide bond formation to two other peptide tags,40 and so in future work it should be possible to construct more orthogonal isopeptide-based linkages from streptavidin subunits.
Streptavidin tetramers have previously been linked together through chemical coupling but without control over subunit identity or full characterization of the multimers obtained.4 Peptide–MHC complexes can also be pentamerized with coiled coils or covalently coupled to dextran (although with heterogeneous valency).4 Such multimers have shown enhanced sensitivity compared to tetramers for staining of low affinity T cell receptors, notably those against MHC class I bound to self-peptides in autoimmune disease and antitumor responses.4 Therefore, eicosamers may have value in identification, isolation for immunotherapy, or depletion of such T cells.4,41 The use of desthiobiotinylated MHC should also allow this multimerization to be reversible, using free biotin as a competitor.42 Multivalent recognition using eicosamers may also help to identify binding partners for other fast off-rate interactions, including protein–protein interactions in leukocyte adhesion43 or malarial erythrocyte invasion,44 and protein–carbohydrate interactions in cancer metastasis.45
It is still an active area of investigation how engagement of the T cell receptor leads to initiation of the tyrosine kinase cascade and activation of the T cell.46,47 In the absence of a cell–cell contact, it is known that peptide–MHC tetramers can activate T cell signaling, whereas single peptide–MHC molecules are inactive.48 Therefore, local regions of T cell receptor clustering, giving local exclusion of transmembrane phosphatases, may be sufficient for signal activation. The extra clustering with eicosamers may be able to prolong surface interactions with the T cell receptor and also generate a larger area of phosphatase exclusion, leading to more efficient downstream signal transduction. In future work it will be valuable to explore this nanoscale control of signal activation,49 to enhance the cellular response to a range of biotinylated macromolecular and small-molecule stimuli.
Acknowledgments
M.F., D.M., and M.H. were funded by the Biotechnology and Biological Sciences Research Council (BBSRC). G.V. was funded by the Medical Research Council (MRC) and Merton College Oxford. M.L. was supported by a Doctoral Training Centre Systems Biology studentship from the Engineering and Physical Sciences Research Council (EPSRC). O.D. was supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (Grant Number 098363). P.A.vdM. was funded by the Wellcome Trust (Grant reference 101799). J.Y. was funded by the MRC. C.V.R. was funded by the Royal Society.
Supporting Information Available
Supporting Information consists of Figures S1–S4. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare the following competing financial interest(s): M.H. is an author on patent applications for monovalent streptavidin compositions (USPTO 20070099248), traptavidin (UK 0919102.4), and isopeptide bond-forming peptides (UK 1002362.0).
Supplementary Material
References
- Cheng M. M.; Cuda G.; Bunimovich Y. L.; Gaspari M.; Heath J. R.; Hill H. D.; Mirkin C. A.; Nijdam A. J.; Terracciano R.; Thundat T.; Ferrari M. Curr. Opin. Chem. Biol. 2006, 10, 11. [DOI] [PubMed] [Google Scholar]
- Laitinen O. H.; Hytonen V. P.; Nordlund H. R.; Kulomaa M. S. Cell. Mol. Life Sci. 2006, 63, 2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooldridge L.; Lissina A.; Cole D. K.; van den Berg H. A.; Price D. A.; Sewell A. K. Immunology 2009, 126, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J.; Dojcinovic D.; Guillaume P.; Luescher I. Front. Immunol. 2013, 4, 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman J. D.; Moss P. A.; Goulder P. J.; Barouch D. H.; McHeyzer-Williams M. G.; Bell J. I.; McMichael A. J.; Davis M. M. Science 1996, 274, 94. [PubMed] [Google Scholar]
- Korndorfer I. P.; Skerra A. Protein Sci. 2002, 11, 883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch J. D.; Eslamizar L.; Filanoski B. J.; Malekzadeh N.; Haugland R. P.; Beechem J. M.; Haugland R. P. Anal. Biochem. 2002, 308, 343. [DOI] [PubMed] [Google Scholar]
- Srisawat C.; Engelke D. R. RNA 2001, 7, 632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber P. C.; Pantoliano M. W.; Simons D. M.; Salemme F. R. J. Am. Chem. Soc. 1994, 116, 2717. [Google Scholar]
- Lamla T.; Erdmann V. A. Protein Expr. Purif. 2004, 33, 39. [DOI] [PubMed] [Google Scholar]
- Keefe A. D.; Wilson D. S.; Seelig B.; Szostak J. W. Protein Expr. Purif. 2001, 23, 440. [DOI] [PubMed] [Google Scholar]
- Reznik G. O.; Vajda S.; Cantor C. R.; Sano T. Bioconjugate Chem. 2001, 12, 1000. [DOI] [PubMed] [Google Scholar]
- Chilkoti A.; Schwartz B. L.; Smith R. D.; Long C. J.; Stayton P. S. Biotechnology (N.Y.) 1995, 13, 1198. [DOI] [PubMed] [Google Scholar]
- Zakeri B.; Fierer J. O.; Celik E.; Chittock E. C.; Schwarz-Linek U.; Moy V. T.; Howarth M. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, E690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.; Fierer J. O.; Rapoport T. A.; Howarth M. J. Mol. Biol. 2014, 426, 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivers C. E.; Crozat E.; Chu C.; Moy V. T.; Sherratt D. J.; Howarth M. Nat. Methods 2010, 7, 391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth M.; Chinnapen D. J.; Gerrow K.; Dorrestein P. C.; Grandy M. R.; Kelleher N. L.; El Husseini A.; Ting A. Y. Nat. Methods 2006, 3, 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairhead M.; Krndija D.; Lowe E. D.; Howarth M. J. Mol. Biol. 2014, 426, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu J.; March P. E.; Lee R.; Tillett D. Nucleic Acids Res. 2004, 32, e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett D.; Kovaleva E.; Schatz P. J. Protein Sci. 1999, 8, 921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer J. O.; Veggiani G.; Howarth M. Proc. Natl. Acad. Sci. U.S.A. 2014, 111, E1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth M.; Ting A. Y. Nat. Protoc. 2008, 3, 534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Callaghan C. A.; Byford M. F.; Wyer J. R.; Willcox B. E.; Jakobsen B. K.; McMichael A. J.; Bell J. I. Anal. Biochem. 1999, 266, 9. [DOI] [PubMed] [Google Scholar]
- Bush M. F.; Hall Z.; Giles K.; Hoyes J.; Robinson C. V.; Ruotolo B. T. Anal. Chem. 2010, 82, 9557. [DOI] [PubMed] [Google Scholar]
- Hernandez H.; Robinson C. V. Nat. Protoc. 2007, 2, 715. [DOI] [PubMed] [Google Scholar]
- Aleksic M.; Dushek O.; Zhang H.; Shenderov E.; Chen J. L.; Cerundolo V.; Coombs D.; van der Merwe P. A. Immunity 2010, 32, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivers C. E.; Koner A. L.; Lowe E. D.; Howarth M. Biochem. J. 2011, 435, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofblom J.; Feldwisch J.; Tolmachev V.; Carlsson J.; Stahl S.; Frejd F. Y. FEBS Lett. 2010, 584, 2670. [DOI] [PubMed] [Google Scholar]
- Li J.; Lundberg E.; Vernet E.; Larsson B.; Hoiden-Guthenberg I.; Graslund T. Biotechnol. Appl. Biochem. 2010, 55, 99. [DOI] [PubMed] [Google Scholar]
- Wikman M.; Steffen A. C.; Gunneriusson E.; Tolmachev V.; Adams G. P.; Carlsson J.; Stahl S. Protein Eng. Des. Sel. 2004, 17, 455. [DOI] [PubMed] [Google Scholar]
- McKay A. R.; Ruotolo B. T.; Ilag L. L.; Robinson C. V. J. Am. Chem. Soc. 2006, 128, 11433. [DOI] [PubMed] [Google Scholar]
- Altan-Bonnet G.; Germain R. N. PLoS Biol. 2005, 3, e356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J. L.; Heuff R.; Cramb D. T. Biophys. J. 2006, 90, 1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buranda T.; Jones G. M.; Nolan J. P.; Keij J.; Lopez G. P.; Sklar L. A. J. Phys. Chem. B 1999, 103, 3399. [Google Scholar]
- Rajendran A.; Endo M.; Katsuda Y.; Hidaka K.; Sugiyama H. J. Am. Chem. Soc. 2011, 133, 14488. [DOI] [PubMed] [Google Scholar]
- Shih W. M.; Lin C. X. Curr. Opin. Struct. Biol. 2010, 20, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher J. M.; Harniman R. L.; Barnes F. R. H.; Boyle A. L.; Collins A.; Mantell J.; Sharp T. H.; Antognozzi M.; Booth P. J.; Linden N.; Miles M. J.; Sessions R. B.; Verkade P.; Woolfson D. N. Science 2013, 340, 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradisar H.; Bozic S.; Doles T.; Vengust D.; Hafner-Bratkovic I.; Mertelj A.; Webb B.; Sali A.; Klavzar S.; Jerala R. Nat. Chem. Biol. 2013, 9, 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove T. Z.; Forster J.; Pimienta G.; Dufresne E.; Regan L. Biopolymers 2012, 97, 508. [DOI] [PubMed] [Google Scholar]
- Zakeri B.; Howarth M. J. Am. Chem. Soc. 2010, 132, 4526. [DOI] [PubMed] [Google Scholar]
- Davis M. M.; Altman J. D.; Newell E. W. Nat. Rev. Immunol. 2011, 11, 551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaume P.; Baumgaertner P.; Angelov G. S.; Speiser D.; Luescher I. F. J. Immunol. 2006, 177, 3903. [DOI] [PubMed] [Google Scholar]
- Jiang L.; Barclay A. N. Immunology 2010, 129, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholdson S. J.; Crosnier C.; Bustamante L. Y.; Rayner J. C.; Wright G. J. Cell. Microbiol. 2013, 15, 1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauselmann I.; Borsig L. Front. Oncol. 2014, 4, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z.; Discher D. E.; Finkel T. H. Front Immunol. 2012, 3, 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Merwe P. A.; Dushek O. Nat. Rev. Immunol. 2011, 11, 47. [DOI] [PubMed] [Google Scholar]
- Ge Q.; Stone J. D.; Thompson M. T.; Cochran J. R.; Rushe M.; Eisen H. N.; Chen J.; Stern L. J. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 13729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiessling L. L.; Gestwicki J. E.; Strong L. E. Angew. Chem., Int. Ed. 2006, 45, 2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.