Abstract
In addition to the small and large ribosomal subunits, aminoacyl-tRNAs, and an mRNA, cellular protein synthesis is dependent on translation factors. The eukaryotic translation initiation factor 5A (eIF5A) and its bacterial ortholog elongation factor P (EF-P) were initially characterized based on their ability to stimulate methionyl-puromycin (Met-Pmn) synthesis, a model assay for protein synthesis; however, the function of these factors in cellular protein synthesis has been difficult to resolve. Interestingly, a conserved lysine residue in eIF5A is post-translationally modified to hypusine and the corresponding lysine residue in EF-P from at least some bacteria is modified by the addition of a βlysine moiety. In this review, we provide a summary of recent data that have identified a novel role for the translation factor eIF5A and its hypusine modification in the elongation phase of protein synthesis and more specifically in stimulating the production of proteins containing runs of consecutive proline residues.
Keywords: translation elongation, EF-P, polyproline, elongation factor, deoxyhypusine synthase (DHS), deoxyhypusine hydroxylase (DOHH)
Introduction
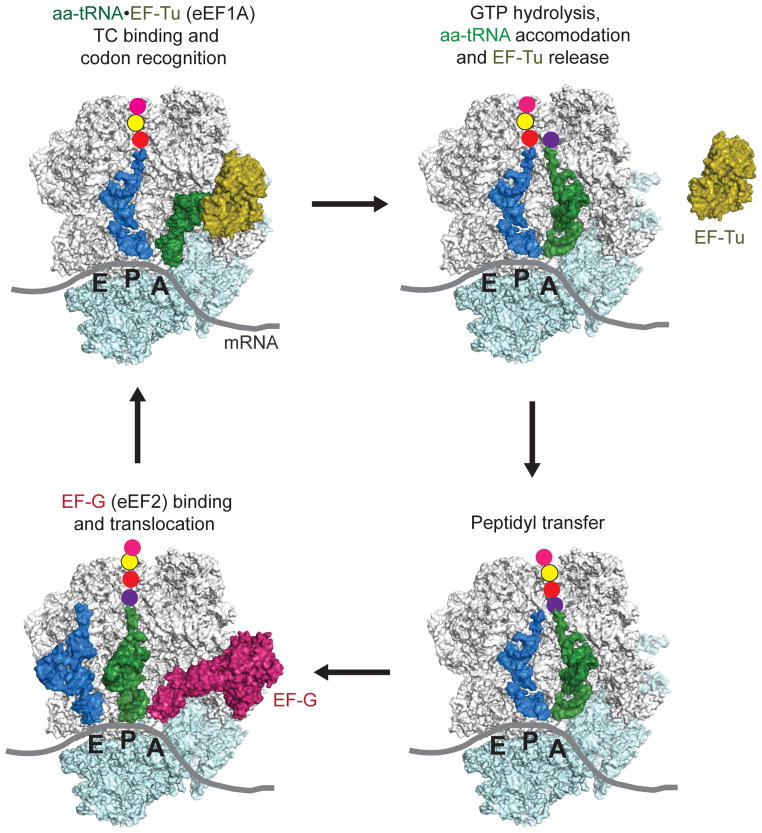
Translation factors assist all four stages of protein synthesis: initiation, elongation, termination and recycling. During the initiation phase of protein synthesis the small ribosomal subunit associates with the mRNA and locates to the start codon for the open reading frame. Upon start codon selection, the large ribosomal subunit joins and a functional ribosome is assembled. Whereas bacteria require only three translation initiation factors (IFs) (reviewed in Simonetti et al., 2009), more than ten eukaryotic translation initiation factors (eIFs) assist in binding the initiator methionyl-tRNA to the 40S subunit, binding the resulting 43S complex to an mRNA, ensuring the fidelity of start codon selection, and joining of the 60S subunit to form an elongation-competent 80S ribosome (reviewed in Hinnebusch and Lorsch, 2012). These dramatic differences in factor requirements for translation initiation in eukaryotes versus bacteria are due in part to the direct targeting of the small subunit to the translation start site in bacteria via the Shine-Delgarno sequence versus the scanning mechanism of translation initiation in eukaryotes. In contrast to the different mechanisms of translation initiation, the mechanism and factor requirements for translation elongation are well conserved between bacteria and eukaryotes (Figure 1) (Dever and Green, 2012). The elongation factor Tu (EF-Tu) in bacteria and its homolog eukaryotic elongation factor 1A (eEF1A) form ternary complexes with GTP and aminoacyl-tRNAs and then bind the aminoacyl-tRNA to the A site of the ribosome in a codon-dependent manner. The ribosome then catalyzes peptide bond formation between the peptidyl-tRNA in the P site and the aminoacyl-tRNA in the A site. Next, the factor EF-G in bacteria or eEF2 in eukaryotes promotes translocation of the tRNAs and mRNA on the ribosome resulting in a deacylated tRNA in the E site, the elongated peptidyl-tRNA in the P site and a vacant A site awaiting delivery of the next aminoacyl-tRNA in complex with EF-Tu/eEF1A (Schmeing and Ramakrishnan, 2009). When a termination codon enters the A site, release factors promote release of the completed polypeptide chain, and then ribosome recycling factor in bacteria, or structurally and mechanistically distinct factors in eukaryotes, promote dissociation of the ribosome, tRNA and mRNA (Dever and Green, 2012). Thus by interacting with ribosomes and other components of the protein synthesis machinery, translation factors play central roles in promoting the various stages of protein synthesis. In addition, some factors, rather than stimulating general protein synthesis, have specialized roles to enhance the translation of specific mRNAs or the incorporation of specific amino acids, such as selenocysteine (Lobanov et al., 2010).
Figure 1. Schematic of the translation elongation cycle.
(Top left) An EF-Tu(eukaryotic eEF1A)–GTP–aminoacyl(aa)-tRNA ternary complex (TC) binds aa-tRNA to the A site of the 70S (eukaryotic 80S) ribosome in a codon-dependent manner. (Top right) Following release of EF-Tu–GDP, the aa-tRNA is accommodated into the A site. Peptide bond formation (lower right) is followed by binding of EF-G (eEF2)–GTP (lower left), which promotes translocation of the deacylated tRNA and peptidyl tRNA into the canonical E and P sites, respectively. Following release of EF-G–GDP, the ribosome is ready for the next cycle of elongation. A color version of the figure is available online.
The protein eIF5A was initially characterized as a translation factor and stimulator of methionyl-puromycin (Met-Pmn) synthesis; however, other biochemical and genetic studies implicated eIF5A in nucleocytoplasmic transport (Bevec et al., 1996; Hofmann et al., 2001; Ruhl et al., 1993) and in mRNA stability (Schrader et al., 2006; Zuk and Jacobson, 1998). In this review we focus on the recent linkage of eIF5A to translation elongation and more specifically on its identification as a factor required to stimulate translation of proteins containing polyproline sequences. Based on the clarified role of eIF5A in translation elongation, we propose to rename the protein as a eukaryotic translation elongation factor (eEF).
The early history of eIF5A and EF-P: from discovery to hypusine
During the initial isolation of eukaryotic translation factors, model assays were used as tools to identify and isolate stimulatory factors from fractionated rabbit reticulocyte lysates. The factor IF-M2Bα, which was later renamed eIF-4D and then eIF5A (from this point onward, we use the name eIF5A), was found to enhance the yield of Met-Pmn synthesis in assays containing 80S ribosomes, the known complement of initiation factors and the trinucleotide 5′-AUG-3′ mRNA (Benne, Brown-Luedi, and Hershey, 1978; Benne and Hershey, 1978; Kemper, Berry, and Merrick, 1976; Schreier, Erni, and Staehelin, 1977). In this assay 80S initiation complexes are assembled with initiator methionyl-tRNA in the P site, and the assay monitors transfer of the methionine to puromycin, a partial mimic of aminoacyl-tRNA that binds to the ribosomal A site. Notably, eIF5A did not stimulate other model assays of translation initiation, such as binding of initiator methionyl-tRNA or mRNA to ribosomes or formation of the 80S product of translation initiation (Benne and Hershey, 1978; Schreier, Erni, and Staehelin, 1977). This inability of eIF5A to stimulate assays monitoring various steps in the translation initiation pathway, combined with its specific ability to stimulate Met-Pmn synthesis, raised the possibility that eIF5A functions late in the translation initiation pathway to facilitate formation of the first peptide bond and promote the transition from the initiation to the elongation phase of protein synthesis (Benne and Hershey, 1978). However, it is also relevant to note that Met-Pmn synthesis is a translation elongation step and that this assay also reports on elongation activity. Moreover, puromycin, an aminonucleoside antibiotic, is an imperfect substrate due to its poor positioning in the ribosome active site (Wohlgemuth et al., 2008; Youngman et al., 2004). Thus, the eIF5A stimulation of Met-Pmn synthesis could indicate roles for the factor in first peptide bond synthesis, general translation elongation, and/or the reactivity of poor substrates in the ribosome active site.
Along with its ability to stimulate Met-Pmn synthesis, eIF5A was found to stimulate the yield of polyphenylalanine (polyPhe) synthesized by 80S ribosomes at low Mg2+ concentrations (Kemper, Berry, and Merrick, 1976). When assayed at 10 mM Mg2+, poly(U)-directed synthesis of polyPhe requires only translation elongation factors; however, at low Mg2+ concentrations this assay also requires the initiation factors eIF1A, eIF2A (now called eIF2D), eIF5B, and eIF5A (Merrick, 1979), though the role of initiation factors in this assay is not clear. The 2–4–fold stimulatory activity of eIF5A in the polyPhe and Met-Pmn synthesis assays, which both monitor the formation of a peptide bond, is consistent with the notion that eIF5A functions in translation elongation. However, at odds with this interpretation, addition of eIF5A failed to stimulate assays monitoring the synthesis of globin protein chains. While this apparent lack of activity might be attributed to the presence of eIF5A in the post-ribosomal supernatant and crude aminoacyl-tRNA synthetase preps used for the globin synthesis assays (Merrick, 1979; Thomas et al., 1979), the addition of eIF5A did not stimulate the yield of globin synthesis in reconstituted assays using purified aminoacyl-tRNAs and thus lacking this proposed eIF5A contamination (Schreier, Erni, and Staehelin, 1977). Interestingly, further analysis by Schreier et al. revealed that addition of eIF5A lowered the Mg2+ optimum for globin synthesis in assays lacking spermine (Schreier, Erni, and Staehelin, 1977). Based on these findings, the authors proposed that, rather than functioning in translation initiation, eIF5A acts like polyamines and has a general stimulatory effect on protein synthesis.
Concurrent with the initial characterization of eIF5A, its bacterial ortholog EF-P was likewise identified by its biochemical activity to stimulate formylmethionyl-puromycin (fMet-Puro) peptide synthesis in an assay consisting of 70S ribosomes, [35S]fMet-tRNAfMet, mRNA, and puromycin. Based upon this initial observation, EF-P, which was predominantly isolated from the postribosomal supernatant (>90%), was proposed to be an elongation factor that functions to promote peptide bond synthesis on the ribosome (Glick and Ganoza, 1975). In a subsequent study, EF-P was found to differentially stimulate dipeptide synthesis between fMet-tRNA and cytidyl-aminoacyladenosine (C-A) analogs of the acceptor ends of various aminoacyl-tRNAs. Interestingly, the EF-P stimulatory activity was most pronounced for the A-site substrates C-A-Gly, puromycin, and C-A-Ala, leading to the hypothesis that EF-P promoted the activity of poor substrates (smaller and/or less hydrophobic side chains) for peptidyl transfer (Glick, Chladek, and Ganoza, 1979; Ganoza and Aoki, 2000).
As will be discussed in greater detail below, eIF5A is the sole cellular protein to contain the amino acid hypusine (Nε-(4-amino-2-hydroxybutyl)lysine) (Park, Cooper, and Folk, 1981). Hypusine was initially identified as a free amino acid isolated from bovine brain, and was later reported to be widely distributed in other tissues. Hypusine is highly basic and was named for its component parts: hydroxyputrescine and lysine (Shiba et al., 1971). When cultured mammalian cells were grown in the presence of [3H]spermidine, a single labeled protein was resolved by two-dimensional gel electrophoresis (Cooper, Park, and Folk, 1982; Cooper et al., 1983; Park, Cooper, and Folk, 1981). The size (~17 kDa) and pI (~5.1) of the hypusine-containing protein, as well as the direct correlation of increased hypusine labeling with increased rates of protein synthesis in mammalian cells, led to the hypothesis that hypusine plays a critical role in cellular protein synthesis. The detection of hypusine in acid hydrolysates of highly purified eIF5A confirmed its identity as the sole hypusine-containing protein (Cooper et al., 1983). Finally, cloning of eIF5A and comparison of the predicted amino acid sequence from the cDNA with the observed sequence based on Edman degradation revealed that Lys50 was the site of the hypusine modification in mammalian eIF5A (Park et al., 1986; Smit-McBride et al., 1989). Further genome sequencing efforts and spermidine labeling studies revealed that the hypusine (or at least deoxyhypusine) modification is conserved in all eukaryotic eIF5A and archaeal aIF5A proteins (Gordon et al., 1987; Bartig et al., 1992; Bartig, Schumann, and Klink, 1990; Park, Lee, and Joe, 1997). Moreover, despite the absence of hypusine in bacteria, amino acid sequence comparisons revealed that bacterial EF-P, eukaryotic eIF5A, and archaeal aIF5A are homologs (Kyrpides and Woese, 1998). Given their common ability to stimulate Met-Pmn synthesis assays, it was reasonably proposed that eIF5A and EF-P perform similar functions in cellular protein synthesis (Saini et al., 2009).
Structural analysis and ribosome-binding of eIF5A and EF-P
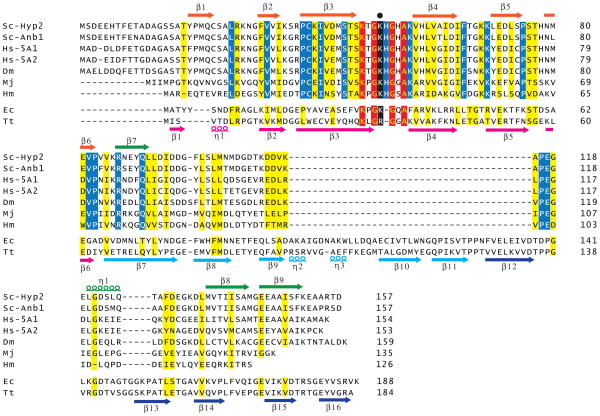
Both yeast and humans express two eIF5A proteins, and the two forms of eIF5A within each organism share 80–90% amino acid sequence identify. Moreover, the eIF5A proteins in yeast, humans, and fruit fly consist of 153–159 residues, and they share >50% amino acid sequence identity with the greatest concentration of identical residues near the site of hypusine modification (Figure 2). The archaeal homolog of eIF5A, called aIF5A, shows strong similarity to the eukaryotic factor, though archaeal aIF5A is truncated by around 10 residues at the N-terminus compared to the eukaryotic factor. Bacterial EF-P is around 30 residues longer than eIF5A (Figure 2), and based on multiple sequence alignments these extra residues are inserted in the C-terminal half of the protein. It is noteworthy that the sequence flanking the site of hypusine/deoxyhypusine modification K-x-G-Hyp-H-G-x-A-K is conserved among eukaryotes and archaea. Four of the residues in this motif (Lys, both Gly, and Ala) are also conserved in EF-P. However, the His residue is not present in EF-P, and the Lys residue that is modified to hypusine is conserved in EF-P from E. coli but substituted by Arg in T. thermophilus EF-P (Figure 2).
Figure 2. Sequence alignment of eIF5A and EF-P.
Multiple sequence alignment of yeast (Saccharomyces cerevisiae, Sc) Hyp2 (Tif51A; eIF5A1) and Anb1 (Tif51B, eIF5A2), human (Hs) eIF5A1 and eIF5A2, fly (Drosophila melanogaster, Dm) eIF5A, archaeal (Methanococcus jannaschii, Mj, and Haloarcula marismortui, Hm) aIF5A, and bacterial (Escherichia coli, Ec, and Thermus thermophilus, Tt) EF-P was performed using Clustal X (v2.0). Residues in red are identical in all eIF5A and EF-P proteins; residues in blue are identical only in eIF5A and aIF5A; and residues in yellow are conserved in eIF5A and EF-P. Secondary structure elements are shown at the top (eIF5A) and bottom (EF-P) of the sequences, and colored based on the protein domain and scheme described in Figure 3. The lysine residue that is modified to hypusine in eIF5A and aIF5A and β-lysinylated in E. coli EF-P is colored black and denoted by the black dot above the alignment. A color version of the figure is available online.
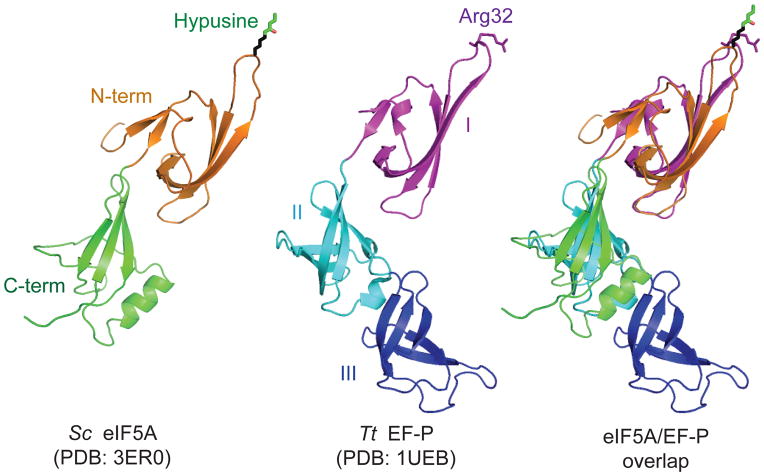
The crystal structures of eIF5A and EF-P from a variety of eukaryotes, archaea and bacteria have been solved and the proteins show significant structural similarity. The aIF5A from M. jannaschii (Kim et al., 1998), P. aerophilum (Peat et al., 1998), and P. horikoshii (Yao et al., 2003), and the eIF5A from human (Tong et al., 2009) and yeast (pdb code 3ER0) fold into a two domain structure with residues ~1–83 forming the N-terminal domain. The N-terminal domain consists of six β-strands that fold into a partially open β-barrel (Kim et al., 1998), while the C-terminal domain, consisting of 3–5 β-strands and 0–2 α-helices, resembles an OB-fold type of β-barrel domain (Kim et al., 1998; Peat et al., 1998; Yao et al., 2003) (Figure 3). In all structures the highly conserved residues flanking the site of hypusine modification are in a long unstructured loop (the hypusine loop) between strands β3 and β4 that protrudes from the N-terminal domain (Figure 3).
Figure 3. Comparison of eIF5A and EF-P structures.
Structures of Saccharomyces cerevisiae eIF5A (Left, PDB: 3ER0) and Thermus thermophilus EF-P (middle, PDB: 1UEB (Hanawa-Suetsugu et al., 2004)) are superimposed (right) using PyMOL software (PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC). Colors are used to highlight the domains of eIF5A, N-terminal domain (orange) and C-terminal domain (green), and EF-P, domain I (magenta), domain II (cyan), and domain III (blue). The same color schemes are used in all figures. The modeled hypusine residue of eIF5A and corresponding Arg32 residue of EF-P are shown in stick presentation. A color version of the figure is available online.
In contrast to the two-lobed structure of eIF5A, EF-P is composed of three domains (Figure 3) (Hanawa-Suetsugu et al., 2004). The N-terminal domain of EF-P can be superimposed on the N-terminal domain of eIF5A, while domains II and III of EF-P share structural similarity with one another and with the C-terminal domain of eIF5A (Figure 3). The three domains of EF-P are arranged in an L-shaped configuration that resembles the structure of tRNA (Hanawa-Suetsugu et al., 2004). In support of this structural resemblance, the co-crystal structure of EF-P bound to the T. thermophilus 70S ribosome revealed that EF-P binds adjacent to the P site tRNA in a location that partially overlaps the E site of the ribosome (Blaha, Stanley, and Steitz, 2009). Domain I of EF-P binds near the acceptor stem of the P-site tRNA and contacts the large ribosomal subunit, while domain III of EF-P is located adjacent to the anticodon of the P-site tRNA and partially overlaps the E site of the small ribosomal subunit. Based on the extensive overlap of domain III of EF-P with the position of an E-site tRNA on the small subunit, EF-P binding is predicted to preclude binding of tRNA to the E site.
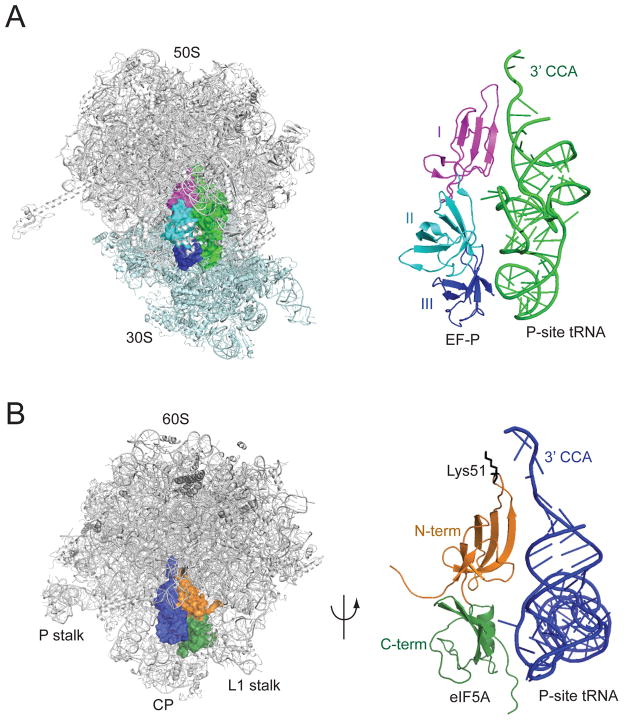
While no structures of eIF5A binding to the ribosome are available, the results of directed hydroxyl radical probing experiments indicate that eIF5A binds to a comparable site on the eukaryotic ribosome as EF-P binds to the bacterial ribosome (Figure 4). Hydroxyl radicals generated by iron linked to the hypusine loop of eIF5A resulted in cleavages to the acceptor stem of the P-site tRNA and to rRNA at the top of the E and P sites including the peptidyl transferase center (PTC) of the ribosome (Gutierrez et al., 2013). In addition, hydroxyl radicals generated by iron tethered to the body of eIF5A yielded cleavages on the side of the P-site tRNA that faces the E site. Consistent with the smaller size of eIF5A compared to EF-P, and the lack of domain III in eIF5A, there was no evidence that eIF5A contacts the small ribosomal subunit. Thus, when bound to the ribosome, eIF5A is predicted to lie adjacent to the P-site tRNA in a position that partially overlaps the E site with the N-terminal domain of eIF5A lying close to the acceptor stem of the P-site tRNA (Figure 4B). Interestingly, when the hypusine modification on eIF5A is modeled into this structure, the hypusine side chain extends into the peptidyl-transferase center of the ribosome (Blaha, Stanley, and Steitz, 2009).
Figure 4. Docking model of eIF5A–60S ribosome complex.
(A) EF-P–70S ribosome complex (PDB: 3HUW and 3HUX, (Blaha, Stanley, and Steitz, 2009)). 50S subunit is colored gray and 30S subunit is colored light cyan, EF-P is colored as described in Figure 3, and tRNAfMet in P site is colored green. (Right) Magnified view of EF-P and P-site tRNAfMet structures.
(B) Yeast eIF5A (PDB: 3ER0) was docked between the P and E sites of the yeast 60S ribosome (PDB: 3O30, (Ben-Shem et al., 2010)) by aligning with the structure of the EF-P–70S ribosome complex (panel A) and adjusting according to the results of hydroxyl radical cleavage data (Gutierrez et al., 2013). Positions of L1 stalk, P stalk and central protuberance (CP) on the 60S subunit are indicated; tRNAiMet is colored blue, and eIF5A is colored as described in Figure 3 with Lys51, the site of hypusine modification, colored black. (Right) Magnified view of docked eIF5A and P site tRNAiMet structures rotated 180°. A color version of the figure is available online.
Post-translational modification of eIF5A and EF-P
Hypusination of eIF5A
As discussed above, eIF5A is the only known protein that contains the amino acid hypusine (Park, Cooper, and Folk, 1981). When cells are grown in the presence of [3H]spermidine, eIF5A is the only protein to be labeled. The hypusine modification in eIF5A appears to occur shortly after eIF5A synthesis with no pools of unmodified eIF5A except when the protein is synthesized in cells depleted of spermidine (Park, 1987; Murphey and Gerner, 1987). Moreover, hypusine formation in eIF5A appears to be constitutive with no evidence of turnover or reversion of the protein to the unmodified state (Gordon et al., 1987; Duncan and Hershey, 1986). Interestingly, in spermidine-dependent mutants of yeast hypusine modification of eIF5A appears to be limiting for cell growth, indicating that hypusine formation is a critical role of spermidine in cells (Chattopadhyay, Park, and Tabor, 2008).
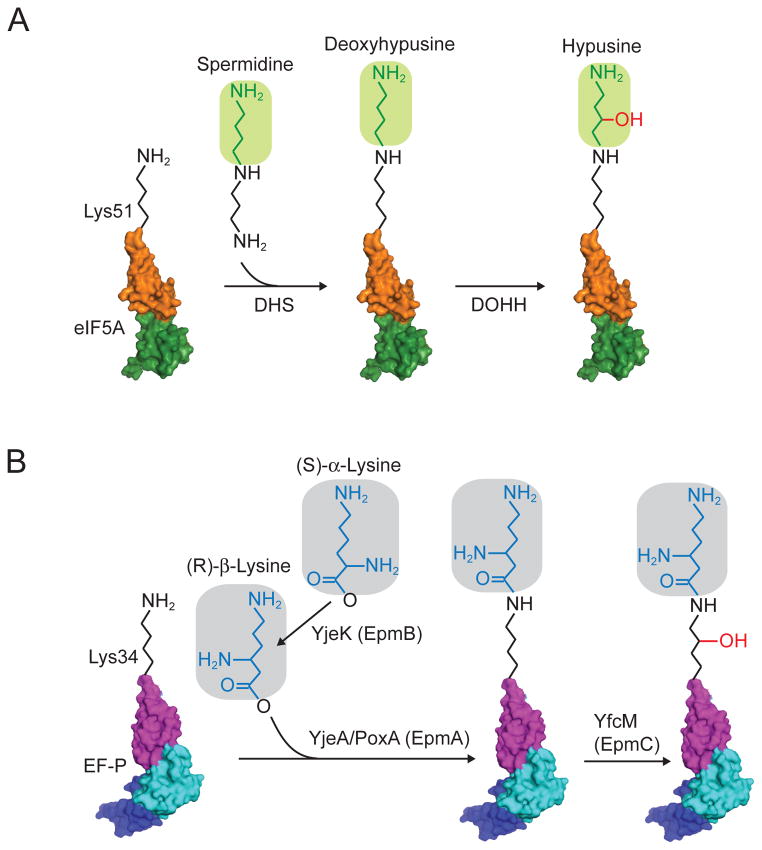
Hypusine is formed by the post-translational modification of a specific lysine residue in eIF5A (Lys50 in human eIF5A and Lys51 in yeast) by the enzymes deoxyhypusine synthase (DHS) and deoxyhypusine hydroxylase (DOHH) (Figure 5A). First, an n-butyl amine group is transferred from spermidine to the ε-amino group of the specific lysine residue by DHS. In a second step, DOHH converts the deoxyhypusine residue to hypusine by addition of a hydroxyl group (Figure 5A) (Park, Cooper, and Folk, 1981; Park et al., 2010). Both genetic and biochemical studies indicate that the hypusine modification is critical for eIF5A function. Substitution of the modified Lys residue by Arg blocked the ability of yeast or human eIF5A to complement the lethal phenotype of a yeast mutant lacking eIF5A (Schnier et al., 1991; Magdolen et al., 1994). Consistent with these in vivo results, unmodified forms of eIF5A that were produced in bacteria or purified from a Chinese hamster ovary (CHO) cell line grown in the presence of 2-difluoromethylornithine, an inhibitor of deoxyhypusine synthase activity, failed to stimulate Met-Pmn synthesis in vitro (Park, 1989; Park et al., 1991; Smit-McBride et al., 1989). Likewise, purified eIF5A-K51R did not stimulate Met-Phe-Phe tripeptide synthesis in a reconstituted yeast translation assay (Saini et al., 2009).
Figure 5. Post-translational modification pathways for eIF5A and EF-P.
(A) In eukaryotes and archaea, an n-butylamine moiety is transferred from spermidine to the ε-amino group of the specific lysine residue (Lys50 in human eIF5A and Lys51 in yeast) by the enzyme deoxyhypusine synthase (DHS). Next, deoxyhypusine hydroxylase (DOHH) adds a hydroxyl group to convert deoxyhypusine to hypusine.
(B) In bacteria, β-lysinylation of EF-P occurs in three steps catalyzed by YjeK (EpmB), YjeA/PoxA (EpmA), and YfcM (EpmC). First, YjeK, a lysine aminomutase, converts lysine to β-lysine. Next, YjeA/PoxA catalyzes the addition of β-lysine onto the specific lysine residue (Lys34 on E. coli EF-P). Finally, YfcM hydroxylates the lysine residue. Domains of eIF5A and EF-P are colored as described in Figure 3. A color version of the figure is available online.
In contrast to the lack of function of unmodified eIF5A, partially modified deoxyhypusine eIF5A is functional. Purified DHS was readily able to add deoxyhypusine to recombinant eIF5A, but not to recombinant eIF5A-K51R (Park et al., 1991), indicating that Arg is not an acceptor for deoxyhypusine and consistent with the failure of the K51R mutant to function in vivo. The deoxyhypusinated recombinant eIF5A stimulated Met-Pmn synthesis, though not as robustly as native eIF5A purified from HeLa cells (Park et al., 1991). While this result could indicate that full hypusine modification is required for eIF5A function, more recent experiments using recombinant eIF5A produced in bacteria co-expressing DHS or DHS plus DOHH revealed nearly equivalent abilities of the deoxyhypusine and hypusine forms of eIF5A to stimulate Met-Pmn synthesis (Park et al., 2011). Consistent with this latter result, it is noteworthy that yeast strains lacking the LIA1 gene encoding yeast DOHH are viable and grow only slightly slower than wild-type strains (Park et al., 2006; Park, 2006).
The enzymes for hypusine synthesis, DHS and DOHH, are conserved in eukaryotes and show high specificity for eIF5A (Park, Lee, and Joe, 1997). The DHS enzyme from yeast was identified based on the sequence of tryptic peptides from purified rat DHS (Kang et al., 1995). The cloned DYS1 gene in yeast encodes a protein of 387 amino acids that possesses DHS activity, and biochemical studies revealed yeast DHS to be a homotetramer of the ~43 kDa subunits (Kang et al., 1995). Consistent with the deoxyhypusine requirement for eIF5A stimulation of Met-Pmn and peptide synthesis, and with the inability of eIF5A-K51R to function in vivo, the yeast DYS1 gene encoding DHS is essential for viability (Sasaki, Abid, and Miyazaki, 1996; Park, Joe, and Kang, 1998). Deoxyhypusine synthesis proceeds through a multistep process including spermidine cleavage, a DHS-imine intermediate, and then transfer of the butylamine moiety from the enzyme to eIF5A (Wolff, Park, and Folk, 1990; Wolff et al., 1995; Wolff, Folk, and Park, 1997). Interestingly, the DHS reaction requires NAD (Chen and Dou, 1988; Wolff, Park, and Folk, 1990), and the NAD cofactor as well as GC7, a spermidine analog and inhibitor of DHS, were observed in crystal structures of human DHS (Liao et al., 1998; Umland et al., 2004).
Yeast DOHH was initially identified based on its interaction with eIF5A in a yeast two-hybrid assay (Thompson, Cano, and Valentini, 2003). The protein was named Lia1 to signify that it was a Ligand of eIF5A. Subsequently, Park and colleagues screened a yeast GST-ORF expression library for DOHH activity and identified Lia1 (Park et al., 2006). Consistent with this biochemical assignment, deletion of the LIA1 gene blocked the production of hypusine, but not deoxyhypusine in yeast (Park et al., 2006). Sequence analysis of the DOHH enzyme suggests that the 325 amino acid protein contains eight HEAT repeats with conserved His-Glu motifs present in four of the repeats (Park et al., 2006). DOHH contains 2 molecules of iron, thought to be present in a diiron active site, per enzyme (Kim et al., 2006), and mutation of the conserved His-Glu motifs impaired Fe and/or eIF5A binding to DOHH (Kang et al., 2007). In contrast to the non-essential nature of DOHH in budding and fission yeast (Park et al., 2006; Weir and Yaffe, 2004), loss of DOHH was recessively lethal in C. elegans (Sievert et al., 2014; Sugimoto, 2004), Drosophila (Patel et al., 2009), and mouse (Sievert et al., 2014), suggesting perhaps that the hydroxyl group on hypusine is more critical for eIF5A function in metazoan cells. While the function of the hydroxyl group on hypusine is unknown, it is noteworthy that the yield of hypusinated eIF5A in bacteria co-expressing eIF5A, DHS and DOHH is greater than the yield of deoxyhypusinated eIF5A in bacteria co-expressing eIF5A and DHS (Park et al., 2011). As the DHS reaction is reversible (Park et al., 2003), it is possible that hydroxylation of deoxyhypusine to hypusine blocks this back reaction and thus stabilizes the hypusine modification. Consistent with this idea, the knock-out of DOHH in mouse was accompanied by the loss of both the hypusine and deoxyhypusine forms of eIF5A (Sievert et al., 2014), though it is worth noting again that in yeast deoxyhypusine eIF5A is present at expected levels in cells lacking DOHH (Park et al., 2006). Interestingly, DOHH orthologs have not been identified in archaea (Park et al., 2006). This lack of DOHH is consistent with the presence of deoxyhypusine rather than hypusine in some archaea (Bartig, Schumann, and Klink, 1990). Moreover, the presence of hypusine in a few archaeal species that grow in the absence of oxygen indicates that a distinct mechanism of hydroxylation has been adopted (Bartig, Schumann, and Klink, 1990).
Genetic and biochemical studies have provided insights into the recognition of eIF5A by DHS and DOHH. In vitro assays with purified DHS and truncated derivatives of human eIF5A identified the fragment from residues 10–90 as the smallest efficient substrate, while the highly conserved region consisting of residues Phe30 to Asp80 was the minimal fragment required for deoxyhypusine biosynthesis (Joe and Park, 1994). Point mutations in the conserved hypusine loop sequence Gly-Hypusine-His-Gly-His-Lys immediately flanking the site of hypusine formation impaired DHS modification of human or yeast eIF5A expressed in yeast cells (Cano et al., 2008; Dias et al., 2008). Taken together, these results indicate that DHS recognizes the N-terminal domain and residues flanking the modified Lys residue when forming hypusine on eIF5A. The DOHH recognition of deoxyhypusinated eIF5A is likewise mediated through the N-terminal domain of eIF5A. Residues 20–90 of eIF5A are sufficient for binding to DOHH and for hydroxylation in vitro (Kang et al., 2007); and similar to the requirements for deoxyhypusine synthesis, mutation of Lys47 and His51 flanking the Lys50 site of modification impairs hydroxylation of human eIF5A expressed in yeast (Cano et al., 2008). Thus, modification of eIF5A by both DHS and DOHH is dependent on recognition of the N-terminal domain of the factor and specific residues within the hypusine loop.
Despite evidence supporting the essential nature of the eIF5A Lys residue that is modified to hypusine, the precise function of hypusine in protein synthesis remains unknown. As stated above, Arg cannot substitute for the modified Lys residue in eIF5A; however, partially modified eIF5A containing deoxyhypusine retained the ability to stimulate Met-Pmn synthesis (Park, 1989) and maintained yeast viability (Park et al., 2006). Interestingly, a structural mimic of eIF5A containing homodeoxyhypusine [Nε-(5-aminopentyl)lysine] was unable to stimulate Met-Pmn synthesis (Park et al., 1991). In contrast to hypusine, which contains an N-butylamine group from spermidine, homodeoxyhypusine contains an aminopentyl moiety from N-(3-aminopropyl)-cadaverine (Park et al., 1991). Thus, the side chain of homodeoxyhypusine is one methyl group longer than the side chain of hypusine. The observation that homodeoxyhypusine-modified eIF5A is unable to stimulate Met-Pmn synthesis indicates a precise length restriction on the hypusine side chain, perhaps reflecting a critical contact between hypusine and other components in the translation complex. However, as will be discussed in more detail below, some forms of bacterial EF-P are post-translationally modified on a Lys residue corresponding to the site of hypusine modification in eIF5A. The EF-P modification contains a β-lysine residue, and thus like homodeoxyhypusine is one methyl group longer than hypusine. As modified EF-P readily promotes polyproline and Met-Pmn synthesis (Bullwinkle et al., 2013; Doerfel et al., 2013; Peil et al., 2013; Ude et al., 2013), the proposed length restriction may be specific to eukaryotic ribosomes or to eIF5A.
Phosphorylation of eIF5A
In addition to its hypusine modification, yeast eIF5A is also phosphorylated. The phosphorylation was mapped to the Ser2 residue of the protein (Klier et al., 1993). As mutation of this residue to Ala did not affect yeast cell growth and the phosphorylated and unphosphorylated forms of the factor were equally active in stimulating Met-Pmn synthesis in assays with mammalian factors and ribosomes, the phosphorylation of eIF5A, unlike its hypusine modification, does not appear to be important for the factor’s function (Klier et al., 1993; Kang, Schwelberger, and Hershey, 1993).
β-lysinylation (lysylation) of EF-P
Alignment of the EF-P sequence from E. coli with the sequence of eIF5A showed conservation of the Lys residue that is modified to hypusine in eIF5A (Figure 2). However, the absence of DHS and DOHH homologs in E. coli (Park et al., 2011), the absence of spermidine labeling of proteins in bacteria, and the substitution of this Lys residue in EF-P by Arg in several bacteria including Thermus thermophilus led to the notion that EF-P was not modified like eIF5A. However, several studies demonstrated that EF-P, at least in some bacteria, is modified. Mass spectrometry analysis of tryptic peptides from native EF-P purified from E. coli revealed that Lys34 was modified, and that the modification contributed an extra mass of ~144 Da (Aoki et al., 2008). Based on comparative genomics analyses including gene clustering and phylogenetic conservation it was hypothesized that the E. coli genes yjeA and yjeK, encoding a truncated lysyl-tRNA synthetase-related protein and a protein related to lysine aminomutase, respectively, function in a pathway to modify EF-P (Bailly and de Crecy-Lagard, 2010). Consistent with the hypothesis that these genes were acting in the same pathway, mutation of yjeA(also known as poxA, genX, and epmA) and yjeK (epmB) in Salmonella typhimurium had equivalent and non-additive negative impacts on S. typhimurium virulence and resistance to antibiotics (Navarre et al., 2010). Biochemical studies demonstrated that YjeA/PoxA could lysinylate (lysylate) EF-P in vitro (Navarre et al., 2010; Yanagisawa et al., 2010), and this activity was dependent on the conserved Lys34 residue (which corresponds to the site of hypusine modification in eIF5A) (Navarre et al., 2010). Notably, Lys34 was also reported to be modified on native EF-P from E. coli (Aoki et al., 2008; Peil et al., 2012; Roy et al., 2011). In vitro and co-expression studies revealed that YjeA/PoxA could add a Lys residue to EF-P, increasing the mass by ~128 Da (Navarre et al., 2010; Park et al., 2012; Yanagisawa et al., 2010). It is notable that this mass is less than the ~144 Da modification detected by Aoki et al., (2008). In the co-expression studies, enhanced levels of EF-P modification were observed in the presence of yjeK, encoding a homolog of lysine-2,3-aminomutase that catalyzes the isomerization of α-lysine to βlysine, and it was thus concluded that EF-P is modified to a β-lysyl-lysine form (Figure 5B) (Yanagisawa et al., 2010).
The initial studies on EF-P modification did not determine whether YjeK acted before or after YjeA/PoxA added an extra molecule of lysine to the Lys34 side chain in EF-P. However, further biochemical analysis of the lysinylation reaction revealed that βlysine was a much-preferred substrate for YjeA/PoxA than was α-lysine (Km decreased 40-fold and kcat increased 2-fold with β-lysine) (Roy et al., 2011). Thus, the EF-P modification pathway is predicted to proceed first with conversion of α-lysine to β-lysine by YjeK and then with addition of the β-lysine to EF-P by YjeA/PoxA to form β-lysyl-lysine (Figure 5B). Analysis of Met-Pmn synthesis in vitro using recombinant forms of unmodified and α-versus β-lysinylated EF-P demonstrated the critical importance of the β-lysyl-lysine modification for EF-P function (Bullwinkle et al., 2013; Park et al., 2012).
Whereas β-lysinylation is predicted to increase the mass of EF-P by 128 Da, analysis of native EF-P indicated that the modification increased EF-P mass by 144 Da (Aoki et al., 2008; Peil et al., 2012). This discrepancy was resolved when Peil et al demonstrated that YfcM (EpmC) hydroxylates β-lysinylated EF-P (Peil et al., 2012). Moreover, their mass spectrometry analyses established that β-lysine is attached to the εamino group of Lys34 in E. coli EF-P (Peil et al., 2012). Thus, similar to the conversion of deoxyhypusine to hypusine by DOHH, YfcM adds a hydroxyl group to complete the post-translational modification of EF-P (Figure 5B). However, in contrast to eIF5A, where the hydroxyl group is added to the spermidine-derived portion of the modification, the hydroxylation of EF-P is directed to the γor δcarbons of the Lys34 sidechain (and not to the added β-lysine) (Peil et al., 2012). Interestingly, similar to the minimal effect of LIA1 deletion in yeast, loss of yfcM in E. coli did not affect bacterial growth or antibiotic sensitivity (Bullwinkle et al., 2013). Thus, the role of the hydroxyl modification in EF-P is not known. Currently, it is also not known whether the EF-P is modified in bacterial species that lack Lys at residue 34, such as T. thermophilus in which Lys34 is substituted by Arg.
Yeast studies reveal a role for eIF5A in translation elongation
Molecular genetic and biochemical studies of yeast eIF5A have provided novel insights into the function of the factor. Using the human eIF5A cDNA as a probe, Schnier et al. identified two genes encoding eIF5A in the yeast Saccharomyces cerevisiae (Schnier et al., 1991). The genes HYP2 and ANB1 encode 157-residue proteins that share 90% amino acid sequence identity with one another and >60% identity with human eIF5A (Figure 2) (Schnier et al., 1991). Further studies by several laboratories revealed that expression of HYP2 and ANB1 is reciprocally regulated by oxygen. Whereas ANB1, also known as HYP1 or TIF51B, is expressed exclusively under anaerobic conditions, HYP2, also known as TIF51A, is preferentially expressed under aerobic conditions (Lowry et al., 1983; Mehta et al., 1990; Schwelberger, Kang, and Hershey, 1993; Magdolen et al., 1994). Consistent with these expression patterns, HYP2 is essential for growth under aerobic conditions, but dispensable for anaerobic growth (Schnier et al., 1991; Schwelberger, Kang, and Hershey, 1993; Wohl et al., 1993). In contrast, ANB1 is dispensable for growth under aerobic conditions (Schnier et al., 1991). These growth phenotypes can be attributed strictly to the expression patterns of HYP2 and ANB1 and not to functional differences between the protein isoforms, as expression of Hyp2 or Anb1 protein from a heterologous promoter was able to restore growth of a yeast strain lacking the HYP2 gene (Magdolen et al., 1994; Schwelberger, Kang, and Hershey, 1993; Wohl et al., 1993). Moreover, expression of human, alfalfa, or slime mold eIF5A protein was able to complement the growth defect in the strain lacking HYP2, demonstrating the functional equivalence of eIF5A across eukaryotes (Magdolen et al., 1994; Schwelberger, Kang, and Hershey, 1993; Wohl et al., 1993).
To gain further insights into eIF5A function, several groups have examined the impact of depleting eIF5A in yeast. By placing Hyp2 expression under the control of a regulated promoter, eIF5A expression was turned off by changing growth conditions. In initial experiments, depletion of eIF5A resulted in a significant inhibition of cell growth but only a 30% reduction in total protein synthesis rates and a correspondingly modest change in polysome profiles (Kang and Hershey, 1994). This discrepancy between the impact of eIF5A depletion on cell growth versus protein synthesis raised the possibility that eIF5A is required for the translation of only a subset of mRNAs whose protein products are critical for cell growth. Further studies with the same eIF5A degron revealed a more substantial impairment in general translation with up to a 4-fold decrease in translation rates upon depletion of eIF5A in rich medium (Henderson and Hershey, 2011). Consistent with this latter study, analysis of an independent eIF5A degron revealed a pronounced defect in total protein synthesis upon depletion of eIF5A (Saini et al., 2009). Likewise, total protein synthesis was impaired when an eIF5A temperature-sensitive mutant was shifted to the restrictive temperature (Dias et al., 2008).
While these eIF5A degron studies revealed that eIF5A promotes general protein synthesis, the role of the factor in translation was not resolved. Polysome profile analyses are commonly used to assess translation function in yeast. In this technique, cells are treated with cycloheximide to freeze ongoing translation elongation and crude extracts are fractionated on sucrose gradients. While one study reported a loss of larger polysomes and accumulation of smaller polysomal species and 80S monosomes upon depletion of eIF5A, indicative of a translation initiation defect (Henderson and Hershey, 2011), this polysome loss was not detected in other studies. Zanelli et al. reported that polysomes were maintained upon shifting two different eIF5A temperature-sensitive (ts−) mutants to the restrictive temperature (Zanelli et al., 2006). Moreover, polysome analyses performed by Saini and co-workers indicated a defect in translation elongation upon depletion or inactivation of eIF5A (Saini et al., 2009). Whereas omission of cycloheximide during analysis of strains expressing wild-type eIF5A resulted in the expected loss of polysomes due to continued translation elongation and release of the ribosomes from mRNAs during extract preparation, polysomes were maintained in the absence of cycloheximide upon depletion of eIF5A (Saini et al., 2009). This retention of polysomes in the absence of cycloheximide in the eIF5A mutant mimicked the effect of cycloheximide addition to the wild-type strain. Thus loss of eIF5A, like addition of cycloheximide, impaired translation elongation.
Though the extended incubations required to fully deplete eIF5A in the degron strains might allow secondary effects resulting in the apparent elongation defect, similar polysome retention in the absence of cycloheximide was observed in an eIF5A ts− mutant after shifting for a short time to the restrictive temperature (Saini et al., 2009) or when grown continuously at a semi-permissive temperature (Gutierrez et al., 2013). Interestingly, polysome analyses in Drosophila and mammalian cells likewise indicated a role for eIF5A in translation elongation. Knockdown of eIF5A expression in DrosophilaS2 cells resulted in the accumulation of polysomes (Patel et al., 2009), and whereas treatment of mammalian cells with arsenite caused impaired translation initiation and polysome loss, knockdown of eIF5A was reported to block this disassembly of polysomes in arsenite-treated cells (Li et al., 2010). In further support of the notion that loss or inactivation of eIF5A impairs translation elongation, two studies reported that eIF5A mutations cause an increase in the average ribosomal transit time, as measured by the time it takes for a ribosome following initiation to synthesize and release a completed polypeptide (Saini et al., 2009; Gregio et al., 2009). It is noteworthy that both the polysome profile analyses and ribosomal transit time measurements assess global translation. Thus, these data indicate that loss of eIF5A impairs translation elongation on a substantial fraction of the mRNAs in yeast cells.
Consistent with the results of the yeast genetic studies, in vitro assays have provided additional evidence that directly link yeast eIF5A to translation elongation. First, the translation defect in crude extracts prepared from eIF5A-depleted or mutant extracts was rescued by addition of purified eIF5A in a hypusine-dependent manner (Saini et al., 2009; Henderson and Hershey, 2011). Moreover, hypusine-modified eIF5A stimulated the rate of tri-peptide synthesis in reconstituted in vitro translation assays employing recombinant or highly purified translation factors and ribosomes (Saini et al., 2009). In further support of these studies that directly link eIF5A to translation elongation, several studies reported eIF5A association with ribosomes or polysomes in whole-cell extracts from yeast (Saini et al., 2009; Zanelli et al., 2006; Jao and Chen, 2006) or mammalian cells (Sievert et al., 2012) and eIF5A was reported to genetically interact with elongation factor eEF2 (Dias et al., 2012). Taken together, the results of these various in vivo and in vitro studies indicate that eIF5A is a translation elongation factor.
EF-P and eIF5A promote production of polyproline proteins
Recent studies have provided new insights into the function of EF-P and eIF5A, and solidified a role for these factors in translation elongation. In concurrently published studies, Ude and colleagues (Ude et al., 2013) and Doerfel and co-workers (Doerfel et al., 2013) showed that EF-P enhanced the synthesis of proteins containing runs of consecutive proline codons. Coming from a genetics perspective, Ude et al. (Ude et al., 2013) found that production of CadC, a transcriptional regulator in E. coli, was impaired in cells lacking EF-P or its modifying enzymes YjeA/PoxA or YjeK. The sensitivity of cadC mRNA translation to defects in EF-P function was mapped to a cluster of proline codons within the cadC open reading frame. Mutating these proline codons substantially restored cadC expression in cells lacking EF-P. Moreover, expression of reporter constructs containing three or more consecutive proline codons was diminished in cells lacking EF-P, and this effect was specific for proline as runs of five consecutive codons for each of the other 19 amino acids did not confer sensitivity to loss of EF-P function (Ude et al., 2013). Based in part on previous findings that Gly and Pro are poor substrates for peptide bond formation (Pavlov et al., 2009; Tanner et al., 2009), Doerfel and coworkers used in vitro translation elongation assays to examine EF-P stimulation of peptide bond formation. Whereas EF-P did not stimulate Met-Pro-Phe synthesis, Met-Pro-Gly and Met-Pro-Pro synthesis was simulated 8- and 16-fold, respectively, by addition of EF-P. Synthesis of peptides containing longer stretches of proline residues showed practically an absolute dependence on EF-P (Doerfel et al., 2013). This requirement for EF-P was also observed when translating derivatives of the PrmC protein with the sequence Pro-Pro-Pro or Pro-Pro-Gly inserted after the 19th codon or when translating native E. coli proteins containing polyproline sequences (Doerfel et al., 2013). Finally, Doerfel et al. and Ude et al. showed that the β-lysyl-lysine modification, independent of its hydroxylation, enhanced the ability of EF-P to stimulate polyproline synthesis (Doerfel et al., 2013; Ude et al., 2013). Consistent with these studies, polyproline sequences were found to cause ribosome stalling in bacteria in a manner that could be alleviated by addition of EF-P in vitro (Woolstenhulme et al., 2013). Taken together, this work establishes a role for EF-P in the specialized translation of polyproline sequences, and demonstrates that EF-P function is not limited to first peptide bond synthesis.
Two studies have further examined the sequence motifs that impose a requirement for EF-P. Both studies employed SILAC to differentially label proteins expressed in bacteria that express or lack EF-P (Hersch et al., 2013; Peil et al., 2013), and Peil et al. also examined cells lacking yjeK, yjeA, or yfcM (Peil et al., 2013). Bioinformatic analysis of the data revealed down-regulation of proteins containing polyproline sequences especially the motifs Pro-Pro-Pro and Pro-Pro-Gly (Hersch et al., 2013; Peil et al., 2013). Systematic analyses of reporter expression containing various X-Pro-Pro or Pro-Pro-Y motifs revealed strong EF-P dependence when X was Pro, Asp or Ala and Y was Pro, Gly, Trp, Asp, or Asn (Peil et al., 2013). Interestingly, Hersh et al. reported that loss of EF-P impaired expression of several non-proline containing motifs when inserted into the fourth codon position of a GFP reporter (Hersch et al., 2013). However, when Peil et al. inserted these motifs into a lacZ reporter they observed no EF-P dependence either in vivo or in vitro (Peil et al., 2013). Thus, it remains to be determined whether EF-P contributes to the translation of non-proline containing motifs. Finally, while these recent studies indicated that polyproline sequences confer the greatest dependency on EF-P, the chemical nature of the other amino acids that when juxtaposed with diproline impose EF-P dependency may provide new insights into the peptidyl-transferase center of the ribosome and help reveal how the β-lysyl-lysine modification of EF-P promotes peptide bond formation.
Consistent with the reports on EF-P, Gutierrez and coworkers reported that eIF5A likewise plays a critical role in translation of polyproline motifs (Gutierrez et al., 2013). A set of dual-luciferase reporter constructs in which the two luciferase ORFs were separated by ten consecutive codons for each of the twenty amino acids were introduced into wild type and eIF5A mutant strains of yeast. Interestingly, expression from the reporter containing the run of proline codons was specifically impaired in the eIF5A mutant (Gutierrez et al., 2013). This 3–5–fold impact of the eIF5A mutation on polyproline reporter expression was observed when the polyproline insert was reduced from 10 to 8 or 6 codons, and was still apparent, albeit with weaker impact, when only 3 or 4 proline codons were inserted in the reporter. Similar to this in vivo requirement for eIF5A stimulation of polyproline synthesis, polyproline peptide synthesis using a reconstituted yeast in vitro translation system was also dependent on eIF5A. Addition of eIF5A modestly (<2-fold) stimulated Met-Pro-Lys or Met-Phe-Phe tripeptide synthesis, suggesting a basic enhancement of peptidyl transferase activity that is not specific for proline. In contrast, synthesis of a peptide containing two consecutive Pro residues was stimulated ~4-fold by addition of eIF5A, and the in vitro synthesis of a peptide containing three consecutive Pro residues showed absolute dependence on eIF5A (Gutierrez et al., 2013). Just as EF-P stimulation of polyproline peptide synthesis was dependent on the β-lysyl-lysine modification (Doerfel et al., 2013), the stimulation of Met-Pro-Pro-Pro-Lys peptide synthesis by eIF5A was dependent on the hypusine modification (Gutierrez et al., 2013). Thus EF-P and eIF5A share a similar function in promoting the ability of the ribosome to translate polyproline sequences.
Further analysis of ribosomes translating polyproline sequences has provided insight into requirement for EF-P or eIF5A. In the in vitro bacterial system, ribosomes translating the sequence Pro-Pro-Gly in the absence of EF-P stalled with diproline bound to the P-site tRNA and Gly-tRNA in the A site (Doerfel et al., 2013). Likewise, toe-printing studies of ribosomes stalled in vivo or in vitro on polyproline sequences indicated that proline is bound to the P-site tRNA and Pro-tRNA is in the A site (Woolstenhulme et al., 2013). Interestingly, Woolstenhulme et al. also found that both the length and amino acid sequence preceding the polyproline stretch affects the efficiency of stalling (Woolstenhulme et al., 2013), consistent with the notion that the conformation of the peptide in the active site and exit tunnel of the ribosome influences the requirement for EF-P. Toe-printing studies of eukaryotic ribosomes translating polyproline sequences in the absence of eIF5A likewise revealed stalling with the second Pro codon in the P site (diproline bound to the P-site tRNA) and a Pro codon (Pro-tRNA) in the A site (Gutierrez et al., 2013). Thus EF-P and eIF5A appear to rescue similarly stalled complexes.
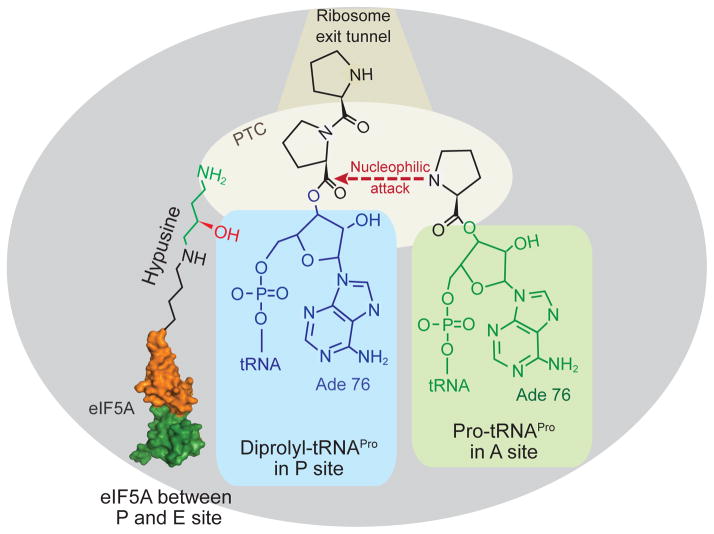
The common appearance of diproline immediately joined to the P-site tRNA raises the possibility that the cis-trans conformation or the rigidity of the Pro-Pro bond (Saha and Shamala, 2012) coupled with a poor A-site substrate (Pavlov et al., 2009) may contribute to the requirement for EF-P and eIF5A for polyproline synthesis. While the precise function of the hypusine or β-lysyl-lysine modification of eIF5A or EF-P, respectively, is unclear, we consider several possibilities. Binding of eIF5A adjacent to the P-site tRNA and insertion of hypusine into the peptidyl-transferase center of the ribosome may sterically restrict the position of the prolines on the P-site tRNA and allow proper alignment of the substrates for peptide bond synthesis (Figure 6). Relatedly, perhaps eIF5A functions as a prolyl cis-trans isomerase to fix the geometry of di-proline on the P-site tRNA, such that the prolines properly enter the ribosomal exit tunnel and enable peptide bond formation with the A-site substrate. Future structural and biochemical studies on elongating ribosomes containing eIF5A will help elucidate the function of eIF5A and its hypusine modification.
Figure 6. Model of eIF5A promoting polyproline peptide bond formation.
Cartoon presentation of the large ribosomal subunit stalled with diprolyl-tRNAPro in the P site and Pro-tRNAPro in the A site. Binding of eIF5A between the P and E sites places the hypusine side chain near the diprolyl-tRNAPro in the peptidyl transferase center (PTC) of the ribosome where it facilitates peptide bond formation. Note that figure is not drawn to scale. A color version of the figure is available online.
Examination of the yeast proteome revealed that 549 out of 5886 proteins contain at least one triproline motif. The number of hits increases to 749 if Pro-Pro-Gly motifs are included as well, and the number of hits increases to nearly 2000 if all poor Pro-Pro-X motifs, as defined for EF-P by Peil et al. (Peil et al., 2013), are included in the search. In humans, proline accounts for ~6% of the total encoded residues (Morgan and Rubenstein, 2013). Among ~18,000 human proteins, there are roughly 10,000 motifs consisting of 3 or more consecutive proline residues (Morgan and Rubenstein, 2013). In their analysis of yeast protein expression, Gutierrez et al. (Gutierrez et al., 2013) reported that levels of the polyproline containing proteins Ldb17, Eap1, and Vrp1 were reduced in eIF5A mutant strains. Moreover, the expression of Ldb17 was restored when the polyproline sequence in this protein was mutated to polyalanine (Gutierrez et al., 2013). Interestingly, while polyproline sequences are found in a variety of proteins in yeast, it is notable that several proteins involved in the actin cytoskeleton contain polyproline motifs. Of particular interest, the protein Bni1, which functions in polarized cell growth, contains a polyproline sequence and when overexpressed partially suppresses the slow-growth phenotype of an eIF5A mutant yeast strain (Zanelli and Valentini, 2005). Based on this finding it seems reasonable to propose that the impacts of eIF5A mutants on the yeast cell cycle, cytoskeleton, and PKC pathways (Chatterjee et al., 2006; Frigieri et al., 2008; Galvao et al., 2013; Valentini et al., 2002) as well as the association of eIF5A with oncogenesis, and paradoxically tumor suppression, in mammals (Scuoppo et al., 2012; Zender et al., 2008; Silvera, Formenti, and Schneider, 2010) may be due to impaired expression of specific proteins containing polyproline sequences.
Conclusions
Since its discovery nearly 40 years ago eIF5A has been an enigma. While the factor was initially linked to translation initiation based on its stimulation of Met-Pmn synthesis, recent studies on eIF5A, and the related bacterial factor EF-P, have established its role in translation elongation and specifically in promoting the translation of polyproline motifs. Based on its clarified role in protein synthesis, we feel that it would be appropriate to rename the protein as an elongation factor. Accordingly, we propose the name eEF5 for eukaryotic elongation factor 5. Future studies will help elucidate the full range of amino acids and sequence motifs that require eIF5A for their efficient translation. Moreover, it is of particular interest to learn how the hypusine modification on eIF5A interacts with the ribosome or diprolyl-tRNA in the ribosomal P site to promote peptide bond formation.
Footnotes
Declaration of interest
This work was supported by the Intramural Research Program of the National Institutes of Health, NICHD. The authors report no conflict of interest.
References
- Aoki H, Xu J, Emili A, Chosay JG, Golshani A, Ganoza MC. Interactions of elongation factor EF-P with the Escherichia coli ribosome. FEBS J. 2008;275:671–81. doi: 10.1111/j.1742-4658.2007.06228.x. [DOI] [PubMed] [Google Scholar]
- Bailly M, de Crecy-Lagard V. Predicting the pathway involved in post-translational modification of elongation factor P in a subset of bacterial species. Biol Direct. 2010;5:3. doi: 10.1186/1745-6150-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartig D, Lemkemeier K, Frank J, Lottspeich F, Klink F. The archaebacterial hypusine-containing protein. Structural features suggest common ancestry with eukaryotic translation initiation factor 5A. Eur J Biochem. 1992;204:751–8. doi: 10.1111/j.1432-1033.1992.tb16690.x. [DOI] [PubMed] [Google Scholar]
- Bartig D, Schumann H, Klink F. The unique posttranslational modification leading to deoxyhypusine or hypusine is a general feature of the archaebacterial kingdom. Syst Appl Microbiol. 1990;13:112–16. [Google Scholar]
- Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–9. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- Benne R, Brown-Luedi ML, Hershey JWB. Purification and characterization of protein synthesis initiation factors eIF-1, eIF-4C, eIF-4D, and eIF-5 from rabbit reticulocytes. J Biol Chem. 1978;253:3070–77. [PubMed] [Google Scholar]
- Benne R, Hershey JWB. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J Biol Chem. 1978;253:3078–87. [PubMed] [Google Scholar]
- Bevec D, Jaksche H, Oft M, Wohl T, Himmelspach M, Pacher A, Schebesta M, Koettnitz K, Dobrovnik M, Csonga R, Lottspeich F, Hauber J. Inhibition of HIV-1 replication in lymphocytes by mutants of the Rev cofactor eIF-5A. Science. 1996;271:1858–60. doi: 10.1126/science.271.5257.1858. [DOI] [PubMed] [Google Scholar]
- Blaha G, Stanley RE, Steitz TA. Formation of the first peptide bond: the structure of EF-P bound to the 70S ribosome. Science. 2009;325:966–70. doi: 10.1126/science.1175800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullwinkle TJ, Zou SB, Rajkovic A, Hersch SJ, Elgamal S, Robinson N, Smil D, Bolshan Y, Navarre WW, Ibba M. (R)-β-Lysine-modified elongation factor P functions in translation elongation. J Biol Chem. 2013;288:4416–23. doi: 10.1074/jbc.M112.438879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano VS, Jeon GA, Johansson HE, Henderson CA, Park JH, Valentini SR, Hershey JW, Park MH. Mutational analyses of human eIF5A-1--identification of amino acid residues critical for eIF5A activity and hypusine modification. FEBS J. 2008;275:44–58. doi: 10.1111/j.1742-4658.2007.06172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee I, Gross SR, Kinzy TG, Chen KY. Rapid depletion of mutant eukaryotic initiation factor 5A at restrictive temperature reveals connections to actin cytoskeleton and cell cycle progression. Mol Genet Genomics. 2006;275:264–76. doi: 10.1007/s00438-005-0086-4. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay MK, Park MH, Tabor H. Hypusine modification for growth is the major function of spermidine in Saccharomyces cerevisiae polyamine auxotrophs grown in limiting spermidine. Proc Natl Acad Sci USA. 2008;105:6554–9. doi: 10.1073/pnas.0710970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KY, Dou QP. NAD+ stimulated the spermidine-dependent hypusine formation on the 18 kDa protein in cytosolic lysates derived from NB-15 mouse neuroblastoma cells. FEBS Lett. 1988;229:325–8. doi: 10.1016/0014-5793(88)81149-9. [DOI] [PubMed] [Google Scholar]
- Cooper HL, Park MH, Folk JE. Posttranslational formation of hypusine in a single major protein occurs generally in growing cells and is associated with activation of lymphocyte growth. Cell. 1982;29:791–97. doi: 10.1016/0092-8674(82)90441-x. [DOI] [PubMed] [Google Scholar]
- Cooper HL, Park MH, Folk JE, Safer B, Braverman R. Identification of the hypusine-containing protein hy+ as translation initiation factor eIF-4D. Proc Natl Acad Sci USA. 1983;80:1854–57. doi: 10.1073/pnas.80.7.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Green R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb Perspect Biol. 2012;4:a013706. doi: 10.1101/cshperspect.a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias CA, Cano VS, Rangel SM, Apponi LH, Frigieri MC, Muniz JR, Garcia W, Park MH, Garratt RC, Zanelli CF, Valentini SR. Structural modeling and mutational analysis of yeast eukaryotic translation initiation factor 5A reveal new critical residues and reinforce its involvement in protein synthesis. FEBS J. 2008;275:1874–88. doi: 10.1111/j.1742-4658.2008.06345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias CA, Gregio AP, Rossi D, Galvao FC, Watanabe TF, Park MH, Valentini SR, Zanelli CF. eIF5A interacts functionally with eEF2. Amino Acids. 2012;42:697–702. doi: 10.1007/s00726-011-0985-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013;339:85–8. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- Duncan RF, Hershey JW. Changes in eIF-4D hypusine modification or abundance are not correlated with translational repression in HeLa cells. J Biol Chem. 1986;261:12903–6. [PubMed] [Google Scholar]
- Frigieri MC, Joao Luiz MV, Apponi LH, Zanelli CF, Valentini SR. Synthetic lethality between eIF5A and Ypt1 reveals a connection between translation and the secretory pathway in yeast. Mol Genet Genomics. 2008;280:211–21. doi: 10.1007/s00438-008-0357-y. [DOI] [PubMed] [Google Scholar]
- Galvao FC, Rossi D, Silveira Wda S, Valentini SR, Zanelli CF. The deoxyhypusine synthase mutant dys1-1 reveals the association of eIF5A and Asc1 with cell wall integrity. PLoS One. 2013;8:e60140. doi: 10.1371/journal.pone.0060140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganoza MC, Aoki H. Peptide bond synthesis: function of the efp gene product. Biol Chem. 2000;381:553–9. doi: 10.1515/BC.2000.071. [DOI] [PubMed] [Google Scholar]
- Glick BR, Chladek S, Ganoza MC. Peptide bond formation stimulated by protein synthesis factor EF-P depends on the aminoacyl moiety of the acceptor. Eur J Biochem. 1979;97:23–8. doi: 10.1111/j.1432-1033.1979.tb13081.x. [DOI] [PubMed] [Google Scholar]
- Glick BR, Ganoza MC. Identification of a soluble protein that stimulates peptide bond synthesis. Proc Natl Acad Sci USA. 1975;72:4257–60. doi: 10.1073/pnas.72.11.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon ED, Mora R, Meredith SC, Lee C, Lindquist SL. Eukaryotic initiation factor 4D, the hypusine-containing protein, is conserved among eukaryotes. J Biol Chem. 1987;262:16585–9. [PubMed] [Google Scholar]
- Gordon ED, Mora R, Meredith SC, Lindquist SL. Hypusine formation in eukaryotic initiation factor 4D is not reversed when rates or specificity of protein synthesis is altered. J Biol Chem. 1987;262:16590–5. [PubMed] [Google Scholar]
- Gregio AP, Cano VP, Avaca JS, Valentini SR, Zanelli CF. eIF5A has a function in the elongation step of translation in yeast. Biochem Biophys Res Commun. 2009;380:785–90. doi: 10.1016/j.bbrc.2009.01.148. [DOI] [PubMed] [Google Scholar]
- Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, Dever TE. eIF5A promotes translation of polyproline motifs. Mol Cell. 2013;51:35–45. doi: 10.1016/j.molcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawa-Suetsugu K, Sekine S, Sakai H, Hori-Takemoto C, Terada T, Unzai S, Tame JR, Kuramitsu S, Shirouzu M, Yokoyama S. Crystal structure of elongation factor P from Thermus thermophilus HB8. Proc Natl Acad Sci USA. 2004;101:9595–600. doi: 10.1073/pnas.0308667101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson A, Hershey JW. Eukaryotic translation initiation factor (eIF) 5A stimulates protein synthesis in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2011;108:6415–9. doi: 10.1073/pnas.1008150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch SJ, Wang M, Zou SB, Moon KM, Foster LJ, Ibba M, Navarre WW. Divergent protein motifs direct elongation factor P-mediated translational regulation in Salmonella enterica and Escherichia coli. MBio. 2013;4:e00180–13. doi: 10.1128/mBio.00180-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG, Lorsch JR. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb Perspect Biol. 2012;4:a011544. doi: 10.1101/cshperspect.a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann W, Reichart B, Ewald A, Muller E, Schmitt I, Stauber RH, Lottspeich F, Jockusch BM, Scheer U, Hauber J, Dabauvalle MC. Cofactor requirements for nuclear export of Rev response element (RRE)- and constitutive transport element (CTE)-containing retroviral RNAs. An unexpected role for actin. J Cell Biol. 2001;152:895–910. doi: 10.1083/jcb.152.5.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao DL, Chen KY. Tandem affinity purification revealed the hypusine-dependent binding of eukaryotic initiation factor 5A to the translating 80S ribosomal complex. J Cell Biochem. 2006;97:583–98. doi: 10.1002/jcb.20658. [DOI] [PubMed] [Google Scholar]
- Joe YA, Park MH. Structural features of the eIF-5A precursor required for posttranslational synthesis of deoxyhypusine. J Biol Chem. 1994;269:25916–21. [PubMed] [Google Scholar]
- Kang HA, Hershey JW. Effect of initiation factor eIF-5A depletion on protein synthesis and proliferation of Saccharomyces cerevisiae. J Biol Chem. 1994;269:3934–40. [PubMed] [Google Scholar]
- Kang HA, Schwelberger HG, Hershey JW. Translation initiation factor eIF-5A, the hypusine-containing protein, is phosphorylated on serine in Saccharomyces cerevisiae. J Biol Chem. 1993;268:14750–6. [PubMed] [Google Scholar]
- Kang KR, Kim YS, Wolff EC, Park MH. Specificity of the deoxyhypusine hydroxylase-eukaryotic translation initiation factor (eIF5A) interaction: identification of amino acid residues of the enzyme required for binding of its substrate, deoxyhypusine-containing eIF5A. J Biol Chem. 2007;282:8300–8. doi: 10.1074/jbc.M607495200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang KR, Wolff EC, Park MH, Folk JE, Chung SI. Identification of YHR068w in Saccharomyces cerevisiae chromosome VIII as a gene for deoxyhypusine synthase. Expression and characterization of the enzyme. J Biol Chem. 1995;270:18408–12. doi: 10.1074/jbc.270.31.18408. [DOI] [PubMed] [Google Scholar]
- Kemper WM, Berry KW, Merrick WC. Purification and properties of rabbit reticulocyte protein synthesis initiation factors M2Bα and M2Bβ. J Biol Chem. 1976;251:5551–57. [PubMed] [Google Scholar]
- Kim KK, Hung LW, Yokota H, Kim R, Kim SH. Crystal structures of eukaryotic translation initiation factor 5A from Methanococcus jannaschii at 1.8 A resolution. Proc Natl Acad Sci USA. 1998;95:10419–24. doi: 10.1073/pnas.95.18.10419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Kang KR, Wolff EC, Bell JK, McPhie P, Park MH. Deoxyhypusine hydroxylase is a Fe(II)-dependent, HEAT-repeat enzyme. Identification of amino acid residues critical for Fe(II) binding and catalysis. J Biol Chem. 2006;281:13217–25. doi: 10.1074/jbc.M601081200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klier H, Wohl T, Eckerskorn C, Magdolen V, Lottspeich F. Determination and mutational analysis of the phosphorylation site in the hypusine-containing protein Hyp2p. FEBS Lett. 1993;334:360–4. doi: 10.1016/0014-5793(93)80712-4. [DOI] [PubMed] [Google Scholar]
- Kyrpides NC, Woese CR. Universally conserved translation initiation factors. Proc Natl Acad Sci USA. 1998;95:224–28. doi: 10.1073/pnas.95.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CH, Ohn T, Ivanov P, Tisdale S, Anderson P. eIF5A promotes translation elongation, polysome disassembly and stress granule assembly. PLoS One. 2010;5:e9942. doi: 10.1371/journal.pone.0009942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao DI, Wolff EC, Park MH, Davies DR. Crystal structure of the NAD complex of human deoxyhypusine synthase: an enzyme with a ball-and-chain mechanism for blocking the active site. Structure. 1998;6:23–32. doi: 10.1016/s0969-2126(98)00004-5. [DOI] [PubMed] [Google Scholar]
- Lobanov AV, Turanov AA, Hatfield DL, Gladyshev VN. Dual functions of codons in the genetic code. Crit Rev Biochem Mol Biol. 2010;45:257–65. doi: 10.3109/10409231003786094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry CV, Weiss JL, Walthall DA, Zitomer RS. Modulator sequences mediate oxygen regulation of CYC1 and a neighboring gene in yeast. Proc Natl Acad Sci U S A. 1983;80:151–5. doi: 10.1073/pnas.80.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magdolen V, Klier H, Wohl T, Klink F, Hirt H, Hauber J, Lottspeich F. The function of the hypusine-containing proteins of yeast and other eukaryotes is well conserved. Mol Gen Genet. 1994;244:646–52. doi: 10.1007/BF00282755. [DOI] [PubMed] [Google Scholar]
- Mehta KD, Leung D, Lefebvre L, Smith M. The ANB1 locus of Saccharomyces cerevisiae encodes the protein synthesis initiation factor eIF-4D. J Biol Chem. 1990;265:8802–07. [PubMed] [Google Scholar]
- Merrick WC. Assays for eukaryotic protein synthesis. Methods Enzymol. 1979;60:108–23. doi: 10.1016/s0076-6879(79)60011-3. [DOI] [PubMed] [Google Scholar]
- Merrick WC. Purification of protein synthesis initiation factors from rabbit reticulocytes. Methods Enzymol. 1979;60:101–8. doi: 10.1016/s0076-6879(79)60010-1. [DOI] [PubMed] [Google Scholar]
- Morgan AA, Rubenstein E. Proline: the distribution, frequency, positioning, and common functional roles of proline and polyproline sequences in the human proteome. PLoS One. 2013;8:e53785. doi: 10.1371/journal.pone.0053785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphey RJ, Gerner EW. Hypusine formation in protein by a two-step process in cell lysates. J Biol Chem. 1987;262:15033–6. [PubMed] [Google Scholar]
- Navarre WW, Zou SB, Roy H, Xie JL, Savchenko A, Singer A, Edvokimova E, Prost LR, Kumar R, Ibba M, Fang FC. PoxA, yjeK, and elongation factor P coordinately modulate virulence and drug resistance in Salmonella enterica. Mol Cell. 2010;39:209–21. doi: 10.1016/j.molcel.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Aravind L, Wolff EC, Kaevel J, Kim YS, Park MH. Molecular cloning, expression, and structural prediction of deoxyhypusine hydroxylase: a HEAT-repeat-containing metalloenzyme. Proc Natl Acad Sci USA. 2006;103:51–6. doi: 10.1073/pnas.0509348102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Dias CA, Lee SB, Valentini SR, Sokabe M, Fraser CS, Park MH. Production of active recombinant eIF5A: reconstitution in E. coli of eukaryotic hypusine modification of eIF5A by its coexpression with modifying enzymes. Protein Eng Des Sel. 2011;24:301–9. doi: 10.1093/protein/gzq110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Johansson HE, Aoki H, Huang BX, Kim HY, Ganoza MC, Park MH. Post-translational modification by β-lysylation is required for activity of Escherichia coli elongation factor P (EF-P) J Biol Chem. 2012;287:2579–90. doi: 10.1074/jbc.M111.309633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Wolff EC, Folk JE, Park MH. Reversal of the deoxyhypusine synthesis reaction. Generation of spermidine or homospermidine from deoxyhypusine by deoxyhypusine synthase. J Biol Chem. 2003;278:32683–91. doi: 10.1074/jbc.M304247200. [DOI] [PubMed] [Google Scholar]
- Park MH. Regulation of biosynthesis of hypusine in Chinese hamster ovary cells. Evidence for eIF-4D precursor polypeptides. J Biol Chem. 1987;262:12730–4. [PubMed] [Google Scholar]
- Park MH. The essential role of hypusine in eukaryotic translation initiation factor 4D (eIF-4D). Purification of eIF-4D and its precursors and comparison of their activities. J Biol Chem. 1989;264:18531–5. [PubMed] [Google Scholar]
- Park MH. The post-translational synthesis of a polyamine-derived amino acid, hypusine, in the eukaryotic translation initiation factor 5A (eIF5A) J Biochem. 2006;139:161–9. doi: 10.1093/jb/mvj034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH, Cooper HL, Folk JE. Identification of hypusine, an unusual amino acid, in a protein from human lymphocytes and of spermidine as its biosynthetic precursor. Proc Natl Acad Sci USA. 1981;78:2869–73. doi: 10.1073/pnas.78.5.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH, Joe YA, Kang KR. Deoxyhypusine synthase activity is essential for cell viability in the yeast Saccharomyces cerevisiae. J Biol Chem. 1998;273:1677–83. doi: 10.1074/jbc.273.3.1677. [DOI] [PubMed] [Google Scholar]
- Park MH, Lee YB, Joe YA. Hypusine is essential for eukaryotic cell proliferation. Biol Signals. 1997;6:115–23. doi: 10.1159/000109117. [DOI] [PubMed] [Google Scholar]
- Park MH, Liu TY, Neece SH, Swiggard WJ. Eukaryotic initiation factor 4D. Purification from human red blood cells and the sequence of amino acids around its single hypusine residue. J Biol Chem. 1986;261:14515–9. [PubMed] [Google Scholar]
- Park MH, Nishimura K, Zanelli CF, Valentini SR. Functional significance of eIF5A and its hypusine modification in eukaryotes. Amino Acids. 2010;38:491–500. doi: 10.1007/s00726-009-0408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH, Wolff EC, Smit-McBride Z, Hershey JW, Folk JE. Comparison of the activities of variant forms of eIF-4D. The requirement for hypusine or deoxyhypusine. J Biol Chem. 1991;266:7988–94. [PubMed] [Google Scholar]
- Patel PH, Costa-Mattioli M, Schulze KL, Bellen HJ. The Drosophila deoxyhypusine hydroxylase homologue nero and its target eIF5A are required for cell growth and the regulation of autophagy. J Cell Biol. 2009;185:1181–94. doi: 10.1083/jcb.200904161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov MY, Watts RE, Tan Z, Cornish VW, Ehrenberg M, Forster AC. Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc Natl Acad Sci USA. 2009;106:50–4. doi: 10.1073/pnas.0809211106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peat TS, Newman J, Waldo GS, Berendzen J, Terwilliger TC. Structure of translation initiation factor 5A from Pyrobaculum aerophilum at 1.75 Å resolution. Structure. 1998;6:1207–14. doi: 10.1016/s0969-2126(98)00120-8. [DOI] [PubMed] [Google Scholar]
- Peil L, Starosta AL, Lassak J, Atkinson GC, Virumae K, Spitzer M, Tenson T, Jung K, Remme J, Wilson DN. Distinct XPPX sequence motifs induce ribosome stalling, which is rescued by the translation elongation factor EF-P. Proc Natl Acad Sci USA. 2013;110:15265–70. doi: 10.1073/pnas.1310642110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peil L, Starosta AL, Virumae K, Atkinson GC, Tenson T, Remme J, Wilson DN. Lys34 of translation elongation factor EF-P is hydroxylated by YfcM. Nat Chem Biol. 2012;8:695–7. doi: 10.1038/nchembio.1001. [DOI] [PubMed] [Google Scholar]
- Roy H, Zou SB, Bullwinkle TJ, Wolfe BS, Gilreath MS, Forsyth CJ, Navarre WW, Ibba M. The tRNA synthetase paralog PoxA modifies elongation factor-P with (R)-β-lysine. Nat Chem Biol. 2011;7:667–9. doi: 10.1038/nchembio.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhl M, Himmelspach M, Bahr GM, Hammerschmid F, Jaksche H, Wolff B, Aschauer H, Farrington GK, Probst H, Bevec D, et al. Eukaryotic initiation factor 5A is a cellular target of the human immunodeficiency virus type 1 Rev activation domain mediating trans-activation. J Cell Biol. 1993;123:1309–20. doi: 10.1083/jcb.123.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha I, Shamala N. Investigating diproline segments in proteins: occurrences, conformation and classification. Biopolymers. 2012;97:54–64. doi: 10.1002/bip.21703. [DOI] [PubMed] [Google Scholar]
- Saini P, Eyler DE, Green R, Dever TE. Hypusine-containing protein eIF5A promotes translation elongation. Nature. 2009;459:118–21. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Abid MR, Miyazaki M. Deoxyhypusine synthase gene is essential for cell viability in the yeast Saccharomyces cerevisiae. FEBS Lett. 1996;384:151–4. doi: 10.1016/0014-5793(96)00310-9. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–42. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- Schnier J, Schwelberger HG, Smit-McBride Z, Kang HA, Hershey JWB. Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:3105–14. doi: 10.1128/mcb.11.6.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader R, Young C, Kozian D, Hoffmann R, Lottspeich F. Temperature-sensitive eIF5A mutant accumulates transcripts targeted to the nonsense-mediated decay pathway. J Biol Chem. 2006;281:35336–46. doi: 10.1074/jbc.M601460200. [DOI] [PubMed] [Google Scholar]
- Schreier MH, Erni B, Staehelin T. Initiation of mammalian protein synthesis: purification and characterization of seven initiation factors. J Mol Biol. 1977;116:727–53. doi: 10.1016/0022-2836(77)90268-6. [DOI] [PubMed] [Google Scholar]
- Schwelberger HG, Kang HA, Hershey JW. Translation initiation factor eIF-5A expressed from either of two yeast genes or from human cDNA. Functional identity under aerobic and anaerobic conditions. J Biol Chem. 1993;268:14018–25. [PubMed] [Google Scholar]
- Scuoppo C, Miething C, Lindqvist L, Reyes J, Ruse C, Appelmann I, Yoon S, Krasnitz A, Teruya-Feldstein J, Pappin D, Pelletier J, Lowe SW. A tumour suppressor network relying on the polyamine-hypusine axis. Nature. 2012;487:244–8. doi: 10.1038/nature11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba T, Mizote H, Kaneko T, Nakajima T, Kakimoto Y. Hypusine, a new amino acid occurring in bovine brain. Isolation and structural determination. Biochim Biophys Acta. 1971;244:523–31. doi: 10.1016/0304-4165(71)90069-9. [DOI] [PubMed] [Google Scholar]
- Sievert H, Pallmann N, Miller KK, Hermans-Borgmeyer I, Venz S, Sendoel A, Preukschas M, Schweizer M, Bottcher S, Janiesch PC, Streichert T, Walther R, Hengartner MO, Manz MG, Brummendorf TH, Bokemeyer C, Braig M, Hauber J, Duncan KE, Balabanov S. A novel mouse model for inhibition of DOHH mediated hypusine modification reveals crucial function for embryonic development, proliferation and oncogenic transformation. Dis Model Mech. 2014 doi: 10.1242/dmm.014449. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievert H, Venz S, Platas-Barradas O, Dhople VM, Schaletzky M, Nagel CH, Braig M, Preukschas M, Pallmann N, Bokemeyer C, Brummendorf TH, Portner R, Walther R, Duncan KE, Hauber J, Balabanov S. Protein-protein-interaction network organization of the hypusine modification system. Mol Cell Proteomics. 2012;11:1289–305. doi: 10.1074/mcp.M112.019059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer. 2010;10:254–66. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- Simonetti A, Marzi S, Jenner L, Myasnikov A, Romby P, Yusupova G, Klaholz BP, Yusupov M. A structural view of translation initiation in bacteria. Cell Mol Life Sci. 2009;66:423–36. doi: 10.1007/s00018-008-8416-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit-McBride Z, Dever TE, Hershey JW, Merrick WC. Sequence determination and cDNA cloning of eukaryotic initiation factor 4D, the hypusine-containing protein. J Biol Chem. 1989;264:1578–83. [PubMed] [Google Scholar]
- Smit-McBride Z, Schnier J, Kaufman RJ, Hershey JW. Protein synthesis initiation factor eIF-4D. Functional comparison of native and unhypusinated forms of the protein. J Biol Chem. 1989;264:18527–30. [PubMed] [Google Scholar]
- Sugimoto A. High-throughput RNAi in Caenorhabditis elegans: genome-wide screens and functional genomics. Differentiation. 2004;72:81–91. doi: 10.1111/j.1432-0436.2004.07202004.x. [DOI] [PubMed] [Google Scholar]
- Tanner DR, Cariello DA, Woolstenhulme CJ, Broadbent MA, Buskirk AR. Genetic identification of nascent peptides that induce ribosome stalling. J Biol Chem. 2009;284:34809–18. doi: 10.1074/jbc.M109.039040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A, Goumans H, Amesz H, Benne R, Voorma HO. A comparison of the initiation factors of eukaryotic protein synthesis from ribosomes and from the postribosomal supernatant. Eur J Biochem. 1979;98:329–37. doi: 10.1111/j.1432-1033.1979.tb13192.x. [DOI] [PubMed] [Google Scholar]
- Thompson GM, Cano VS, Valentini SR. Mapping eIF5A binding sites for Dys1 and Lia1: in vivo evidence for regulation of eIF5A hypusination. FEBS Lett. 2003;555:464–8. doi: 10.1016/s0014-5793(03)01305-x. [DOI] [PubMed] [Google Scholar]
- Tong Y, Park I, Hong BS, Nedyalkova L, Tempel W, Park HW. Crystal structure of human eIF5A1: insight into functional similarity of human eIF5A1 and eIF5A2. Proteins. 2009;75:1040–5. doi: 10.1002/prot.22378. [DOI] [PubMed] [Google Scholar]
- Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013;339:82–5. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- Umland TC, Wolff EC, Park MH, Davies DR. A new crystal structure of deoxyhypusine synthase reveals the configuration of the active enzyme and of an enzyme•NAD•inhibitor ternary complex. J Biol Chem. 2004;279:28697–705. doi: 10.1074/jbc.M404095200. [DOI] [PubMed] [Google Scholar]
- Valentini SR, Casolari JM, Oliveira CC, Silver PA, McBride AE. Genetic interactions of yeast eukaryotic translation initiation factor 5A (eIF5A) reveal connections to poly(A)-binding protein and protein kinase C signaling. Genetics. 2002;160:393–405. doi: 10.1093/genetics/160.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BA, Yaffe MP. Mmd1p, a novel, conserved protein essential for normal mitochondrial morphology and distribution in the fission yeast Schizosaccharomyces pombe. Mol Biol Cell. 2004;15:1656–65. doi: 10.1091/mbc.E03-06-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohl T, Klier H, Ammer H, Lottspeich F, Magdolen V. The HYP2 gene of Saccharomyces cerevisiae is essential for aerobic growth: characterization of different isoforms of the hypusine-containing protein Hyp2p and analysis of gene disruption mutants. Mol Gen Genet. 1993;241:305–11. doi: 10.1007/BF00284682. [DOI] [PubMed] [Google Scholar]
- Wohlgemuth I, Brenner S, Beringer M, Rodnina MV. Modulation of the rate of peptidyl transfer on the ribosome by the nature of substrates. J Biol Chem. 2008;283:32229–35. doi: 10.1074/jbc.M805316200. [DOI] [PubMed] [Google Scholar]
- Wolff EC, Folk JE, Park MH. Enzyme-substrate intermediate formation at lysine 329 of human deoxyhypusine synthase. J Biol Chem. 1997;272:15865–71. doi: 10.1074/jbc.272.25.15865. [DOI] [PubMed] [Google Scholar]
- Wolff EC, Lee YB, Chung SI, Folk JE, Park MH. Deoxyhypusine synthase from rat testis: purification and characterization. J Biol Chem. 1995;270:8660–6. doi: 10.1074/jbc.270.15.8660. [DOI] [PubMed] [Google Scholar]
- Wolff EC, Park MH, Folk JE. Cleavage of spermidine as the first step in deoxyhypusine synthesis. The role of NAD. J Biol Chem. 1990;265:4793–9. [PubMed] [Google Scholar]
- Woolstenhulme CJ, Parajuli S, Healey DW, Valverde DP, Petersen EN, Starosta AL, Guydosh NR, Johnson WE, Wilson DN, Buskirk AR. Nascent peptides that block protein synthesis in bacteria. Proc Natl Acad Sci USA. 2013;110:E878–87. doi: 10.1073/pnas.1219536110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa T, Sumida T, Ishii R, Takemoto C, Yokoyama S. A paralog of lysyl-tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P. Nat Struct Mol Biol. 2010;17:1136–43. doi: 10.1038/nsmb.1889. [DOI] [PubMed] [Google Scholar]
- Yao M, Ohsawa A, Kikukawa S, Tanaka I, Kimura M. Crystal structure of hyperthermophilic archaeal initiation factor 5A: a homologue of eukaryotic initiation factor 5A (eIF-5A) J Biochem. 2003;133:75–81. doi: 10.1093/jb/mvg011. [DOI] [PubMed] [Google Scholar]
- Youngman EM, Brunelle JL, Kochaniak AB, Green R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004;117:589–99. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- Zanelli CF, Maragno AL, Gregio AP, Komili S, Pandolfi JR, Mestriner CA, Lustri WR, Valentini SR. eIF5A binds to translational machinery components and affects translation in yeast. Biochem Biophys Res Commun. 2006;348:1358–66. doi: 10.1016/j.bbrc.2006.07.195. [DOI] [PubMed] [Google Scholar]
- Zanelli CF, Valentini SR. Pkc1 acts through Zds1 and Gic1 to suppress growth and cell polarity defects of a yeast eIF5A mutant. Genetics. 2005;171:1571–81. doi: 10.1534/genetics.105.048082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender L, Xue W, Zuber J, Semighini CP, Krasnitz A, Ma B, Zender P, Kubicka S, Luk JM, Schirmacher P, McCombie WR, Wigler M, Hicks J, Hannon GJ, Powers S, Lowe SW. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135:852–64. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk D, Jacobson A. A single amino acid substitution in yeast eIF-5A results in mRNA stabilization. EMBO J. 1998;17:2914–25. doi: 10.1093/emboj/17.10.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]