Background: The guanine nucleotide exchange factor Tiam1 regulates the activity of the small GTPase Rac1, a crucial regulator of cell adhesion, proliferation, and survival.
Results: The SCFβTrCP ubiquitin ligase in cooperation with CK1 targets Tiam1 for proteasome-dependent degradation.
Conclusion: Tiam1 degradation is required to terminate the mTOR-S6K signaling pathway.
Significance: Tiam1 degradation controls the duration of mTOR-S6K signaling in response to mitogens.
Keywords: E3 Ubiquitin Ligase, Proteasome, Proteolysis, Signal Transduction, Ubiquitin, Tiam1, betaTrCP, mTOR-S6K Signaling
Abstract
Tiam1 (T-cell lymphoma invasion and metastasis 1) is a guanine nucleotide exchange factor that specifically controls the activity of the small GTPase Rac, a key regulator of cell adhesion, proliferation, and survival. Here, we report that in response to mitogens, Tiam1 is degraded by the ubiquitin-proteasome system via the SCFβTrCP ubiquitin ligase. Mitogenic stimulation triggers the binding of Tiam1 to the F-box protein βTrCP via its degron sequence and subsequent Tiam1 ubiquitylation and proteasomal degradation. The proteolysis of Tiam1 is prevented by βTrCP silencing, inhibition of CK1 and MEK, or mutation of the Tiam1 degron site. Expression of a stable Tiam1 mutant that is unable to interact with βTrCP results in sustained activation of the mTOR/S6K signaling and increased apoptotic cell death. We propose that the SCFβTrCP-mediated degradation of Tiam1 controls the duration of the mTOR-S6K signaling pathway in response to mitogenic stimuli.
Introduction
Tiam1 is a ubiquitously expressed guanine nucleotide exchange factor that specifically activates the Rho-like GTPase Rac1 in response to growth factors and cell-substrate interaction (1–3). It was originally identified in a retroviral insertional mutagenesis screen for genes that confer invasive properties to T-lymphoma cells (1). Thereafter, a plethora of studies have demonstrated that Tiam1 controls different Rac-dependent cellular processes, such as cell polarity, cell-matrix adhesion, cell-cell adhesion, cell survival, and cell growth and proliferation. Hence, it does not come as a surprise that Tiam1 plays a critical role in cancer development. Tiam1 deficiency in mice protects against Ras-induced skin carcinogenesis and correlates with increased apoptosis and reduced proliferation and growth in keratinocytes (4). This is in agreement with in vitro studies reporting that Tiam1 associates with GTP-bound Ras through a Ras-binding domain and functions as an effector that mediates Ras activation of Rac (4). However, the fewer skin tumors developing in Tiam1 knock-out mice progress more frequently to malignancy, indicating that loss of Tiam1 stimulates malignant conversion (4). Indeed, in vitro, Tiam1 is known to promote the formation of adherens junctions and the induction of an epithelial-like phenotype in mesenchymal cells (5, 6) and to be required for both the formation and maintenance of cadherin-based adhesions (7).
Despite the importance of Tiam1 in cancer, little is known about the regulation of Tiam1 protein levels in normal and cancer cells. Here, we report that in response to mitogenic stimulation, Tiam1 is targeted for proteasome-dependent degradation by the SCFβTrCP ubiquitin ligase in cooperation with casein kinase 1 and the MEK/ERK pathway. We also show that ubiquitin-dependent degradation of Tiam1 is required to terminate the activation of mTOR-S6K signaling in response to mitogenic stimulation.
EXPERIMENTAL PROCEDURES
Cell Culture and Drug Treatments
HEK293, HEK293T, T98G, and MDCK2 cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) with 10% FCS and 100 units/ml penicillin-streptomycin. The following drugs were used: MG132 (Peptide Institute; 10 μm), D4476 (Sigma-Aldrich; 50 μm), IC261 (Sigma-Aldrich; 50 μm), cycloheximide (Sigma-Aldrich; 100 μm), PD0325901 (Sigma-Aldrich; 10 μm), U0126 (Millipore; 10 μm), Rac inhibitor III EHop-016 (Millipore; 50 μm), and phorbol 12-myristate 13-acetate (Sigma-Aldrich; 10 ng/ml). 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assays were carried according to the manufacturer's protocol.
Transient Transfections
Cells were transfected by the polyethylenimine or liposome-mediated (Lipofectamine 2000; Invitrogen) methods
Plasmids
Full-length and N-terminally truncated (C1199) human Tiam1 carrying a C-terminal HA tag cDNAs were cloned by PCR into the EcoRV and NotI sites of pcDNA3. GFP-tagged Tiam1 was kindly provided by F. Zwartkruis. The full-length constructs were used as template to generate the Tiam1(S370A/S374A), Tiam1(S414A/S418A), Tiam1(S329A/S334A), and Tiam1 (S329A/S334A/T340A) mutants using the QuikChange site-directed mutagenesis kit (Stratagene) according to manufacturer's directions. The oligonucleotides used were as follows: Tiam1(S370A/S373A) forward primer, 5′-CTTTGTGGGCAGCGACGCCGGCAGCAGCGCCACCGGGGATGCGGCTC-3′; Tiam1(S370A/S373A) reverse primer, 5′-GAGCCGCATCCCCGGTGGCGCTGCTGCCGGCGTCGCTGCCCACAAAG-3′; Tiam1(S414A/S418A) forward primer 5′-CAGCGATGAGCAGAGCGCCGGCACCCTGGCCTCTCCGGGCCAAGTCGGAC-3′; Tiam1(S414A/S418A) reverse primer, 5′-GTCCGACTTGGCCCGGAGAGGCCAGGGTGCCGGCGCTCTGCTCATCGCTG-3′;Tiam1(S329A/S334A) forward primer, 5′-GGCGAGGGCGCTGAGTTTGCAGACGCTGGGATTGAAGGG-3′; Tiam1(S329A/S334A) reverse primer, 5′-CCCTTCAATCCCAGCGTCTGCAAACTCAGCGCCCTCGCC-3′; Tiam1(S329A/S334A/T340A) forward primer, 5′-GAGTTTGCAGACGCTGGGATTGAAGGGGCCGCTACCGACACG-3′; and Tiam1(S329A/S334A/T340A) reverse primer, 5′-CGTGTCGGTAGCGGCCCCTTCAATCCCAGCGTCTGCAAACTC-3′. All constructs were verified by sequencing.
Biochemical Methods
Extract preparation, immunoprecipitation, and immunoblotting were previously described (8). Mouse monoclonal antibodies were from Invitrogen (Cul1), Sigma-Aldrich (FLAG), Santa Cruz Biotechnology (Actin, p70-S6K), Covance (HA), Cell Signaling Technology (phospho-Erk1/2(Thr202/Tyr204)), and BD Biosciences (Rac1). Rabbit polyclonal antibodies were from Santa Cruz Biotechnology (Tiam1), Cell Signaling Technology (βTrCP1, Erk1/2, phospho-p70-S6K(Thr421/Ser424), phospho-p70-S6K(Thr389), cleaved caspase-3(Asp175)), Sigma-Aldrich (FLAG), and Torrey Pines Biolabs (GFP).
In Vitro Binding Assay
In vitro translated 35S-labeled βTrCP1 and FBXW5 were incubated with protein G-Sepharose beads precoupled with the following Tiam1 synthetic peptides: 327EGSEFADSGIEGAT340, 330EFADSGIEGATTDT343, 327EGpSEFADpSGIEGAT340, or 330EFADpSGIEGApTTDT343 for 2 h at 4 °C. The beads were washed four times with lysis buffer (50 mm Tris-HCl, pH 7.4, 1 mm EDTA, 250 mm NaCl, 0.1% Triton X-100, 50 mm NaF, 1 mm DTT, 0.1 mm Na3VO4). Proteins were eluted with Laemmli buffer for 5 min at 95 °C and subjected to SDS-PAGE followed by autoradiography.
In Vitro Ubiquitylation Assay
Tiam1 ubiquitylation was performed in a volume of 10 μl containing SCFβTrCP-Tiam1 immunocomplexes, 50 mm Tris, pH 7.6, 5 mm MgCl2, 0.6 mm DTT, 2 mm ATP, 1.5 ng/μl E1 (Boston Biochem), 10 ng/μl Ubc3, 2.5 μg/μl ubiquitin (Sigma-Aldrich), and 1 μm ubiquitin aldehyde. The reactions were incubated at 30 °C for 60 min and analyzed by immunoblotting.
Purification of βTrCP2 Interactors
HEK293T cells grown in 15-cm dishes were transfected with pcDNA3–2×FLAG-2×HA-βTrCP2 and treated with 10 μm MG132 for 5 h. Cells were harvested and subsequently lysed in lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40, 2 mm β-glycerophosphate plus protease and phosphatase inhibitors). βTrCP2 was immunopurified with mouse anti-FLAG M2-agarose resin (Sigma-Aldrich). After washing, proteins were eluted by competition with FLAG peptide (Sigma-Aldrich). The eluate was then subjected to a second immunopurification with anti-HA resin (12CA5 monoclonal antibody cross-linked to protein G-Sepharose; Invitrogen) prior to elution in Laemmli sample buffer. The final eluate was separated by SDS-PAGE, and proteins were visualized by colloidal Coomassie Blue. Bands were sliced out from the gels and subjected to in-gel digestion. Gel pieces were then reduced, alkylated, and digested according to a published protocol (9). For mass spectrometric analysis, peptides recovered from in-gel digestion were separated with a C18 column and introduced by nano-electrospray into the LTQ Orbitrap XL (Thermo Fisher) with a configuration as described (10). Peak lists were generated from the MS/MS spectra using MaxQuant build 1.0.13.13 and then searched against the IPI Human database (version 3.37, 69164 entries) using Mascot search engine (Matrix Science). Carbaminomethylation (+57 Da) was set as fixed modification and protein N-terminal acetylation and methionine oxidation as variable modifications. Peptide tolerance was set to 7 ppm, and fragment ion tolerance was set to 0.5 Da, allowing two missed cleavages with trypsin enzyme. Finally, Scaffold (version Scaffold_3.6.1; Proteome Software Inc.) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if their Mascot scores exceeded 20.
Gene Silencing by Small Interfering RNA
The oligonucleotides designed for targeting both βTrCP1 and βTrCP2 are as follows: sense sequence, 5′-GTGGAATTTGTGGAACATC dTdT-3′; and antisense sequence, 5′-GATGTTCCACAAATTCCACdTdT-3′. Cells were transfected with the oligonucleotides twice (24 and 48 h after plating) using Oligofectamine (Invitrogen) according to the manufacturer's recommendations. Forty-eight hours after the last transfection, lysates were prepared and analyzed by immunoblotting.
Phosphorylation Analysis by Mass Spectrometry
Samples were reduced with 10 mm DTT for 30 min at 60 °C, followed by 30-min incubation with iodoacetamide (20 mm) in the dark at room temperature. The first digestion was performed using Lys-C for 4 h at 37 °C. Subsequently, the digest was diluted 5-fold using 50 mm ammonium bicarbonate to a final urea concentration of less than 2 m, and a second digestion with trypsin was performed overnight at 37 °C. Finally, the digestion was quenched by the addition of formic acid to a final concentration of 0.1% (v/v). The resulting solution was desalted using 200-mg Sep-Pak C18 cartridges (Waters Corporation), lyophilized, and reconstituted in 10% formic acid. LC-MSMS was performed with higher energy collision dissociated in a Q-Exactive Plus Orbitrap instrument. MS spectra to peptide sequence assignment were performed with Proteome Discoverer version 1.4 and MASCOT version 2.4 as search engine. The localization of phosphorylated sites was evaluated with PhosphoRS version 3 (11).
Rac Activity Assay
The abundance of GTP-bound Rac1 was determined as follows. Cells were lysed in lysis buffer (10 mm Tris, 150 mm NaCl, 50 mm MgCl2, 0.5% Nonidet P-40) containing 2 μg of biotinylated p21-binding domain of Pak1. The cleared supernatant was then incubated with streptavidin beads for 30 min at 4 °C. Beads were washed, and GTP-bound Rac1 was analyzed by immunoblotting with an anti-Rac1 antibody (Upstate Biotechnology Inc.).
RESULTS
Tiam1 Interacts with βTrCP
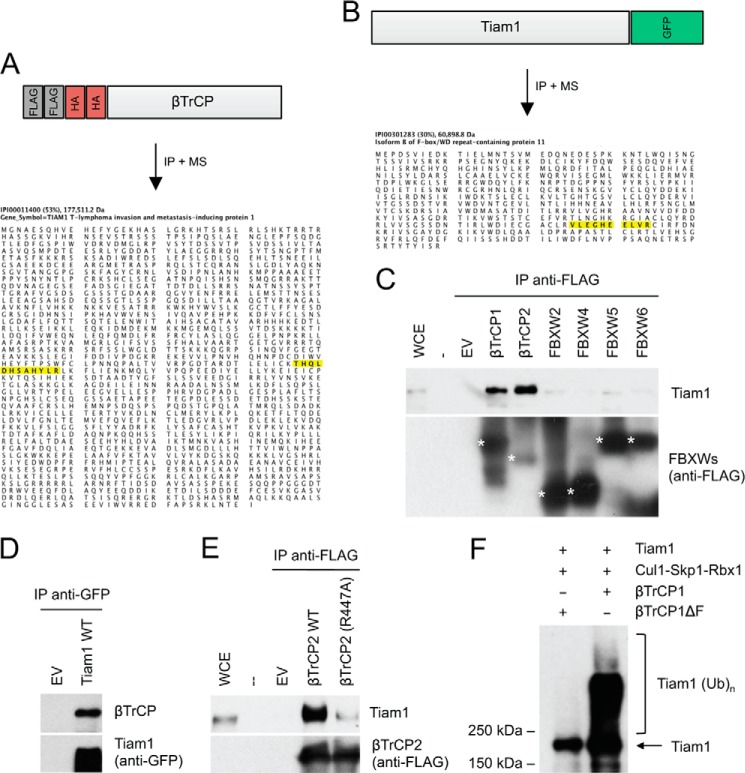
In an attempt to identify new targets of the SCFβTrCP ubiquitin ligase, we expressed βTrCP with an N-terminal FLAG-HA epitope tag in HEK293T cells and purified FLAG-HA-βTrCP immunoprecipitates, which were then analyzed by mass spectrometry. In two independent immunopurifications, peptides corresponding to Tiam1 (T-cell lymphoma invasion and metastasis 1) were recovered (Fig. 1A). Conversely, when we immunopurified Tiam1 from HEK293T cells, we identified peptides corresponding to βTrCP (Fig. 1B). To confirm the binding between βTrCP and Tiam1 and test its specificity, we overexpressed different FLAG-tagged F-box proteins (all containing WD40 repeats) in HEK293T cells. We then carried out FLAG immunoprecipitations to examine the interaction with endogenous Tiam1. βTrCP1 and its paralog βTrCP2 were the only proteins able to coimmunoprecipitate with Tiam1 (Fig. 1C). The coimmunoprecipitation of endogenous βTrCP with exogenously expressed Tiam1 was confirmed by immunoprecipitation coupled to immunoblotting (Fig. 1D).
FIGURE 1.
Tiam1 interacts with and is ubiquitylated by the SCFβTrCP ubiquitin ligase. A, peptide coverage of Tiam1 in the mass spectrometry analysis of βTrCP2 immunopurification. Amino acid sequences of detected Tiam1 peptides are highlighted in yellow. B, peptide coverage of βTrCP2 (also known as FBXW11) in the mass spectrometry analysis of Tiam1 immunopurification. Amino acid sequences of detected βTrCP2 peptides are highlighted in yellow. C, the indicated FLAG-tagged F-box proteins containing WD40 repeats (FBXWs) or an empty vector (EV) were expressed in HEK293T cells. Forty-eight hours after transfection, cells were treated for 5 h with the proteasome inhibitors MG132, then harvested, and lysed. Whole cell extracts (WCE) were immunoprecipitated (IP) with anti-FLAG antibody and immunoblotted with antibodies specific for the indicated proteins. D, HEK293T cells were transfected with GFP-tagged wild type Tiam1 or an empty vector. Cells were collected and lysed. Tiam1 was immunoprecipitated from whole cell extracts, and immunocomplexes were analyzed by immunoblotting with antibodies specific for the indicated proteins. E, arginine 447 in the WD40 repeat of βTrCP2 is required for the interaction with Tiam1. HEK293T cells were transfected as indicated and analyzed as in C. F, Tiam1, Skp1, Cul1, and Rbx1 were expressed in HEK293T in the absence or presence of βTrCP1 or an inactive βTrCP1-ΔF-box mutant. After immunopurification with anti-FLAG resin, an in vitro ubiquitylation assay of Tiam1 was performed. Samples were analyzed by immunoblotting with an anti-Tiam1 antibody. The bracket indicates a ladder of bands corresponding to polyubiquitylated Tiam1.
It has been shown that substitution to alanine of Arg474 in βTrCP1 (or Arg447 in βTrCP2) within its WD40 β-propeller structure disrupts βTrCP interaction with the substrate (8, 12). To assess whether the interaction between Tiam1 and βTrCP is mediated by the βTrCP WD40 β-propeller, we immunoprecipitated FLAG-tagged wild type βTrCP2 and the βTrCP2(R447A) mutant from HEK293T cells and examined their binding to endogenous Tiam1. As shown in Fig. 1E, wild type βTrCP coimmunoprecipitated with Tiam1, whereas the βTrCP2(R447A) mutant did not.
These results suggest that Tiam1 is a substrate of SCFβTrCP. To test whether SCFβTrCP targets Tiam1 for ubiquitin conjugation, we reconstituted the ubiquitylation of Tiam1 in vitro. βTrCP1, but not an inactive βTrCP1(ΔF box) mutant, was indeed able to efficiently ubiquitylate Tiam1 (Fig. 1F).
The Interaction of Tiam1 with βTrCP Requires a Conserved Phosphodegron
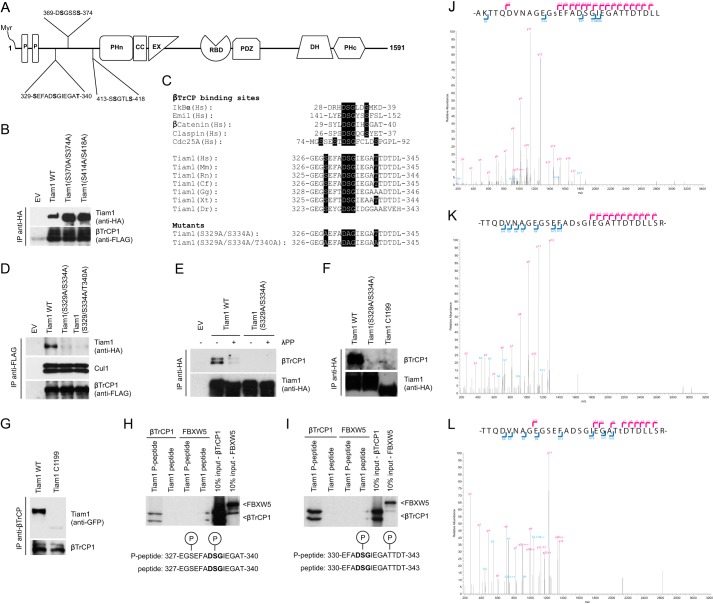
Substrates of SCFβTrCP share a conserved DSGXX(X)S degron that is bound by βTrCP (13–16). Two potential degron motifs are present at the N terminus of human Tiam1, namely, 369DSGSSS374 and 413SSGTLS418 (Fig. 2A). To test which of these motifs is responsible for the interaction of Tiam1 with βTrCP, we generated a number of mutants in which either Ser370/Ser374 or Ser414/Ser418 were replaced by alanine and evaluated the ability of these mutants to coimmunoprecipitate with βTrCP. Fig. 2B shows that wild type Tiam1, Tiam1(S370A/S374A), and Tiam1(S414A/S418A) were all able to bind βTrCP, indicating that these serine residues do not mediate the interaction of Tiam1 with βTrCP. We then tested the requirement of a third putative Tiam1 degron motif, i.e. 329SEFADSGIEGAT340 (Fig. 2A), for the binding of Tiam1 to βTrCP. The upstream Ser329 resembles the serine residue in Cdc25A (Ser79 preceding the conserved aspartate) whose phosphorylation is required for Cdc25A binding to βTrCP (Fig. 2C and Refs. 17–19). We generated Tiam1 mutants in which Ser329 and Ser334 residues, alone or in combination with Thr340, were replaced by alanine. These Tiam1 mutants were not able to pull down exogenous (Fig. 2D) or endogenous (Fig. 2E) βTrCP in immunoprecipitation experiments. Of note, treatment of Tiam1 immunocomplexes with λ-phosphatase disrupted its binding to βTrCP (Fig. 2E). Moreover, a Tiam1 mutant truncated at the N terminus (C1199) lacking the βTrCP degron (5) did not coimmunoprecipitate with endogenous βTrCP (Fig. 2, F and G). Taken together, these results demonstrate that the association of Tiam1 with βTrCP requires the conserved degron motif SEFADSGIEGAT.
FIGURE 2.
Phosphorylation of Ser329, Ser334, and Thr340 in Tiam1 is required for its interaction with βTrCP1. A, schematic representation of Tiam1 functional domains and three putative βTrCP-binding motifs. B, HEK293T cells were transfected with FLAG-tagged βTrCP1 and either HA-tagged wild type Tiam1, HA-tagged Tiam1(S370A/S374A), HA-tagged Tiam1(S414A/S418A), or an empty vector (EV). Forty-eight hours after transfection, cells were harvested and lysed. Whole cell extracts were subjected to immunoprecipitation (IP) with anti-HA resin, followed by immunoblotting with antibodies specific for the indicated proteins. C, alignment of the amino acid regions corresponding to the βTrCP-binding motifs in Tiam1 orthologs and previously reported βTrCP substrates, i.e. IκBα, Emi1, β-catenin, Claspin, and Cdc25A. The amino acidic sequences of the phosphodegron mutants are shown below. D, HEK293T cells were transfected with FLAG-tagged βTrCP1 and either HA-tagged wild type Tiam1, HA-tagged Tiam1(S329A/S334A), HA-tagged Tiam1(S329A/S334A/T340A), or an empty vector. Forty-eight hours after transfection, the cells were harvested and lysed. Whole cell extracts were subjected to immunoprecipitation with anti-FLAG resin, followed by immunoblotting with antibodies specific for the indicated proteins. E, HEK293T cells were transfected with an empty vector, HA-tagged wild type Tiam1, or HA-tagged Tiam1(S329A/S334A). Forty-eight hours after transfection, cells were harvested and lysed. Whole cell extracts were subjected to immunoprecipitation with anti-HA resin and then immunoblotted with antibodies specific for the indicated proteins. When indicated, immunocomplexes were incubated with lambda phosphatase (λPP) for 30 min and then washed. F, HEK293T cells were transfected with HA-tagged wild type Tiam1, HA-tagged Tiam1(S329A/S334A), or HA-tagged Tiam1-C1199. Whole cell extracts were subjected to immunoprecipitation and immunoblotting as in E. G, HEK293T cells were transfected with GFP-tagged wild type Tiam1 or GFP-tagged Tiam1-C1199. Cells were collected and lysed. βTrCP was immunoprecipitated from whole cell extracts, and immunocomplexes were analyzed by immunoblotting with antibodies specific for the indicated proteins. H and I, 35S-βTrCP1 and 35S-FBXW5 were transcribed/translated in vitro and incubated with beads coupled to peptides spanning the Tiam1 degron. Beads were washed with Triton-X buffer, and bound proteins were eluted and subjected to electrophoresis and autoradiography. The last two lanes correspond to 10% of the in vitro translated protein inputs. Peptide sequence spanning the Tiam1 degron is shown on the left. J–L, FLAG-HA-tagged Tiam1 was immunopurified from HEK293T cells and analyzed by mass spectrometry. Ion fragmentation spectra are shown. The indicated Tiam1 tryptic peptides spanning the phosphodegron were found phosphorylated on Ser329 (J), Ser334 (K), or Thr341 (L). As shown, peptide sequences can be explained by their respective collision-induced dissociation MS/MS spectra including Ser(P)329, Ser(P)334, and Thr(P)341.
As a second approach to examine whether phosphorylation of Tiam1 is required for the interaction with βTrCP, we employed an in vitro binding assay using immobilized synthetic phosphopeptides containing the βTrCP recognition domain of Tiam1. Peptides containing phosphorylated residues at positions Ser329 and Ser334 (Fig. 2H) or at positions Ser334 and Thr340 (Fig. 2I) efficiently associated with in vitro-translated βTrCP (but not with FBXW5), whereas the unphosphorylated peptides did not. These results indicate that phosphorylation of Ser329, Ser334, and Thr340 directly mediates the interaction of Tiam1 with βTrCP.
To test whether the Tiam1 degron is phosphorylated in cells, FLAG-HA epitope-tagged Tiam1 immunoprecipitated from HEK293T cells was subjected to tryptic digestion followed by phosphopeptide enrichment by Ti4+-IMAC and LC-MS/MS analysis. We recovered phosphopeptides containing Ser(P)329 (Fig. 2J), Ser(P)334 (Fig. 2K), and Thr(P)341 (Fig. 2L), indicating that the Tiam1 degron is phosphorylated in cultured cells.
Mitogens Induce the Degradation of Tiam1
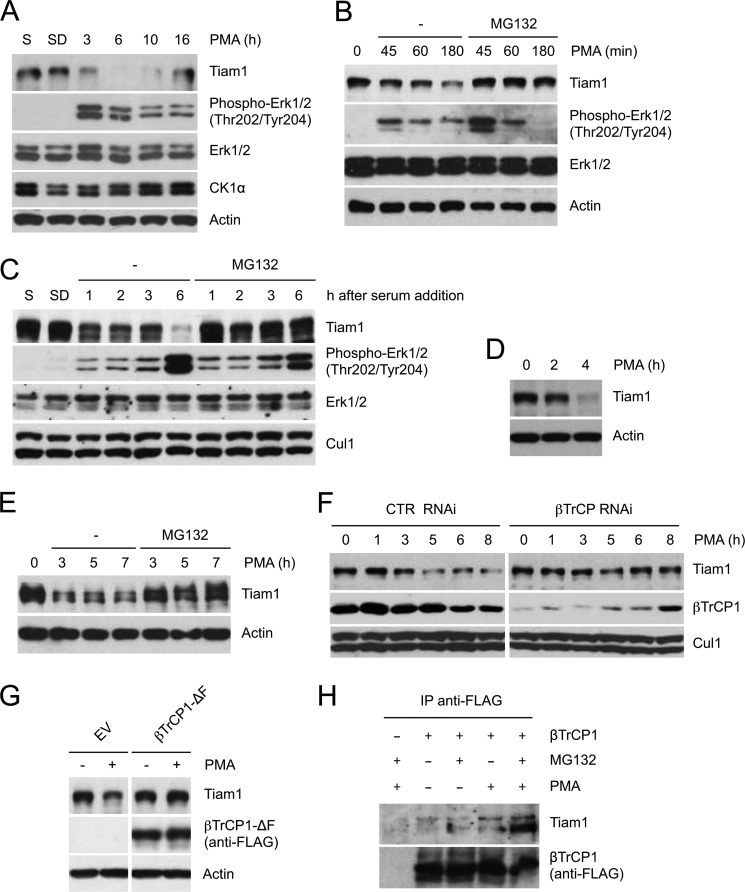
Tiam1-Rac signaling controls a wide range of signaling pathways regulated by extracellular signals. While testing the effect of diverse stimuli on the abundance of Tiam1, we observed a decrease of Tiam1 levels in HEK293 cells treated with the potent mitogen phorbol 12-myristate 13-acetate (PMA; Fig. 3A). Mitogen-induced down-regulation of Tiam1 is transient because Tiam1 abundance goes back approximately to the initial levels 10–16 h after PMA treatment. The reduction in Tiam1 levels was prevented when cells were treated with the proteasome inhibitor MG132, indicating that Tiam1 degradation in response to PMA is mediated by the proteasome (Fig. 3B). Similarly, Tiam1 protein levels decreased in a proteasome-dependent manner in HEK293 cells that were first deprived of serum for 48 h and then stimulated by the readdition of serum (Fig. 3C). The degradation of Tiam1 in response to mitogenic stimulation was observed also in other cell types, namely T98G cells (Fig. 3D) and MDCK cells (Fig. 3E).
FIGURE 3.
Proteasome- and βTrCP-dependent degradation of Tiam1 in response to mitogens. A, HEK293 cells, cultured in low serum for 48 h, were treated with PMA. Cells were collected at the indicated times and lysed. Whole cell extracts were subjected to immunoblotting with antibodies specific for the indicated proteins. Actin is shown as a loading control. B, HEK293 cells were treated with PMA as in A with or without the proteasome inhibitor MG132. Whole cell extracts were analyzed as in A. C, HEK293 cells were either cultured in presence of serum (S) or deprived of serum for 48 h (SD). After serum readdition, the cells were collected at the indicated times. When indicated, the proteasome inhibitor MG132 was added. Protein extracts were analyzed by immunoblotting with antibodies specific for the indicated proteins. Cul1 is shown as a loading control. D and E, T98G cells (D) and MDCK cells (E) were treated with PMA as in A and with MG132 when indicated. Whole cell extracts were analyzed as in A. F, HEK293 cells were transfected with the indicated siRNA oligonucleotides and treated as in A. Cul1 is shown as loading control (CTR). G, HEK293 cells were transfected with FLAG-tagged βTrCP1-ΔF or an empty vector (EV) and treated with PMA for 2 h. Cells were collected and lysed. Whole cell extracts were analyzed as in A. H, HEK293 were transfected with an empty vector or FLAG-tagged βTrCP1. Forty-eight hours after transfection, cells were treated with MG132 and PMA for 5 h (when indicated), then harvested, and lysed. Whole cell extracts were immunoprecipitated (IP) with anti-FLAG resin and immunoblotted with anti-Tiam1 and anti-FLAG antibodies.
Next, we examined whether the observed mitogen-induced degradation of Tiam1 is mediated by βTrCP. We silenced by RNAi the expression of both βTrCP1 and βTrCP2 (20–24) in HEK293 cells, which were then treated with PMA. As shown in Fig. 3F, βTrCP knockdown inhibited the PMA-induced degradation of Tiam1. Ectopic expression of βTrCP with a deleted F-box motif (βTrCP-ΔF), which has been shown to function as a dominant negative mutant (25–29), also prevented the degradation of Tiam1 (Fig. 3G). Moreover, the binding of βTrCP to endogenous Tiam1 was enhanced by PMA treatment (Fig. 3H). Altogether, these results indicate that mitogenic stimulation of cells triggers the βTrCP-mediated proteasomal degradation of Tiam1.
Casein Kinase 1 and MAPK Signaling Control the βTrCP-mediated Degradation of Tiam1
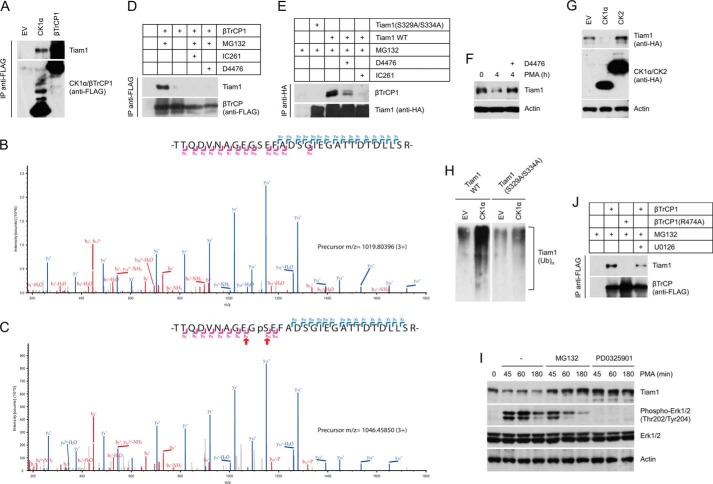
In the Tiam1 immunopurification described above, we also retrieved three peptides (DIKPDNFLMGIGR, MSTPVEVLCK, and AAQQAASSSGQGQQAQTPTGK) corresponding to casein kinase 1α (CK1α) and one peptide (HPQLHIESK) corresponding to casein kinase 1ϵ (CK1ϵ). First, we confirmed the interaction between Tiam1 and CK1α (Fig. 4A). Second, to test whether CK1α is able to phosphorylate the Tiam1 degron and pinpoint the specific Tiam1 site targeted by CK1α, immunopurified, dephosphorylated Tiam1 was subjected to an in vitro kinase assay in the presence or absence of purified CK1α prior to mass spectrometry analysis. We identified phosphopeptides containing Ser(P)329 in CK1α-treated Tiam1 samples (Fig. 4, B and C). These phosphopeptides were not found in Tiam1 treated with other kinases or when no kinase was added to the reaction. Next, we assessed the possible involvement of CK1 in the βTrCP-dependent degradation of Tiam1. We employed the CK1 pharmacological inhibitors D4476 and IC261 to inactivate CK1 in HEK293T cells. As shown in Fig. 4 (D and E), the Tiam1-βTrCP binding was abolished by both D4476 and IC261, suggesting that CK1 is required for the βTrCP-dependent degradation of Tiam1. Indeed, pharmacological inhibition of CK1 prevented the degradation of Tiam1 in response to PMA treatment (Fig. 4F). Accordingly, in cultured cells, Tiam1 degradation (Fig. 4G) and ubiquitylation (Fig. 4H) were stimulated by the ectopic expression of CK1α.
FIGURE 4.
CK1 and the MAPK pathway control the βTrCP-mediated degradation of Tiam1. A, HEK293T cells were transfected with FLAG-tagged CK1α, FLAG-tagged βTrCP1, or an empty vector. Forty-eight hours after transfection, cells were treated with MG132 for 5 h. Cells were then harvested and lysed. Whole cell extracts were immunoprecipitated (IP) with anti-FLAG resin and immunoblotted with antibodies specific for the indicated proteins. B and C, Tiam1 was immunopurified and dephosphorylated with lambda phosphatase prior to an in vitro kinase assay in the presence of purified CK1α. A mock reaction (no kinase) was used as a negative control. Individual mixes were subsequently trypsinized and analyzed by mass spectrometry. The Tiam1 tryptic peptide TTQDVNAGEGSEFADSGIEGATTDTDLLSR was not found to be phosphorylated in the negative control (B). When CK1α was used in the in vitro kinase assay, the same peptide was found to be phosphorylated on Ser329 (C). As shown, both peptide sequences can be clearly explained by their respective HCD MSMS spectrum (Mascot score 139 and 92), including the Ser(P)329 site (PhosphoRS site probability = 100.0%). In these figures, pS denotes phosphorylated serine, b denotes b ions, and y denotes y ions. D, HEK293T cells were transfected with FLAG-tagged βTrCP1. Forty-eight hours after transfection, cells were treated with MG132 for 5 h (when indicated) in the presence or absence of the indicated kinase inhibitors. Cells were then harvested and lysed. Whole cell extracts were immunoprecipitated with anti-FLAG resin and immunoblotted with antibodies specific for the indicated proteins. E, HEK293T cells were transfected with HA-tagged wild type Tiam1 or HA-tagged Tiam1(S329A/S334A). Forty-eight hours after transfection, cells were treated as in D. Whole cell extracts were processed as in D except that an anti-HA resin was used for immunoprecipitation. F, cells were treated with PMA for 4 h with or without the CK1 inhibitor D4476. Whole cell extracts were analyzed by immunoblotting. G, HEK293 cells were transfected with plasmids expressing HA-tagged wild type Tiam1 along with plasmids expressing the indicated kinases. Twenty-four hours after transfection, cells were collected and lysed. Whole cells extracts were analyzed by immunoblotting with antibodies specific for the indicated proteins. H, HEK293T cells were transfected with either HA-tagged wild type Tiam1 or HA-tagged Tiam1(S329A/S334A) along with Myc-tagged ubiquitin with or without CK1α. Cells were harvested and lysed in 0.1% Triton X-100 lysis buffer. Whole cell extracts were denatured by adding 1% SDS and boiling for 10 min. SDS was quenched and diluted. Whole cell extracts were then immunoprecipitated with anti-HA resin and immunoblotted with anti-Myc antibodies. The bracket indicates a ladder of bands corresponding to polyubiquitylated Tiam1. I, HEK293 cells were treated with PMA in the absence or presence of the indicated compounds. Cells were collected at the indicated times and lysed. Whole cell extracts were analyzed by immunoblotting. J, HEK293T cells were transfected with FLAG-tagged βTrCP1 or FLAG-tagged βTrCP1(R474A). Forty-eight hours after transfection, cells were treated with MG132 for 5 h in the presence or absence of the MEK inhibitor U1026. Cells were then harvested and lysed. Whole cell extracts were immunoprecipitated with anti-FLAG resin and immunoblotted with antibodies specific for the indicated proteins. EV, empty vector.
The finding that mitogens induce the CK1- and βTrCP-mediated degradation of Tiam1 prompted us to test whether MAPKs control Tiam1 proteolysis. Fig. 4I shows that pharmacological inhibition of MEK1 and MEK2, two protein kinases of the MAPK cascade known to be activated by mitogenic stimuli, blocked the PMA-induced degradation of Tiam1 to the same extent of MG132. Accordingly, inhibition of MEK1 and MEK2 reduced the interaction between βTrCP and Tiam1 (Fig. 4J).
Tiam1 Degradation Is Required for the Termination of mTOR-S6K Signaling
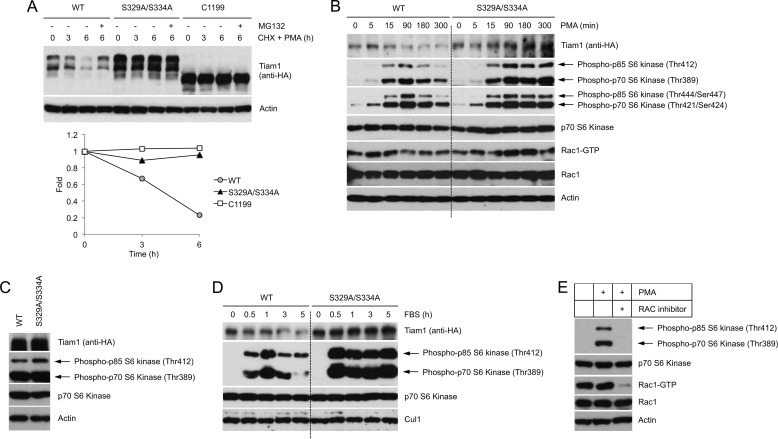
To investigate the biological significance of Tiam1 degradation, physiological levels of wild type Tiam1 and the Tiam1(S239A/S334A) mutant were expressed in HEK293 cells. Tiam1(S239A/S334A) and the Tiam1 mutant truncated at the N terminus (C1199) lacking the βTrCP binding domain were resistant to the degradation induced by mitogenic stimulation, as indicated by the half-life experiment shown in Fig. 5A.
FIGURE 5.
Tiam1 degradation controls the duration of the mitogen-induced mTOR-S6K signaling pathway. A, HEK293 cells expressing HA-tagged wild type Tiam1, HA-tagged Tiam1(S329A/S334A), or HA-tagged Tiam1-C1199 were incubated in low serum for 24 h and then treated with PMA and cycloheximide (CHX) for the indicated times. Cells were collected and lysed. Whole cell lysates were analyzed by immunoblotting with an anti-HA antibody. Actin is shown as a loading control. The graph shows the quantification of Tiam1 abundance relative to the amount at time 0. B, HEK293 cells transduced with retroviruses expressing HA-tagged wild type Tiam1 or HA-tagged Tiam1(S329A/S334A) were incubated in low serum for 48 h and then treated with PMA. At the indicated times, cells were collected and lysed. Whole cell extracts were analyzed by immunoblotting with antibodies for the indicated proteins. Levels of GTP-loaded Rac1 were analyzed in a pulldown assay as described under “Experimental Procedures.” Actin is shown as loading control. To facilitate comparison, a dotted line separates samples from cells expressing wild type Tiam1 and samples from cells expressing Tiam1(S329A/S334A). C, whole cell lysates of asynchronously growing HEK293 cells expressing HA-tagged wild type Tiam1 or HA-tagged Tiam1(S329A/S334A) were analyzed by immunoblotting with antibodies for the indicated proteins. D, as in B except that serum (instead of PMA) was used as mitogen. Cul1 is shown as loading control. E, HEK293 cells were incubated in low serum for 48 h and then treated with PMA. Six hours after PMA treatment, cells were analyzed by immunoblotting. Rac1 activity was assessed as in B.
Because it has been shown that both Tiam1 and Rac1 control the mTOR-S6K signaling pathway, which mediates the mitogenic response in cells (30–32), we analyzed the activation of mTOR-S6K signaling in response to mitogenic stimulation in cell expressing wild type Tiam1 and the nondegradable Tiam1(S239A/S334A) mutant. As shown in Fig. 5B, mitogenic stimulation of cells expressing wild type Tiam1 resulted in an increase of the mTOR-mediated phosphorylation of p70-S6K (Thr389) and its isoform p85-S6K (Thr412), which then rapidly decreased. In contrast, cells expressing the nondegradable Tiam1(S239A/S334A) mutant displayed a sustained (at least up to 5 h after PMA treatment) phosphorylation of p70-S6K (Thr389) and p85-S6K (Thr412) in response to mitogenic stimulation. No remarkable effect on the phosphorylation of p70-S6K (Thr389) and p85-S6K (Thr412) was observed in asynchronously growing cells (Fig. 5C). The expression of the degradation-resistant Tiam1(S239A/S334A) mutant had a similar effect on the mitogen-induced activating phosphorylation of additional residues of p70-S6K (Thr421/Ser424) and p85-S6K (Thr444/Ser447) (Fig. 5B). Similar results were obtained when serum was used to stimulate cells expressing wild type Tiam1 or the Tiam1(S239A/S334A) mutant (Fig. 5D). In agreement with previous reports (30), pharmacological inhibition of Rac activity prevented the induction of S6K phosphorylation following mitogenic stimulation, suggesting that the mitogen-induced activation of the mTOR-S6K signaling pathway is dependent on Rac activity (Fig. 5E). Taken together, these results demonstrate that failure to degrade Tiam1 in response to mitogens results in prolonged activation of the mTOR/S6K signaling.
Failure to Degrade Tiam1 Results in Loss of Cell Viability and Increased Apoptosis
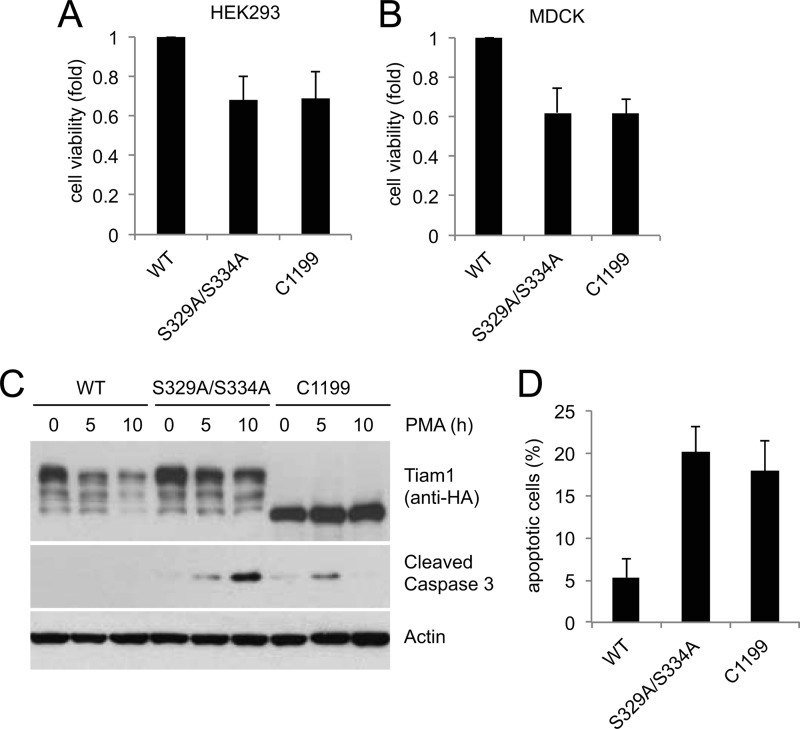
Finally, we investigated the effect of defective degradation of Tiam1 on cell growth and survival. After mitogenic stimulation, cells expressing Tiam1(S239A/S334A) or Tiam1-C1199 displayed reduced cell viability when compared with cells expressing wild type Tiam1 (Fig. 6, A and B). Moreover, failure to degrade Tiam1 resulted in apoptotic cell death as shown by the induction of the cleaved active form of Caspase 3 (Fig. 6, C and D).
FIGURE 6.
Inhibition of Tiam1 degradation results in loss of cell viability and increased apoptosis. A and B, HEK293 cells (A) or MDCK cells (B) transfected with HA-tagged wild type Tiam1, HA-tagged Tiam1(S329A/S334A), or HA-tagged Tiam1-C1199 were incubated in low serum for 24 h and then treated with PMA for 6 h. Cell viability was analyzed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. The results are the averages of three independent experiments. The error bars represent S.D. C, MDCK cells as in B were incubated in low serum for 24 h and then treated with PMA for the indicated times. The cells were collected, lysed, and analyzed by immunoblotting with antibodies for the indicated proteins. D, as in C except that cells were fixed, stained for cleaved caspase 3, and analyzed by flow cytometry. The results are the averages of three independent experiments. The error bars represent S.D.
DISCUSSION
Here we have shown that Tiam1 is a novel substrate of the SCFβTrCP ubiquitin ligase, identified a conserved phosphodegron at the N terminus of Tiam1 that is required for Tiam1 binding to βTrCP, and demonstrated that SCFβTrCP targets Tiam1 for proteasome-dependent degradation in response to mitogenic stimulation. We have also found that the mitogen-induced degradation of Tiam1 is mediated by CK1α, which phosphorylates a residue within the Tiam1 phosphodegron. Tiam1 destruction is required for terminating the activation of the mTOR-S6K signaling pathway following mitogenic stimulation. Indeed, expression of a Tiam1 mutant unable to bind βTrCP results in sustained phosphorylation of S6K, a substrate of mTOR. Failure to degrade Tiam1 leads to loss of cell viability caused by increased apoptosis. Interestingly, it has been recently reported that constitutive mTOR-S6K signaling sensitizes cells to p53-dependent cell death in response to stress conditions (33). In agreement with these studies, a recent report has shown that cells expressing the degradation-resistant Tiam1-C1199 mutant are more sensitive to DNA-damaging drug-induced apoptosis (34).
We have shown that the βTrCP-mediated degradation of Tiam1 is blocked by pharmacological inhibition of MEK. Accordingly, the Malliri group (35) has demonstrated that ERK1 and ERK2 associate with Tiam1 and that ERK activation is required for the v-Src-induced degradation of Tiam1. The involvement of the MEK-ERK pathway in the destruction of Tiam1 via βTrCP also suggests that the MEK-ERK-dependent phosphorylation of Tiam1 could prime the phosphorylation of the Tiam1 degron by CK1, as has been shown for other βTrCP substrates (16) (36, 37). Phospho-specific antibodies against the Tiam1 degron motif (329pSEFADpSGIEGApT340) are required to test (both in vitro and in cultured cells) whether Tiam1 is phosphorylated by CK1 on its degron following initial phosphorylation by MEK-ERK on different sites. In conclusion, the finding that the degradation of Tiam1, induced by CK1 in cooperation with the MAPK cascade, is needed to terminate mTOR-S6K signaling suggests a new mechanism by which the MAPK cascade controls the timing of activation of the mTOR-S6K signaling pathway.
Acknowledgments
We thank R. Lim, S. Rahmy for contributions; S. Ellenbroek, J. den Hertog, H. Ovaa, and F. Zwartkruis for reagents; and A. Malliri for suggestions and reagents.
This work was supported by funds from the Royal Dutch Academy of Arts and Sciences (KNAW), the Dutch Cancer Society (KWF), the Cancer Genomics Centre, and the European Union under Marie Curie Actions (to D. G.) and from the Netherlands Proteomics Center (to T. Y. L. and A. J. R. H.).
- MDCK
- Madin-Darby canine kidney
- PMA
- phorbol 12-myristate 13-acetate
- CK
- casein kinase.
REFERENCES
- 1. Habets G. G., Scholtes E. H., Zuydgeest D., van der Kammen R. A., Stam J. C., Berns A., Collard J. G. (1994) Identification of an invasion-inducing gene, Tiam-1, that encodes a protein with homology to GDP-GTP exchangers for Rho-like proteins. Cell 77, 537–549 [DOI] [PubMed] [Google Scholar]
- 2. Mertens A. E., Roovers R. C., Collard J. G. (2003) Regulation of Tiam1-Rac signalling. FEBS Lett. 546, 11–16 [DOI] [PubMed] [Google Scholar]
- 3. Michiels F., Habets G. G., Stam J. C., van der Kammen R. A., Collard J. G. (1995) A role for Rac in Tiam1-induced membrane ruffling and invasion. Nature 375, 338–340 [DOI] [PubMed] [Google Scholar]
- 4. Lambert J. M., Lambert Q. T., Reuther G. W., Malliri A., Siderovski D. P., Sondek J., Collard J. G., Der C. J. (2002) Tiam1 mediates Ras activation of Rac by a PI3K-independent mechanism. Nat. Cell Biol. 4, 621–625 [DOI] [PubMed] [Google Scholar]
- 5. Hordijk P. L., ten Klooster J. P., van der Kammen R. A., Michiels F., Oomen L. C., Collard J. G. (1997) Inhibition of invasion of epithelial cells by Tiam1-Rac signaling. Science 278, 1464–1466 [DOI] [PubMed] [Google Scholar]
- 6. Malliri A., Collard J. G. (2003) Role of Rho-family proteins in cell adhesion and cancer. Curr. Opin. Cell Biol. 15, 583–589 [DOI] [PubMed] [Google Scholar]
- 7. Malliri A., van Es S., Huveneers S., Collard J. G. (2004) The Rac exchange factor Tiam1 is required for the establishment and maintenance of cadherin-based adhesions. J. Biol. Chem. 279, 30092–30098 [DOI] [PubMed] [Google Scholar]
- 8. Kruiswijk F., Yuniati L., Magliozzi R., Low T. Y., Lim R., Bolder R., Mohammed S., Proud C. G., Heck A. J., Pagano M., Guardavaccaro D. (2012) Coupled activation and degradation of eEF2K regulates protein synthesis in response to genotoxic stress. Sci. Signal. 5, ra40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 10. Raijmakers R., Berkers C. R., de Jong A., Ovaa H., Heck A. J., Mohammed S. (2008) Automated online sequential isotope labeling for protein quantitation applied to proteasome tissue-specific diversity. Mol. Cell. Proteomics 7, 1755–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Taus T., Köcher T., Pichler P., Paschke C., Schmidt A., Henrich C., Mechtler K. (2011) Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res. 10, 5354–5362 [DOI] [PubMed] [Google Scholar]
- 12. D'Annibale S., Kim J., Magliozzi R., Low T. Y., Mohammed S., Heck A. J., Guardavaccaro D. (2014) Proteasome-dependent degradation of transcription factor AP4 (TFAP4) controls mitotic division. J. Biol. Chem. 289, 7730–7737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Frescas D., Pagano M. (2008) Deregulated proteolysis by the F-box proteins SKP2 and β-TrCP: tipping the scales of cancer. Nat. Rev. Cancer 8, 438–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu G., Xu G., Schulman B. A., Jeffrey P. D., Harper J. W., Pavletich N. P. (2003) Structure of a β-TrCP1-Skp1-β-catenin complex: destruction motif binding and lysine specificity of the SCF(β-TrCP1) ubiquitin ligase. Mol. Cell 11, 1445–1456 [DOI] [PubMed] [Google Scholar]
- 15. Cardozo T., Pagano M. (2004) The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5, 739–751 [DOI] [PubMed] [Google Scholar]
- 16. Magliozzi R., Low T. Y., Weijts B. G., Cheng T., Spanjaard E., Mohammed S., van Veen A., Ovaa H., de Rooij J., Zwartkruis F. J., Bos J. L., de Bruin A., Heck A. J., Guardavaccaro D. (2013) Control of epithelial cell migration and invasion by the IKKβ- and CK1α-mediated degradation of RAPGEF2. Dev. Cell 27, 574–585 [DOI] [PubMed] [Google Scholar]
- 17. Busino L., Donzelli M., Chiesa M., Guardavaccaro D., Ganoth D., Dorrello N. V., Hershko A., Pagano M., Draetta G. F. (2003) Degradation of Cdc25A by β-TrCP during S phase and in response to DNA damage. Nature 426, 87–91 [DOI] [PubMed] [Google Scholar]
- 18. Jin J., Shirogane T., Xu L., Nalepa G., Qin J., Elledge S. J., Harper J. W. (2003) SCFβ-TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes Dev. 17, 3062–3074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Melixetian M., Klein D. K., Sørensen C. S., Helin K. (2009) NEK11 regulates CDC25A degradation and the IR-induced G2/M checkpoint. Nat. Cell Biol. 11, 1247–1253 [DOI] [PubMed] [Google Scholar]
- 20. Mailand N., Bekker-Jensen S., Bartek J., Lukas J. (2006) Destruction of Claspin by SCFβTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol. Cell 23, 307–318 [DOI] [PubMed] [Google Scholar]
- 21. Margottin-Goguet F., Hsu J. Y., Loktev A., Hsieh H. M., Reimann J. D., Jackson P. K. (2003) Prophase destruction of Emi1 by the SCF(βTrCP/Slimb) ubiquitin ligase activates the anaphase promoting complex to allow progression beyond prometaphase. Dev. Cell 4, 813–826 [DOI] [PubMed] [Google Scholar]
- 22. Guardavaccaro D., Frescas D., Dorrello N. V., Peschiaroli A., Multani A. S., Cardozo T., Lasorella A., Iavarone A., Chang S., Hernando E., Pagano M. (2008) Control of chromosome stability by the β-TrCP-REST-Mad2 axis. Nature 452, 365–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mantovani F., Banks L. (2003) Regulation of the discs large tumor suppressor by a phosphorylation-dependent interaction with the β-TrCP ubiquitin ligase receptor. J. Biol. Chem. 278, 42477–42486 [DOI] [PubMed] [Google Scholar]
- 24. Dorrello N. V., Peschiaroli A., Guardavaccaro D., Colburn N. H., Sherman N. E., Pagano M. (2006) S6K1- and βTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science 314, 467–471 [DOI] [PubMed] [Google Scholar]
- 25. Yaron A., Hatzubai A., Davis M., Lavon I., Amit S., Manning A. M., Andersen J. S., Mann M., Mercurio F., Ben-Neriah Y. (1998) Identification of the receptor component of the IκBα-ubiquitin ligase. Nature 396, 590–594 [DOI] [PubMed] [Google Scholar]
- 26. Spencer E., Jiang J., Chen Z. J. (1999) Signal-induced ubiquitination of IKB by the F-box protein Slimb/β-TrCP. Genes Dev. 13, 284–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu K., Fuchs S. Y., Chen A., Tan P., Gomez C., Ronai Z., Pan Z. Q. (2000) The SCF(HOS/β-TRCP)-ROC1 E3 ubiquitin ligase utilizes two distinct domains within CUL1 for substrate targeting and ubiquitin ligation. Mol. Cell. Biol. 20, 1382–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Winston J. T., Strack P., Beer-Romero P., Chu C. Y., Elledge S. J., Harper J. W. (1999) The SCFb-TRCP ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev. 13, 270–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fuchs S. Y., Chen A., Xiong Y., Pan Z. Q., Ronai Z. (1999) HOS, a human homolog of Slimb, forms an SCF complex with Skp1 and Cullin1 and targets the phosphorylation-dependent degradation of IκB and β-catenin. Oncogene 18, 2039–2046 [DOI] [PubMed] [Google Scholar]
- 30. Chou M. M., Blenis J. (1996) The 70 kDa S6 kinase complexes with and is activated by the Rho family G proteins Cdc42 and Rac1. Cell 85, 573–583 [DOI] [PubMed] [Google Scholar]
- 31. Saci A., Cantley L. C., Carpenter C. L. (2011) Rac1 regulates the activity of mTORC1 and mTORC2 and controls cellular size. Mol. Cell 42, 50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Buchsbaum R. J., Connolly B. A., Feig L. A. (2003) Regulation of p70 S6 kinase by complex formation between the Rac guanine nucleotide exchange factor (Rac-GEF) Tiam1 and the scaffold spinophilin. J. Biol. Chem. 278, 18833–18841 [DOI] [PubMed] [Google Scholar]
- 33. Lee C. H., Inoki K., Karbowniczek M., Petroulakis E., Sonenberg N., Henske E. P., Guan K. L. (2007) Constitutive mTOR activation in TSC mutants sensitizes cells to energy starvation and genomic damage via p53. EMBO J. 26, 4812–4823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhu G., Fan Z., Ding M., Mu L., Liang J., Ding Y., Fu Y., Huang B., Wu W. (2014) DNA damage induces the accumulation of Tiam1 by blocking β-TrCP-dependent degradation. J. Biol. Chem. 289, 15482–15494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Woodcock S. A., Rooney C., Liontos M., Connolly Y., Zoumpourlis V., Whetton A. D., Gorgoulis V. G., Malliri A. (2009) SRC-induced disassembly of adherens junctions requires localized phosphorylation and degradation of the rac activator tiam1. Mol. Cell 33, 639–653 [DOI] [PubMed] [Google Scholar]
- 36. Dehan E., Bassermann F., Guardavaccaro D., Vasiliver-Shamis G., Cohen M., Lowes K. N., Dustin M., Huang D. C., Taunton J., Pagano M. (2009) βTrCP- and Rsk1/2-mediated degradation of BimEL inhibits apoptosis. Mol. Cell 33, 109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Duan S., Skaar J. R., Kuchay S., Toschi A., Kanarek N., Ben-Neriah Y., Pagano M. (2011) mTOR generates an auto-amplification loop by triggering the βTrCP- and CK1α-dependent degradation of DEPTOR. Mol. Cell 44, 317–324 [DOI] [PMC free article] [PubMed] [Google Scholar]