Abstract
Background
Squamous cell carcinoma (cSCC) and basal cell carcinomas are the two most common cutaneous carcinomas. Molecular profiles predicting metastasis of these cancers have not been identified.
Methods
Epigenetic profiles of 37 primary cases of cSCC and BCC were quantified via the Illumina Goldengate Cancer Panel. Differential protein expression by metastatic potential was analyzed in 110 total cases by immunohistochemical staining.
Results
Unsupervised hierarchical clustering analysis revealed that metastatic BCCs had a methylation profile resembling cSCCs. Metastatic cSCCs were found to be hypermethylated at FRZB (median methylation: 46.7% vs 4.7%; p=4×10−5). Metastatic BCCs were found to be hypomethylated at MYCL2 (median methylation: 3.8% vs 83.4%, p=1.9×10−6). Immunohistochemical staining revealed few differences between metastatic and non-metastatic cancers.
Conclusions
Metastatic primary BCCs and cSCCs had distinct epigenetic profiles when compared to their non-metastatic counterparts. Epigenetic profiling may prove useful in future diagnosis and prevention of advanced non-melanoma skin cancers.
Keywords: DNA methylation, metastasis, cutaneous carcinoma, epigenetic, squamous, basal
Introduction
Cutaneous squamous cell carcinoma (cSCC) and basal cell carcinoma (BCC) are the two most common cutaneous malignancies in the United States. Most non-melanoma skin cancers (NMSC) remain localized and non-invasive, however, there is a risk of loco-regional and distant spread of disease with both cSCC and BCC. Cutaneous squamous cell carcinoma has a risk of metastasis, which has been reported between 2% and 9.9% (1-7). Despite the high incidence of BCC, metastatic transformation is extremely rare with an estimated rate of metastasis between 0.0028% and 0.55% (8-11). In recent decades, much has been learned about the metastatic potential of cutaneous squamous cell carcinoma. Risk factors such as primary location, tumor size, depth of invasion, immunosuppression, and perineural invasion have all been associated with increased risk of metastasis (12-16). Risk factors associated with the metastasis of BCC have been reported despite the rarity of its occurrence. Metastatic BCC tends to arise preferentially from tumors in the head and neck region, which account for 70%-80% of reported cases in the literature, with ear, cheek, and forehead predominating (8, 10-11). The size of the primary tumor appears to be an important risk factor with large, neglected tumors comprising many of the reported cases in the literature. Snow et al. in their series of metastatic BCC found that 75% of metastatic tumors occurred in tumors greater than 2 cm and/or with invasion of bone or other deep structures (10).
Epigenetic modifications, including DNA methylation, have been shown to play an integral role in carcinogenesis, cancer progression, and metastasis. Compared to normal tissues, tumors tend to be hypomethylated at intergenic distal regulatory regions and repetitive elements, which generally results in genomic instability, and hypermethylated at gene promoters, resulting in gene silencing (17). Additionally, epigenetic modifications are known to regulate gene expression patterns and affect cell fate decisions (18). Thus, quantification of epigenetic profiles across the genome can establish similarities or differences in tumor cell type of origin and can provide insight as to whether rare metastatic basal cancers actually represent a basal-like differentiation of squamous cell carcinomas. Epigenetic biomarkers of metastatic potential have previously been identified in malignant melanoma (19), however, it is unknown if primary cSCCs and BCCs with metastatic potential have a distinct epigenetic profile. The high incidences of these cancers make the identification of novel biomarkers of metastasis in these tumors of high clinical utility.
Despite numerous reports citing risk factors associated with metastatic skin cancers, many patients with high-risk features such as large size and invasion of bone do not develop regional or distant metastases. A central question is how to more precisely identify the patients with cSCC or BCC that have a high potential to metastasize. To address this question, we undertook a study of the epigenetic profiles of a cohort of patients from the University of Michigan Cancer Center with documented metastasis of cSCC or BCC and compared their epigenetic signatures to an age-matched control group of non-metastatic cSCC or BCC specimens, respectively, to determine unique patterns that might be associated with metastatic disease in each malignancy.
Materials and Methods
Identification of Cases
Institutional Review Board approval at the University of Michigan was prospectively obtained (HUM00028920) and cases of metastatic cSCC and BCC were identified in a systematic chart review of all surgical cases performed in the Department of Otolaryngology-Head and Neck Surgery at the University of Michigan between 1990 and 2010. Patient records for these patients were examined and pertinent data were collected regarding patient demographics, tumor pathology and course of disease. Surgical specimens were collected to confirm histopathologic characteristics and presence of cSCC or BCC in primary and metastatic tumor by an experienced head and neck pathologist (JBM). Patient records and specimens were scrutinized according to the criteria set forth by Lattes and Kessler in 1951 (20), and adapted by Cotran's review(21) in 1961, which states that an accurate diagnosis of metastatic BCC must satisfy the following criteria: (1) the primary lesion must be of cutaneous origin; (2) metastases must be distinguished from growth by direct extension of the original tumor; and (3) the primary and metastatic tissue must share similar histologic subtypes. These criteria were similarly applied for patients with metastatic cSCC.
FFPE tissue, DNA isolation, and bisulfite conversion
Paraffin blocks containing pathologic tissue from both primary and metastatic cSCC and BCC tumors were obtained from the University of Michigan Pathology Core. In order to provide an experimental control group for patients with metastatic cSCC or BCC, paraffin blocks from age-matched patients that did not metastasize were randomly selected in a 1:2 ratio. Tumor blocks were recut for uniform histopathologic review and microdissection, with the first and last slides of a series of 12 reviewed by a qualified pathologist (JM) to confirm the original diagnosis and to circle areas for DNA extraction. Percent cellularity was estimated for each tumor, and areas with >70% cancer cellularity were designated for use in the analyses.
Tumor regions that were identified for DNA extraction were cored from the formalin fixed paraffin embedded (FFPE) tissue blocks using an 18 gauge needle. DNA was isolated from the cored tissue samples using the QIAamp DNA FFPE Tissue Kit (Qiagen, Valencia, CA) with a modification to the manufacturer's recommended lysis protocol (incubation overnight at 56°C in lysis buffer). DNA concentration and purity was confirmed via NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA). We performed sodium bisulfite modification on 500ng to 1μg of DNA using the EZ DNA Methylation kit (Zymo Research, Orange, CA) following the manufacturer's recommended protocol.
Tissue Microarray (TMA)
Formalin-fixed, paraffin-embedded tissue blocks of 110 cases were obtained from the files of the Department of Pathology, University of Michigan Medical Center, Ann Arbor, MI. The University of Michigan Institutional Review Board provided a waiver of informed consent to obtain these samples. After pathological review, a tissue microarray was constructed from the most representative area using the methodology of Nocito et al. (22). Each case was represented by two 1 mm diameter cores obtained from the most representative, non-necrotic area of the tumor.
Immunohistochemistry
Immunohistochemical staining was performed on the DAKO Autostainer (DAKO, Carpinteria, CA) using diaminobenzadine as the chromogen. Serial sections of de-paraffinized TMA sections were labeled with the antibodies listed in Supplementary Table 1. Appropriate negative (no primary antibody) and positive controls were stained in parallel with each set of tumors studied. The immunoreactivity was scored on a three-tier scale (negative, low- (1+) and high-positive (2+)). Following titration of each antibody, and subsequent immunohistochemical staining of the two TMA slides, slides were taken to an experienced pathologist (JBM) for review and scoring.
Scoring of each stained core on the TMA slides was carried out using a 2-variable system. The percentage of malignant cells with positive staining was represented by a number from 0-100. The intensity of staining was scored on a scale from 0 to 4, where 0 = negative, 1+ = 0-25%, 2+ = 26-50%, 3+ = 51-75% and 4+ = 76-100%. As each tumor core was represented on the TMA in duplicate, the numerical scores of the two cores were averaged separately by percentage and intensity. The resultant values were then analyzed statistically using chi-square contrast tests in a linear regression model to test differences across groups.
Bead Array Methods
The Illumina Goldengate Methylation Cancer Panel was utilized to detect DNA methylation patterns in tumor samples. The Cancer Panel quantifies methylation at 1505 CpG sites located in known CpG islands across 807 genes related to cancer, including oncogenes, tumor suppressor genes, imprinted genes, and genes involved in cell cycle regulation, DNA repair, apoptosis and metastasis. Bead arrays were run at the University of Michigan DNA Sequencing Core Facility according to the manufacturer's protocol. Briefly, bisulfite converted tumor DNA was hybridized to the bead array as described previously (23), and bead arrays were imaged using Illumina Cancer Panel Reader software. Raw bead array fluorescence data was initially analyzed using Illumina BeadStudio Methylation software, which converts fluorescence values of the methylated (Cy5) and unmethylated (Cy3) alleles into an average methylation value at a specific probe using the formula β = [Max(Cy5,0)]/[Max(Cy5,0) + Max(Cy3,0) + 100] with β ∊ (0,1).
Methylation at specific CpG probes on the Goldengate Cancer Panel has been previously shown to be biased by probe thermodynamic properties (24). Known biases include probe length and GC content, which can affect the melting temperature of the probes as well as probe fluorescence intensities. Thus, we used the method proposed by Kuan et al. to normalize our average β values based on probe length and GC content (24).
Statistical Methods
Unsupervised hierarchical clustering of the methylation array data was performed using the Euclidean distance metric and the Ward clustering method in the hclust package in R version 2.10.1.(25). To minimize sex-specific effects, we excluded CpG sites on the sex chromosomes, and included the top 50% of CpG sites with the greatest variance in methylation across samples. For survival analyses, death was considered an “event”; survival time was censored at 3 years (1095 days). The log-rank test was used to test differences in survival distributions using the R Survival package.
Methylation Scores
We calculated a tumor suppressor gene methylation score using our previously described methodology (26). Briefly, we identified genes associated with tumor suppression by conducting a simply query of the National Center for Biotechnology Information (NCBI) Gene database for “tumor suppressor” and extracted Gene IDs. A total of 444 individual CpG sites across 237 genes on the Goldengate Cancer Panel were linked to tumor suppression. Next, for each study participant, the methylation score was defined as the number of tumor suppressor gene-associated CpG sites with normalized average β values above 0.5. Thus, the methylation score ranged from 0 to 444. We infer that a higher methylation score is associated with higher levels of DNA methylation of promoter regions of genes associated with tumor suppression and cell cycle regulation.
We also calculated a Polycomb Repressor Complex 2 (PRC2) gene occupancy methylation score for each study participant. PRC2 target genes were identified in human embryonic stem cells (27), and are known to be important regulators of development (28) and dysregulated in cancer (29). A total of 232 sites across 107 genes on the Cancer Panel were identified as PRC2 targets, as described by Lee et al. A PRC2 occupancy methylation score was calculated by summing the number of PRC2-associated CpG sites with normalized average β values above 0.5. The PRC2 occupancy methylation scores ranged from 0 to 232. Twenty-six genes were found in both the PRC2 occupancy gene list and the tumor suppressor gene list.
Site Specific Analyses
To determine whether specific sites of the genome may be differentially methylated in metastatic cancers, we calculated associations between DNA methylation and metastatic potential. The association between metastasis and individual CpG site DNA methylation at the 1505 CpG sites measured on the Goldengate Cancer Panel were examined using the Limma package in R 2.10.1 (30), adjusting for sex and age. Sample weights based on detection p-values across samples were used in the lmFit function from the Limma package to downweight samples with higher detection p-values. An empirical Bayes method (using the eBayes function in Limma) was used to shrink standard errors to a common value and to rank CpG sites in order of differential methylation. Multiple comparisons were accounted for using the q-value method previously described (31).
Results
Clinical Findings of Cohort
Our cSCC study cohort (Table 1) included 46 patients diagnosed with metastatic cSCC of the head and neck, including tumors from 41 men and 5 women. The mean age at diagnosis of metastatic disease was 73 years with a shorter time interval from primary diagnosis to metastasis of 8.5 months, as compared to BCC. The most common primary site resulting in regional metastasis was periauricular (14/46; 30%) followed by the frontotemporal region (8/46; 18%). Adverse pathological features included perineural invasion (16/46; 35%), perivascular invasion (7/46; 15%), and bone involvement (3/46; 6%). Sixty percent of patients (28/46) were either current or former smokers. The medial overall survival for patients with metastatic cSCC was 4.4 years with no statistical difference at 3 years between metastatic and non-metastatic patients. There was however a trend at 2 years for worse survival in the metastatic cSCC cohort (p=0.08). The demographics of patients with metastatic cSCC are consistent with other reports in the literature (13-16). The majority of patients were male (89%) and the most common primary sites involved with regional metastasis were from the ear (30%) or the frontotemporal region (18%). Perineural invasion was present in 35% of patients, perivascular invasion in 15%, and 18 of 46 patients (39%) of those with metastatic disease were poorly differentiated on histopathology, compared with 0% of the specimens in the non-metastatic group. The median survival for patients with metastatic cSCC was 4.4 years, similar to that reported elsewhere (13-16). There was a trend at 2 years for worse survival in the metastatic cSCC cohort (p=0.08).
Table 1.
Clinical features of patients with metastatic BCC or cSCC.
| Clinical characteristics of patients with metastatic disease | BCC Patients (N = 5) | SCC Patients (N=46) |
|---|---|---|
| Male : Female | 4 M : 1 F | 41 M : 5 F |
| Mean age at dx of metastasis (S.D.) | 65 (8) | 73 (11) |
| Median interval between primary and met | 7 years | 8.5 months |
| Perineural Invasion | 5 | 16 |
| Perivascular Invasion | 2 | 7 |
| Bone Involvement | 5 | 3 |
| Smoking Status | 3 current; 2 former | 1 current; 27 former |
Our BCC study cohort included five patients diagnosed with verified metastatic basal cell carcinoma (Table 1). The group consisted of 4 males and 1 female. The mean age at the time of primary BCC diagnosis was 55.0 while the mean age at the diagnosis of metastatic disease was 65 (range: 52-75). Sixty percent of the patients had multiply recurrent tumors prior to the diagnosis of a metastasis. The median interval between the development of the primary lesion and progression to clinically evident metastatic disease was 7 years. Eighty percent of primary sites originated in the facial region including temple, ear, chin, and forehead. Primary lesions ranged in size from 1.3 cm2 to 70 cm2. Each of these cases developed metastatic disease in the neck: one patient with Level 1 disease, 3 patients with Level 2 disease, and the lower back primary had Level IV and V disease along with extensive axillary disease (13/17 positive nodes). Histopathologic examination confirmed the presence of BCC in all metastatic tumor specimens, which were identical in subtype to the corresponding primary pathology. All specimens were representative of an aggressive growth pattern, and subtypes included nodular, infiltrative, metatypical and infiltrating keratotic, with several cases including mixed features. Perineural invasion was present in all cases and perivascular invasion was evident in 2/5 patients. Only one of the five patients received radiation prior to development of metastasis, due to extent of the primary tumor. The other four received radiation following resection of the metastases, and only one of those underwent adjuvant chemotherapy. The mean overall survival time after the development of regional metastases for the cohort was 4.8 years. Two patients are still alive, but one has distant metastatic disease (lung). For all the deceased patients, the cause of death is unknown but each of the three was disease free at the date of the last documented follow-up. The BCC case cohort of five patients is largely representative of the published literature on age, gender and ethnic demographics(8-11); we had a male predominance, with the primary diagnosis in the fifth decade and an extended interval before clinically apparent metastases were diagnosed. All patients had T4 disease based on current AJCC criteria (bone involvement) and were treated with radical surgical resection of both the primary disease and regional metastases. Four of these patients underwent adjuvant radiation, one of whom also received post-chemoradiotherapy (cisplatin). Despite the co-morbidities of these procedures, as well as the reported median survival of 8 months (11), the shortest observed survival following surgery was 18 months, and the median overall survival from the time of definitive surgical management was almost 5 years.
Methylation Patterns
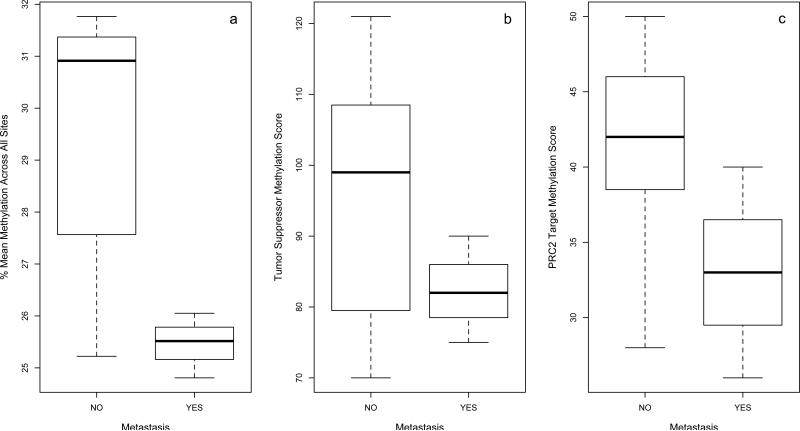
Methylation patterns across all of the 1505 CpG sites assayed on the Goldengate Cancer Panel were compared between primary BCC and SCC tissues that did and did not metastasize. When the top 50% most variable sites were included in an unsupervised hierarchical clustering analysis, non-metastatic BCCs tended to cluster separately from metastatic BCCs and SCCs (Figure 1). For SCCs, there was no overall difference in mean methylation across all sites observed between the two groups (Wilcoxon p=0.18), while for BCCs, primary tumors that metastasized were more likely to be hypomethylated (Wilcoxon p=0.07; Figure 2). Additionally, comparing tumor suppressor methylation score as well as Polycomb Repressive Complex 2 occupancy score across metastatic and non-metastatic primary BCCs revealed a strong trend that had a similar pattern to that seen with mean methylation, with both tumor suppressor genes and PRC2 target genes hypomethylated in metastatic tumors, however, this did not reach statistical significance (Figure 2). For SCCs, there was no significant difference in tumor suppressor methylation score comparing primary tumors that metastasized to those that did not (Wilcoxon p=0.83). Nor was any difference observed between the two groups when comparing PRC2 gene occupancy methylation score (Wilcoxon p=0.30).
Figure 1.
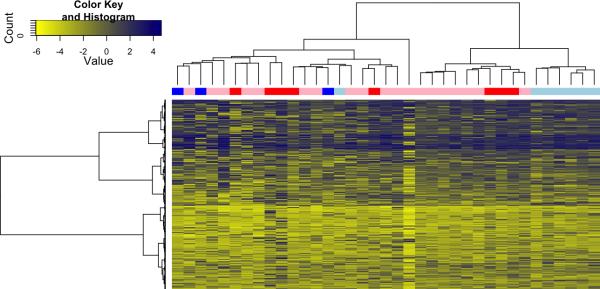
Unsupervised hierarchical cluster analysis of DNA methylation identified nonmetastatic BCCs (light blue) as having a distinct methylation pattern when compared to metastatic BCCs (dark blue), non-metastatic SCCs (pink), and metastatic cSCCs (red).
Figure 2.
(a) Mean methylation across 1505 CpG sites, (b) tumor suppressor methylation score, and (c) PRC2 target methylation score compared between non-metastatic (n=7) and metastatic (n=3) primary basal cell carcinomas.
Site Specific Methylation Analyses – Cutaneous Squamous Cell Carcinomas
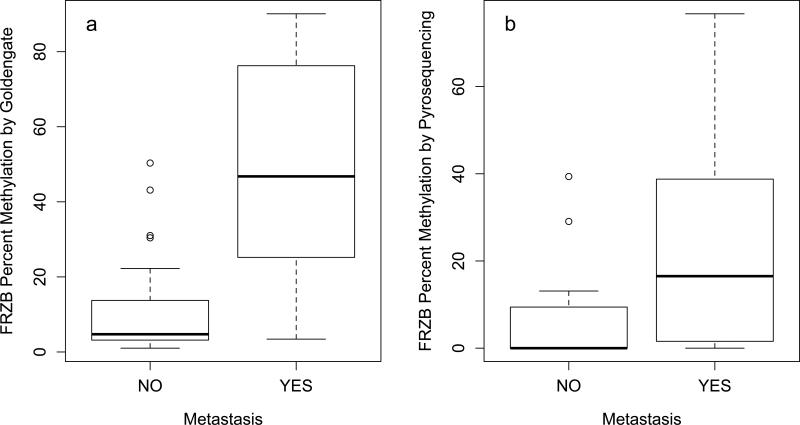
To quantify site-specific epigenetic differences between primary cSCCs that metastasized and those that did not, we measured the associations between each of the 1505 CpG sites on the Goldengate Cancer Panel and metastatic potential of the tumor. The ten most differentially methylated sites that were identified are listed in Table 2. The top hit, a CpG site in the promoter region of FRZB, a modulator of Wnt signaling known to be involved in regulation of bone development, was found to be hypermethylated in metastatic (median methylation: 46.7%) compared to non-metastatic (median methylation: 4.7%) primary SCCs (p=4×10−5,q=0.14). Additionally, CpG sites associated with the developmental transcription factors TFAP2C and ASCL2 were also found to be hypermethylated in metastatic primary SCCs, while CpG sites associated with the actin protein ACTG2 were found to be hypomethylated in metastatic primary SCCs. We also compared DNA methylation of 4 sites in the FRZB promoter sequence in a subset of the samples analyzed on the Goldengate Cancer Panel as well as 10 additional primary SCCs and observed a similar pattern of methylation based on metastatic potential as measured on the array (Wilcoxon p = 0.08) (Figure 3).
Table 2.
The ten most differentially methylated CpG sites associated with metastatic potential of the primary cSCC, adjusting for patient age and sex.
| Gene | Chromosome | CpG Coordinate | Log Fold Change | T Value | P Value | Q Value |
|---|---|---|---|---|---|---|
| FRZB | 2 | 183439557 | 2.632 | 4.612 | 0.0001 | 0.14 |
| ASCL2 | 11 | 2249118 | 1.635 | 4.050 | 0.0004 | 0.29 |
| TFAP2C | 20 | 54638025 | 1.115 | 3.840 | 0.0007 | 0.29 |
| GSTM2 | 1 | 110012367 | 2.501 | 3.717 | 0.0010 | 0.29 |
| GSTM1 | 1 | 110031602 | −2.489 | −3.699 | 0.0010 | 0.29 |
| TFF1 | 21 | 42659893 | −2.570 | −3.623 | 0.0012 | 0.29 |
| RARRES1 | 3 | 159933026 | −1.883 | −3.587 | 0.0014 | 0.29 |
| ACTG2 | 2 | 73973146 | −1.565 | −3.539 | 0.0015 | 0.29 |
| ACTG2 | 2 | 73973699 | −1.943 | −3.350 | 0.0025 | 0.41 |
| NGFB | 1 | 115682393 | 1.085 | 3.154 | 0.0040 | 0.46 |
Figure 3.
In cSCC, methylation of FRZB promoter quantified by (a) the Goldengate methylation panel or (b) pyrosequencing of 4 CpG sites in the FRZB promoter.
Site Specific Methylation Analyses – Basal Cell Carcinomas
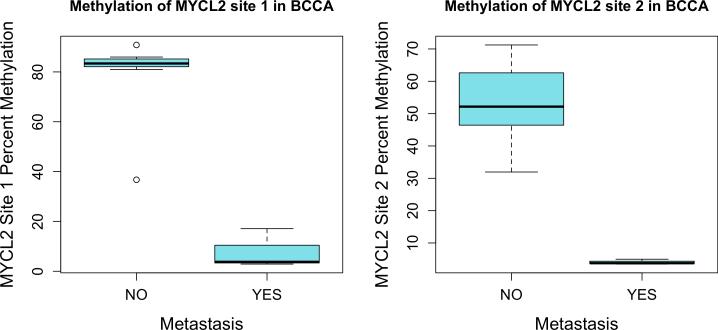
To quantify specific epigenetic differences seen in metastatic and non-metastatic primary basal cell carcinomas, we measured the associations between methylation at each of the 1505 CpG sites on the GoldenGate array to assess the possible markers of metastatic potential in primary tumors. Two CpG sites, both associated with the putative tumor suppressor gene MYCL2 (Figure 4), were found to be significantly hypomethylated in metastatic tumors compared to control primary BCC that did not metastasize (p=1.9×10−6, q=0.002 and p=3.7×10−5, q=0.028 respectively). While not reaching statistical significance when adjusting for multiple comparisons (q<0.05), a number of other genes were also found to be differentially methylated (p<0.05) in metastatic BCCs (Supplementary Table 2). The majority of these sites were found to be hypomethylated, similar to the global measures of methylation presented above, including sites associated with EGF, GNG7, GRB10, and FGF9.
Figure 4.
Two CpG sites associated with MYCL2 were found to be significantly hypomethylated in BCCs that became metastatic (p=1.9×10−6, q=0.002 and p=3.7×10−5, q=0.028 respectively).
Immunohistochemical Staining
During preliminary quantification of epigenetic differences between metastatic and nonmetastatic samples, numerous candidate genes were identified as potentially associated with metastatic potential. To investigate whether this differential methylation was reflected in protein expression, immunohistochemical (IHC) staining was performed on a subset of the proteins for which IHC antibodies are commercially available (Supplementary Table 1).
Following the construction of a tissue microarray (TMA) that included pathological specimens from both cSCC and BCC groups, tissue from metastatic and non-metastatic tumors were used for IHC staining. Staining scores were then compared between groups, however, a significant difference in protein expression that differentiated metastatic from nonmetastatic primary tumors was discovered in only one case. When comparing the intensity of staining across cSCC samples, a neurotrophin receptor kinase, TrkB (also known as NTRK2), was identified by this process as being more highly expressed in SCC primaries that went on to metastasize compared to those that did not . Notably, this data reflects the highly elevated staining intensity of only 3 specimens out of 26 stained in the metastatic SCC group, all of which scored 2 on a 0-4 scale. This, when compared to the 6 cores of 70 with positive staining in the nonmetastatic group, all of which received an intensity score of 1, achieved statistical significance in our model. IHC staining of BCC samples did not yield any significant differences between metastatic and non-metastatic tissues.
Discussion
In addition to reviewing our clinical experience with metastatic cSCC and BCC, we undertook a study of the epigenetic signatures in primary tumors that developed regional or distant metastatic disease to determine how to identify patients who might be at high risk of developing metastatic cSCC or BCC. We identified epigenetic predictors of metastatic potential in both cSCC and BCC, with metastatic cSCC primaries hypermethylated at FRZB and metastatic BCC primaries hypomethylated at MYCL2. Additionally, we identified that metastatic primary cSCCs were more likely to stain positive for TrkB, with a higher staining intensity, although this relationship was driven by a small number of cases.
Despite a small sample size, our comprehensive experimental analysis of epigenetic regulation in metastatic cSCC and BCC was adequately powered to detect significant differences in methylation signatures between groups. When comparing epigenetic profiles between metastatic vs. non-metastatic primary cSCCs, we did not see widespread differences in methylation patterns, but did identify differential methylation at a CpG site in the promoter region of FRZB. The protein product of FRZB is secreted and involved in regulation of bone morphogenesis during development (32-33) through antagonism of Wnt signaling (34-35). Previous studies of the role of FRZB in cancer have identified that overexpression of FRZB is associated with a more differentiated tumor phenotype in gastric cancer (36), as well as a less invasive phenotype in prostate cancer (37). Additionally, promoter methylation of FRZB was associated with a higher grade in non-invasive bladder cancer cases (38). Similarly, in this study, we observed increased promoter methylation in primary cSCC tumors that metastasized. Our results, combined with previous findings, suggest that FRZB methylation could serve as a biomarker of tumor aggressiveness or metastatic potential at multiple cancer sites.
We observed widespread methylation differences between metastatic and non-metastatic primary BCCs. Metastatic BCCs were found to be hypomethylated across the genome, as well as at tumor suppressor genes and PRC2 target genes (Figure 2). Additionally, we found that in an unsupervised hierarchical cluster analysis, non-metastatic primary BCCs clustered separately from the rest of tumors, suggesting a unique methylation profile (Figure 1). In gene specific analyses, MYCL2 was the only gene to show a significantly different methylation score across metastatic and non-metastatic primaries, and this was observed at both loci included on the array. This gene localizes to the X-chromosome, has a poorly characterized function, and has previously been described as highly and specifically expressed in testes (39). MYCL2 shares sequence homology with the tumor suppressor genes MYCL1 and MYC. In these samples, MYCL2 was unmethylated in metastatic BCCA and methylated in non-metastatic BCCA, suggesting that methylation at these loci could be used as biomarkers of metastatic potential. Of interest in the gene list with borderline significance, GNG7, involved in transmembrane signaling, has been shown to be hypermethylated in head and neck squamous cell carcinoma (40) and fibroblast growth factor 9 (FGF9) is upregulated in cutaneous squamous cell carcinomas (41).
Immunohistochemical staining for a range of proteins, including EGFR and RUNX3, did not reveal significant differences in expression when comparing primary tumors that metastasized and those that did not. Interestingly, despite the differences observed in methylation of FRZB in metastatic cSCCs, we did not observe a significant difference in protein expression between metastatic and non-metastatic primaries. We did, however, observe a significant increase in staining intensity of TrkB in metastatic cSCCs. TrkB has been described previously as an important oncogene in head and neck cancer, medullary thyroid carcinoma, myelomas, prostate cancer, and lymphoid tumors (42-45). The small number of metastatic cases that stained positively for TrkB suggests that there may be a subset of cSCCs where metastasis is driven, in part, by TrkB overexpression.
There exists an ongoing debate as to whether BCC truly metastasizes, with some arguing that it is rather the cells of a primary lesion with squamous cell differentiation that initiate invasion and spread of disease, acting as a vehicle for BCC cells to disseminate. However, the pathology of our BCC specimens, both primary and metastatic, consistently demonstrates the features of basal cell carcinoma on histologic analysis. Interestingly, however, the primary BCCs that developed metastases had a methylation pattern that strongly clustered with cSCC (Figure 1). This cluster profile was shared with both primary cSCC that metastasized and did not metastasize, which would suggest that metastatic BCCs, at least when viewed from a gene methylation perspective, more closely resembles cSCC. Perhaps, analysis of the gene methylation profiles of high risk BCC might offer clues to those primaries at greatest risk of regional and distant metastases and could offer the possibility of increased surveillance or the use of adjuvant treatments such as the hedgehog pathway inhibitor (vismodegib) for high risk lesions.
This study has elucidated definitive differences in epigenetic signatures among the metastatic forms of cutaneous squamous cell carcinoma and basal cell carcinoma. FRZB and MYCL2 may be putative biomarkers of the metastatic phenotype and useful in future diagnosis and prevention of advanced disease. However, our data highlight the need for additional research on this subject; mechanistic and mutational data remains elusive due to the relative rare nature of this process. However, epigenetic modification of functionally important genes may hold special significance in the improved management of metastatic cutaneous disease.
Supplementary Material
Acknowledgements
This study was funded as a part of the University of Michigan Head and Neck Specialized Program of Research Excellence, NIH/NCI CA097248, as well as through NIH/NCI R01CA158286. Support for JAC was provided by a Rackham Predoctoral Fellowship from the University of Michigan and Institutional Training Grants from the National Institute of Environmental Health Sciences (NIEHS) (T32 ES007062) and the National Human Genome Research Institute (NHGRI) (T32 HG00040).
References
- 1.Chuang TY, Popescu NA, Su WP, Chute CG. Squamous cell carcinoma. A population-based incidence study in Rochester. Minn. Arch Derm. 1990;126:185–8. doi: 10.1001/archderm.126.2.185. [DOI] [PubMed] [Google Scholar]
- 2.Dinehart SM, Pollack SV. Metastases from squamous cell carcinoma of the skin and lip. An analysis of twenty-seven cases. J Am Acad Derm. 1989;21:241–8. doi: 10.1016/s0190-9622(89)70168-7. [DOI] [PubMed] [Google Scholar]
- 3.Epstein E, Epstein NN, Bragg K, Linden G. Metastases from squamous cell carcinomas of the skin. Arch Derm. 1968;97:245–51. [PubMed] [Google Scholar]
- 4.Katz AD, Urbach F, Lilienfeld AM. The frequency and risk of metastases in squamous-cell carcinoma of the skin. Cancer. 1957;10:1162–6. doi: 10.1002/1097-0142(195711/12)10:6<1162::aid-cncr2820100612>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 5.Lund HZ. How often does squamous cell carcinoma of the skin metastasize? Arch Derm. 1965;92:635–7. [PubMed] [Google Scholar]
- 6.Rowe DE, Carroll RJ, Day CL., Jr. Prognostic factors for local recurrence, metastasis, and survival rates in squamous cell carcinoma of the skin, ear, and lip. Implications for treatment modality selection. J Am Acad Derm. 1992;26:976–90. doi: 10.1016/0190-9622(92)70144-5. [DOI] [PubMed] [Google Scholar]
- 7.Tavin E, Persky M. Metastatic cutaneous squamous cell carcinoma of the head and neck region. The Laryngoscope. 1996;106:156–8. doi: 10.1097/00005537-199602000-00009. [DOI] [PubMed] [Google Scholar]
- 8.Farmer ER, Helwig EB. Metastatic basal cell carcinoma: a clinicopathologic study of seventeen cases. Cancer. 1980;46:748–57. doi: 10.1002/1097-0142(19800815)46:4<748::aid-cncr2820460419>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 9.Lo JS, Snow SN, Reizner GT, Mohs FE, Larson PO, Hruza GJ. Metastatic basal cell carcinoma: report of twelve cases with a review of the literature. J Am Acad Derm. 1991;24:715–9. doi: 10.1016/0190-9622(91)70108-e. [DOI] [PubMed] [Google Scholar]
- 10.Snow SN, Sahl W, Lo JS, Mohs FE, Warner T, Dekkinga JA, et al. Metastatic basal cell carcinoma. Report of five cases. Cancer. 1994;73:328–35. doi: 10.1002/1097-0142(19940115)73:2<328::aid-cncr2820730216>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 11.von Domarus H, Stevens PJ. Metastatic basal cell carcinoma. Report of five cases and review of 170 cases in the literature. J Am Acad Derm. 1984;10:1043–60. doi: 10.1016/s0190-9622(84)80334-5. [DOI] [PubMed] [Google Scholar]
- 12.Weinberg AS, Ogle CA, Shim EK. Metastatic cutaneous squamous cell carcinoma: an update. Derm Surg. 2007;33:885–99. doi: 10.1111/j.1524-4725.2007.33190.x. [DOI] [PubMed] [Google Scholar]
- 13.Turner SJ, Morgan GJ, Palme CE, Veness MJ. Metastatic cutaneous squamous cell carcinoma of the external ear: a high-risk cutaneous subsite. J Laryngol Otol. 2010;124:26–31. doi: 10.1017/S0022215109991101. [DOI] [PubMed] [Google Scholar]
- 14.Veness MJ, Palme CE, Morgan GJ. High-risk cutaneous squamous cell carcinoma of the head and neck: results from 266 treated patients with metastatic lymph node disease. Cancer. 2006;106:2389–96. doi: 10.1002/cncr.21898. [DOI] [PubMed] [Google Scholar]
- 15.Veness MJ, Porceddu S, Palme CE, Morgan GJ. Cutaneous head and neck squamous cell carcinoma metastatic to parotid and cervical lymph nodes. Head Neck. 2007;29:621–31. doi: 10.1002/hed.20576. [DOI] [PubMed] [Google Scholar]
- 16.Veness MJ. High-risk cutaneous squamous cell carcinoma of the head and neck. J Biomed Biotechnol. 2007;2007:80572. doi: 10.1155/2007/80572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–13. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 18.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–36. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanemura A, Terando AM, Sim MS, van Hoesel AQ, de Maat MF, Morton DL, et al. CpG island methylator phenotype predicts progression of malignant melanoma. Clin Cancer Res. 2009;15:1801–7. doi: 10.1158/1078-0432.CCR-08-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LATTES R, KESSLER RW. Metastasizing basal-cell epithelioma of the skin; report of two cases. Cancer. 1951;4:866–78. doi: 10.1002/1097-0142(195107)4:4<866::aid-cncr2820040424>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 21.COTRAN RS. Metastasizing basal cell carcinomas. Cancer. 1961;14:1036–40. doi: 10.1002/1097-0142(196109/10)14:5<1036::aid-cncr2820140518>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 22.Nocito A, Kononen J, Kallioniemi OP, Sauter G. Tissue microarrays (TMAs) for high-throughput molecular pathology research. Int J Cancer. 2001;94:1–5. doi: 10.1002/ijc.1385. [DOI] [PubMed] [Google Scholar]
- 23.Bibikova M, Lin Z, Zhou L, Chudin E, Garcia EW, Wu B, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–93. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuan PF, Wang S, Zhou X, Chu H. A statistical framework for Illumina DNA methylation arrays. Bioinformatics. 2010;26:2849–55. doi: 10.1093/bioinformatics/btq553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qu Y, Li J-f, Cai Q, Wang Y-w, Gu Q-l, Zhu Z-g, et al. Over-expression of FRZB in gastric cancer cell suppresses proliferation and induces differentiation. J Cancer Research Clin Oncology. 2008;134:353–64. doi: 10.1007/s00432-007-0291-0. [DOI] [PubMed] [Google Scholar]
- 26.Colacino JA, Arthur AE, Dolinoy DC, Sartor MA, Duffy SA, Chepeha DB, et al. Pretreatment dietary intake is associated with tumor suppressor DNA methylation in head and neck squamous cell carcinomas. Epigenetics. 2012;7:883–91. doi: 10.4161/epi.21038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–13. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Surface LE, Thornton SR, Boyer LA. Polycomb group proteins set the stage for early lineage commitment. Cell stem cell. 2010;7:288–98. doi: 10.1016/j.stem.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 29.Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nature Reviews Cancer. 2009;9:773–84. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 30.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 31.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoang B, Moos M, Vukicevic S, Luyten FP. Primary structure and tissue distribution of FRZB, a novel protein related to Drosophila frizzled, suggest a role in skeletal morphogenesis. J Biol Chem. 1996;271:26131–7. doi: 10.1074/jbc.271.42.26131. [DOI] [PubMed] [Google Scholar]
- 33.Hoang BH, Thomas JT, Abdul-Karim FW, Correia KM, Conlon RA, Luyten FP, et al. Expression pattern of two Frizzled-related genes, Frzb-1 and Sfrp-1, during mouse embryogenesis suggests a role for modulating action of Wnt family members. Dev Dyn. 1998;212:364–72. doi: 10.1002/(SICI)1097-0177(199807)212:3<364::AID-AJA4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 34.Enomoto-Iwamoto M, Kitagaki J, Koyama E, Tamamura Y, Wu C, Kanatani N, et al. The Wnt antagonist Frzb-1 regulates chondrocyte maturation and long bone development during limb skeletogenesis. Dev Biol. 2002;251:142–56. doi: 10.1006/dbio.2002.0802. [DOI] [PubMed] [Google Scholar]
- 35.Wang S, Krinks M, Lin K, Luyten FP, Moos M. Frzb, a secreted protein expressed in the Spemann organizer, binds and inhibits Wnt-8. Cell. 1997;88:757–66. doi: 10.1016/s0092-8674(00)81922-4. [DOI] [PubMed] [Google Scholar]
- 36.Qu Y, Li JF, Cai Q, Wang YW, Gu QL, Zhu ZG, et al. Over-expression of FRZB in gastric cancer cell suppresses proliferation and induces differentiation. J Cancer Res Clin Oncol. 2008;134:353–64. doi: 10.1007/s00432-007-0291-0. [DOI] [PubMed] [Google Scholar]
- 37.Zi X, Guo Y, Simoneau AR, Hope C, Xie J, Holcombe RF, et al. Expression of Frzb/secreted Frizzled-related protein 3, a secreted Wnt antagonist, in human androgen-independent prostate cancer PC-3 cells suppresses tumor growth and cellular invasiveness. Cancer Res. 2005;65:9762–70. doi: 10.1158/0008-5472.CAN-05-0103. [DOI] [PubMed] [Google Scholar]
- 38.Marsit CJ, Houseman EA, Christensen BC, Gagne L, Wrensch MR, Nelson HH, et al. Identification of methylated genes associated with aggressive bladder cancer. PLoS One. 2010;5:e12334. doi: 10.1371/journal.pone.0012334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robertson NG, Pomponio RJ, Mutter GL, Morton CC. Testis-specific expression of the human MYCL2 gene. Nucleic Acids Res. 1991;19:3129–37. doi: 10.1093/nar/19.11.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hartmann S, Szaumkessel M, Salaverria I, Simon R, Sauter G, Kiwerska K, et al. Loss of protein expression and recurrent DNA hypermethylation of the GNG7 gene in squamous cell carcinoma of the head and neck. J Appl Genet. 2012;53:167–74. doi: 10.1007/s13353-011-0079-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kathpalia VP, Mussak EN, Chow SS, Lam PH, Skelley N, Time M, et al. Genome-wide transcriptional profiling in human squamous cell carcinoma of the skin identifies unique tumor-associated signatures. J Dermatol. 2006;33:309–18. doi: 10.1111/j.1346-8138.2006.00075.x. [DOI] [PubMed] [Google Scholar]
- 42.Kupferman ME, Jiffar T, El-Naggar A, Yilmaz T, Zhou G, Xie T, et al. TrkB induces EMT and has a key role in invasion of head and neck squamous cell carcinoma. Oncogene. 2010;29:2047–59. doi: 10.1038/onc.2009.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee J, Jiffar T, Kupferman ME. A novel role for BDNF-TrkB in the regulation of chemotherapy resistance in head and neck squamous cell carcinoma. PloS one. 2012;7:e30246. doi: 10.1371/journal.pone.0030246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thiele CJ, Li Z, McKee AE. On Trk--the TrkB signal transduction pathway is an increasingly important target in cancer biology. Clin Cancer Res. 2009;15:5962–7. doi: 10.1158/1078-0432.CCR-08-0651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yilmaz T, Jiffar T, de la Garza G, Lin H, Milas Z, Takahashi Y, et al. Theraputic targeting of Trk supresses tumor proliferation and enhances cisplatin activity in HNSCC. Cancer Bio Therapy. 2010;10:644–53. doi: 10.4161/cbt.10.6.12782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.