Summary
Reasons for performing study
Biological treatments for osteoarthritis (OA) are an important component of disease control. Understanding the expression of catabolic and anabolic genes during osteoarthritis progression should help to identify the major mediators of the disease.
Objective
To compare the cytokine and anabolic marker concentrations in synovium, synovial fluid, and cartilage between normal and osteoarthritic joints.
Methods
Equine carpi from horses age 2–11 years were used. Tissues were harvested at the time of surgery or euthanasia, and RNA was isolated for RT-PCR analysis. Tumor necrosis factor alpha (TNFα), interleukin-1beta (IL-1β), aggrecanase 1 (ADAMTS-4), aggrecanase 2 (ADAMTS-5), matrix metalloproteinase-13 (MMP-13), interleukin 17 (IL-17), and collagen I alpha 1(Col-1) expression was determined in synovium. TNFα, IL-1β, ADAMTS-4, ADAMTS-5, MMP-13, IL-17, collagen IIB (Col-2B), and aggrecan expression was determined in cartilage. TNFα concentration in the synovial fluid was determined by enzyme-linked immunosorbent assay (ELISA).
Results
Expression of TNFα, ADAMTS-5, and MMP-13 was significantly increased in synovial tissue from OA joints. Synovial membrane IL-1β abundance showed only moderate elevations in OA, without reaching significant levels. Cytokine expression was increased in OA cartilage samples, particularly for TNFα (p=0.0007), IL-1β (p<0.0001), ADAMTS-4 (p=0.0011), and MMP-13 (p<0.0001). Collagen type I expression was significantly increased in synovial tissues from OA groups. Collagen type II message was diminished in mild and moderate stages of OA, but rebounded to significant elevations in severely degenerate joints. Conversely, aggrecan levels significantly declined in all OA cartilage groups (p<0.001). Synovial fluid TNFα peptide concentration was significantly increased in severe OA cases (p=0.021).
Conclusion
TNFα was significantly increased in all degrees of equine OA, and was abundantly expressed in synovial membrane and cartilage. Similarly, IL-1β was overexpressed in OA cartilage, but not to a significant extent in synovium. ADAMTS-4 was more abundant in OA cartilage while ADAMTS-5 predominated in OA synovium. IL-17 expression was not observed in osteoarthritic equine synovium nor cartilage.
Potential relevance
Control of TNFα should be considered further as a target in the treatment of OA. ADAMTS-4 may be the primary aggrecanase causing cartilage breakdown in OA.
Keywords: horse, osteoarthritis, cytokine, TNFα, IL-1, cartilage, aggrecanase, MMP
Introduction
The importance of different inflammatory molecules in the development of osteoarthritis (OA) is a critical factor in the development of biomedical therapies. Intraarticular steroid administration is one of the most commonly used pharmaceuticals for human and equine osteoarthritis. Steroids have been shown to decrease a variety of cytokines including tumor necrosis factor (TNFα) and interleukin 17 (IL-17) (Alex et al. 2007). Biologic treatments such as tumor necrosis factor alpha antibody and decoy receptors, and interleukin-1beta receptor antagonist protein (IRAP) are increasingly used to control rheumatoid and osteoarthritis, respectively, in human patients (Augustsson et al. 2009; Chevalier et al. 2009). Enhanced application of biomolecular controls may provide attenuation of catabolic processes while better preserving existing matrix formation, compared to steroids.
Tumor necrosis factor-α and interleukin 1 beta (IL-1β) are increased in osteoarthritic joints (Furuzawa-Carballeda et al. 2009). They bind at different receptors but cause a similar increase in NFκB, a transcription factor that binds regulatory elements on DNA to increase transcriptional activity, and JNK, a kinase which controls the nuclear and mitochondrial activation of other molecules (Brennan et al. 2002; Yoshizawa et al. 2008). These intermediates then increase the transcription of matrix metalloproteinases (MMPs), cycloxygenase-2 (COX-2), and inducible nitric oxide synthase (iNOS). The end result is increased cartilage degradation and prostaglandin release leading to joint swelling and pain (Kunisch 2009).
IL-1β has been called the master cytokine in human osteoarthritis (Berenbaum 2007). The IL-1β receptor is more concentrated in the cartilage surrounding OA lesions when compared to areas in the same joint that are more remote to the lesion (Schlopov. et al. 2000). Further, the innate IRAP to IL-1β ratio in synovium decreases as the severity of OA increases (Smith et al. 1997). This allows the IL-1β to bind more of its receptors leading to an increase in several different MMPs and aggrecanases. These enzymes are proteolytic and cause cartilage matrix destruction. Furthermore, in vitro treatment of cartilage with IL-1β has been shown to decrease the anabolic profile in chondrocytes, including suppression of both aggrecan and collagen elaboration (Stove et al. 2000; Venkatesan et al. 2004). Despite this, stable IL-1β expression has an important role in cartilage homeostasis. A recent study in IL-1β knockout mice, using a collateral ligament transection and meniscectomy model to create OA, found that IL-1β knockout mice had on average a higher degree of OA than normal mice (Clements et al. 2003).
TNFα has also been implicated in joint degeneration. Suppression of TNFα led to decreased cartilage damage in OA joints compared to untreated OA joints (Fernandez et al. 2002). Like IL-1β, the TNFα receptor is also increased in OA cartilage. Additionally, the enzyme that converts TNFα to its active form is increased in OA (Amin 1999), suggesting the ubiquity of the TNF system in OA.
Based on these data, we sought better to understand which joint tissues have increased levels of cytokines and to what degree different cytokines increase in equine OA joints as compared to normal joints. We hypothesized that an array of cytokines, including TNFα, IL-1β, ADAMTS-4, ADAMTS-5, MMP-13, and IL-17, would be significantly increased in OA synovium and cartilage. We also hypothesized that anabolic matrix markers such as Col-1, or Col-2B and aggrecan, would be decreased in OA synovium or cartilage, respectively. This study aimed to identify the important mediators in the progression of equine OA as the basis for treatment programs that inhibit these degradatory mediators.
Materials and methods
Inclusion criteria and case analysis
Horses included in this study were patients of the Cornell University Hospital for Animals, horses used in other research projects, or donations made to the hospital. Owners of clinical patients signed waivers for their samples to be used in this study, and this study was completed under IACUC guidelines. All horses ranged in age from 2 to 11 years and were primarily Thoroughbred, with several other breeds including Warmbloods (2), Quarter Horses (1), and Standardbreds (4). All OA joints had a radiographic examination performed prior to surgery or euthanasia. Carpal joints were scored and grouped as normal, or as mild, moderate, or severe OA according to radiographic analysis of osteophytes, enthesiophytes, osteoproliferation, joint space narrowing, or chronic fracture lines. Similarly, a score was assigned at gross or arthroscopic examination to grade severity of cartilage disruption and subchondral bone loss. An assessment sheet for each case was completed by the authors to include the location of the lesion, the extent and type of of articular cartilage involvement, and the extent of progression into subchondral bone at surgery or gross examination. The arthroscopic/gross exam score varied from 0= no involvement; 1=fibrillation or fragmentation not extending more than 5mm from the fracture line; 2=degeneration extending more than 5mm but less than 30% of the articular surface; 3=loss of 50% or more of the articular surface. The radiographic category was primarily used to assign OA extent, with additional corroborating information derived from the arthroscopic/gross scoring and histologic sections in cases which fell between categories.
Thirty-six horses with OA joints and 22 horses with normal joints were included in the study, and all had synovial tissue cytokine gene expression assessed. Of these, 21 horses with OA joints and 8 horses with normal joints also had cartilage cytokine expression levels determined, and synovial fluid was examined in 31 horses with OA and 22 horses with normal joints. There were 10 mild OA cases, 18 moderate cases, and 8 severe cases.
Tissue harvesting
All carpal joints were clipped and aseptically prepared. Synovial fluid samples were aspirated from the affected or normal joint using an 18g 1.5 inch needle. Fluid was centrifuged at 1000g for 5 minutes at 5–10°C. The supernatant was aspirated and the pellet was discarded. Synovial samples acquired at the time of carpal arthroscopy were gathered using a biopsy suction punch rongeur (Dyonics1) with a sterile tube extension and 40 micron mesh bag attached to the fluid output. Cartilage samples were collected using Ferris Smith ronguers. Cartilage immediately surrounding or on the osteochondral fragment and directly involved with the lesion was used for RNA isolation. For necropsy cases, tissues were collected by aseptically opening the affected or normal joint. Synovium and cartilage were removed using a #10 blade and forceps. Joint capsule was excluded from samples for RNA isolation, but included for histologic assessment. Bone slabs were retrieved using an oscillating saw. These samples were placed in 4% paraformaldehyde at 4°C. All samples for RNA isolation and synovial fluid were frozen at −80°C until their time of use.
Synovium and cartilage histology
All samples were kept in paraformaldehyde for five days. The synovium samples were then placed in 70% alcohol until they could be batch processed and embedded into paraffin blocks. Sections were then cut 6μm in width and stained with hematoxylin and eosin (H&E). The bone slabs were decalcified in a citrate buffered formic acid or a formic and hydrochloric acid mixture. Slabs took from 5 to 7 days to decalcify. Decalcification was monitored by radiographic examination. Sections were cut again at 6μm and stained with H&E and toluidine blue.
Histologic grading was performed in order to better characterize the severity groups. Synovium was scored according to the following parameters: villus architecture, villus subintimal fibrosis, intimal layer thickening, vasculature, and inflammatory cell infiltrate. Cartilage was scored according to the following parameters: chondrocyte cloning, structure, cell type, and toluidine blue staining. Scores from 0 (normal) to 3 (severe) were assigned to each category. Scores were then used to better categorize the samples.
Tissue homogenization
All synovium samples and cartilage collected at surgery were homogenized in chaotropic lysis buffer at 10,000rpm for 3 repetitions of 10 second homogenization followed by a 20 second cool down period. A high speed homogenizer (Powergen 700D2) was used, and samples were kept on ice at all times. A freezer mill (SPEX CertiPrep 6750 Freezer/Mill3) was used to homogenize normal cartilage. Four cycles of 30 second pulverization followed by one minute rests were completed in liquid nitrogen in order to powder the cartilage for RNA isolation.
RNA Isolation and Quantitative RT-PCR
The homogenized tissue was placed on ice, and 100mg was weighed and separated for RNA isolation using a kit (Versagene’s 5-Prime RNA Isolation Kit4). RNAse free procedures and filtered pipettes were used throughout the isolation procedure. The nanodrop spectrophotometer (ND-1000 Spectrophotometer5) was used to analyze the amount and purity of the RNA product. Concentrations ranged from 3.8–51.8ng/ul, and one joint (mild OA) was excluded as its RNA concentration was less than 3.0ng/ul and its 260/280 wavelength ratio was <1.6.
Gene expression was quantified through the use of quantitative real-time PCR (ABI PRISM 7900 Sequence Detection System 6). Primers and labeled probes (6-FAM as the 5′ reporter label and TAMRA as the 3′ quenching label) were created using commercial software (ABI Primer Express Software 2.0b8a6). Sequences for these primers and probes were generated from Genbank7 or available from clones within the laboratory of Dr Alan Nixon. Copy number was quantified for the mRNA of TNF-α, IL-1β, aggrecanase 1 (ADAMTS-4), aggrecanase 2 (ADAMTS-5), MMP-13, and IL-17 in both the synovium and the cartilage. Collagen type I alpha 1(Col-1) was quantified in synovium, and collagen type IIB (Col-2B) and aggrecan were quantified in cartilage. The housekeeping gene used was 18S, and the 18S copy number was similar for all tissues used. Data used for statistical analysis was copy number per microgram.
Enzyme Linked Immunosorbent Assay (ELISA)
Spun synovial fluid samples were diluted to 50% with phosphate buffered saline (PBS) for TNFα analysis. TNFα concentration was quantified using a commercial ELISA (Thermo Fisher Scientific Equine TNFα ELISA2). The samples for MMP-13 ELISA were used undiluted (Biotrak human MMP-13 ELISA8). This MMP-13 ELISA has not been validated in the horse, but had been successfully used anecdotally (verbal communication, L Fortier, 2009). An equine IL-1β monoclonal antibody (R&D Equine Monoclonal Antibody9) was used at different concentrations and with different marker antibodies to develop an equine IL-1β ELISA. The TNFα and MMP-13 assays were performed using the normal and OA synovial fluid samples to bind to the antibody-coated plates. The assays were completed according to manufacturers directions, and the plates were read on a visible/fluor plate reader (Tecan Safire Absorbance Reader using the Magellan 3.11 analysis program10). Absorbance was converted to concentration using the standard curve derived from serially diluted human peptide standards.
Statistical Analysis
Cytokine expression data and protein concentration in normal joints, and mild OA, moderate OA, or severe OA groups were compared. A two-way analysis of variance (ANOVA) and Tukey’s and LSD post-hoc tests were performed on the qRT-PCR values using a software package (Statistix 9.0 Analytical Software11). Log values were used to normalize all data sets, and summary statistics were performed on the untransformed data. Significance was noted as p<0.05.
Results
Cytokine Gene Expression
Synovial membrane from OA joints had significantly increased gene expression for TNFα (p=0.01), ADAMTS-5 (0.0016) and MMP-13 (0.01) (Table 1, Figures 1–2). Despite obvious upregulation with increased OA burden, no significant difference existed in synovial gene expression for IL-1β and ADAMTS-4. Profiling cartilage pro-inflammatory cytokine gene expression showed significant increases for TNFα (p=0.0007), IL-1β (p<0.001), ADAMTS-4 (p=0.0011), and MMP-13 (p<0.001) in OA samples (Table 2, Figures 3–4). Cartilage ADAMTS-5 expression was unchanged in OA.. No expression of IL-17 was seen in either normal or OA synovium and cartilage. In all cases where a significant difference was noted, the moderate OA mean value was the highest of all of the groups, and mean values often decreased slightly from moderate to severe cases.
Table 1.
Catabolic gene expression in synovial membrane from normal and osteoarthritic joints. Mean values and standard errors are listed, and significant groupings of post-hoc Tukeys and LSD evaluation are shown in parentheses
| Mean Transcription Levels | TNFα | IL-1β | ADAMTS-4 | ADAMTS-5 | MMP-13 | Col-1 |
|---|---|---|---|---|---|---|
| Normal | 18.34 ±5.5 (a) | 20.85±3.1 | 12.48±7.25 | 5177.2±756.7 (a) | 3.047 ± 1.43 (a) | 26,786 ±5,157.5 (a) |
| Mild OA | 20.98 ±6.75 (ab) | 23.04±4.1 | 565.3±324.5 | 9664.1 ± 1506.4 (ab) | 24.78 ± 20.1 (ab) | 65,486 ±25,689 (ab) |
| Moderate OA | 35.72 ± 5.13 (b) | 32.05±6.8 | 725.3±423.6 | 12250.4 ±1945.2 (b) | 153.7 ± 69.1 (b) | 154,448 ±48,107 (b) |
| Severe OA | 18.64 ±6.33 (a) | 24.36±5.1 | 309±191.2 | 8291 ± 1546.0(ab) | 24.81 ± 12.7 (ab) | 132,524 ±102,615 (b) |
| P value | 0.01 | 0.7 | 0.23 | 0.0016 | 0.01 | 0.0001 |
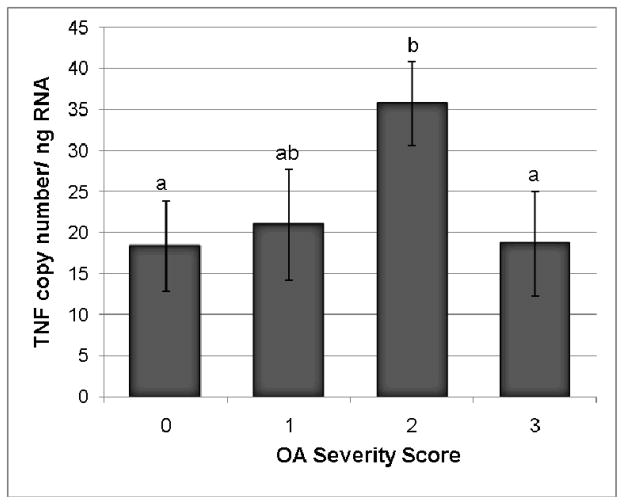
Figure 1.
TNFα transcription levels in synovial membrane from normal and OA joints. Categories were numbered as normal (0), mild OA (1), moderate OA (2), and severe OA (3). TNFα levels were significantly increased in the moderate OA group as compared to the control group. Data presented as Mean ± SE. Differing letters indicate significant differences at p<0.05.
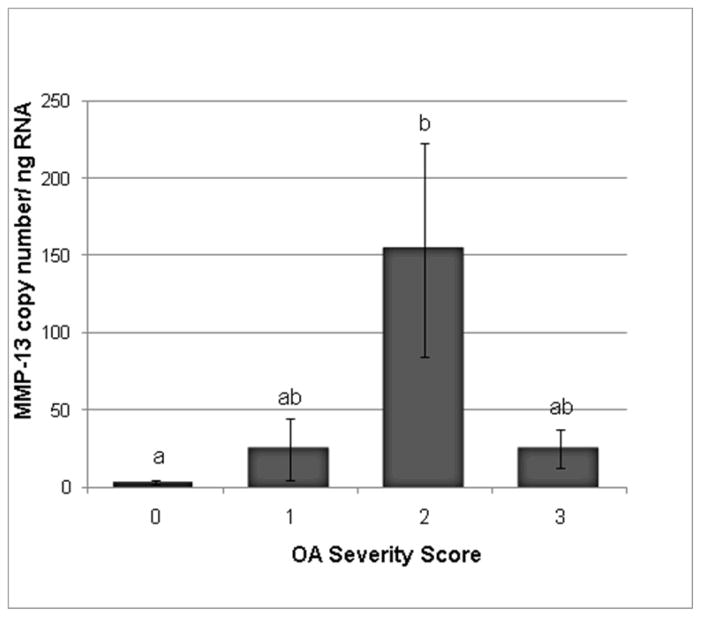
Figure 2.
MMP-13 transcription levels in synovial tissue from normal and OA joints. Categories were numbered as normal (0), mild OA (1), moderate OA (2), and severe OA (3). MMP-13 levels were significantly increased in the moderate OA group as compared to the control group. Data presented as Mean ± SE. Differing letters indicate significant differences at p<0.05.
Table 2.
Catabolic mediator and matrix gene expression in cartilage from normal and arthritic joints. Mean values and standard errors are listed, and significant groupings of post-hoc Tukeys and LSD evaluation are shown by differing letters in parentheses.
| Mean Transcription Levels | TNFα | IL-1β | ADAMSTS-4 | ADAMSTS-5 | MMP-13 | Col-2B | Aggrecan |
|---|---|---|---|---|---|---|---|
| Normal | 0.5205± 0.23(a) | 2.62 ± 0.49(a) | 408.5 ±272(a) | 4579.3±846.8 | 0.69± 0.35(a) | 117,236±43,815(a) | 260,529±42,704(a) |
| Mild OA | 5.167± 3.21(b) | 7.22 ± 1.8(b) | 1209 ± 523(ab) | 4449±1442.0 | 1471.5 ±1030 (b) | 30,562 ±10,778 (a) | 61,518 ±19,688(b) |
| Moderate OA | 15.46± 5.29(b) | 19.95 ± 9.0(c) | 1825 ±522 (b) | 3285.1±721.4 | 2828.5 ± 1066(b) | 44,832 ±35,573 (a) | 17,356± 14,188(c) |
| Severe OA | 15.13± 6.58(b) | 19.17 ± (7.8bc) | 1491.9 ±579 (b) | 3602.9±1067.0 | 1567.6 ± 749(b) | 297,913±81,986(b) | 44,131 ±12,290(b) |
| P value | 0.0007 | <0.001 | 0.0011 | 0.49 | <0.001 | 0.0047 | <0.0010 |
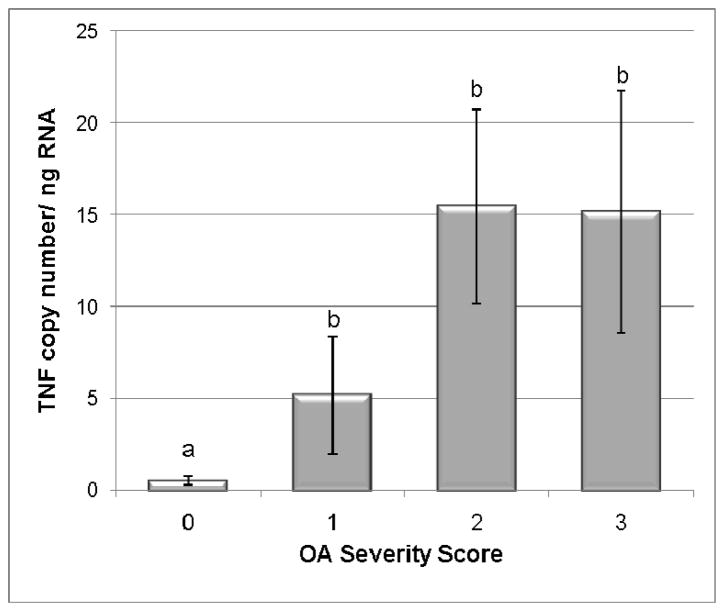
Figure 3.
TNFα transcription levels in normal and OA cartilage. Categories were numbered as normal (0), mild OA (1), moderate OA (2), and severe OA (3). TNFα levels were significantly increased in all OA groups as compared to the control group. Data presented as Mean ± SE. Differing letters indicate significant differences at p<0.05.
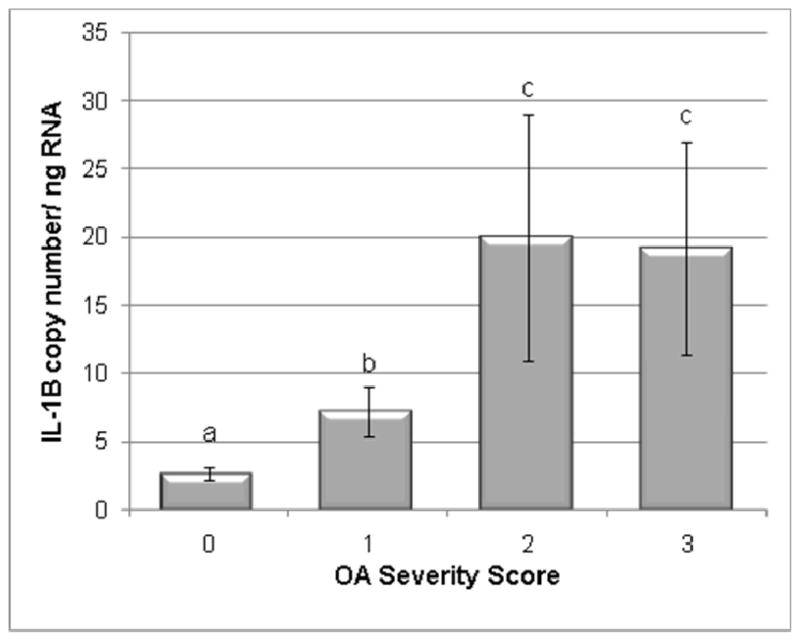
Figure 4.
IL-1β transcription levels in normal and OA cartilage. Categories were numbered as normal (0), mild OA (1), moderate OA (2), and severe OA (3). IL-1β levels were significantly increased in all OA groups as compared to the control group. Data presented as Mean ± SE. Differing letters indicate significant differences at p<0.05.
Anabolic Marker Expression
Collagen type I expression in synovium was significantly increased in OA groups (p=0.0001) (Table 1). For cartilage, the severe OA group had a significant increase in Col-2B compared to other OA groups and the normals (p=0.0047) (Table 2). Aggrecan levels significantly decreased in the OA cartilage groups (p<0.0001).
Synovial Fluid Protein Concentration
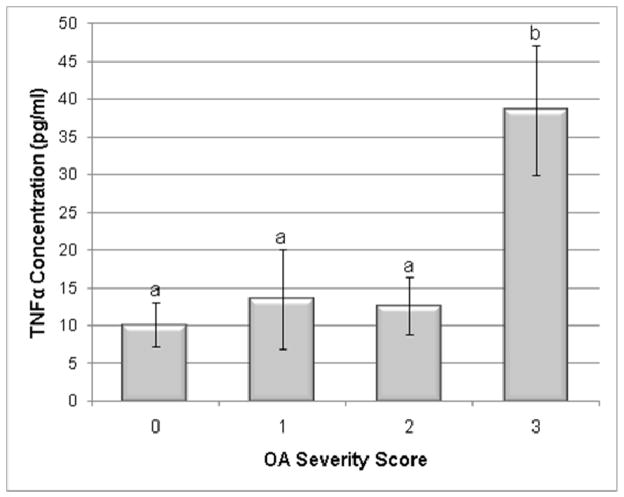
Synovial fluid TNFα peptide concentration was significantly increased in severe OA cases (p=0.021) (Fig 5). The other OA groups did not have significantly different TNF concentration compared to the normal group. However, the means of all OA groups were higher than the normal group. The MMP-13 kit and the IL-1β trials using an equine monoclonal antibody did not yield concentrations above the lower limit of sensitivity. It is likely that the human MMP-13 ELISA did not cross-react adequately with equine in vivo derived samples. Commercially available antibodies targeting equine IL-1β are not yet sensitive enough for the low concentration found in joint fluid.
Figure 5.
Synovial fluid TNFα cytokine concentration in normal and OA joints. Categories were numbered as normal (0), mild OA (1), moderate OA (2), and severe OA (3). TNFα concentration was significantly increased in the severe OA group compared to the control group. Data presented as Mean ± SE. Differing letters indicate significant differences at p<0.05.
Discussion
This study served to identify the key inflammatory mediators in equine osteoarthritis. The data from 36 OA and 22 normal joints indicated that gene expression of TNFα, ADAMTS-5, and MMP-13 were significantly increased in the synovium of OA joints. Similarly, TNFα, IL-1β, ADAMTS-4, and MMP-13 transcripts were significantly increased in the cartilage of 22 OA joints compared to 8 normal joints. TNFα protein concentrations were significantly increased in the synovial fluid of 31 OA joints when compared to normal joints.
The role of TNFα and IL-1B
TNFα is thought to be most prominent cytokine in the acute stages of osteoarthritis while IL-1β remains high throughout all stages (Toncheva et al. 2009). Toncheva et al. 2009 used the serum from human patients with and without knee OA to show that TNFα was significantly increased in active OA patients compared to normal patients, while more chronic, inactive OA patients did not have significantly different concentration of TNFα compared to normals. In our study, TNFα gene expression in the synovium decreased significantly from moderate to severe cases, but expression in cartilage remained significantly increased in all OA groups, particularly in the moderate and severe OA groups. Additionally, the TNFα concentration in the synovial fluid increased significantly during progression from moderate to severe OA groups. Although systemic and articular levels of TNFα should not be compared, this study tends to contradict earlier studies that suggest TNFα is less significant in cases that have progressed beyond the acute stages of OA.
Of all the inflammatory mediators examined in this study, only TNFα and MMP-13 expression were significantly increased in both the synovium and cartilage. The increase in TNFα in both synovium and cartilage may signify that it can not only damage cartilage but also lead to increased secretion of other cytokines from the synovium. MMP-13 is a downstream mediator that is upregulated by TNFα and its concurrent increase with TNF seems predictable (Burrage et al, 2006).
IL-1β also stimulates increased levels of MMP-13, but IL-1β transcription was not significantly increased in OA synovial tissue compared to normals. No equine IL-1β ELISA is currently available that reads below the 1ng/ml that is necessary for detection in synovial fluid. Lacking this information, we were unable to conclude if the increased IL-1β transcription in cartilage can cause an increase in IL-1β in synovial fluid and therefore have a generalized effect on the joint rather than just a local effect on the cartilage.
The Aggrecanases
Whether aggrecanase 1 or aggrecanase 2 is of greater importance in osteoarthritis has been in contention lately (Huang and Wu 2008). Aggrecan organized through hyaluronan chains into the complex of the matrix is particularly susceptible to degradation at specific G1-G2 sites by the two aggrecanases (Aggrecanase 1 or ADAMTS-4 and Aggrecanase 2 or ADAMTS-5), which cleave the aggrecan and thereby decrease biomechanical integrity of the cartilage. As aggrecan makes up a significant portion of the extracellular matrix of cartilage, this cleavage decreases the ability of the cartilage to resist compressive forces.
Previous in vitro studies using synoviocytes have shown that ADAMTS-4, but not ADAMTS-5, is inducible by TNFα and IL-1β (Bondeson et al. 2006). Although our study does not directly correlate expression of ADAMTS to TNF or IL-1, the co-expression data appears to counter Bondeson et al., in that ADAMTS-5 but not ADAMTS-4 co-expression was significantly increased in the synovium when TNFα was concurrently increased.
Increased pro-inflammatory cytokine expression in equine OA cartilage does support previous studies examining chondrocyte transcriptional changes in OA. A human study using chondrocytes showed that ADAMTS-4 was dramatically induced by TNFα and IL-1β, while ADAMTS-5 increased only slightly after exposure to the same cytokines (Song et al. 2007). Our in vivo study shows that chondrocyte expression of ADAMTS-4, but not ADAMTS-5, was increased in chondrocytes from OA joints. The complete lack of transcriptional change of ADAMTS-5 in OA cartilage is an important finding, as it shows that ADAMTS-5 is not increased by IL-1β flux in equine chondrocytes. Additionally, OA treatment modalities may better target ADAMTS-5 in the synovium of arthritic joints, and ADAMTS-4 should be abrogated in the cartilage.
Interleukin-17
Although IL-17 has been documented to be increased in human rheumatoid arthritis, little is known of its action in OA (Gaffen et al. 2004). Published data has shown that IL-17 can increase NO and MMP production to cause cartilage breakdown and can be used in conjunction with other cytokines to create an OA model (Gaffen et al. 2004). However, there is little evidence of its increase in expression in vivo in OA joints compared to normal joints (Malemud et al. 2004). Our data shows very little expression of IL-17, in either the synovium or the cartilage in OA joints. E. coli and LPS stimulated monocytes were used to test the sensitivity of our quantitative RT-PCR probes and primers. High levels of expression were seen in these equine leukocytes, verifying the accuracy of our assay. Fluctuation in IL-17 expression seems to have little relevance in equine OA.
Anabolic Markers
The increased collagen type II transcription in severe OA cartilage was unexpected, as IL-1β suppresses type II expression during in vitro experiments (Toegel et al. 2008), and previous studies suggest collagen type II content is reduced in more advanced OA samples (Squires et al, 2003). Our data shows collagen type II expression decreases from normal to mild and moderate OA cartilage, but then transcription rebounds in the severe group. Previous gene expression data help explain the difference in collagen type II expression levels, where the deeper cartilage zones are more capable of rebound collagen overexpression when cartilage is deeply eroded but not eburnated (Aigner et al, 1997; Brew et al, 2008). The severely affected joints in our study had cartilage that was deeply eroded. Aggrecan mRNA levels followed a more predictable decline, with significant decreases in the OA cartilage when compared to normals. Collagen type I transcription was increased in the synovium, most likely as a consequence of the proliferation of synovium and subintimal fibrosis in OA joints.
Conclusion
This in vivo analysis of articular cytokine and matrix genes shows that TNFα expression is significantly increased through all stages of OA, and is increased in both the synovium and the cartilage. It appears that ADAMTS-4 is more significant than ADAMTS-5 in cartilage destruction in OA joints, while ADAMTS-5 may have more of a role at the synovial level. IL-17 is not increased in OA synovium or cartilage and seems less likely to be a major contributor to OA progression than previously described. On the other hand, TNFα is a more significant player in equine OA than previously believed. Indeed the gene expression data presented here shows that TNFα is more consistently increased in equine OA synovium and cartilage than IL-1β.
Acknowledgments
The assistance of Dr Hussni Mohammed with statistical analysis is gratefully acknowledged. Funding for this project was provided by the Harry M. Zweig Memorial Fund for Equine Research and NIH grant 5R01-AR055373-01. This project was supported by Award Number R01AR055373 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Arthritis and Musculoskeletal and Skin Diseases or the National Institutes of Health.
Footnotes
Dyonics 5.2, Smith and Nephew, London, UK.
Thermo Fisher Scientific, Waltham, Massachusetts, USA.
SPEX CertiPrep, Metuchen, New Jersey, USA.
Versagene, Carlsbad, California, USA.
Nanodrop, Wilmington, Deleware, USA.
Applied Biosystems, Foster City, California, USA.
National Institutes of Health, Bethesda, Maryland, USA.
GE Amersham, Piscataway, New Jersey, USA.
R and D Systems, Minneapolis, Minnesota, USA.
Tecan Group Ltd, Mannedorf, Switzerland.
Analytical Software, Tallahassee, FL, USA.
References
- Aigner T, Vornehm SI, Zeiler G, Dudhia J, von der MK, Bayliss MT. Suppression of cartilage matrix gene expression in upper zone chondrocytes of osteoarthritic cartilage. Arthritis Rheum. 1997;40:562–569. doi: 10.1002/art.1780400323. [DOI] [PubMed] [Google Scholar]
- Alex P, Szodoray P, Arthur E, Willis L, Hynd R, Flinn D, Centola M. Influence of intraarticular corticosteroid administration on serum cytokines in rheumatoid arthritis. Clin Rheumatol. 2007;26:845–848. doi: 10.1007/s10067-006-0419-7. [DOI] [PubMed] [Google Scholar]
- Amin AR. Regulation of tumor necrosis factor-alpha and tumor necrosis factor converting enzyme in human osteoarthritis. Osteoarth Cart. 1999;7:392–4. doi: 10.1053/joca.1998.0221. [DOI] [PubMed] [Google Scholar]
- Augustsson J, Neovius M, Cullinane-Carli C, Eksborg S, van Vollenhoven RF. Patients with rheumatoid arthritis treated with tumour necrosis factor antagonists increase their participation in the work-force: potential for significant long-term indirect cost gains (data from a population-based registry) Ann Rheum Dis. 2010;69:126–131. doi: 10.1136/ard.2009.108035. [DOI] [PubMed] [Google Scholar]
- Berenbaum F. The quest for the Holy Grail: a disease-modifying osteoarthritis drug. Arthritis Res Ther. 2007;9:111. doi: 10.1186/ar2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondeson J, Wainwright SD, Lauder S, et al. The role of synovial macrophages and macrophage-produced cytokines in driving aggrecanases, matrix metalloproteinases, and other destructive and inflammatory responses in osteoarthritis. Arthritis Res Ther. 2006;8:R187. doi: 10.1186/ar2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan FM, Hayes AL, Ciesielski CJ, Green P, Foxwell BM, Feldmann M. Evidence that rheumatoid arthritis synovial T cells are similar to cytokine-activated T cells: involvement of phosphatidylinositol 3-kinase and nuclear factor kappaB pathways in tumor necrosis factor alpha production in rheumatoid arthritis. Arthritis Rheum. 2002;46:31–4. doi: 10.1002/1529-0131(200201)46:1<31::AID-ART10029>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Brew CJ, Clegg PD, Boot-Handford RP, Andrews G, Hardingham T. Gene expression in human chondrocytes in late OA is changed in both fibrillated and intact cartilage without evidence of generalised chondrocyte hypertrophy. Ann Rheum Dis. 2010;69:234–240. doi: 10.1136/ard.2008.097139. [DOI] [PubMed] [Google Scholar]
- Burrage PS, Mix KS, Brinckerhoff CE. Matrix metalloproteinases: role in arthritis. Front Biosci. 2006;11:529–43. doi: 10.2741/1817. [DOI] [PubMed] [Google Scholar]
- Chevalier X, Goupille P, Beaulieu AD, Burch FX, Bensen WG, Conrozier T, Loeuille D, Kivitz AJ, Silver D, Appleton BE. Intraarticular injection of anakinra in osteoarthritis of the knee: a multicenter, randomized, double-blind, placebo-controlled study. Arthritis Rheum. 2009;61:344–52. doi: 10.1002/art.24096. [DOI] [PubMed] [Google Scholar]
- Clements KM, Price JS, Chambers MG, Visco DM, Poole AR, Mason RM. Gene deletion of either Interleukin-1β, Interleukin-1β–Converting Enzyme, Inducible Nitric Oxide Synthase, or Stromelysin 1 accelerates the development of knee osteoarthritis in mice after surgical transection of the medial collateral ligament and partial medial meniscectomy. Arthritis Rheum. 2003;48:3452–3463. doi: 10.1002/art.11355. [DOI] [PubMed] [Google Scholar]
- Fernandes JC, Martel-Pelletier J, Pelletier JP. The role of cytokines in osteoarthritis pathophysiology. Biorheology. 2002;39:237–46. [PubMed] [Google Scholar]
- Furuzawa-Carballeda J, Macip-Rodríguez PM, Cabral AR. Osteoarthritis and rheumatoid arthritis pannus have similar qualitative metabolic characteristics and pro-inflammatory cytokine response. Clin Rheumatol. 2009;28:749–56. [PubMed] [Google Scholar]
- Gaffen SL. Biology of recently discovered cytokines: Interleukin-17–a unique inflammatory cytokine with roles in bone biology and arthritis. Arthritis Res The. 2004;6:240–247. doi: 10.1186/ar1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Wu LD. Aggrecanase and Aggrecan Degradation in Osteoarthritis: a Review. J International Med Res. 2008;36:1149–1160. doi: 10.1177/147323000803600601. [DOI] [PubMed] [Google Scholar]
- Kunisch E, Jansen A, Kojima F, Loffler I, Kapoor M, Kawai S, Rubio I, Crofford LJ, Kinne RW. Prostaglandin E2 differentially modulates proinflammatory/prodestructive effects of TNF-a on synovial fibroblasts via specific E prostanoid receptors/cAMP. J Immunol. 2009;183:1328–1336. doi: 10.4049/jimmunol.0900801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malemud CJ. Cytokines as therapeutic targets for osteoarthritis. Biodrugs. 2004;18:23–35. doi: 10.2165/00063030-200418010-00003. [DOI] [PubMed] [Google Scholar]
- Rai MF, Rachakonda PS, Manning K, Vorwerk B, Brunnberg L, Kohn B, Schmidt MFG. Quantification of cytokines and inflammatory mediators in a three-dimensional model of inflammatory arthritis. Cytokine. 2010;42:8–17. doi: 10.1016/j.cyto.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Shlopov BV, Gumanovskaya ML, Hasty KA. Autocrine regulation of collagenase 3 (matrix metalloproteinase 13) during osteoarthritis. Arthritis Rheum. 2000;43:195–205. doi: 10.1002/1529-0131(200001)43:1<195::AID-ANR24>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Smith MD, Triantafillou S, Parker A, Youssef PP, Coleman M. Synovial membrane inflammation and cytokine production in patients with early osteoarthritis. J Rheumatol. 1997;24:365–71. [PubMed] [Google Scholar]
- Song RH, Tortorella MD, Malfait AM, et al. Aggrecan degradation in human articular cartilage explants is mediated by both ADAMTS-4 and ADAMTS-5. Arthritis Rheum. 2007;56:575–585. doi: 10.1002/art.22334. [DOI] [PubMed] [Google Scholar]
- Stove J, Huch K, Gunther KP, Scharf HP. Interleukin-1s induces different gene expression of stromelysin, aggrecan and tumor necrosis factor-stimulated gene 6 in human osteoarthritic chondrocytes in vitro. Pathobiology. 2000;68:144–9. doi: 10.1159/000055915. [DOI] [PubMed] [Google Scholar]
- Squires GR, Okouneff S, Ionescu M, Poole AR. The pathobiology of focal lesion development in aging human articular cartilage and molecular matrix changes characteristic of osteoarthritis. Arthritis Rheum. 2003;48:1261–1270. doi: 10.1002/art.10976. [DOI] [PubMed] [Google Scholar]
- Toegel S, Wu SQ, Piana C, Unger FM, Wirth M, Goldring MB, Gabor F, Viernstein H. Comparison between chondroprotective effects of glucosamine, curcumin, and diacerein in IL-1beta-stimulated C-28/I2 chondrocytes. Osteoarth Cart. 2000;16:1205–12. doi: 10.1016/j.joca.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Toncheva A, Remichkova M, Ikonomova K, Dimitrova P, Ivanovska N. Inflammatory response in patients with active and inactive osteoarthritis. Rheumatol Int. 2009;29:1197–1203. doi: 10.1007/s00296-009-0864-0. [DOI] [PubMed] [Google Scholar]
- Venkatesan N, Barre L, Benani A. Stimulation of proteoglycan synthesis by glucuronosyltransferase-I gene delivery: a strategy to promote cartilage repair. Proc Natl Acad Sci. 2004;101:18087–92. doi: 10.1073/pnas.0404504102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa T, Hammaker D, Sweeney SE, Boyle DL, Firestein GS. Synoviocyte innate immune responses: I. Differential regulation of interferon responses and the JNK pathway by MAPK kinases. J Immunol. 2008;181:3252–8. doi: 10.4049/jimmunol.181.5.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]