Abstract
The intrinsic ability to display self-organizing morphogenetic properties in ex vivo culture may represent a general property of tissue stem cells. Here we show that single luminal stem/progenitor cells can generate prostate organoids in a three-dimensional culture system in the absence of stroma. Organoids generated from CARNs (castration-resistant Nkx3.1-expressing cells) or normal prostate epithelium exhibit tissue architecture containing luminal and basal cells, undergo long-term expansion in culture, and display functional androgen receptor signaling. Lineage-tracing demonstrates that luminal cells are favored for organoid formation, and generate basal cells in culture. Furthermore, tumor organoids can initiate from CARNs after oncogenic transformation, and from mouse models of prostate cancer, and can facilitate analyses of drug response. Finally, we provide evidence supporting the feasibility of organoid studies of human prostate tissue. Our studies underscore the progenitor properties of luminal cells, and identify in vitro approaches for studying prostate biology.
Despite the apparent simplicity of cell types in the prostate epithelium, there has long been a dearth of suitable cell culture-based systems for investigating prostate biology1. In the normal prostate, there are three epithelial cell types, corresponding to: 1) luminal cells, which are columnar cells expressing cytokeratin (CK) 8, CK18, and high levels of androgen receptor (AR); 2) basal cells, which express CK5 and p63; and 3) rare neuroendocrine cells2. During prostate tumorigenesis, basal cells undergo progressive loss in pre-neoplastic lesions known as prostatic intraepithelial neoplasia (PIN), and are essentially absent in prostate adenocarcinoma, which typically has a luminal phenotype3, 4.
Historically, prostate luminal cells have been difficult to grow in culture, which has hindered the establishment of cell lines from normal or transformed prostate epithelium. One approach to circumvent this limitation has been culture of three-dimensional “prostaspheres” containing epithelial cells explanted from primary mouse or human prostate tissue5–8. Such prostaspheres can be serially passaged and used in assays for prostate epithelial stem cells and tumor-initiating cells9, 10. However, prostaspheres typically originate from basal epithelial cells and fail to display complete luminal differentiation in the presence of androgens9, 11–13. Notably, prostaspheres fail to demonstrate strong nuclear AR expression in the presence of androgens or a functional response to androgen-deprivation6, 9.
Recent work has described alternative explant approaches for three-dimensional culture of epithelial cells in the absence of stroma. Such “organoid” culture systems contain similar extracellular matrix components as often used in sphere assays, but also utilize conditions that enhance the survival, proliferation, and/or differentiation of stem/progenitor populations14. In particular, cultured stem cells of the mouse small intestine and colon15, 16 can form organoids that display normal epithelial architecture and serve as the basis for tissue repair17, while tumor organoids can be established from transformed colon as a model of colon adenocarcinoma18, 19. Additional studies of organoids from intestine20, stomach21, liver22, and pancreas23, 24 have demonstrated the general feasibility of this approach.
In previous studies, we identified a luminal epithelial stem/progenitor population known as CARNs (castration-resistant Nkx3.1-expressing cells), which are also cells of origin for prostate cancer25. We also showed that single CARNs can reconstitute prostate ducts in a renal grafting assay25. Below, we introduce an ex vivo culture system that can support the growth and serial passaging of epithelial organoids derived from CARNs or more generally from normal prostate epithelium. We show that these prostate organoids are primarily derived from luminal epithelial cells, and display functional AR activity in culture. We demonstrate that mouse tumor organoids can model tumor phenotypes and drug response, and show that organoids can be established from benign human prostate tissue and a luminal prostate cancer cell line. Consequently, we propose that organoid culture represents an excellent system for investigating prostate biology and cancer.
Results
Establishment of prostate epithelial organoids from CARNs
Previously, we identified a rare luminal epithelial population in the regressed prostate epithelium that has stem cell properties in vivo and in tissue reconstitution assays25. To pursue further analyses of these CARNs, we sought to establish conditions for their isolation and successful propagation in culture. For this purpose, we surgically castrated adult male Nkx3.1CreERT2/+; R26R-YFP/+ mice to induce androgen-deprivation, followed by tamoxifen induction to lineage-mark CARNs (Fig 1a). Following dissociation of prostate tissue into a single-cell suspension, we used flow-sorting to isolate CARNs based on their yellow fluorescent protein (YFP) expression (Fig. 1b).
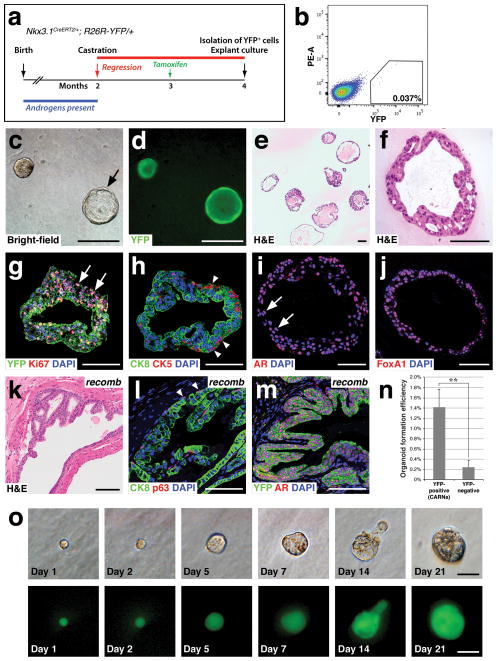
Figure 1.
Generation of prostate epithelial organoids from lineage-marked CARNs. (a) Time course of lineage-marking of CARNs in Nkx3.1CreERT2/+; R26R-YFP/+ mice. (b) Isolation of YFP-positive lineage-marked CARNs by flow cytometry. (c,d) Bright-field (c) and epifluorescent (d) views of CARN-derived organoids that are filled or hollow (arrow). (e,f) Hematoxylin-eosin (H&E) staining of CARN organoids at low-power (e) showing range of phenotypes, and high-power (f). (g) Uniform YFP expression with Ki67 immunostaining (arrows). (h) The basal marker CK5 is expressed on the exterior (arrowheads), while the luminal marker CK8 is expressed internally. (i,j) Strong nuclear expression of AR (arrows, i) and Foxa1 (j). (k–m) Renal grafts generated by tissue recombination of CARN-derived organoids with rat embryonic urogenital mesenchyme (k) display normal stratification of basal (arrowheads, l) and luminal cells (l), and uniform YFP and nuclear AR immunostaining (m); note that the slightly atypical histology in (k) likely reflects the heterozygous phenotype of Nkx3.1 mutants40, 41. (n) Efficiency of organoid formation by lineage-marked CARNs (YFP-positive cells from tamoxifen-induced and castrated Nkx3.1CreERT2/+; R26R-YFP/+ mice; n=4 experiments) and non-CARNs (YFP-negative cells from the same mice; n=3 experiments). Source data are provided in Supplementary Table 1. Error bars represent one standard deviation; the difference between CARNs and non-CARNs is statistically significant (p = 0.002, two-tailed Student’s t-test). (o) Generation of organoids from single CARNs. Time course of paired images shown under bright-field (top) and epifluorescent (bottom) illumination shows organoid growth from isolated single CARN. Scale bars in o correspond to 25 microns, in f–j,l,m to 50 microns, and in c–e,k to 100 microns.
To culture CARNs, we developed a protocol based in part on the importance of Matrigel in three-dimensional culture of prostate and mammary epithelium26, 27, hepatocyte medium for prostate epithelial cell culture28, and ROCK inhibitor to improve the survival of dissociated epithelial cells29–31. The resulting protocol involves low-percentage Matrigel floating culture in the presence of epidermal growth factor, heat-inactivated charcoal-stripped fetal bovine serum (FBS), which lacks androgens, and supplementation with dihydrotestosterone (DHT) (see Methods). Under these conditions, isolated CARNs formed epithelial “organoids” that could grow for at least 3–4 weeks in culture (Fig 1c), displaying a range of morphologies, and varying in size from 15 microns in diameter to greater than 0.5 mm in diameter. Importantly, most organoids were homogeneously composed of YFP-expressing cells, indicating their derivation from lineage-marked CARNs (Fig. 1d), and lacked stroma (Fig. 1e,f). Consistent with their growth in culture, many cells within organoids were positive for Ki67 (Fig. 1g). CARN-derived organoids typically displayed an outer rim of cells positive for the basal marker cytokeratin 5 (CK5), and internal cells positive for the luminal marker cytokeratin 8 (CK8) (Fig. 1h); few “intermediate” cells that co-express basal and luminal markers were observed. Notably, the organoids expressed nuclear AR (Fig. 1i), as well as nuclear Foxa1, a transcription factor that is essential for prostate organogenesis32 (Fig. 1j). Thus, lineage-marked CARNs are able to generate basal cells in organoid culture, similar to their ability in vivo and in tissue reconstitution assays.
To confirm that these organoids retained properties of prostate epithelium, we performed tissue reconstitution assays13. CARN-derived organoids were recombined with urogenital mesenchyme from rat embryos, followed by implantation under the kidney capsule of immunodeficient mice. The resulting grafts displayed prostate ductal structures (Fig. 1k) and expressed both basal (p63) and luminal (CK8) markers (Fig 1l). Furthermore, the epithelial cells were completely YFP-positive and expressed nuclear AR (Fig. 1m), indicating that the CARN-derived organoids could successfully reconstitute prostate tissue.
Establishment of prostate organoids from single CARNs
To determine the efficiency of organoid formation, we assessed the number of organoids formed after 7 days of culture. We found that the average efficiency of organoid formation by lineage-marked CARNs was 1.42% (Fig. 1n; Supplementary Table 1). For comparison, we also assayed non-YFP expressing epithelial cells from the same mice used to isolate the lineage-marked CARNs. These non-YFP expressing cells could also form organoids in culture, but at a nearly 6-fold lower average frequency of 0.24% (Fig. 1n; Supplementary Table 1; Methods), suggesting that non-CARNs can also form organoids, but at a reduced efficiency.
Given their stem cell properties, we examined whether individual lineage-marked CARNs could form an organoid. To isolate single cells, we used flow cytometry to purify lineage-marked CARNs, and then mouth-pipetted single fluorescent cells into individual wells of a 96-well plate. We imaged each well to confirm plating of single cells, and followed their potential growth every other day (Fig. 1o). We found that organoids formed from 5 out of 300 individual lineage-marked CARNs, with an overall frequency (1.67%) similar to that after plating of CARNs as a population (1.42%).
Organoid formation by prostate epithelial cells from hormonally-intact mice
Since both CARNs and non-CARNs could form organoids, we investigated whether normal prostate epithelial cells could initiate organoid formation. For this purpose, we performed flow-sorting of dissociated prostate cells to remove non-epithelial EpCAM−E-cadherin− cells (Fig. 2a). The resulting organoids displayed variable morphology and growth rates, suggesting heterogeneity in the starting population (Fig. 2b,c). Many organoids had a ductal structure resembling that of normal prostates, with a bi-layered epithelium surrounding a lumen (Fig. 2d), while other organoids contained multi-layered masses of cells (Fig. 2e). The organoids contained proliferating cells (Fig. 2f), and showed stratification into an outer basal layer and an internal luminal layer (Fig. 2g–i), with intermediate cells co-expressing luminal and basal markers rarely observed; neuroendocrine cells have not been detected to date. Furthermore, the organoids displayed nuclear expression of AR and Foxa1 (Fig. 2i,j), and could reconstitute prostate tissue in grafts (Fig. 2k). Importantly, organoids derived from normal prostate epithelium could be grown for at least 13 passages, with no apparent alterations in growth rate or phenotype (Supplementary Fig. 1a–c), and could be frozen and thawed with no loss of viability. After passaging, organoids continued to express luminal and basal markers, as well as nuclear AR, and were indistinguishable from low-passage number organoids (Supplementary Fig. 1d,e).
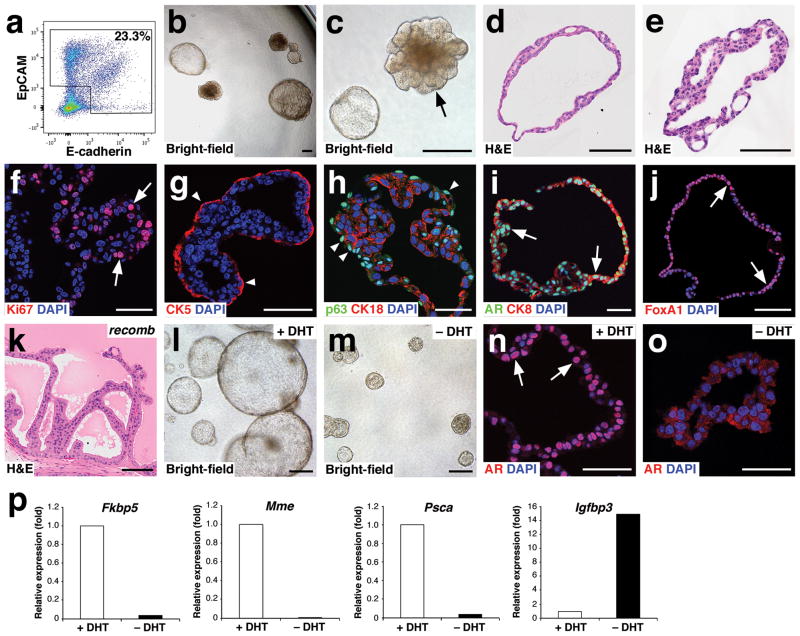
Figure 2.
Growth and androgen-responsiveness of prostate organoids from normal prostate epithelium. (a) Flow-sorting strategy to eliminate EpCAM−E-cadherin− cells from dissociated prostate tissue for organoid culture. (b) Low-power view of organoids at 20 days after plating, showing heterogeneity of phenotype. (c) Higher-power view showing hollow and filled budding organoid (arrow). (d,e) H&E staining of sections from a hollow organoid (d) and a multi-layered organoid (e). (f) Many proliferating cells are detectable by Ki67 immunostaining (arrows). (g) Organoids have an outer layer that expresses the basal marker CK5 (arrowheads). (h) Outer cells express the basal marker p63 (arrowheads), while interior cells are positive for the luminal marker CK18. (i) Nuclear immunostaining of AR (arrows) in organoids cultured in standard conditions with DHT. (j) Nuclear immunostaining for Foxa1 (arrows). (k) Tissue recombination of normal organoids with rat embryonic urogenital mesenchyme followed by renal grafting results in reconstitution of prostate tissue. (l,m) Organoids at passage 4 were passaged as single-cell suspensions and plated in the presence of DHT (l) or absence of DHT (m). (n,o) Strong nuclear AR immunostaining in the presence of DHT (n) and weak cytoplasmic AR immunostaining in the absence of DHT (o). (p) qPCR analysis of expression of AR downstream genes in organoids cultured in the presence or absence of DHT. Results are from a single experiment representative of 2 independent experiments. All assays were performed using three technical replicates and normalized to GAPDH expression; Scale bars in e–j,n,o correspond to 50 microns, and in b–d,k,l,m to 100 microns.
Interestingly, the efficiency of organoid formation from normal hormonally-intact prostate epithelium was 0.30%, significantly lower than from lineage-marked CARNs (Supplementary Table 1), which are isolated from the androgen-deprived regressed prostate. We also found that the efficiency of organoid formation from wild-type regressed prostate epithelium was 0.49%, which is not significantly different than that of hormonally-intact epithelium. (Supplementary Table 1). These efficiencies are also similar to that of YFP-negative cells in the CARNs lineage-marking experiment (Fig. 1n), suggesting that cells distinct from CARNs can form organoids, but at a lower efficiency.
To examine the effects of androgen-deprivation, organoids established from normal prostate epithelium were cultured and passaged in the presence or absence of DHT. We found that organoids could still form in the absence of DHT, but were reduced in size (Fig. 2l,m). Notably, AR immunostaining was nuclear in the presence of DHT, but was weaker and mostly cytoplasmic in the absence of DHT (Fig. 2n,o). To assess the molecular response to androgen-deprivation, we performed quantitative RT-PCR (qPCR) analysis of the expression of several known AR-regulated genes at passage 1. We found that Fkbp5, Mme, and Psca were down-regulated in organoids after DHT withdrawal, whereas expression of Igfbp3 was up-regulated, as expected for decreased AR activity (Fig. 2p). These findings indicate that organoids are highly responsive to androgen-deprivation.
Lineage-tracing demonstrates the preferential origin of organoids from luminal cells
We next used lineage-tracing to investigate which epithelial cell type(s) can give rise to organoids (Fig. 3a). To mark basal cells, we used the tamoxifen-inducible CK5-CreERT2 transgene13 in combination with the R26R-YFP reporter allele33. For marking of luminal cells, we used the CK8-CreERT2 or CK18-CreERT2 transgenes34, 35, either in combination with the R26R-YFP reporter or R26R-Tomato reporter36. Notably, these inducible Cre drivers were highly specific in marking basal or luminal epithelial cells in vivo at efficiencies similar to those previously observed13, 35 (Supplementary Fig. 2; Supplementary Table 2).
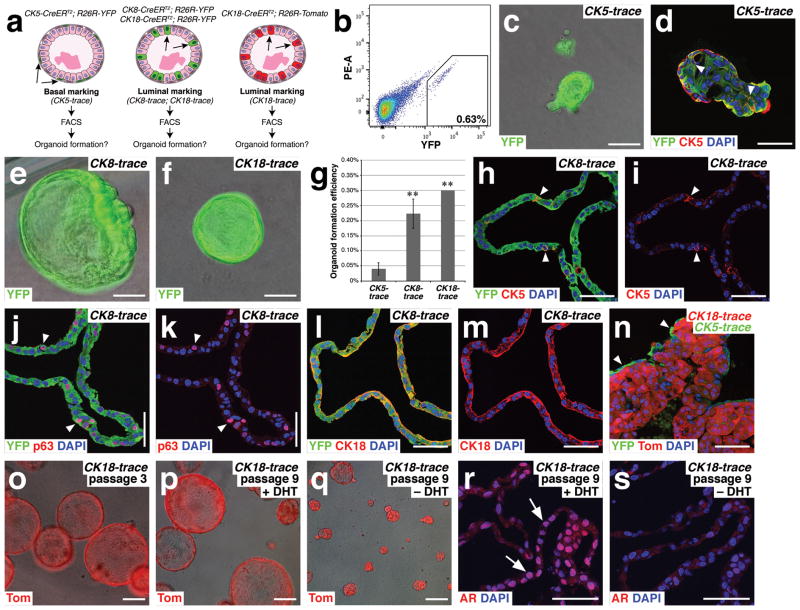
Figure 3.
Lineage-tracing shows that luminal cells are favored for generation of prostate organoids. (a) Strategy for lineage-marking of basal and luminal epithelial cells for organoid culture. (b) Isolation of YFP-positive luminal cells from CK8-CreERT2; R26R-YFP (CK8-trace) mice by flow cytometry. (c) CK5-trace organoid. (d) Many cells within a CK5-trace organoid are CK5-positive, including internal cells (arrowheads). (e) CK8-trace organoid. (f) CK18-trace organoid. (g) Efficiency of organoid formation from YFP-positive CK5-trace (n=4 experiments), CK8-trace (n=3 experiments), and CK18-trace (n=2 experiments) epithelial cells. The differences in efficiency between CK5-trace and CK8-trace (p=0.001) and between CK5-trace and CK18-trace (p=0.0009) are statistically significant (**) using a two-tailed Student’s t-test; error bars correspond to one standard deviation. Source data are provided in Supplementary Table 1. (h,i) Expression of the basal marker CK5 (arrowheads) in a CK8-trace organoid, shown with (h) and without (i) YFP overlay. (j,k) Expression of the basal marker p63 (arrowheads) in a CK8-trace organoid, shown with (j) and without (k) YFP overlay. (l,m) Expression of the luminal marker CK18 in a CK8-trace organoid, shown with (l) and without (m) YFP overlay. (n) Organoid generated from mixing of red CK18-trace cells and green CK5-trace cells shows green cells on the exterior, consistent with the localization of basal cells. (o) Serial passaging of CK18-trace organoids at passage 3. (p,q) CK18-trace organoids at passage 9 cultured in the presence (p) and absence (q) of DHT. (r,s) AR immunostaining is nuclear in CK18-trace organoids in the presence of DHT (r), but is weakly cytoplasmic in the absence of DHT (s). Scale bars in c–f,h–n correspond to 50 microns, and in o–s to 100 microns.
Using tamoxifen-induced CK5-CreERT2; R26R-YFP mice (which we term CK5-trace), we isolated YFP-positive cells by flow cytometry for organoid culture (Fig. 3b). We found that the isolated CK5-trace cells were extremely inefficient at organoid formation (0.04% efficiency) (Supplementary Table 1). Moreover, when organoids did form, they were often heterogeneous, containing regions derived from non-YFP expressing cells; for example, such organoids could arise from doublets containing a YFP-expressing and a non-expressing cell after flow sorting. The few homogeneously YFP-expressing CK5-trace organoids were small and contained both CK5-expressing and non-expressing cells (Fig. 3c,d).
In contrast, YFP-positive cells from tamoxifen-induced CK8-CreERT2; R26R-YFP mice (CK8-trace) or CK18-CreERT2; R26R-YFP mice (CK18-trace) gave rise to hollow organoids with large lumens (Fig. 3e,f), most of which were homogeneously YFP-positive. Interestingly, the efficiency of organoid formation by luminal CK8-trace cells (0.22%) and CK18-trace cells (0.30%) was significantly higher than that of basal CK5-trace cells (Fig. 3g; Supplementary Table 1). In addition, the efficiency of organoid formation by CK8-trace or CK18-trace cells from castrated mice was similar (0.34%), consistent with the enhanced efficiency of CARNs relative to other luminal cells in the regressed prostate (Supplementary Table 1). Thus, both basal and luminal cells can give rise to organoids, potentially explaining the heterogeneity of organoids from normal prostate epithelium (Fig. 2b,c), but luminal cells are favored for organoid formation.
Notably, luminal cells could generate basal cells in organoid culture, as CK8-trace organoids with homogeneous YFP expression contained cells expressing basal markers (CK5, p63) (Fig. 3h–m). These basal cells were typically found on the outer layer of the organoids, as for normal organoids, but displayed an irregular morphology that might suggest incomplete basal differentiation. To assess whether luminal cells would give rise to basal cells in the presence of normal basal cells, we mixed green CK5-trace cells from CK5-CreERT2; R26R-YFP mice with red CK8-trace cells isolated from CK18-CreERT2; R26R-Tomato mice. In the resulting cultures, we found organoids with an outer layer of green cells and inner red cells (Fig. 3n), suggesting that both basal and luminal cells are preferentially lineage-restricted, consistent with lineage-tracing analyses in vivo13, 37, 38.
We further investigated the properties of luminal-derived organoids generated from lineage-marked CK18-CreERT2; R26R-Tomato mice (CK18-trace). These organoids could be serially passaged at least 9 times without apparent loss of viability (Fig. 3o,p), suggesting that the normal luminal compartment contains a stem/progenitor population that can propagate organoids in culture. Moreover, following androgen-deprivation after passaging, these luminal-derived organoids were decreased in size and lacked nuclear AR expression (Fig. 3q–s). Thus, lineage-marked luminal cells generate organoids that recapitulate key properties of organoids cultured from the bulk prostate epithelium.
Establishment of tumor organoids from single transformed CARNs
Since CARNs are a cell of origin for prostate cancer in vivo25, we investigated organoid formation from CARNs that had undergone oncogenic transformation in the context of a model of aggressive lethal prostate cancer39. In particular, Nkx3.1CreERT2/+; Ptenflox/flox; KrasLSL-G12D/+; R26R-YFP/+ mice (termed NPK) were castrated and induced with tamoxifen, so that combined Pten deletion, KrasG12D activation, and YFP expression occurred specifically in CARNs (Fig. 4a). Transformed lineage-marked CARNs were isolated by flow cytometry on the basis of their YFP expression, and used for organoid culture. The resulting NPK-CARN tumor organoids grew rapidly and displayed extensive budding and branching (Fig. 4b). Notably, these NPK-CARN organoids displayed histological phenotypes resembling PIN (Fig. 4c), and contained many proliferating cells (Fig. 4d). Immunostaining of NPK-CARN organoids showed membrane-localized phospho-Akt (pAkt) (Fig. 4e), as well as patchy expression of phospho-Erk (pErk) (Fig. 4f). Consistent with a tumor phenotype, the organoids displayed strong luminal features, with relatively few cells expressing the basal markers p63 and CK5 (Fig. 4g,h); in addition, the NPK-CARN organoids showed nuclear Foxa1 expression (Fig. 4i). Importantly, the organoids displayed nuclear AR in the presence of DHT, but mostly cytoplasmic AR in the absence of DHT (Fig. 4j,k). Furthermore, these tumor organoids could be frozen and thawed, and passaged at least 10 times without apparent loss of viability. Finally, these tumor organoids could be used to generate renal grafts that displayed a high-grade PIN phenotype (Fig. 4l), and contained proliferating cells (Fig. 4m). These grafts displayed membrane-localized phospho-Akt, patchy pErk expression, and nuclear AR, and were uniformly YFP-positive (Fig. 4n–p), indicating their phenotypic similarity to donor tumors in vivo39.
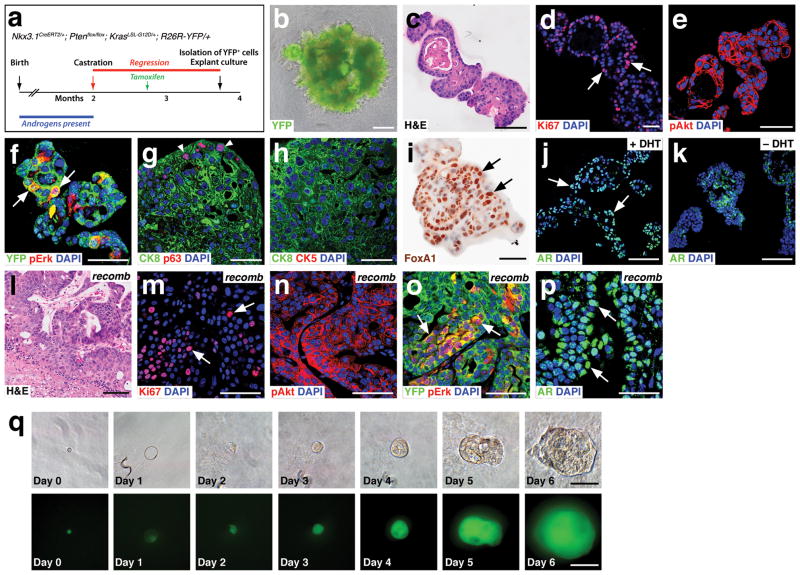
Figure 4.
Tumor organoids can be generated from single transformed CARNs. (a) Time course for generation of transformed CARNs in Nkx3.1CreERT2/+; Ptenflox/flox; KrasLSL/+; R26R-YFP/+ (NPK) mice. (b) NPK-CARN organoid shows extensive budding. (c) H&E staining of NPK-CARN organoids. (d–f) NPK-CARN organoids display Ki67 immunostaining (arrows, d), membrane localization of phospho-Akt (e), and patchy expression of phospho-Erk (arrows, f). (g,h) Luminal phenotype of NPK-CARN organoids, with limited expression of p63 (arrowheads, g) or CK5 (h). (i) Nuclear expression of Foxa1 (arrows). (j,k) AR expression is nuclear in the presence of DHT (arrows, j), but is cytoplasmic in the absence of DHT (k). (l–p) Renal grafts generated by recombination of NPK-CARN organoids with rat embryonic urogenital mesenchyme display high-grade PIN/carcinoma histological phenotypes (l), abundant Ki67 immunostaining (arrows, m), membrane-localized pAkt (n), patchy pErk (arrows, o), and nuclear AR (arrows, p). (q) Generation of organoids from single NPK CARNs. Time course of paired images shown under bright-field (top) and epifluorescent (bottom) illumination shows organoid growth from isolated single NPK CARN. Scale bars in q correspond to 25 microns, in d–h,m–p to 50 microns, and in b,c,i–l to 100 microns.
Given the tumor phenotype of NPK-CARN organoids, we investigated whether organoids could be derived from single transformed CARNs. We used flow-sorting to isolate transformed YFP-positive cells from Nkx3.1CreERT2/+; Ptenflox/flox; KrasLSL-G12D/+; R26R-YFP/+ mice that were castrated and induced with tamoxifen, and mouth-pipetted individual cells into a 96-well plate. We found that 6/80 (7.5%) of single transformed CARNs could form organoids after ten days of culture (Fig. 4q). Thus, single NPK-CARNs can initiate organoid formation at a frequency significantly higher than that of untransformed CARNs.
Tumor organoids can model prostate cancer phenotypes in culture
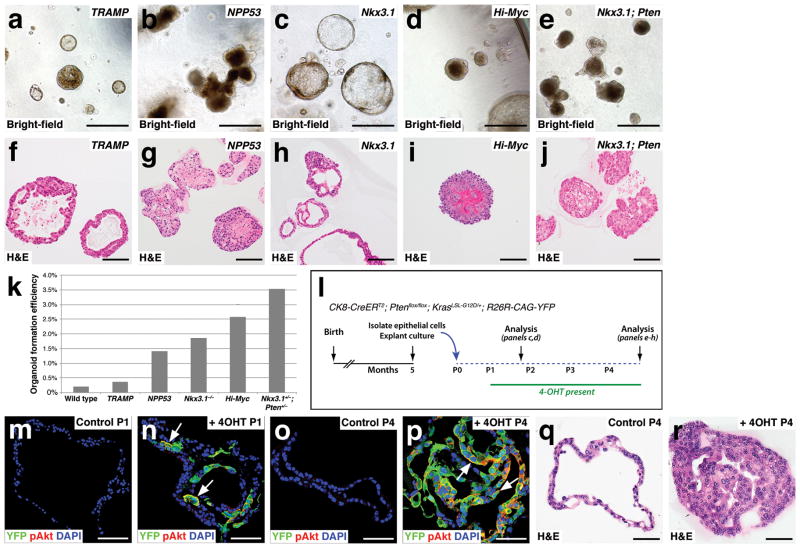
These findings suggest that organoid culture might represent a suitable system for modeling of tumor phenotypes and drug treatment responses. To test this idea, we first investigated whether tumor organoids could be generated from a range of well-studied mouse models of prostate cancer, namely: 1) Nkx3.1−/− null mutants40, 41; 2) Nkx3.1+/−; Pten+/− double mutants42; 3) TRAMP transgenic mice43, 44; 4) Hi-Myc transgenic mice45; and 5) tamoxifen-induced Nkx3.1CreERT2/+; Ptenflox/flox; p53flox/flox (NPP53) mice46 (Fig. 5a–j). Interestingly, many of these organoids displayed filled morphologies consistent with oncogenic transformation; in contrast, the Nkx3.1−/− organoids displayed a more normal morphology, consistent with the low-grade PIN phenotype of Nkx3.1 mutant mice41, 47. Moreover, all of these mouse models displayed significantly enhanced efficiencies of organoid formation (Fig. 5k).
Figure 5.
Modeling tumor phenotypes in organoid culture. (a–j) Formation of organoids from mouse models of prostate cancer, shown in bright-field (a–e) and H&E stained sections (f–j). (a,f) Organoids generated from TRAMP mice at 22 weeks of age. (b,g) Organoids from Nkx3.1CreERT2/+; Ptenflox/flox; p53flox/flox (NPP53) mice induced with tamoxifen at two months of age and assayed at 10 months. (c,h) Organoids from Nkx3.1− − null mutant mice at 14 months of age. (d,i) Organoids from Hi-Myc transgenic mice at 9 months. (e,j) Organoids from Nkx3.1+/−; Pten+/− mice at 10 months. (k) Organoid formation efficiency from the indicated mouse models (data are from 10 technical replicates). (l–r) Induction of tumor phenotypes in culture by tamoxifen treatment of organoids derived from CK8-CreERT2; Ptenflox/flox; KrasLSL-G12D/+; R26R-CAG-YFP mice. (l) Time course of induction experiment. (m–p) Immunostaining for YFP and pAkt in control untreated organoids (m,o) or 4-hydroxy-tamoxifen (4OHT) treated organoids (n,p) at passage 1 (m,n) and at passage 4 (o,p). (q,r) H&E staining of control (q) and 4OHT-treated organoids (r) at passage 4. Scale bars in a–j correspond to 100 microns, and in m–r to 50 microns.
We also examined whether organoid culture could be used for the rapid induction of tumor phenotypes, using tamoxifen-inducible organoids from CK8-CreERT2; Ptenflox/flox; KrasLSL-G12D/+; R26R-CAG-YFP mice (Fig. 5l). Alhough these organoids had normal phenotypes, they displayed YFP expression and membrane-localized phospho-Akt after induction in culture with 4-hydroxy-tamoxifen (4-OHT) (Fig. 5m,n). Following serial passaging in the absence of 4-OHT, the control organoids retained a hollow morphology without any detectable YFP expression. In contrast, in the presence of 4-OHT, the organoids were mostly YFP and pAkt-positive, and displayed PIN-like phenotypes (Fig. 5o–r).
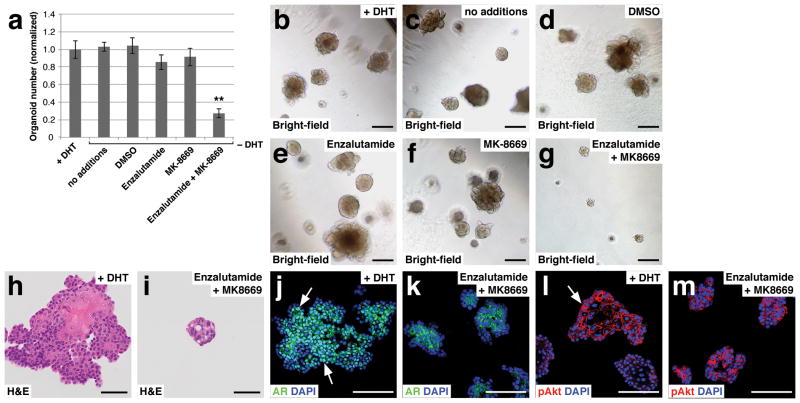
We next determined whether tumor organoids could be utilized to assess drug response, using organoids from Nkx3.1CreERT2/+; Ptenflox/flox; R26R-YFP/+ (NP) mice, which were previously used to analyze therapeutic response in vivo48. Although NP mice initially form castration-sensitive prostate tumors, they eventually develop castration-resistant disease that is sensitive to combined treatment with the Akt inhibitor MK-2206 and the mTOR inhibitor MK-8669 (ridaforolimus)48. To assess therapeutic response, we isolated YFP-positive prostate cells from tamoxifen-induced NP mice for organoid culture, and subsequently dissociated organoids at the third passage to single cell suspensions, followed by plating at 1,000 cells/well embedded within Matrigel/culture medium. Control cultures were established in the presence of DHT, while treatment cultures were established without DHT. Treatment with the DMSO solvent control had no effect, as expected, while either the AR antagonist enzalutamide or MK-8669 had minimal effects on organoid formation (Fig. 6a–f,h). In contrast, combined treatment with enzalutamide and MK-8669 inhibited organoid formation (Fig. 6a,g,i), consistent with the known synergistic activities of AR and PI3K signaling in human prostate cancer49. Interestingly, these effects were not simply due to inhibition of AR and PI3K pathway activities, as combined treatment with enzalutamide and MK-8669 could greatly reduce nuclear AR expression (Fig. 6j,k), but had no effect on phospho-Akt (Fig. 6l,m).
Figure 6.
Modeling drug treatment response in organoid culture. (a) Efficiency of organoid formation using organoids from Nkx3.1CreERT2/+; Ptenflox/flox; R26R-YFP/+ (NP) mice. Passaged organoids were treated with the indicated compounds (n=3 samples analyzed per treatment condition); source data are provided in Supplementary Table 1. (b–g) Bright-field images of treated NP organoids. (h,i) H&E sections from control +DHT organoids (h) and enzalutamide + MK8669 treated organoids (i). (j,k) AR expression in control +DHT organoids (arrows, j) and enzalutamide treated organoids (k). (l,m) pAkt expression in control +DHT organoids (arrow, l) and enzalutamide + MK8669 treated organoids (m). Scale bars in h–m correspond to 50 microns, and in b–g correspond to 100 microns. Error bars represent one standard deviation; ** p < 0.01.
Culture of human prostate organoids
Finally, we examined whether organoids could be established from human prostate tissue and cell lines. Therefore, we obtained tissue samples from three radical prostatectomies, confirmed that they contained benign glands, and isolated epithelial cells by flow-sorting for EpCAM and E-cadherin. All three patient-derived samples could establish organoids over the course of three weeks and could be passaged successfully, with organoids displaying cystic morphologies and containing proliferative cells (Supplementary Fig. 3a–c). These organoids displayed outer p63+CK8+ double-positive intermediate cells, and inner CK8+ luminal cells (Supplementary Fig. 3d), with nearly all of the cells expressing AR and CK18 (Supplementary Fig. 3e). Thus, benign human organoids resemble normal mouse organoids, except that outer cells mostly co-express basal and luminal markers.
We also examined whether organoid culture could be used to propagate the VCaP human prostate cancer cell line, which has a luminal phenotype50. VCaP organoids displayed a relatively solid morphology, contained proliferating cells, and were successfully passaged (Supplementary Fig. 3f–h). Notably, VCaP organoids expressed luminal markers including AR, but not the basal marker p63 (Supplementary Fig. 3i,j). Consequently, our organoid culture conditions could readily maintain the VCaP luminal phenotype, and may therefore be suitable for studies of human prostate cancer.
Discussion
The establishment of self-organizing organoids in ex vivo culture has become an emerging paradigm for the study of tissue stem cells51. We have shown that organoids derived from normal mouse prostate can self-renew, generate differentiated basal and luminal cells, and display long-term expansion of prostate epithelial progenitors for at least 13 passages. In addition, luminal cells are favored for organoid formation, and at least some luminal cells display bipotentiality in culture. Moreover, organoids can reconstitute either normal or transformed prostate tissue in renal grafts, depending upon the starting material. These findings indicate that prostate organoids represent an excellent system for investigating prostate biology.
AR signaling represents a central theme in studies of prostate development and cancer. Although there have been many efforts to develop culture systems for prostate epithelium, functional AR activity has not been unambiguously demonstrated in previous studies27, 29, 52–54. For instance, prostaspheres exhibit low or absent AR expression in the presence of DHT6, 9, while other spheroid culture methods show nuclear AR but not alterations of growth or AR-regulated gene expression after androgen withdrawal27, 54. In prostate organoids, however, AR protein is localized to the nucleus in the presence of DHT, while DHT withdrawal affects organoid growth, AR subcellular localization, and expression of AR-regulated genes. These responses to androgen withdrawal suggest that molecular mechanisms of castration-resistance in prostate tumors can be effectively investigated in organoid culture.
Interestingly, despite the importance of epithelial-mesenchymal interactions in prostate organogenesis and regeneration55, 56, much of the stromal requirement for prostate epithelial self-renewal and differentiation can apparently be replaced by soluble factors in the presence of extracellular matrix components found in Matrigel, such as collagen IV and laminin. Furthermore, prostate organoids generated from normal tissue appear to have unlimited potential for expansion of epithelial progenitors, similar to organoids established from other tissue types. Since prostate epithelium in vivo is generally quiescent13, our organoid culture conditions may contain potent proliferative signals and/or lack anti-proliferative signals derived from the adult stroma in vivo. In this regard, we note that our culture conditions are distinct from conditions employed in other organoid studies16, 22, 23, 57 that utilize defined media containing EGF, the BMP inhibitor Noggin, and the canonical Wnt pathway activator R-spondin, in contrast with our serum-containing media. Future analyses may yield insights into improved culture protocols, and optimization through approaches such as epithelial-stromal co-culture.
Our studies provide important insights into luminal progenitors in the prostate epithelium. Although lineage-tracing studies have reported that luminal cells in the hormonally-intact prostate epithelium do not display bipotentiality in vivo37, 38, 58, luminal cells can generate basal cells in organoid culture. This bipotentiality resembles that of CARNs in the regressed (androgen-deprived) epithelium during prostate regeneration. Notably, CARNs have a six-fold higher efficiency of organoid formation than that of non-CARNs. However, since CARNs represent less than 1% of epithelial cells in the regressed prostate25, a substantial proportion of organoid-forming ability within the regressed epithelium appears to arise from cells that are not CARNs. One likely interpretation is that luminal progenitors distinct from CARNs exist within the regressed prostate epithelium, and perhaps in the hormonally-intact epithelium as well. Another, non-mutually exclusive possibility is that some prostate luminal progenitors are lineage-restricted in vivo, but can display plasticity in culture, similar to prostate basal cells13.
Since luminal cells are favored for organoid formation, our culture conditions should be suitable for analyses of prostate tumor initiation and progression, as prostate adenocarcinoma has a luminal phenotype. Indeed, prostate tumor organoids can be established from genetically-engineered mouse models ranging from relatively indolent (Nkx3.1 null) to highly aggressive (Hi-Myc, NPP53). Moreover, tumor phenotypes can be experimentally induced in phenotypically normal organoids in culture, indicating that stromal cells are not required for oncogenic transformation. The ability to passage organoids as single cells suggests that manipulations such as lentiviral infection and CRISPR/Cas9 targeting should be feasible for genetic-engineering of tumor phenotypes in vitro, as shown for intestinal organoids59, 60. Furthermore, the ability to recapitulate treatment responses observed in human prostate cancer49 suggests that organoid culture can be used for drug screens and mechanistic studies of therapeutic response and resistance61, 62.
Finally, we have shown that organoids can be established from benign human prostate tissue as well as a luminal human prostate cancer cell line. Given the presence of intermediate cells in the benign human organoids, further optimization of our culture conditions may be advantageous. Nonetheless, a logical next step is to establish organoid cultures from human prostate tumor samples. Organoid culture may provide an alternative to tissue slice cultures63, which are short-lived and display varying androgen responsiveness, and to patient-derived xenografts64, 65, which are laborious and require large numbers of immunodeficient mice. In particular, patient-derived organoids established from primary tumors or metastases might be suitable for generation of a cryopreserved tumor organoid bank, and could potentially be used for prospective drug screening. Therefore, the continuing development of organoid culture systems may ultimately lead to advances in personalized medicine.
Methods
Mouse strains and genotyping
The Nkx3.1CreERT2 allele (Nkx3-1tm4(cre/ERT2)Mms) has been previously described25. The CK8-CreERT2 (Tg(Krt8-cre/ERT2)17Blpn/J, #017947)34, CK18-CreERT2 (Tg(KRT18-cre/ERT2)23Blpn/J, #017948)66, R26R-YFP (B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J, #006148)33, R26R-CAG-YFP (B6.Cg-Gt(ROSA)26Sortm3(CAG-EYFP)Hze/J, #007903)36, R26R-Tomato (B6;129S6-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J, #007908)36, and conditional Ptenflox (B6.129S4-Ptentm1Hwu/J, #006440)67 strains were obtained from the Jackson Laboratory Induced Mutant Resource. The inducible KrasLSL-G12D (B6.129-Krastm4Tyj/Nci)68 allele was obtained from the National Cancer Institute Mouse Models of Human Cancer Consortium Repository. CK5-CreERT2 (Krt5tm1.1(cre/ERT2)Blh) mice were generously provided by Brigid Hogan69. Animals were maintained on a congenic C57BL/6N background. Genotyping was performed using the primers listed in Supplementary Table 3.
For lineage-marking of CARNs, Nkx3.1CreERT2/+; R26R-YFP/+ males were castrated after 8 weeks of age and allowed to regress for 4 weeks, then treated with tamoxifen (Sigma) (9 mg/40 g body weight in corn oil) by daily oral gavage for 4 consecutive days, followed by a chase period as previously described25. For lineage-marking with the CK5-CreERT2, CK8-CreERT2, and CK18-CreERT2 drivers, 8 week old hormonally-intact males were treated with tamoxifen as previously described13. To generate androgen-deprived males with lineage-marked cells using CK8-CreERT2 and CK18-CreERT2 drivers, tamoxifen-treated animals were castrated and allowed to regress for 2–3 weeks. Lineage marking efficiencies were calculated using sections from anterior prostate lobes. No statistical methods were used to pre-determine sample size, and experiments were not randomized; investigators were not blinded to allocation during experiments and outcome assessment. All animal experiments received approval from the Institutional Animal Care and Use Committee at Columbia University Medical Center.
Sample acquisition for human organoids
Benign human organoids were derived from radical prostatectomy samples obtained from patients undergoing surgery at Columbia University Medical Center. All patients gave informed consent under the auspices of an Institutional Review Board approved protocol. Candidate benign regions were dissected and transported to the laboratory in DMEM/F12 (Gibco #10565) supplemented with 5% FBS for tissue dissociation. Benign pathology was initially confirmed by H&E analyses of adjacent rapid frozen sections, and was further confirmed by immunostaining of paraffin sections from the primary sample used for organoid establishment for p63, high-molecular weight cytokeratins, and α-methyl acyl coenzyme-A racemase (AMACR) using the PIN-4 cocktail70 (Biocare Medical #PPM 225 DSAA). The VCaP cell line was purchased from the American Type Culture Collection (CRL-2876).
Tissue dissociation and isolation of prostate epithelial cells
For tissue dissociation, dissected mouse prostate tissue (all lobes combined) from male mice at two to six months of age (five to fourteen months for tumor models) were dissected away from other urogenital tissues in cold phosphate buffered saline (PBS) and minced with scissors. Human tissue was prepared by mincing with scalpels and washing three times in PBS with 4 mg/ml Gentamycin (Gibco #15750-060). Prostate tissues were then incubated in 1.5 ml of DMEM/F12 (Gibco #10565), supplemented with 5% FBS and 1:10 dilution of collagenase/hyaluronidase (STEMCELL Technologies #07912) at 37°C for 3 hrs. Dissociated tissues were then spun down at 350 g for 5 min, and resuspended in 1.5 ml of ice-cold 0.25% trypsin-EDTA (STEMCELL Technologies #07901), followed by incubation at 4°C for 1 hr. Trypsinization was then stopped by addition of 3 ml Modified Hank’s Balanced Salt Solution (HBSS) (STEMCELL Technologies #37150) medium supplemented with 2% FBS, followed by centrifugation at 350 g. The cell pellet was resuspended with 1 ml pre-warmed 5 mg/ml dispase (STEMCELL Technologies #07913) supplemented with 1:10 dilution of 1 mg/ml DNase I (STEMCELL Technologies #07900). The sample was triturated vigorously for 1 min, followed by addition of 5 ml HBSS/2% FBS to neutralize dispase activity, and passed through a 40 μm cell strainer (Corning #352340). Dissociated cells were spun down again and resuspended in HBSS/2% FBS. Dissociation of human tissue was performed using the same protocol with 10-fold reagent volume and overnight digestion in collagenase/hyaluronidase solution.
Flow cytometry
For isolation of normal and transformed prostate epithelial cells, single-cell suspensions were stained using fluorescent-tagged EpCAM (BioLegend #118214 for mouse and #324208 for human) and E-cadherin (eBioscience #46-3249-82) antibodies on ice for 25 min. The stained cells were spun down, and the cell pellet washed with HBSS/2% FBS, followed by resuspension in HBSS/2% FBS with 10 μM Y-27632 (ROCK inhibitor; STEMCELL Technologies #07171) and a 1:1,000 dilution of 0.5 mg/ml DAPI to exclude dead cells during sorting. For flow cytometry, unstained cells as well as cells stained with fluorescent-tagged EpCAM or E-cadherin were used for compensation. Both side-scatter pulse width (SSC-W) vs. area (SSC-A) and forward side-scatter pulse area (FSC-A) vs. heights (FSC-H) were used to isolate single dissociated cells. For normal prostate epithelium, cells expressing either EpCAM and/or E-cadherin were isolated. For isolation of lineage-marked CARNs and transformed CARNs, as well as lineage-marked basal or luminal populations, cells were sorted based on their YFP or Tomato expression; non-YFP expressing cells were obtained by sorting EpCAM and/or E-cadherin positive but YFP-negative cells. Sorted cells were plated in low-attachment 96-well plates at densities ranging from 100 (for CARNs) to 10000 cells/well. For single cell experiments, sorted YFP-positive cells were picked by mouth-pipetting using an inverted microscope, followed by re-plating in wells of 96-well low attachment plates.
Organoid culture
We used two methods for three-dimensional culture of prostate organoids from isolated prostate epithelial cells, corresponding to flotation on top of Matrigel or embedding within Matrigel; the embedding method was used for drug treatment experiments and is described below. For the floating method, prostate epithelial cells were resuspended in prostate organoid culture medium, consisting of: hepatocyte medium supplemented with 10 ng/ml epidermal growth factor (EGF) (Corning #355056), 10 μM Y-27632 (STEMCELL Technologies #07171), 1x glutamax (Gibco #35050), 5% Matrigel (Corning #354234), and 5% charcoal-stripped FBS (Gibco #12676), which had been heat-inactivated at 55°C for 1 hr. After resuspension in prostate organoid medium, 100 – 10,000 dissociated cells were plated into wells of ultra low-attachment 96 well plates (Corning #3474) in the presence of 100 nM DHT for mouse or 10 nM DHT for human (Sigma A-8380). 100 μl of fresh organoid medium was added to the wells every four days, and the medium changed every 12 days for up to one month.
For serial passaging experiments, organoids were passaged at a 1:4 dilution every 1–2 weeks with 0.25% trypsin for 5 minutes at 37°C, followed by mechanical dissociation to nearly single-cell suspensions. Organoids were frozen in complete media with 50% FBS and 10% DMSO. The efficiency of organoid formation was calculated by averaging the number of organoids visible in each well after 7 days of growth using a 10x objective. For statistical analyses, efficiency percentages were arcsin converted to perform unpaired two-tailed Student’s t-tests.
For analyses of androgen withdrawal, organoids were passaged and then cultured for 7–10 days in culture medium in the presence or absence of DHT. For induction of Cre recombinase activity in culture, epithelial cells from un-induced CK8-CreERT2; Ptenflox/flox; KrasLSL-G12D/+; R26R-CAG-YFP mice were sorted based on EpCAM and E-cadherin expression, and cultured until organoid formation was evident. The resulting organoids were passaged, followed by addition of 1 μM 4-OHT on the day after passaging to induce Cre recombination.
A detailed protocol for organoid establishment and culture will be provided on Nature Protocol Exchange immediately following publication.
Drug treatments
The embedding method was used to culture organoids for drug treatment experiments. Organoids were dissociated by digestion with 0.25% trypsin-EDTA (STEMCELL Technologies #07901) and passed through a 40 μm cell strainer. 40 μl of the resulting cell suspension containing 500 – 3,000 dissociated cells were mixed with 60 μl of Matrigel, and the mixture pipetted around the rim of wells in a 24 well plate. The mixture was allowed to solidify for 30 minutes at 37°C, prior to addition of 400 μl organoid culture medium to each well, with or without supplementation with 100 nm DHT in the presence or absence of drugs. The culture medium was changed every other day, and organoids were counted after 8 days. Drugs were dissolved in DMSO to generate a final concentration of 0.1% in all drug-treated groups. Drug concentrations were as follows: 100 nM DHT, no additions, 0.1% DMSO, 10 μM enzalutamide (provided by Charles Sawyers, Memorial Sloan-Kettering Cancer Center), 1 nM MK-8669 (provided by Cory Abate-Shen, Columbia University Medical Center).
Tissue recombination and renal grafting
For tissue recombination, organoids from one well of a 96-well plate were mixed with 250,000 dissociated rat urogenital mesenchyme cells from embryonic day 18.5 rat embryos and resuspended in 12 μl of 9:1 collagen:setting buffer solution (10x Earle’s Balanced Salt Solution (Life Technologies), 0.2 M NaHCO3 and 50 mM NaOH). The recombinants were cultured overnight in DMEM with 10% FBS and 100 nM DHT, followed by grafting under the kidney capsules of male NOD.Cg-Prkdcscid Il2rgtm1Sug/JicTac (NOG) mice (Taconic). Renal grafts were harvested for analysis at 8 weeks after grafting.
Histology and immunostaining
Tissues were processed for cryosections or paraffin sectioning using standard protocols. For cryosections, organoids and tissues were fixed in 4% paraformaldehyde in PBS at 4°C for 1 hr, placed in 30% sucrose in PBS, and embedded in OCT (Tissue-Tek). For paraffin sectioning, organoids were fixed in 10% formalin for 1 hr and placed in Histogel (Thermo Scientific) prior to tissue processing and embedding.
For immunostaining, sections underwent antigen-retrieval by boiling in citrate acid-based antigen unmasking solution (Vector Labs) for 10 min. Primary antibodies were applied to sections and incubated at 4°C overnight in a humidified chamber. Alexa Fluors (Life Technologies) were used for secondary antibodies. Tyramide amplification (Life Technologies) or ABC Elite (Vector Labs) kits were used for signal detection. For lineage-tracing experiments, analysis of marked basal or luminal cells was performed by manual counting of cells from confocal images taken with a 40x objective. Details on antibodies used are provided in Supplementary Table 4.
Quantitative real-time PCR analysis
For RNA extraction, 4–6 wells of organoids were pooled, pelleted, and dissolved in Trizol reagent prior to processing by the MagMAX 96 Total RNA Isolation Kit (Ambion, Life Technologies). 300–500ng of RNA was used for cDNA synthesis using the Superscript First Strand Synthesis System (Invitrogen). Quantitative real-time PCR was carried out using SYBR green master mix reagent (QIAGEN) in the Realplex2 instrument (Eppendorf). cDNA samples were diluted 1:5 to 1:10 for all analyses, which were performed in triplicate. Expression values were obtained using the ΔΔCT method and normalized to GAPDH expression; average values are shown as the mean ± standard deviation (SD). Primer sequences are provided in Supplementary Table 3.
Repeatability of experiments
For histological and immunofluorescence analyses of organoids and tissue recombination experiments, representative staining patterns were confirmed in at least 3 samples from at least 2 independent experiments. All DHT withdrawal experiments were repeated at least twice. Data shown for quantitative real-time PCR analysis are from a single experiment that was representative of 2 independent experiments. The drug treatment experiment was repeated at a different passage and gave similar results and statistical significance.
Supplementary Material
Acknowledgments
We thank Marianna Kruithof-de Julio, Maher Hanoun, and Paul Frenette for initial discussions about organoid culture, Charles Sawyers and Cory Abate-Shen for providing pathway inhibitors, Chenhong Liu and the HICCC Flow Cytometry Shared Resource for flow-sorting, Dajiang Sun for assistance with specimen acquisition, the HICCC Molecular Pathology Shared Resource for organoid sectioning and H&E staining, Flaminia Talos for helpful comments on the culture protocol, David-Emlyn Parfitt for assistance with cell-picking, and Cory Abate-Shen and Flaminia Talos for insightful discussions and comments on the manuscript.
This work was supported by post-doctoral fellowships from the DOD Prostate Cancer Research Program (C.W.C., M.S., and R.T.), by a Residency Research Award from the Urology Care Foundation (L.J.B.), and by grants from the National Institutes of Health (M.M.S.).
Footnotes
Author contributions
C.W.C. and M.L. developed the organoid culture protocol, C.W.C. and M.S. performed analyses of CARN-derived and normal prostate organoids, M.S. performed lineage-tracing studies, M.L. performed analyses of transformed CARN organoids and drug response, R.T. performed analyses of benign human prostate organoids, M.S. and C.W.C. analyzed VCaP organoids, L.J.B. performed studies of tumor organoids from mouse models, and S.J.B. assisted with single-cell experiments and tissue grafts, K.K.B. and J.M.M. provided surgical specimens, and H.H. performed pathological analyses. C.W.C., M.S., M.L., R.T., L.J.B., M.C.B, H.H., and M.M.S. designed experiments, analyzed data, and wrote the manuscript.
References
- 1.Peehl DM. Primary cell cultures as models of prostate cancer development. Endocr Relat Cancer. 2005;12:19–47. doi: 10.1677/erc.1.00795. [DOI] [PubMed] [Google Scholar]
- 2.Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 2010;24:1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grisanzio C, Signoretti S. p63 in prostate biology and pathology. J Cell Biochem. 2008;103:1354–1368. doi: 10.1002/jcb.21555. [DOI] [PubMed] [Google Scholar]
- 4.Humphrey PA. Diagnosis of adenocarcinoma in prostate needle biopsy tissue. J Clin Pathol. 2007;60:35–42. doi: 10.1136/jcp.2005.036442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawson DA, Xin L, Lukacs RU, Cheng D, Witte ON. Isolation and functional characterization of murine prostate stem cells. Proc Natl Acad Sci USA. 2007;104:181–186. doi: 10.1073/pnas.0609684104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xin L, Lukacs RU, Lawson DA, Cheng D, Witte ON. Self-renewal and multilineage differentiation in vitro from murine prostate stem cells. Stem Cells. 2007;25:2760–2769. doi: 10.1634/stemcells.2007-0355. [DOI] [PubMed] [Google Scholar]
- 7.Shi X, Gipp J, Bushman W. Anchorage-independent culture maintains prostate stem cells. Dev Biol. 2007;312:396–406. doi: 10.1016/j.ydbio.2007.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garraway IP, et al. Human prostate sphere-forming cells represent a subset of basal epithelial cells capable of glandular regeneration in vivo. Prostate. 2010;70:491–501. doi: 10.1002/pros.21083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lukacs RU, Goldstein AS, Lawson DA, Cheng D, Witte ON. Isolation, cultivation and characterization of adult murine prostate stem cells. Nat Protoc. 2010;5:702–713. doi: 10.1038/nprot.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo C, Zhang B, Garraway IP. Isolation and characterization of human prostate stem/progenitor cells. Methods Mol Biol. 2012;879:315–326. doi: 10.1007/978-1-61779-815-3_18. [DOI] [PubMed] [Google Scholar]
- 11.Goldstein AS, et al. Trop2 identifies a subpopulation of murine and human prostate basal cells with stem cell characteristics. Proc Natl Acad Sci USA. 2008;105:20882–20887. doi: 10.1073/pnas.0811411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawson DA, et al. Basal epithelial stem cells are efficient targets for prostate cancer initiation. Proc Natl Acad Sci USA. 2010;107:2610–2615. doi: 10.1073/pnas.0913873107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang ZA, et al. Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell-of-origin model for prostate cancer heterogeneity. Nature Cell Biology. 2013;15:274–283. doi: 10.1038/ncb2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 15.Sato T, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 17.Yui S, et al. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5(+) stem cell. Nat Med. 2012;18:618–623. doi: 10.1038/nm.2695. [DOI] [PubMed] [Google Scholar]
- 18.Sato T, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 19.Kondo J, et al. Retaining cell-cell contact enables preparation and culture of spheroids composed of pure primary cancer cells from colorectal cancer. Proc Natl Acad Sci USA. 2011;108:6235–6240. doi: 10.1073/pnas.1015938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ootani A, et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat Med. 2009;15:701–706. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barker N, et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 22.Huch M, et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huch M, et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 2013;32:2708–2721. doi: 10.1038/emboj.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greggio C, et al. Artificial three-dimensional niches deconstruct pancreas development in vitro. Development. 2013;140:4452–4462. doi: 10.1242/dev.096628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, et al. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461:495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo W, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lang SH, et al. Experimental prostate epithelial morphogenesis in response to stroma and three-dimensional matrigel culture. Cell Growth Differ. 2001;12:631–640. [PubMed] [Google Scholar]
- 28.Cano P, Godoy A, Escamilla R, Dhir R, Onate SA. Stromal-epithelial cell interactions and androgen receptor-coregulator recruitment is altered in the tissue microenvironment of prostate cancer. Cancer Res. 2007;67:511–519. doi: 10.1158/0008-5472.CAN-06-1478. [DOI] [PubMed] [Google Scholar]
- 29.Liu X, et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am J Pathol. 2012;180:599–607. doi: 10.1016/j.ajpath.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, et al. ROCK inhibitor Y-27632 suppresses dissociation-induced apoptosis of murine prostate stem/progenitor cells and increases their cloning efficiency. PLoS One. 2011;6:e18271. doi: 10.1371/journal.pone.0018271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Y, et al. Revealing a core signaling regulatory mechanism for pluripotent stem cell survival and self-renewal by small molecules. Proc Natl Acad Sci USA. 2010;107:8129–8134. doi: 10.1073/pnas.1002024107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao N, et al. Forkhead box A1 regulates prostate ductal morphogenesis and promotes epithelial cell maturation. Development. 2005;132:3431–3443. doi: 10.1242/dev.01917. [DOI] [PubMed] [Google Scholar]
- 33.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Keymeulen A, et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- 35.Ousset M, et al. Multipotent and unipotent progenitors contribute to prostate postnatal development. Nat Cell Biol. 2012;14:1131–1138. doi: 10.1038/ncb2600. [DOI] [PubMed] [Google Scholar]
- 36.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature neuroscience. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi N, Zhang B, Zhang L, Ittmann M, Xin L. Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer Cell. 2012;21:253–265. doi: 10.1016/j.ccr.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu TL, et al. Conditionally ablated Pten in prostate basal cells promotes basal-to-luminal differentiation and causes invasive prostate cancer in mice. Am J Pathol. 2013;182:975–991. doi: 10.1016/j.ajpath.2012.11.025. [DOI] [PubMed] [Google Scholar]
- 39.Aytes A, et al. ETV4 promotes metastasis in response to activation of PI3-kinase and Ras signaling in a mouse model of advanced prostate cancer. Proc Natl Acad Sci USA. 2013;110:E3506–3515. doi: 10.1073/pnas.1303558110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhatia-Gaur R, et al. Roles for Nkx3.1 in prostate development and cancer. Genes Dev. 1999;13:966–977. doi: 10.1101/gad.13.8.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim MJ, et al. Nkx3.1 mutant mice recapitulate early stages of prostate carcinogenesis. Cancer Res. 2002;62:2999–3004. [PubMed] [Google Scholar]
- 42.Kim MJ, et al. Cooperativity of Nkx3.1 and Pten loss of function in a mouse model of prostate carcinogenesis. Proc Natl Acad Sci USA. 2002;99:2884–2889. doi: 10.1073/pnas.042688999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greenberg NM, et al. Prostate cancer in a transgenic mouse. Proc Natl Acad Sci USA. 1995;92:3439–3443. doi: 10.1073/pnas.92.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masumori N, et al. A probasin-large T antigen transgenic mouse line develops prostate adenocarcinoma and neuroendocrine carcinoma with metastatic potential. Cancer Res. 2001;61:2239–2249. [PubMed] [Google Scholar]
- 45.Ellwood-Yen K, et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4:223–238. doi: 10.1016/s1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 46.Aytes A, et al. Cross-species analysis of genome-wide regulatory networks identifies a synergistic interaction between FOXM1 and CENPF that drives prostate cancer malignancy. Cancer Cell. 2014;25:638–651. doi: 10.1016/j.ccr.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Irshad S, et al. A molecular signature predictive of indolent prostate cancer. Sci Transl Med. 2013;5:202ra122. doi: 10.1126/scitranslmed.3006408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Floc’h N, et al. Dual targeting of the Akt/mTOR signaling pathway inhibits castration-resistant prostate cancer in a genetically engineered mouse model. Cancer Res. 2012;72:4483–4493. doi: 10.1158/0008-5472.CAN-12-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carver BS, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–586. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Korenchuk S, et al. VCaP, a cell-based model system of human prostate cancer. In Vivo. 2001;15:163–168. [PubMed] [Google Scholar]
- 51.Sasai Y. Next-generation regenerative medicine: organogenesis from stem cells in 3D culture. Cell Stem Cell. 2013;12:520–530. doi: 10.1016/j.stem.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 52.McKeehan WL, Adams PS, Rosser MP. Direct mitogenic effects of insulin, epidermal growth factor, glucocorticoid, cholera toxin, unknown pituitary factors and possibly prolactin, but not androgen, on normal rat prostate epithelial cells in serum-free, primary cell culture. Cancer Res. 1984;44:1998–2010. [PubMed] [Google Scholar]
- 53.Lang SH, et al. Differentiation of prostate epithelial cell cultures by matrigel/stromal cell glandular reconstruction. In vitro cellular & developmental biology. Animal. 2006;42:273–280. doi: 10.1290/0511080.1. [DOI] [PubMed] [Google Scholar]
- 54.Lamb LE, Knudsen BS, Miranti CK. E-cadherin-mediated survival of androgen-receptor-expressing secretory prostate epithelial cells derived from a stratified in vitro differentiation model. J Cell Sci. 2010;123:266–276. doi: 10.1242/jcs.054502. [DOI] [PubMed] [Google Scholar]
- 55.Marker PC, Donjacour AA, Dahiya R, Cunha GR. Hormonal, cellular, and molecular control of prostatic development. Dev Biol. 2003;253:165–174. doi: 10.1016/s0012-1606(02)00031-3. [DOI] [PubMed] [Google Scholar]
- 56.Cunha GR. Mesenchymal-epithelial interactions: past, present, and future. Differentiation. 2008;76:578–586. doi: 10.1111/j.1432-0436.2008.00290.x. [DOI] [PubMed] [Google Scholar]
- 57.Stange DE, et al. Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell. 2013;155:357–368. doi: 10.1016/j.cell.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu J, et al. Regenerated luminal epithelial cells are derived from preexisting luminal epithelial cells in adult mouse prostate. Mol Endocrinol. 2011;25:1849–1857. doi: 10.1210/me.2011-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koo BK, et al. Controlled gene expression in primary Lgr5 organoid cultures. Nature methods. 2012;9:81–83. doi: 10.1038/nmeth.1802. [DOI] [PubMed] [Google Scholar]
- 60.Schwank G, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 61.Sachs N, Clevers H. Organoid cultures for the analysis of cancer phenotypes. Curr Opin Genet Dev. 2014;24C:68–73. doi: 10.1016/j.gde.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 62.Ranga A, Gjorevski N, Lutolf MP. Drug discovery through stem cell-based organoid models. Advanced drug delivery reviews. 2014;69–70C:19–28. doi: 10.1016/j.addr.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 63.Centenera MM, Raj GV, Knudsen KE, Tilley WD, Butler LM. Ex vivo culture of human prostate tissue and drug development. Nat Rev Urol. 2013;10:483–487. doi: 10.1038/nrurol.2013.126. [DOI] [PubMed] [Google Scholar]
- 64.Toivanen R, et al. A preclinical xenograft model identifies castration-tolerant cancer-repopulating cells in localized prostate tumors. Sci Transl Med. 2013;5:187ra171. doi: 10.1126/scitranslmed.3005688. [DOI] [PubMed] [Google Scholar]
- 65.Lin D, et al. High fidelity patient-derived xenografts for accelerating prostate cancer discovery and drug development. Cancer Res. 2014;74:1272–1283. doi: 10.1158/0008-5472.CAN-13-2921-T. [DOI] [PubMed] [Google Scholar]
- 66.Van Keymeulen A, et al. Epidermal progenitors give rise to Merkel cells during embryonic development and adult homeostasis. J Cell Biol. 2009;187:91–100. doi: 10.1083/jcb.200907080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lesche R, et al. Cre/loxP-mediated inactivation of the murine Pten tumor suppressor gene. Genesis. 2002;32:148–149. doi: 10.1002/gene.10036. [DOI] [PubMed] [Google Scholar]
- 68.Jackson EL, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rock JR, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci USA. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Giannico GA, Ross HM, Lotan T, Epstein JI. Aberrant expression of p63 in adenocarcinoma of the prostate: a radical prostatectomy study. Am J Surg Pathol. 2013;37:1401–1406. doi: 10.1097/PAS.0b013e31828d5c32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.