Background: miR-21 is an anti-apoptotic microRNA, and its role in alcoholic liver disease (ALD) is unknown.
Results: miR-21, increased in ALD and regulated by IL-6/Stat3, is essential for transformation, survival, and liver fibrosis.
Conclusion: miR-21 plays a protective role against alcoholic hepatitis through the anti-extrinsic apoptotic pathway.
Significance: Understanding the recovery function of miR-21 may have important implications for patients with ALD.
Keywords: Alcohol, Interleukin 6 (IL-6), Liver Injury, MicroRNA (miRNA), STAT3, DR5, FASLG, Alcoholic Liver Diseases
Abstract
IL-6/Stat3 is associated with the regulation of transcription of key cellular regulatory genes (microRNAs) during different types of liver injury. This study evaluated the role of IL-6/Stat3 in regulating miRNA and miR-21 in alcoholic liver disease. By microarray, we identified that ethanol feeding significantly up-regulated 0.8% of known microRNAs in mouse liver compared with controls, including miR-21. Similarly, the treatment of normal human hepatocytes (N-Heps) and hepatic stellate cells (HSCs) with ethanol and IL-6 significantly increased miR-21 expression. Overexpression of miR-21 decreased ethanol-induced apoptosis in both N-Heps and HSCs. The expression level of miR-21 was significantly increased after Stat3 activation in N-Heps and HSCs, in support of the concept that the 5′-promoter region of miR-21 is regulated by Stat3. Using real time PCR, we confirmed that miR-21 activation is associated with ethanol-linked Stat3 binding of the miR-21 promoter. A combination of bioinformatics, PCR array, dual-luciferase reporter assay, and Western blot analysis revealed that Fas ligand (TNF superfamily, member 6) (FASLG) and death receptor 5 (DR5) are the direct targets of miR-21. Furthermore, inhibition of miR-21 by specific Vivo-Morpholino and knock-out of IL-6 in ethanol-treated mice also increased the expression of DR5 and FASLG in vivo during alcoholic liver injury. The identification of miR-21 as an important regulator of hepatic cell survival, transformation, and remodeling in vitro, as well as its upstream modulators and downstream targets, will provide insight into the involvement of altered miRNA expression in contributing to alcoholic liver disease progression and testing novel therapeutic approaches for human alcoholic liver diseases.
Introduction
Alcohol abuse in the United States contributes, either directly or indirectly, to 5% of deaths annually. Although alcohol in moderation is thought to offer beneficial effects in the cardiovascular system, chronic excessive alcohol intake confers deleterious effects on nearly all major organ systems. The liver is one of the most critical targets of alcohol-induced damage. Alcoholic liver diseases (ALDs)4 encompass a broad spectrum of clinical features, including alcoholic fatty liver, alcoholic steatohepatitis and cirrhosis, and increased risk of hepatocellular carcinoma (1). Although the toxic effects of alcohol likely result from complex interactions between genes and the environment, the molecular mechanisms of alcohol-induced liver damage remains undefined. Thus, a better understanding of the mechanisms regulating hepatic cell injury may lead to more effective therapeutic approaches for ALD.
Recent studies have discovered a genetic switch that regulates critical properties of hepatic cells during liver injury. This regulator, which belongs to a class of noncoding RNAs (ncRNAs) called microRNAs, is indeed a regulatory master switch that controls the hepatic cell proliferation/survival and tissue repair through its ability to switch off specific target genes by either binding to imperfect complementary sites within the 3′-untranslated region (UTR) of their mRNA targets or by targeting specific cleavage of homologous mRNAs (2–6). miRNAs are ncRNAs recently found to down-regulate a large subset of human genes, and evidence suggests that some miRNAs have powerful cellular functions. We have seen that several miRNAs are aberrantly expressed after alcoholic liver injury (7) and partial hepatectomy (8). Also, specific miRNAs, such as miR-34a and miR-181, regulate cell survival, migration, and remodeling properties via repression of target genes and modulation of downstream signaling pathways (9). Therefore, we postulate that alterations in the expression of miRNAs influence cellular behaviors, such as apoptosis, survival, and remodeling by alteration of key cellular targets. In fact, aberrant expression of miRNAs, such as miR-34a, alters the cellular expression of Sirt1 and caspase-2 during ALD. However, the contribution of aberrantly expressed miRNAs to hepatic cell responses in ALD is unclear.
MicroRNA-21 is an established survival factor during liver injury and hepatocellular carcinoma development. The miR-21 gene is located on chromosome 17, close to the location of p53. The regulation of miR-21 by the transcription factor p53 suggests a potential role for miR-21 in the modulation of hepatic cell behavior during liver injury. Normally, p53 inhibits hepatic cell proliferation and stimulates cell death. However, disruption of the p53 pathway promotes liver injury. One pathway by which p53 regulates cell growth is through miRNA. Cellular stress stabilizes p53 that in turn regulates the expression of a set of miRNAs, which may control apoptosis and senescence (10–14). After intercrossing miR-21−/− and Trp53−/− mice, tissues from the Trp53−/− miR-21−/− mice have a higher percentage of apoptotic cells in response to genotoxic agents (15). Although there is no miR-21-binding site in the p53 3′-untranslated region in either humans or mice, there are several reports demonstrating that miR-21 represses the expression of multiple genes in the p53 network (16). Meanwhile, the overexpression of miR-21 during human liver regeneration suggests the presence of additional mechanisms by which miR-21 contributes to hepatic cell survival and regeneration (17). Thus, we assessed the role of aberrant expression of miR-21 in hepatic cell survival during ALD by posing the following questions. (i) Is miR-21 expression altered in ethanol-exposed mice and ALD human liver tissues? (ii) Does modulation of miR-21 alter apoptosis and cell survival in vitro and in animals with ALD? (iii) What is the upstream regulator for miR-21 during ALD? (iv) What downstream targets of miR-21 are involved in ALD?
MATERIALS AND METHODS
Cells and Tissues
Normal human hepatocytes (N-Hep) and human hepatic stellate cells (HSC) were obtained from ScienCell Inc. (San Diego). The human hepatocellular cancer cell line, HepG2, was obtained from the American Type Culture Collection (Manassas, VA) and cultured as recommended by the supplier. All N-Heps and HSCs were purchased from ScienCell Inc. and used in the current project within five passages.
Transfections
Transfections were performed by nuclear electroporation using the Nucleofector system (Amaxa Biosystems, Koln, Germany). Fifty μl of 100 nm microRNA precursor, antisense inhibitor, or controls (Ambion, Austin, TX) were added to 1 × 106 cells suspended in 50 μl of Nucleofector solution at room temperature. The sequences of the microRNA precursors and inhibitors used can be obtained from Ambion. After electroporation, transfected cells were resuspended in culture medium containing 10% fetal bovine serum (FBS) for 48–72 h prior to study. All studies were performed in quadruplicate unless otherwise specified.
miRNA Array Hybridization and Analysis
RNA was extracted using TRIzol reagent (Invitrogen). Total RNA (5 μg) was reverse-transcribed using biotin end-labeled random octamer oligonucleotide primers. Hybridization of biotin-labeled complementary DNA was done using a custom miRNA microarray chip (ncRNA Program at Center for Targeted Therapy, MD Anderson Cancer Center, Houston, TX), containing 627 probes for mature miRNA corresponding to 324 different human miRNAs spotted in quadruplicate. The images were scanned and quantitated using an Axon 4000B scanner (Molecular Devices, Sunnyvale, CA). The scanned images were quantified using GenePix 6.0 software (Molecular Devices). The data from three samples for each tissue type were further analyzed by the BRB-ArrayTools software from NCI, National Institutes of Health (Bethesda, MD). Cluster analysis was carried out using MultiExperiment Viewer software from the Institute of Genomic Research (Rockville, MD).
Real time PCR for Mature miRNA
The expression of mature miRNAs in human hepatic cell lines was analyzed by TaqMan miRNA assay (Applied Biosystems, Foster City, CA). Briefly, single-stranded cDNA was synthesized from 10 ng of total RNA in a 15-μl reaction volume by using the TaqMan MicroRNA reverse transcription kit (Applied Biosystems). The reactions were incubated first at 16 °C for 30 min and then at 42 °C for 30 min. The reactions were inactivated by incubation at 85 °C for 5 min. Each cDNA generated was amplified by quantitative PCR by using sequence-specific primers from the TaqMan microRNA Assays on a MX 3000P PCR Instrument (Stratagene, San Diego). The 20-μl PCR included 10 μl of 2× Universal PCR Master Mix (No AmpErase uracil N-glycosylase), 2 μl of each 10× TaqMan MicroRNA Assay Mix, and 1.5 μl of reverse transcription (RT) product. The reactions were incubated in a 96-well plate at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The threshold cycle (CT) is defined as the fractional cycle number at which the fluorescence passes the fixed threshold.
SuperArray Quantitative PCR Assay and Real Time PCR Analysis
RNA was isolated from liver tissues or cell lysates using TRIzol (Invitrogen) according to the manufacturer's protocol. The RNA was subsequently cleaned using RNeasy kit (Qiagen, Valencia, CA), according to the manufacturer's protocol. The optional on-column DNase treatment was performed. Reverse transcription was done using 1 μg of RNA with RT2 first strand kit according to the manufacturer's protocol (SABiosciences, Frederick, MD). Mouse liver tissue or normal human hepatocyte cDNA was analyzed using SuperArray plates (human apoptosis PCR array (PAHS-012ZA) or mouse apoptosis miScript miRNA PCR array (MIMM-114ZA) (SABiosciences). To validate the translational significance of these gene expression findings, mouse liver or human hepatocyte samples were analyzed using real time PCR. RT2 quantitative PCR primer assays (SABiosciences) or TaqMan miRNA PCR assays were used. Real time PCR was performed usingRT2 SYBR Green/ROX quantitative PCR master mix for a Stratagene Mx3005P real time PCR system according to the manufacturer's protocol (SABiosciences). ROX was used as an endogenous reference, and data were analyzed using the PCR array data analysis template (SABiosciences). The comparative CT method (ΔΔCT) was used for quantification of gene expression. All samples were tested in triplicate, and average values used for quantification.
Animal Experiments
For chronic intragastric alcohol administration, C57BL/6 mice (10 weeks old) were aseptically implanted with gastrostomy catheters as described (18, 19). An increasing dose of ethanol (22.7–35 g/kg per day for mice) or control solution was infused for 4 weeks (18). All animal experiments were performed with age- and sex-matched mice from the same littermates and performed in accordance with the approved Institutional Animal Care and Use Committee protocol, University of Southern California.
C3H/HeOu/J mice, female C57Bl/6J mice, and IL-6 knock-out mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and used for ethanol (EtOH) treatments and studies with miR-21 Vivo-Morpholino (control miRNA morpholino was also utilized). All mice were used at the age of 6 weeks (weight 20–25 g). These and the following animal studies were performed at the Animal Resource Facility, Scott & White Healthcare and Texas A&M University Health Science Center. Animals were housed in standard microisolator cages and fed standard laboratory chow (rodent diet 2918, Harlan-Teklad, Madison, WI) prior to initiation of liquid diet feeding. All animal procedures were approved by Scott & White Healthcare and Texas A&M Health Science Center Institutional Animal Care and Use Committee. To produce EtOH-induced liver injury with microRNA Vivo-Morpholino treatment, the mice were fed a control liquid diet for 2 days, then 1% (v/v) ethanol diet for 2 days, then 2, 3, and 4% (v/v) ethanol diet for 2 days, followed by 5% (v/v) ethanol diet for 4 more weeks. For microRNA Vivo-Morpholino treatment, miR-21 and control miRNA Vivo-Morpholinos were designed and obtained from Gene Tools, LLC (Philomath, OR). After 2 weeks of EtOH feeding, the mice received one intravenous injection of miR-21 Vivo-Morpholino or control miRNA Vivo-Morpholino (30 mg/kg body weight in 0.1 ml per injection) through the tail vein. After 5 weeks, mice were weighed and anesthetized. Blood was collected from the posterior vena cava. Livers were excised, and mice were then euthanized by exsanguination. Livers were weighed and portions fixed in formalin, frozen in Optimal Cutting Temperature medium (Sakura Finetek U.S.A., Torrance, CA), snap-frozen in liquid nitrogen, or stored in RNAlater (Ambion) for further analysis. Plasma was separated from whole blood and stored at −80 °C.
High Frequency Ultrasound
The Visual Sonic Vevo® 2100 system (Toronto, Ontario, Canada) was used for ultrasound imaging. Mice were anesthetized with 1.5–2.5% isoflurane in O2 at 1 liter/min. Mice were maintained under continuous isoflurane anesthesia and placed on the heated Vevo mouse handling table that was mounted on the Vevo 2100 imaging station. Mouse fur was removed with a commercially available depilatory cream. Pre-warmed ultrasound coupling gel (Aquasonic 100, Parker Laboratories, Inc, Fairfield, NJ) was applied directly to the skin. Mice were scanned from the ventral body wall using either the MS250 or the MS55D real time MicroScanTM transducer and the Vevo 2100 imaging system. The livers were imaged in the parasternal long axis view, and three measurements of the portal vein area were obtained as indicated. Echohepatograph acquisition and analysis were performed by an experienced echohepatographer who was blinded to the genotype and treatment.
miRNA Array Hybridization and Analysis
RNA was extracted using the miRVANA isolation kit (Ambion). Hybridization of biotin-labeled complementary DNA was done using a custom miRNA microarray chip (ncRNA Program at Center for Targeted Therapy, MD Anderson Cancer Center, Houston, TX). The data from three samples for each cell type were further analyzed by the BRB-ArrayTools software from NCI, National Institutes of Health.
Primer Sequence for Chromatin Immunoprecipitation
Primer pairs specific for the miR-21 upstream region containing the Stat3 enhancer region were sequenced as follows: sense 5′-CCT CTG AGA AGA GGG GAC AA-3′, antisense 5′-ACC GCT TCC AGC AAA AGA GT-3′; real time PCR primer, pri-miR-21 forward, 5′-CAT TGT GGG TTT TGA AAA GGT TA-3′, and pri-miR-21 reverse, 5′-CCA CGA CTA GAG GCT GAC TTAGA-3′; Stat3 forward, 5′-CTG GCC TTT GGT GTT GAA AT-3′, and Stat3 reverse, 5′-AAG GCA CCC ACA GAA ACA AC-3′; and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward, 5′-GTC AGT GGT GGA CCT GAC CT-3′, and GAPDH reverse, 5′-AGG GGA GAT TCA GTG TGG TG-3′ (20, 30).
Plasmids for Stat3-miR-21 Binding Assay
The miR-21 promoter/enhancer region was amplified by PCR from human genomic DNA using the following primers: 5′-TTT GGT ACC TTG CTA ATG CAT TCT-3′ and 5′-TTT AGA TCT AGT TCA GCT ATG GTA AGA GC-3′; and inserted into the XhoI and BglII sites of pGL3-Basic (Promega). Mutations were introduced into the two Stat3 sites using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) with the oligonucleotides 5′-GTT CAA ACC AGT TCT CGA GGG AAC TAG TGG TGA T-3′ and 5′-GTG ATA AAT GTG GGA TCC CTG AGA AGT CAT TCA-3′ (alterations underlined). For convenience of selection, two novel restriction sites (XhoI and BamHI, respectively) were introduced. Construction of pSUPER-siStat3, encoding siRNA targeting Stat3, and a control vector (pSUPER-scrambled) have been used. A vector, pStat3res, encoding murine Stat3 resistant to siStat3 was generated by introducing silent mutations into the siRNA target sequence by site-directed mutagenesis. Nucleotides 823–841 of murine Stat3 cDNA were changed from 5′-CTT CAG ACC CGC CAA CAA A-3′ (encoding Leu-Gln-Thr-Arg-Gln-Gln) to TTG CAA ACG CGT CAG CAG A-3′ (altered bases underlined). For expression of miR-21 in N-Hep and HSC cells, a DNA fragment containing the precursor miR-21 sequence was amplified from human genomic DNA using the primers 5′-TTT GGA TCC GCC TAC CAT CGT GAC-3′ (sense) and 5′-TTT AAG CTT GGA TGG TCA GAT GAA-3′ (antisense), introducing BamHI and HindIII sites, respectively, at the 5′-ends. The resulting PCR product was cloned into the BglII and HindIII sites of pSUPER, yielding pSUPER-miR-21 (20).
Western Blotting
Cells grown in 100-mm dishes were lysed, and protein content was measured using the Bradford protein assay. Equivalent amounts of protein were resolved by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked and incubated with the specific primary antibody overnight at 4 °C, washed, and incubated with the appropriate IRDye700- and IRDye800-labeled secondary antibodies (Rockland, Gilbertsville, PA) (1:1000) for 1 h. Blots were stripped and re-probed with mouse monoclonal antibodies for β-actin (Sigma) (1:1000) or total Stat3 antibody (1:1000), which was used for normalization. Protein expression was visualized and quantified using the LI-COR Odyssey Infrared Imaging System (LI-COR Bioscience, Lincoln, NE).
Cell Proliferation Assay
Cell proliferation was measured using the CellTiter 96 aqueous assay kit (Promega, Madison, WI). Transfected cells (10,000/well) were plated in 96-well plates (BD Biosciences) and incubated at 37 °C, and cell proliferation was assessed after 72 h as described previously (9). Ethanol IC50 values were calculated using XLfit software (IDBS, Burlington, MA).
Anchorage-independent Hepatic Cell Growth
N-Hep, HSC, and HepG2 cells were seeded in 96-well plates (10,000 per well) in modified DMEM with 10% FBS after miRNA transfection. The final concentration of the bottom and top feeder layers of the agar system was 0.6% and the cell suspension layer was 0.4%. Anchorage-independent transformed cell growth was fluorometrically assayed after 7 days using Alamar Blue (BIOSOURCE), and the SpectraMax® M5 multi-mode microplate Reader (Molecular Devices, Inc. Sunnyvale, CA; excitation 530/25 nm; emission 580/50 nm).
Luciferase Reporter Assay
Intact putative miR-21 recognition sequence from the 3′-UTR of FASLG and DR5 (pMIR-FASLG/DR5-WT-3′-UTR) or with random mutations (pMIR-FASLG/DR5-mut-3′-UTR) were cloned downstream of the firefly luciferase reporter gene. Cells were co-transfected with 1 μg of pMIR-FASLG/DR5-WT or mut-3′-UTR construct and 1 μg of pRL-TK Renilla luciferase expression construct with or without precursor miR-21 using TransIT-siQUEST transfection reagent (Mirus, Madison, WI). Luciferase assays were performed 72 h after transfection using the Dual-Luciferase Reporter Assay system (Promega, Madison, WI).
Immunohistochemistry Analysis
Lobular necrosis was evaluated in liver sections stained with hematoxylin and eosin (H&E). Liver sections were also incubated overnight at 4 °C with specific antibodies (1:50), washed in phosphate-buffered saline (PBS), incubated for 20 min at room temperature with a secondary biotinylated antibody (Dako Cytomation LSAB Plus System-HRP, Glostrup, Denmark), then with Dako ABC for 20 min, and developed with 3–3′-diaminobenzidine (Dako Cytomation Liquid DAB Plus Substrate Chromogen System). For all immunoreactions, negative controls were included. Immunohistochemical observations were taken by BX-51 light microscopy (Olympus, Tokyo, Japan) with a Videocam (Spot Insight; Diagnostic Instrument, Sterling Heights, MI) and processed with an image analysis system.
Chemicals and Reagents
Specific microRNA precursors and inhibitors were obtained from Ambion. Rabbit anti-FASLG, DR5, α-SMA, survivin polyclonal antibodies, as well as IL-6, Stat3, and control siRNA plasmids were obtained from Santa Cruz Biotechnology Inc. (Santa Cruz, CA).
Statistical Analysis
Data are expressed as the means ± S.E. from at least three separate experiments performed in triplicate, unless otherwise noted. The differences between groups were analyzed using a double-sided Student's t test when only two groups were present and analysis of variance when there were more than two groups, and the null hypothesis was rejected at the 0.05 level unless otherwise specified.
RESULTS
Expression of miRNAs in Mouse Liver with Alcoholic Liver Injury
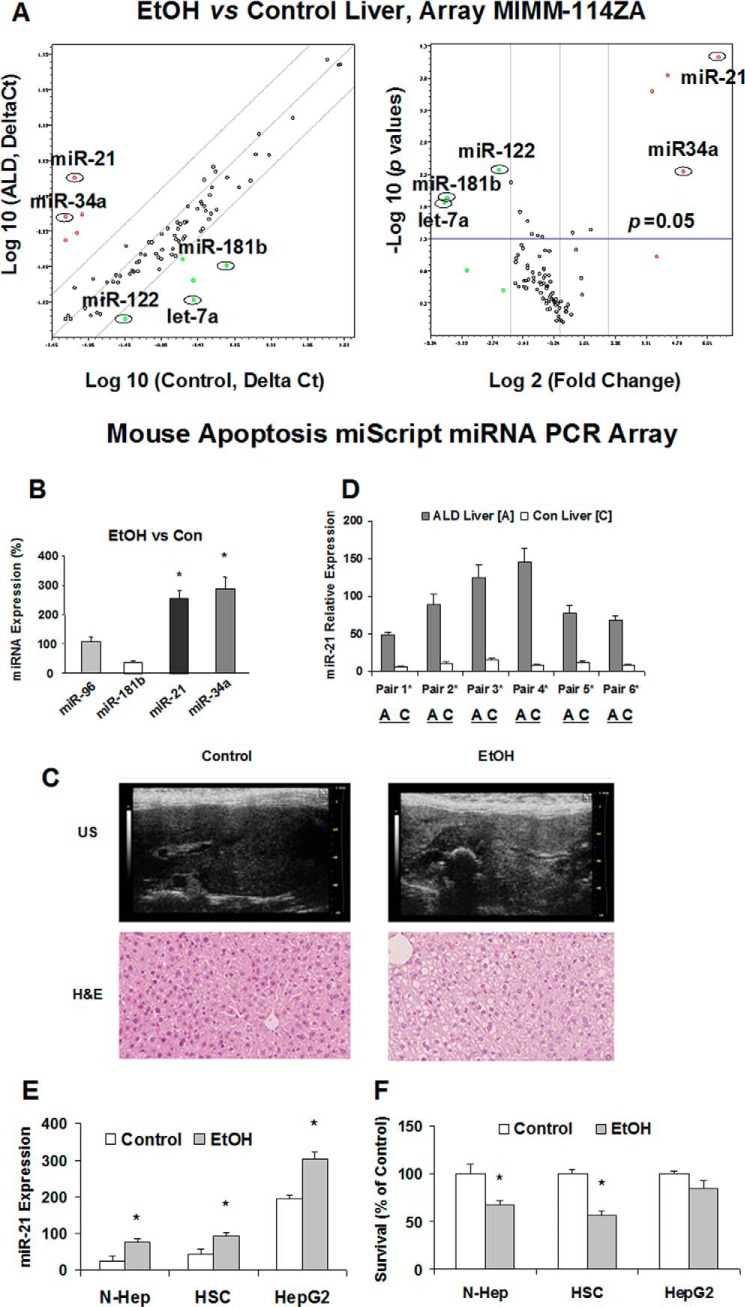
We first isolated and compared the miRNA expression profile with tissues from control mice and mice receiving intragastric EtOH feeding. Four microarray hybridization studies were performed on three different pairs of liver-derived RNA from intragastric EtOH feeding and normal mice (7). The miRNAs differentially overexpressed in livers from EtOH-fed mice included miR-21, miR-34a, miR-882, miR-409-5p, miR-509-3p, and miR-137 (Table 1). The overexpression of miR-21 and miR-34a was also seen in the liver tissues from mice administered 5% (v/v) EtOH after 5 weeks (Fig. 1, A and B). Along with the altered ultrasound liver images (Fig. 1C), there was a significantly increased alanine aminotransferase level and pathology score in these mice (Table 2). The expression of specific microRNAs (miR-21 and miR-34a) is increased significantly, which may contribute to the pathogenesis or phenotypic behavior of alcoholic liver injury (7).
TABLE 1.
Differential expression of miRNAs between intragastric ethanol-feeding mouse liver tissues and control normal mouse liver tissues
| miRNA | -Fold change | p value | Regulation |
|---|---|---|---|
| mmu-miR-34a | 3.95 ± 0.09 | 0.007 | Up |
| mmu-miR-21 | 3.83 ± 0.13 | 0.016 | Up |
| mmu-miR-882 | 2.87 ± 0.12 | 0.019 | Up |
| mmu-miR-409–5p | 2.79 ± 0.14 | 0.026 | Up |
| mmu-miR-509–3p | 2.22 ± 0.15 | 0.031 | Up |
| mmu-miR-137 | 2.08 ± 0.10 | 0.015 | Up |
| mmu-miR-122 | 0.28 ± 0.01 | 0.016 | Down |
| mmu-let-7a | 0.34 ± 0.01 | 0.008 | Down |
| mmu-miR-192 | 0.36 ± 0.04 | 0.021 | Down |
| mmu-miR-181b | 0.39 ± 0.02 | 0.012 | Down |
| mmu-miR-871 | 0.41 ± 0.05 | 0.026 | Down |
| mmu-miR-127 | 0.43 ± 0.07 | 0.042 | Down |
| mmu-miR-181a | 0.45 ± 0.04 | 0.024 | Down |
| mmu-let-7b | 0.46 ± 0.06 | 0.028 | Down |
| mmu-let-7g | 0.49 ± 0.08 | 0.035 | Down |
FIGURE 1.
Aberrant miR-21 expression in EtOH-fed mouse liver. A, expression level of miR-21 is up-regulated in the liver from EtOH-fed mice. Relative miRNA gene expression profile between EtOH mouse liver versus control liver tissues is shown. The expression of a panel of diverse miRNA genes was evaluated by real time PCR using Mouse Apoptosis miScript miRNA PCR array. Gene expression relative to GAPDH was plotted as the Volcano Plots, depicting the relative expression levels (log10) for selected genes in EtOH versus control panels (left). The relative expression levels and p values for each gene in the related samples were also plotted against each other in the scatter plot (right). miR-21 and miR-34a are the greatest up-regulated genes among the microRNAs involved in apoptotic signaling pathways in EtOH mice liver. B, miRNA was isolated from normal or EtOH mouse liver. Three clusters were generated using MultiExperiment Viewer software from the Institute of Genomic Research (Rockville, MD). Cluster 1 is composed of a group of miRNAs that were overexpressed after long term alcohol exposure; cluster 2 is composed of miRNAs that were not significantly altered after ethanol exposure; and cluster 3 is composed of miRNAs that were decreased in expression. The expression of a selected miRNA from each cluster (miR-21 and miR-34a from cluster 1, miR-96 from cluster 2, and miR-181b from cluster 3) was assessed using TaqMan real time PCR assay. Results represent the mean ± S.E. of miRNA expression from four separate determinations. C, mouse liver imaging using high resolution ultrasound in EtOH-treated mice livers with control (top panel). Severe steatosis could be seen by a marked increase in hepatic echogenicity with nonvisualization of hepatic vessels and bile ducts. H&E staining of the same mouse liver sections is also displayed (bottom panel, ×40). D, miR-21 is increased in ALD mice liver. Total RNA was isolated from control mouse liver (C) or ALD mouse liver (A). Real time PCR analysis was performed, and the ratio of miR-21 to U6 small RNA expression in ALD liver samples was determined. The PCR products were verified by 1.8% agarose gel electrophoresis. E, miRNA was isolated from N-Hep, HSC, and HepG2 cell lines with or without ethanol treatment (100 mm, 72 h). The expression of miR-21 was assessed using TaqMan real time PCR assay. Results represent the mean ± S.E. of miRNA expression from four separate determinations. F, ethanol reduces hepatic cell survival. Cell survival was assessed using a viable cell assay, and the survival index was assessed after the treatment of ethanol (100 mm) or PBS for 72 h. *, p < 0.05 relative to controls.
TABLE 2.
Pathophysiological and biochemical characterization of ethanol-exposed mouse model
Data are expressed as mean ± S.E. (n = 5). The following abbreviations are used: BW, body weight; Wt, weight; ALT, alanine aminotransferase; BAL, blood alcohol level. BAL −, <20 mg/dl.
| Control | EtOH | IL-6 KO | IL-6 KO plus EtOH | |
|---|---|---|---|---|
| Final BW (g) | 21.33 ± 0.63 | 23.55 ± 0.57 | 20.08 ± 0.54 | 24.21 ± 0.62 |
| Liver Wt (g) | 1.21 ± 0.05 | 2.38 ± 0.14a | 1.44 ± 0.06 | 2.55 ± 0.17a |
| Liver Wt/body weight (%) | 5.67 ± 0.11 | 10.10 ± 0.46a | 7.17 ± 0.23 | 10.53 ± 0.38a |
| ALT (units/liter) | 68.00 ± 10.23 | 531.60 ± 63.44a | 84.67 ± 15.43 | 954.33 ± 85.62a,b |
| BAL (mg/dl) | − | 234.53 ± 19.23 | − | 227.54 ± 17.69 |
| Total path scores | 0.60 ± 0.32 | 4.30 ± 0.52a | 0.92 ± 0.58 | 6.32 ± 0.75a,b |
| Morbidity (%) | 0 | 0 | 0 | 40 |
a p <0.05 compared with control.
b p <0.05 compared with IL-6 KO. IL-6 KO, IL-6 knockout mice.
miR-21 Is Up-regulated in Hepatic Tissues and Cells after Ethanol Treatment
miR-21 is the most overexpressed miRNA during liver injury and liver cancer (21). To determine whether miR-21 is indeed up-regulated during alcoholic liver injury, real time PCR analysis was performed in EtOH-treated liver tissues and cells. The results showed that among the six ALD mouse liver samples analyzed, miR-21 was up-regulated significantly 4-fold or more (4–15-fold) in all samples compared with control mouse liver tissues (Fig. 1D). In vitro, the expression of miR-21 was also increased in ethanol-treated N-Heps and HSCs with a higher apoptotic rate compared with that of controls (Fig. 1, E and F). The expression of miR-21 was also up-regulated in human malignant hepatocytes (HepG2) relative to that of N-Heps. These results show that miR-21 expression is increased frequently in human alcoholic liver injury.
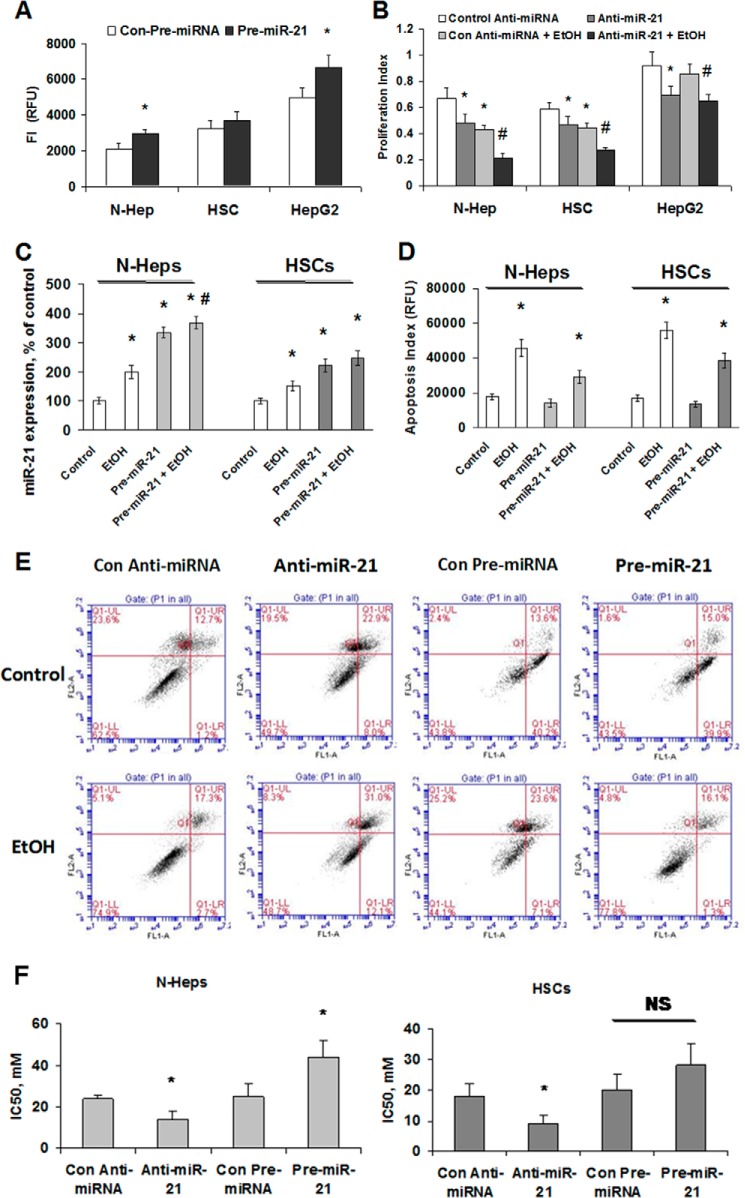
Overexpression of miR-21 Increases Cell Survival and Transformation in Vitro
The role of miR-21 in alcoholic liver injury is unknown. Thus, we performed studies aimed at exploring the possible biological significance of aberrant miR-21 by using a precursor specific to miR-21. The efficacy of transfection as well as the expression of mature miR-21 by real time PCR in N-Heps and HepG2 cells were verified previously (21). A significant change was seen in anchorage-independent growth after the transfections of miR-21 precursor in two out of the three cell lines tested (Fig. 2A). Next, we assessed cell survival after EtOH treatment in N-Hep, HSC, and HepG2 cells. In N-Hep, HSC, and HepG2 cells, transfected with anti-miR-21 inhibitors, cell survival was significantly altered compared with cells transfected with controls with different dosages and durations of EtOH treatment (Fig. 2B). We also quantified miR-21 expression in miR-21-transfected cells before and after EtOH exposure and demonstrated that the levels of miR-21 significantly increased by ∼1.6- and 2.2-fold in N-Hep, HSC, and HepG2 cells, respectively (Fig. 2C). Meanwhile, relatively long term ethanol treatment (1 week, without miR-21 overexpression) induced a 2.1- and 2.5-fold increase in miR-21 expression, respectively, which is higher than that in transfected cells. The anti-apoptotic effect of miR-21 was also verified in EtOH-treated N-Hep and HSC cells by the Apo-ONE® homogeneous caspase-3/7 assay (Fig. 2D) as well as flow cytometric analysis (Fig. 2E). Furthermore, dose responses in N-Heps and HSCs to ethanol treatment (1–200 mm), with and without miR-21 modulation, were also performed; IC50 was calculated using XLfit software. The results show that N-Heps are more resistant to EtOH-induced apoptosis compared with HSCs, and miR-21 plays a more important role in regulating survival in N-Heps (Fig. 2F). Overall these observations indicate a key role for miR-21 in the regulation of survival and transformation of human hepatic cells during alcoholic liver injury.
FIGURE 2.
Overexpression of miR-21 increases hepatic cell transformation, survival, and apoptosis. A, hepatic cells were transfected with pre-miR-21 (■) or with control precursor (□). Cells were plated in agar wells in 96-well plates, and anchorage-independent growth was assessed fluorometrically after 7 days. The pre-miR-21 increased anchorage-independent growth in all three hepatic cell lines tested (pre-miR-21 versus Con-miRNA, N-Hep, p = 0.042; HepG2, p = 0.038). B, hepatic cells were transfected with anti-miR-21 or control miRNA inhibitors, and cell survival against ethanol treatment (100 mm, 72 h) was assessed using a viable cell assay, and the survival index was assessed after 72 h. Anti-miR-21 significantly decreased survival rate in all three hepatic cell lines tested. C, miR-21 expression was assessed by real time PCR in hepatic cells transfected with either control or miR-21 precursors with or without EtOH treatment (100 mm, 72 h). The ability of these constructs and EtOH to modulate miR-21 expression was verified in N-Hep and HSC cells. D, cellular apoptosis was assessed using Apo-ONE® homogeneous caspase-3/7 assay as described under “Materials and Methods” and is expressed as arbitrary fluorescence units (RFU). Normal hepatic cells varied in their ability to against EtOH induced apoptosis. pre-miR-21 decreased cell apoptosis index in both normal hepatic cell lines. E, flow cytometric analysis of control (Con) and miR-21 overexpressed/silenced normal human hepatocytes demonstrate the basis for the gating of viable, apoptotic, and necrotic cells. The lower left quadrant shows the viable cells; the lower right quadrant represents the early apoptotic cells. The upper right quadrant represents nonviable late apoptotic/necrotic cells positive for annexin-V and APOTEST-FITC staining. The upper left quadrant shows nonviable necrotic cells/nuclear fragments. Anti-apoptotic effect of miR-21 was verified in miR-21 overexpressed/silenced N-Heps with or without EtOH treatment. F, N-Heps are more resistant to ethanol-induced cell death than HSCs. After serum starvation of cultured cells overnight, ethanol was added at various concentrations (1–200 mm) with 0.5% FBS, and cell viability was assessed after 72 h. Quantitative data of 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfonylphenyl)-2H-tetrazolium assay in ethanol-treated cells were calculated for IC50 values with XLfit software, which is illustrated in a bar graph. N-Heps are more resistant to ethanol-induced apoptosis than HSCs (IC50, N-Heps, 24.5 ± 2.3; HSCs, 18.1 ± 4.4), and modulation of miR-21 in both ways resulted in significant changes in N-Heps. IC50 results are expressed as the mean ± S.E. of eight different experiments. *, p < 0.05 relative to controls. #, p < 0.05 relative to pre-miR-21 or anti-miR-21controls. NS, no significant difference.
IL-6/Stat3 Activation of miR-21 during Alcoholic Liver Injury
Recent studies have suggested that Stat3, a transcription factor activated by IL-6, directly activates miR-21. Constitutive phosphorylation and activation of Stat3 in hepatic cells after alcoholic liver injury have been shown to be IL-6-dependent. Consistent with these observations, there was an increase in Tyr705-phosphorylated Stat3 (p-Stat3) in EtOH-fed mouse liver tissues and EtOH-treated N-Heps when compared with controls (Fig. 3A). An increase of survivin, the downstream mediator of Stat3 signaling, was also noted in the tissues and cells. We next evaluated the effect of inhibition of IL-6/Stat3 on miR-21. Silencing of IL-6 in ethanol-treated N-Hep, HSC, and HepG2 cells significantly reduced miR-21 expression, along with the reduced activity of phospho-Stat3 and overall survival rate (Fig. 3, B–D). Furthermore, depletion of IL-6 in mice significantly enhanced EtOH-induced liver damage and morbidity (Table 2), along with reduced activation of miR-21 and Stat3 in vivo (Fig. 3, E and F). Stat3 has several well characterized targets that inhibit apoptosis and modulate survival, such as survivin, Bcl-XL, and Mcl-1. Of these, survivin expression can predict prognosis in human liver injury, and Mcl-1 expression can modulate responses to ethanol. Consistent with a role for Stat3 in mediating the survival effects of IL-6 in liver injury, the expression of both survivin and Mcl-1 was decreased in IL-6 knock-out mice liver treated with EtOH in a Stat3-dependent manner (Fig. 3G).
FIGURE 3.
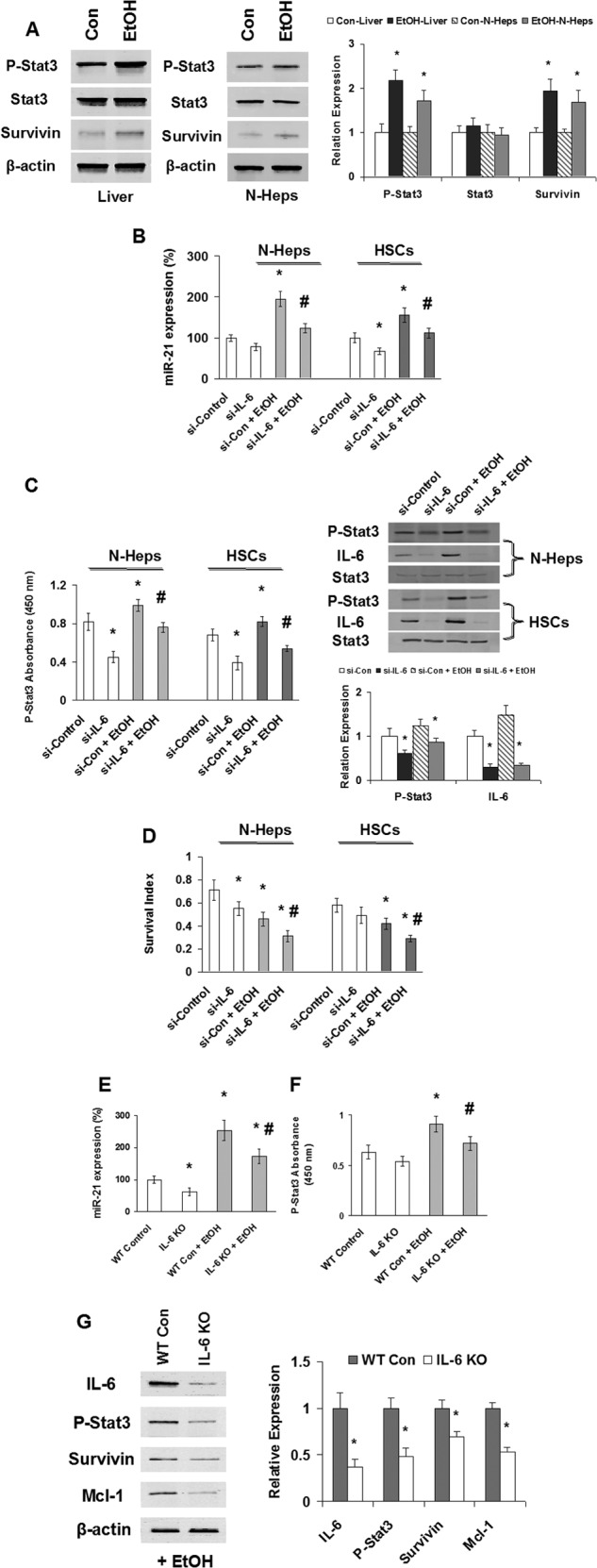
EtOH/IL-6 activation of Stat3 and miR-21. A, Western blots of EtOH mouse liver tissues and N-Hep cell lysates were performed and sequentially probed with antibodies against total-Stat3, p-Stat3, survivin, and β-actin as a loading control in liver tissues or hepatocytes as indicated. Representative immunoblots are shown on the left panel along with quantitative data that show the mean ± S.E. from four separate blots of independent experiments on the right panel. The expression of phospho-Stat3 (Tyr-705) and downstream protein survivin is up-regulated after EtOH treatment in vivo and in vitro. B–D, silencing IL-6 modulates EtOH-dependent miR-21 and Stat3 activation, as well as cell survival in normal human hepatocytes and hepatic stellate cells. B and C, siRNA to IL-6 or control was transfected with or without EtOH treatment (100 mm, 72 h). TaqMan real time assay (B), ELISA (C, left panel), and Western blot analysis (C, right panel) have demonstrated that silencing IL-6 significantly decreased miR-21 expression and Stat3 activation in both cell lines with or without EtOH treatment. C, right panel, representative immunoblots are shown on the top panel along with quantitative data that show the mean ± S.E. from four separate blots of independent experiments on the bottom panel. D, illustrated is the survival index measured by 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfonylphenyl)-2H-tetrazolium assay in EtOH-treated N-Heps and HSCs with or without si-IL-6 treatment. Representative and quantitative data (means ± S.E.) from four separate experiments are shown. E and F, IL-6 knock-out mice (IL-6 KO) or wild type control mice (WT Con) were treated with or without EtOH for 5 weeks. TaqMan real time assay (E) and ELISA (F) have demonstrated that silencing IL-6 in vivo significantly decreased miR-21 expression and Stat3 activation by EtOH treatment in mice liver. G, Western blots of EtOH mice liver tissues (with or without IL-6 knock-out) sequentially probed with antibodies against IL-6, p-Stat3 (Tyr-705), survivin, Mcl-1, and β-actin as a loading control as indicated. Representative immunoblots are shown on the left panel along with quantitative data that show the mean ± S.E. from four separate blots of independent experiments on the right panel. The expression of phospho-Stat3 (Tyr-705) and downstream protein survivin/Mcl-1 is down-regulated after IL-6 knock-out in EtOH-treated mice liver in vivo. *, p < 0.05 relative to si-controls or WT controls. #, p < 0.05 relative to si-controls or WT-controls + EtOH.
Transcriptional Regulation of miR-21 by IL-6/Stat3 Signaling
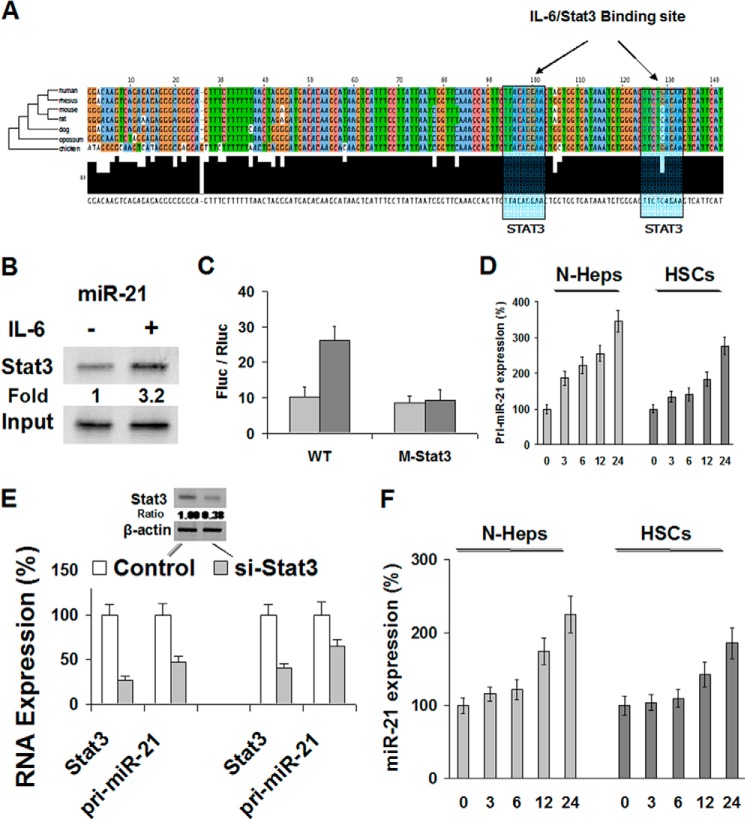
To elucidate whether IL-6/Stat3 is involved in the regulation of the miR-21 gene expression during alcoholic liver injury, we used phylogenetic footprinting to analyze the genomic distribution of potential Stat3-binding sites and their correlation with the miR-21 gene. The putative regulatory region of the miR-21 gene at chromosome 17q23.2 is located within an intron of the overlapping vacuole membrane protein (VMP1) gene, and it contains two consensus Stat3-binding sites ∼800 bp upstream of the transcription start site (Fig. 4A and supplemental File 1). VMP1 is a stress-induced protein that, when overexpressed, promotes formation of intracellular vacuoles followed by cell death. It is evident from these data that the Stat3 sites and the surrounding region, spanning 2,000 bp, are highly conserved in human, mouse, and rat (Fig. 4A and supplemental File 1).
FIGURE 4.
Transcriptional regulation of miR-21 by IL-6/Stat3. A, highly conserved binding sites for IL-6/Stat3-mediated responsiveness of the miR-21 promoter identified by PhastCons track program. 148-bp regions containing two predicted IL-6/Stat3-binding sites upstream of the miR-21 gene of various vertebrate species are aligned. T, G, C, and A nucleotides are colored green, orange, blue, and red, respectively. IL-6/Stat3 sites are highlighted by yellow boxes. B, N-Heps were deprived of IL-6 for 72 h or restimulated with IL-6 (50 ng/ml) for 30 min and subjected to a chromatin immunoprecipitation (ChIP) assay using anti-Stat3 or IgG isotype control. Co-immunoprecipitated DNA was amplified by PCR with primers specific for the miR-21 upstream enhancer. Representative images from four independent experiments are shown. C, dual-luciferase reporter gene assays were performed in N-Heps transfected either with a Fluc luciferase vector driven by the miR-21 promoter/enhancer alone (−), in the presence of a vector encoding a small hairpin RNA silencing Stat3 expression (siStat3), or with a miR-21 promoter/enhancer construct containing point mutations in both Stat3 sites (ΔStat3). The light gray bar represents control, and the dark gray bar represents IL-6-treated group. Values represent the mean Fluc/Rluc luciferase activities ± S.E. of four independent experiments relative to samples from unmanipulated IL-6-treated N-Heps. D, N-Heps and HSCs were either restimulated with IL-6 for the times indicated after cytokine withdrawal for 72 and 12 h, respectively, or continuously cultured with IL-6. Levels of pri-miR-21 were determined by real time PCR assay. Values obtained for cells deprived of IL-6 were set to 1. E, N-Heps and HSCs cultured in the presence of IL-6 were transiently transfected with an expression plasmid for GFP-tagged si-Stat3 or siRNA control. After 48 h, green fluorescent cells were sorted, and Stat3 and pri-miR-21 transcript levels were determined by real time PCR. RNA levels were normalized to GAPDH. Values for the control samples were taken as reference. F, in N-Heps and HSCs, mature miR-21 was quantified by TaqMan real time PCR assay. miR-21 levels were normalized to U6 RNA.
Our chromatin immunoprecipitation (ChIP) studies revealed that Stat3 is recruited to the miR-21 upstream region in N-Heps in response to IL-6 (Fig. 4B). Furthermore, transcriptional regulation of miR-21 by IL-6 through the putative Stat3 enhancer region was confirmed by a luciferase reporter assay (Fig. 4C). IL-6-dependent miR-21 promoter activity was abolished when the Stat3-binding sites were eliminated by site-directed mutagenesis. Meanwhile, knocking down IL-6 by RNA interference inhibited miR-21 promoter activity. These data demonstrate that the transcriptional regulation of miR-21 by IL-6 is through functional Stat3 binding at the 5′-promoter region.
miR-21 Gene Transcription Is Controlled by Ethanol and Requires Stat3
The human microRNA-21 gene is located on the plus strand of chromosome 17q23.2 (55,273,409–55,273,480) within a coding gene vacuole membrane protein (VMP1 as introduced in this study). Despite being located in the intronic regions of a coding gene in the direction of transcription, miR-21 has its own promoter regions and forms an ∼3433-nucleotide-long primary transcript of miR-21 (named pri-miR-21), which is independently transcribed. The stem-loop precursor of miR-21 (named pre-miR-21) resides between nucleotides 2445 and 2516 of pri-miR-21. pri-miR-21 is cut by the endonuclease Drosha in the nucleus to produce pre-miR-21, which is exported into the cytosol. To verify that ethanol induces the transcription of the miR-21 gene, pri-miR-21 was quantified by real time PCR in N-Hep and HSC cells. Ethanol increased pri-miR-21 expression significantly in each cell line (Fig. 4D). Upon Stat3 knockdown, the comparable reductions of Stat3 mRNA and pri-miR-21 levels were observed (Fig. 4E), demonstrating that EtOH-induced miR-21 expression is Stat3-dependent. Induction of miR-21 expression by ethanol is not restricted to hepatocytes, as observed in human hepatic stellate cells as well. Our data demonstrate that miR-21 gene transcription is controlled by ethanol and requires Stat3. In contrast to the rapid induction of pri-miR-21, mature miR-21 levels increased slowly in N-Hep cells, eventually reaching a 2.3-fold increase (Fig. 4F). The same was observed in HSCs, indicating a rather slow processing rate of pri-miR-21.
Identification of FASLG and DR5 as Targets for miR-21
Because miRNAs target mRNA stability and translation, we used a mouse apoptosis PCR array to identify specific target protein levels of miR-21 in pre-miR-21-treated N-Heps. Because the aim of our study was to correlate miR-21 with target transcript expression, we used a cutoff of at least 4-fold difference in our expression analysis. Six genes were initially selected according to this strategy and further screened based on 3′-UTR sequence analysis and prediction algorithms; only two proteins can be potentially targeted by miR-21 (Fig. 5A). Interestingly, the target prediction program Sanger miRBase database indicated the presence of a highly conserved binding site for miR-21 in the 3′-UTR region of FASLG and DR5, the well characterized regulator genes of apoptosis in liver biology. After 1 week of EtOH treatment, FASLG and DR5 were significantly down-regulated in surviving N-Heps and HSCs and were also silenced in HepG2 cells related to normal controls (Fig. 5B). To verify that FASLG and DR5 are bona fide targets of translational regulation by miR-21 in hepatocytes, we performed studies using luciferase reporter constructs containing the miR-21 recognition sequence from the 3′-UTR of FASLG and DR5 inserted downstream of the luciferase gene (Fig. 5C). Transfection with the miR-21 precursor decreased reporter activities of both FASLG and DR5 in normal human hepatocytes. However, when these studies were repeated with reporter constructs that contained random mutations in the recognition sequence, the effects of reporter deactivation by the miR-21 precursor were abolished (Fig. 5, D and E). Moreover, an increase in FASLG and DR5 expression occurred after 3 days in normal human hepatocytes and hepatic stellate cells incubated with an anti-miR-21 inhibitor. Concomitant with enhanced DR5 expression, there was a significant increase in survivin expression, a confirmed downstream mediator of DR5 signaling (Fig. 5F). In contrast, transfection with an inhibitor to miR-122a, which modulates cell survival in hepatic cells, did not alter the expression of FASLG and DR5 with a relative expression of 0.9 ± 0.3- and 1.3 ± 0.4-fold of controls, respectively. Taken together, these findings indicate that FASLG and DR5 are biologically relevant targets of miR-21 in hepatic cells.
FIGURE 5.
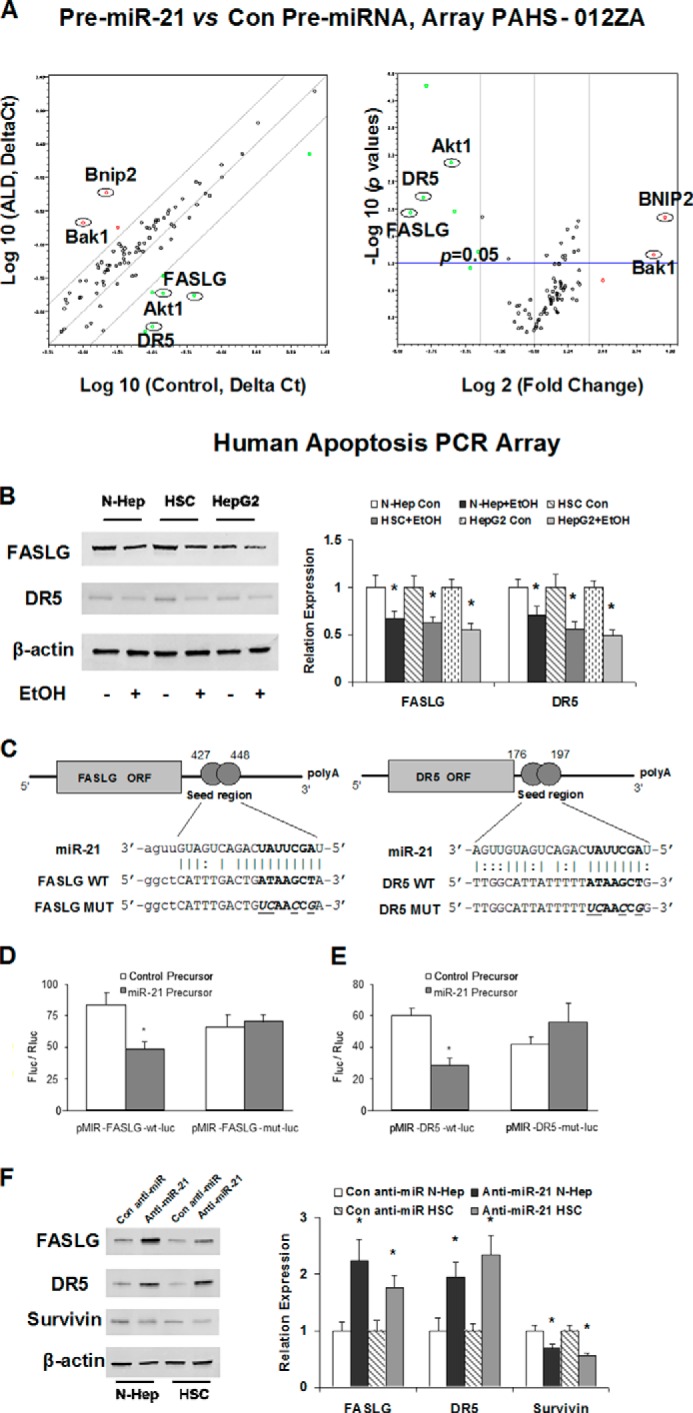
miR-21 regulates expression of FASLG and DR5. A, identification of protein targets modulated by miR-21 in normal human hepatocytes by real time PCR array analysis. N-Hep cells were transfected with pre-miR-21 or control precursor (50 nm) for 72 h. Relative gene expression profile between pre-miR-21 versus control pre-miRNA transfected cells is shown. The expression of a panel of diverse apoptosis-associated genes was evaluated by real time PCR using human apoptosis PCR array (PAHS-012ZA) from SABiosciences. Gene expression relative to glyceraldehyde-3-phosphate dehydrogenase was plotted as the Volcano plots, depicting the relative expression levels (log2) for selected genes in HSC pre-miR-21 versus control pre-miRNA panels (left panel). The relative expression levels and p values for each gene in the related samples were also plotted against each other in the scatter plot (right panel). FASLG and DR5 are the most down-regulated genes among the apoptosis signaling pathways in human hepatocytes. B, miR-21 target proteins are altered in normal hepatocytes after ethanol treatment. Hepatic cells were treated with ethanol (100 mm, right panel) or PBS controls (left panel). Cell lysates were obtained after 7 days, and Western blots performed for FASLG, DR5, and β-actin. Treatment with ethanol down-regulated the expression of FASLG and DR5 in all three hepatic cell lines. Representative immunoblots are shown on the left panel along with quantitative data that show the mean ± S.E. from four separate blots of independent experiments on the right panel. C, schematic of predicted miR-21 site in the 3′-UTR of human FASLG and DR5. D and E, luciferase reporter constructs containing the miR-21 recognition sequence from the 3′-UTR of FASLG and DR5 inserted downstream of the luciferase gene were generated. pMIR-FASLG-WT-luc or pMIR-DR5-WT-luc contains the intact sequence, whereas pMIR-FASLG-mut-luc or pMIR-DR5-mut-luc contains the sequence with random nucleotide changes. Reporter constructs were co-transfected with either miR-21 precursor or control precursor in normal human hepatocytes. The expression of firefly luciferase activity was normalized to that of Renilla luciferase activity for each sample. The decreases in relative firefly luciferase activity in the presence of miR-21 indicate the presence of an miR-21-modulated target sequence in the 3′-UTR of FASLG and DR5. Data represent the means from six separate experiments. *, p < 0.05 relative to control precursor group. F, N-Heps and HScs were transfected with anti-miR-21 or control anti-miRNA. Cell lysates were obtained after 48 h, and Western blots were performed for FASLG, DR5, survivin, and β-actin. Relative ratios normalized with β-actin and control group are displayed below the image. Representative immunoblots are shown on the left panel along with quantitative data that show the mean ± S.E. from four separate blots of independent experiments on the right panel. Inhibition of miR-21 increased the expression of FASLG and DR5, and subsequently decreased the level of survivin, a downstream mediator of Stat3 in normal human hepatocytes and hepatic stellate cells. *, p < 0.05 relative to specific controls.
Regulation of FASLG/DR5 Expression by IL-6 and miR-21 in Vivo during Alcoholic Liver Injury
The cell death and tissue repair process involves a series of death receptor signaling pathways. Alterations of the IL-6/Stat3 have been linked to decreased expression of death receptors and cellular apoptosis. Therefore, we examined the expression of FASLG/DR5 involved in cell apoptosis in normal and EtOH-exposed mouse liver tissues with or without IL-6 depletion/miR-21 inhibition. Compared with control liver tissue, the expression of FASLG/DR5 increased in IL-6 knock-out mice liver with ethanol exposure (Fig. 6, A and B). Enhanced expression of FASLG and DR5 was also associated with the morbidity of IL-6 knock-out mice with ethanol exposure, which was 40% higher than the EtOH control group (n = 5). To confirm the functional effect and relevance of miR-21-dependent modulation of FASLG and DR5 expression in vivo, we assessed the effect of inhibition of miR-21 by specific Vivo-Morpholino on FASLG and DR5 expression in ethanol-exposed mouse liver. The silencing effects on miR-21 by specific Vivo-Morpholino were verified in liver tissues by TaqMan real time PCR analysis (Fig. 6C). Treatment of EtOH-exposed mice with miR-21 Vivo-Morpholino increased hepatic FASLG and DR5 expression (Fig. 6, D and E). Furthermore, the serum alanine aminotransferase level was significantly increased after miR-21 inhibition, along with enhanced α-smooth muscle actin and Sirius red staining in ethanol-exposed mouse liver sections (Fig. 6, F and G). These findings link miR-21 with putative mediators of tissue injury in EtOH-exposed mice and suggest that deregulated expression of miR-21 contributes to liver reconstruction and fibrosis during alcoholic liver injury.
FIGURE 6.
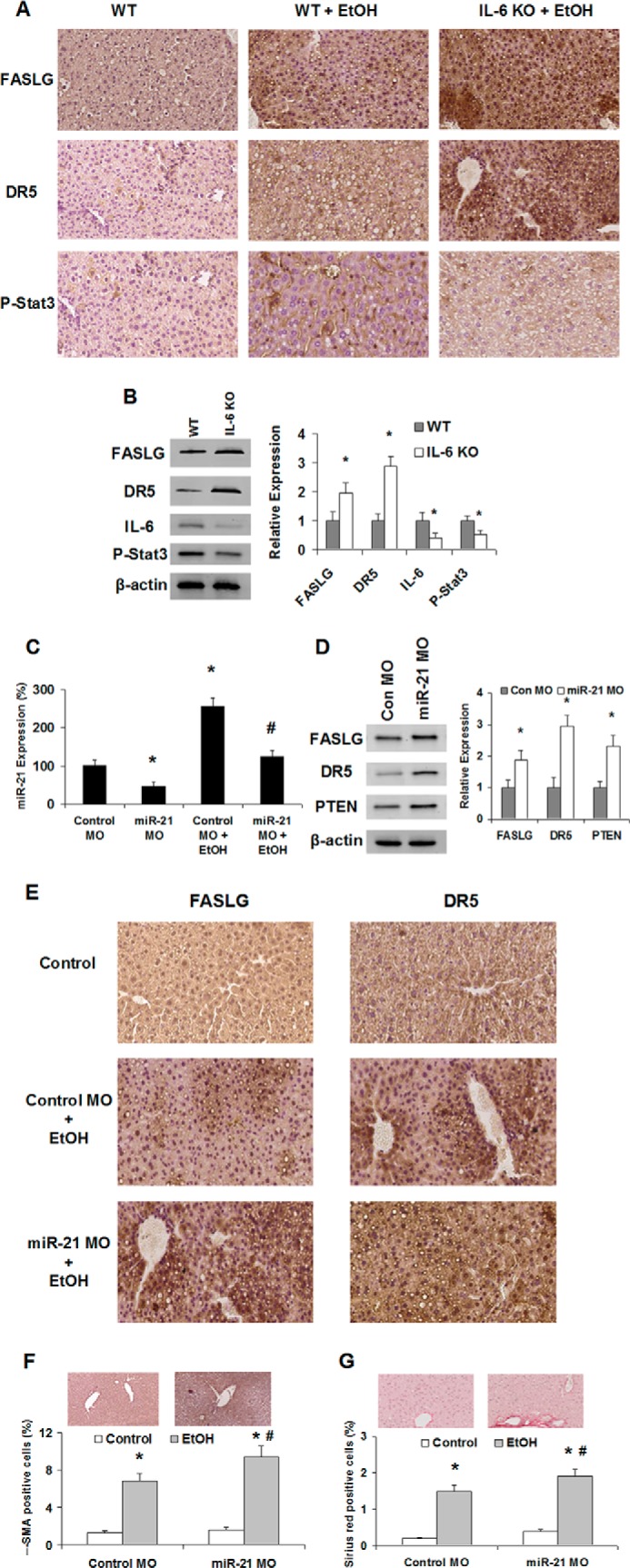
Regulation of FASLG and DR5 by miR-21 Vivo-Morpholino and IL-6 knock-out in EtOH-exposed mice liver. A, steatohepatitis is induced by 5 weeks of ethanol feeding in mouse liver and enhanced by IL-6 knock-out. Liver histology of a regular feeding control mouse and EtOH mice with or without IL-6 knock-out is displayed. For hematoxylin and eosin (H&E) staining, enhanced expression of FASLG and DR5 was seen in EtOH mouse livers. Original magnifications, ×200. B, liver tissue homogenates were obtained from control and IL-6 knock-out mice. Increases of miR-21 target proteins FASLG and DR5, along with the reduction of IL-6 and phospho-Stat3, were verified by Western blot analysis. Representative immunoblots are shown on the left panel along with quantitative data that show the mean ± S.E. from four separate blots of independent experiments on the right panel. C, expression of miR-21 was assessed by TaqMan real time PCR assay in normal liver and EtOH-exposed mouse liver tissues with control or miR-21 Vivo-Morpholino treatment (time and dose). miR-21 was significantly reduced after specific Vivo-Morpholino treatment in vivo. D, liver tissue homogenates were obtained from control and miR-21 Vivo-Morpholino-treated mice. Increases of miR-21 target proteins FASLG, DR5, and PTEN were confirmed by Western blot analysis. Representative immunoblots are shown on the left panel along with quantitative data that show the mean ± S.E. from four separate blots of independent experiments on the right panel. E, liver histology of the regular feeding control mouse and EtOH mouse with miR-21 or control Vivo-Morpholino treatment is displayed. Enhanced expression of FASLG and DR5 was seen in EtOH mouse livers after miR-21 Vivo-Morpholino treatment (E). Original magnifications ×200. F and G, percentages of positive cells stained with fibrotic marker α-smooth muscle actin and Sirius red were counted in miR-21 Vivo-Morpholino-treated mouse livers relative to controls after EtOH exposure. Anti-miR-21 treatment in vivo significantly increased the intensity of liver fibrosis confirmed by enhanced α-SMA and Sirius red expression (n = 3). *, p < 0.05 relative to WT controls or Morpholino-controls. #, p < 0.05 relative to EtOH controls.
DISCUSSION
In this study, we described the role of altered expression of miR-21 by IL-6/Stat3 in the contribution to cellular phenotypic changes that are associated with ALD progression. We have shown that miR-21 is increased in mouse livers with ALD after the activation of IL-6/Stat3 signaling in vivo, and it is overexpressed in ethanol-treated hepatic cell lines compared with controls. We demonstrated that IL-6 contributes to alcoholic liver injury and tissue repairing through miR-21 by modulating cell proliferation, apoptosis, and survival. Some of these effects are mediated through DR5 and FASLG, the well characterized regulator genes of apoptosis that are also involved in ALD. Increased expression of miR-21 was shown by in situ hybridization during liver regeneration, and a similar role for miR-21 has been postulated in cardiovascular injury (17, 22). The concomitant miR-21-dependent activation of survival genes such as survivin in hepatic cells can facilitate tissue recovery. These findings taken together support a functional role for miR-21 in promoting liver tissue repair and against liver fibrosis during the development of ALD.
miRNA-mediated mechanisms are being increasingly implicated in liver injury. Dysregulation of miR-21 can occur as a result of altered IL-6 expression. Likewise, ectopic expression of miR-21 decreases the expression of genes regulated by IL-6 such as cyclin D1, cyclin E1, and RhoB (17, 23). These and other studies (24) support the concept of a cell cycle regulation role for miR-21. miR-21 may mediate an anti-apoptotic response through additive and synergistic effects induced by IL-6. Tissues with low expression of miR-21 lack pro-apoptotic stimulus and acquire the capability to proliferate and grow. Variable expression of miR-21 has been described in different organ systems and diseases states (25). Indeed, in normal and malignant hepatocytes, miR-21 was shown to be a potent contributor of cell proliferation, but miR-21 induction is not sufficient to regulate cell cycle progression during liver regeneration, particularly in ethanol-fed rats (26). These diverse observations necessitate a clear definition of tissue injury-specific expression and function of miR-21 expression. It is likely that targeted therapeutic approaches involving miR-21 may result from defining tissue- and disease state-specific roles of miR-21.
IL-6/Stat3 controls the expression of a number of antiapoptotic proteins at the transcriptional level, including survivin, Bcl-2, and Mcl-1 (27, 28), thereby providing an important fast-track survival signal for hepatic cells during liver injury. IL-6/Stat3-deficient mice are more susceptible to ethanol-induced hepatic steatosis (29). However, some data suggest that this regulatory route does not sufficiently explain the full antiapoptotic potential of Stat3 (30, 31). We have shown that miR-21 induction by Stat3 represents a rather slowly acting yet long-lasting survival stimulus, which appears as an ideal complementation of the immediate induction of antiapoptotic proteins after alcoholic liver injury. Overexpression of miR-21 has been described in several liver injury and regeneration experiments (17, 32), most of which contain constitutively activated Stat3 or even rely on Stat3 with respect to cell survival or growth. Overall, our observations strongly suggest pivotal relevance of miR-21 for the survival/regeneration potential of Stat3 and its involvement in the pathogenesis of alcoholic liver diseases and other related liver disorders.
Members of the TNF family are involved in the pathogenesis of liver diseases. Serum FASLG levels were significantly higher in patients with any form of hepatic fibrosis compared with those without it, and increasing FASLG was a significant predictor of hepatic fibrosis (33). The receptor/ligand pair Fas (CD95)/FASLG (CD95L) regulates the apoptosis of hepatocytes in viral hepatitis and alcoholic liver injury. Actually, Fas/FASLG is strongly overexpressed in hepatocytes in liver specimens from rats treated with an ethanol and high fat diet as well as nonalcoholic steatohepatitis patients, indicating that high Fas/FASLG activity is involved in apoptosis of hepatocytes in NASH (34, 35). Although hepatocytes are constitutively susceptible to FASLG-mediated apoptosis, the expression of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) does not result in apoptosis of hepatocytes in healthy livers (36, 37). Indeed, TRAIL-mediated apoptosis of hepatocytes needs to be triggered through viral infection, and overexpression of TRAIL enables the organism to selectively eliminate virally infected hepatocytes (38). Lack of hepatocyte-specific NF-κB activation through deletion of NEMO (NEMOΔhepa) in mice leads to steatohepatitis, cirrhosis, and hepatocellular carcinoma (39, 40), along with the high expression of the TRAIL receptor DR5 (41). Moreover, bile acids up-regulate DR5 expression (42), and the essential contribution of FASLG in ethanol-induced liver injury has been demonstrated (43), indicating that FASLG and DR5 are the important mediators of chronic liver diseases, including alcoholic liver injury.
Our findings identify a previously unrecognized mechanism for direct regulation of FASLG and DR5, involving ncRNA in ALD. The abusive consumption of alcohol can cause serious cellular injuries that often lead to apoptosis and cell death. Aberrant IL-6/Stat3 signaling has been implicated in many human diseases, including ALDs. Recent developments indicate that ethanol induces alterations of the IL-6/Stat3 signaling pathway, particularly on enhanced expression of Stat3-mediated anti-apoptotic pathways. This has opened up a new area of interest in ethanol research and is providing novel insight into actions of ethanol at the nucleosomal level in relation to gene expression, pathophysiological consequences, and injury recovery/liver regeneration. Although IL-6/Stat3 signaling has been tightly linked to liver injuries and disease outcome in many hepatic disorders, including human ALDs, its application to ethanol-dependent ncRNA expression is novel. A better understanding of how ethanol interacts with specific cytokines to contribute to aberrant ncRNA expression will clearly advance the field and increase our understanding of the mechanisms involved in the development of ALDs. Genomic scanning approaches to identify transcriptional/translational factors and their modified targets in ALD are lacking, but such strategies could identify other novel targets that could be genetically or epigenetically modified in ALD.
With chronic alcohol abuse, early and reversible liver damage occurs in the form of fatty liver long before the onset of clinically symptomatic and irreversible form of hepatitis and cirrhosis. Currently, the mechanisms and regulation of ALDs are poorly understood, and the ALD-associated putative biomarkers and regulatory molecules remain tenuous and unproven. In this study we characterized the role of dysregulated miR-21 in ethanol-induced altered extrinsic apoptotic signaling and its progression to ALD. The data presented here may have direct application to the future translational research for an improved diagnosis and treatment of ALD patients.
Supplementary Material
Acknowledgments
We express our deepest appreciation to Dr. Hidekazu Tsukamoto and the tissue sharing program of the Southern California Research Center for ALPD and Cirrhosis (supported by National Institutes of Health Grant P50AA011999) for their assistance with the animal studies in this project. We also acknowledge Glen Cryer and Gina Dupar in the Publications Department for editing assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants DK58411 and DK07698 (to G. A., S. G., and F. M.). This work was also supported by Veterans Affairs Career Development Award IK2 BX001760 (to H. F. and S. G.), Veterans Affairs Merit Review Grant 1I01BX001724 (to F. M. and G. A.), Veterans Affairs Research Career Award (to G. A.), the Dr. Nicholas C. Hightower Centennial Chair of Gastroenterology from Scott & White (to G. A.), and Scott & White Research Grants Program Project 90190 (to F. M.).

This article contains supplemental File 1.
- ALD
- alcoholic liver disease
- miRNA
- microRNA
- DR5
- death receptor 5
- FASLG
- Fas ligand (TNF superfamily, member 6)
- N-Hep
- normal human hepatocyte
- HSC
- hepatic stellate cell
- ncRNA
- noncoding RNA
- TRAIL
- tumor necrosis factor-related apoptosis-inducing ligand.
REFERENCES
- 1. Mathurin P., Louvet A., Dharancy S. (2008) Acute alcoholic hepatitis. Management practices for 2007. Gastroenterol. Clin. Biol. 32, S179–S181 [DOI] [PubMed] [Google Scholar]
- 2. Ji J., Shi J., Budhu A., Yu Z., Forgues M., Roessler S., Ambs S., Chen Y., Meltzer P. S., Croce C. M., Qin L. X., Man K., Lo C. M., Lee J., Ng I. O., Fan J., Tang Z. Y., Sun H. C., Wang X. W. (2009) MicroRNA expression, survival, and response to interferon in liver cancer. N. Engl. J. Med. 361, 1437–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ruvkun G. (2008) The perfect storm of tiny RNAs. Nat. Med. 14, 1041–1045 [DOI] [PubMed] [Google Scholar]
- 4. Gartel A. L., Kandel E. S. (2008) miRNAs: Little known mediators of oncogenesis. Semin. Cancer Biol. 18, 103–110 [DOI] [PubMed] [Google Scholar]
- 5. Moss E. G. (2007) Heterochronic genes and the nature of developmental time. Curr. Biol. 17, R425–R434 [DOI] [PubMed] [Google Scholar]
- 6. Siomi H., Siomi M. C. (2009) On the road to reading the RNA-interference code. Nature 457, 396–404 [DOI] [PubMed] [Google Scholar]
- 7. Meng F., Glaser S. S., Francis H., Yang F., Han Y., Stokes A., Staloch D., McCarra J., Liu J., Venter J., Zhao H., Liu X., Francis T., Swendsen S., Liu C. G., Tsukamoto H., Alpini G. (2012) Epigenetic regulation of miR-34a expression in alcoholic liver injury. Am. J. Pathol. 181, 804–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Meng F., Francis H., Glaser S., Han Y., DeMorrow S., Stokes A., Staloch D., Venter J., White M., Ueno Y., Reid L. M., Alpini G. (2012) Role of stem cell factor and granulocyte colony-stimulating factor in remodeling during liver regeneration. Hepatology 55, 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Meng F., Glaser S. S., Francis H., Demorrow S., Han Y., Passarini J. D., Stokes A., Cleary J. P., Liu X., Venter J., Kumar P., Priester S., Hubble L., Stoloch D., Sharma J., Liu C. G., Alpini G. (2011) Functional analysis of microRNAs in human hepatocellular cancer stem cells. J. Cell. Mol. Med. 16, 160–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tazawa H., Tsuchiya N., Izumiya M., Nakagama H. (2007) Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc. Natl. Acad. Sci. U.S.A. 104, 15472–15477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. He L., He X., Lim L. P., de Stanchina E., Xuan Z., Liang Y., Xue W., Zender L., Magnus J., Ridzon D., Jackson A. L., Linsley P. S., Chen C., Lowe S. W., Cleary M. A., Hannon G. J. (2007) A microRNA component of the p53 tumour suppressor network. Nature 447, 1130–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tarasov V., Jung P., Verdoodt B., Lodygin D., Epanchintsev A., Menssen A., Meister G., Hermeking H. (2007) Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle 6, 1586–1593 [DOI] [PubMed] [Google Scholar]
- 13. Raver-Shapira N., Marciano E., Meiri E., Spector Y., Rosenfeld N., Moskovits N., Bentwich Z., Oren M. (2007) Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell 26, 731–743 [DOI] [PubMed] [Google Scholar]
- 14. Chang T. C., Wentzel E. A., Kent O. A., Ramachandran K., Mullendore M., Lee K. H., Feldmann G., Yamakuchi M., Ferlito M., Lowenstein C. J., Arking D. E., Beer M. A., Maitra A., Mendell J. T. (2007) Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell 26, 745–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ma X., Choudhury S. N., Hua X., Dai Z., Li Y. (2013) Interaction of the oncogenic miR-21 microRNA and the p53 tumor suppressor pathway. Carcinogenesis 34, 1216–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mathé E., Nguyen G. H., Funamizu N., He P., Moake M., Croce C. M., Hussain S. P. (2012) Inflammation regulates microRNA expression in cooperation with p53 and nitric oxide. Int. J. Cancer 131, 760–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ng R., Song G., Roll G. R., Frandsen N. M., Willenbring H. (2012) A microRNA-21 surge facilitates rapid cyclin D1 translation and cell cycle progression in mouse liver regeneration. J. Clin. Invest. 122, 1097–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tsukamoto H., Mkrtchyan H., Dynnyk A. (2008) Intragastric ethanol infusion model in rodents. Methods Mol. Biol. 447, 33–48 [DOI] [PubMed] [Google Scholar]
- 19. Xiong S., She H., Zhang A. S., Wang J., Mkrtchyan H., Dynnyk A., Gordeuk V. R., French S. W., Enns C. A., Tsukamoto H. (2008) Hepatic macrophage iron aggravates experimental alcoholic steatohepatitis. Am. J. Physiol. Gastrointest. Liver Physiol. 295, G512–G521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Löffler D., Brocke-Heidrich K., Pfeifer G., Stocsits C., Hackermüller J., Kretzschmar A. K., Burger R., Gramatzki M., Blumert C., Bauer K., Cvijic H., Ullmann A. K., Stadler P. F., Horn F. (2007) Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood 110, 1330–1333 [DOI] [PubMed] [Google Scholar]
- 21. Meng F., Henson R., Wehbe-Janek H., Ghoshal K., Jacob S. T., Patel T. (2007) MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133, 647–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hu S., Huang M., Nguyen P. K., Gong Y., Li Z., Jia F., Lan F., Liu J., Nag D., Robbins R. C., Wu J. C. (2011) Novel microRNA prosurvival cocktail for improving engraftment and function of cardiac progenitor cell transplantation. Circulation 124, S27–S34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Parikh V. N., Jin R. C., Rabello S., Gulbahce N., White K., Hale A., Cottrill K. A., Shaik R. S., Waxman A. B., Zhang Y. Y., Maron B. A., Hartner J. C., Fujiwara Y., Orkin S. H., Haley K. J., Barabási A. L., Loscalzo J., Chan S. Y. (2012) MicroRNA-21 integrates pathogenic signaling to control pulmonary hypertension: results of a network bioinformatics approach. Circulation 125, 1520–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. He L., He X., Lowe S. W., Hannon G. J. (2007) MicroRNAs join the p53 network–another piece in the tumour-suppression puzzle. Nat. Rev. Cancer 7, 819–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Olivieri F., Rippo M. R., Monsurrò V., Salvioli S., Capri M., Procopio A. D., Franceschi C. (2013) MicroRNAs linking inflamm-aging, cellular senescence and cancer. Ageing Res. Rev. 12, 1056–1068 [DOI] [PubMed] [Google Scholar]
- 26. Dippold R. P., Vadigepalli R., Gonye G. E., Hoek J. B. (2012) Chronic ethanol feeding enhances miR-21 induction during liver regeneration while inhibiting proliferation in rats. Am. J. Physiol. Gastrointest. Liver Physiol. 303, G733–G743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gao B. (2012) Hepatoprotective and anti-inflammatory cytokines in alcoholic liver disease. J. Gastroenterol. Hepatol. 27, 89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gong J., Zhang J. P., Li B., Zeng C., You K., Chen M. X., Yuan Y., Zhuang S. M. (2013) MicroRNA-125b promotes apoptosis by regulating the expression of Mcl-1, Bcl-w and IL-6R. Oncogene 32, 3071–3079 [DOI] [PubMed] [Google Scholar]
- 29. Zhang X., Tachibana S., Wang H., Hisada M., Williams G. M., Gao B., Sun Z. (2010) Interleukin-6 is an important mediator for mitochondrial DNA repair after alcoholic liver injury in mice. Hepatology 52, 2137–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brocke-Heidrich K., Kretzschmar A. K., Pfeifer G., Henze C., Löffler D., Koczan D., Thiesen H. J., Burger R., Gramatzki M., Horn F. (2004) Interleukin-6-dependent gene expression profiles in multiple myeloma INA-6 cells reveal a Bcl-2 family-independent survival pathway closely associated with Stat3 activation. Blood 103, 242–251 [DOI] [PubMed] [Google Scholar]
- 31. Tumang J. R., Hsia C. Y., Tian W., Bromberg J. F., Liou H. C. (2002) IL-6 rescues the hyporesponsiveness of c-Rel deficient B cells independent of Bcl-xL, Mcl-1, and Bcl-2. Cell. Immunol. 217, 47–57 [DOI] [PubMed] [Google Scholar]
- 32. Chen Y. P., Jin X., Xiang Z., Chen S. H., Li Y. M. (2013) Circulating MicroRNAs as potential biomarkers for alcoholic steatohepatitis. Liver Int. 33, 1257–1265 [DOI] [PubMed] [Google Scholar]
- 33. Page S., Birerdinc A., Estep M., Stepanova M., Afendy A., Petricoin E., Younossi Z., Chandhoke V., Baranova A. (2013) Knowledge-based identification of soluble biomarkers: hepatic fibrosis in NAFLD as an example. PLoS One 8, e56009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Y., Seitz H. K., Wang X. D. (2010) Moderate alcohol consumption aggravates high-fat diet induced steatohepatitis in rats. Alcohol. Clin. Exp. Res. 34, 567–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Feldstein A. E., Canbay A., Angulo P., Taniai M., Burgart L. J., Lindor K. D., Gores G. J. (2003) Hepatocyte apoptosis and fas expression are prominent features of human nonalcoholic steatohepatitis. Gastroenterology 125, 437–443 [DOI] [PubMed] [Google Scholar]
- 36. Walczak H., Miller R. E., Ariail K., Gliniak B., Griffith T. S., Kubin M., Chin W., Jones J., Woodward A., Le T., Smith C., Smolak P., Goodwin R. G., Rauch C. T., Schuh J. C., Lynch D. H. (1999) Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat. Med. 5, 157–163 [DOI] [PubMed] [Google Scholar]
- 37. Ashkenazi A., Pai R. C., Fong S., Leung S., Lawrence D. A., Marsters S. A., Blackie C., Chang L., McMurtrey A. E., Hebert A., DeForge L., Koumenis I. L., Lewis D., Harris L., Bussiere J., Koeppen H., Shahrokh Z., Schwall R. H. (1999) Safety and antitumor activity of recombinant soluble Apo2 ligand. J. Clin. Invest. 104, 155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mundt B., Kühnel F., Zender L., Paul Y., Tillmann H., Trautwein C., Manns M. P., Kubicka S. (2003) Involvement of TRAIL and its receptors in viral hepatitis. FASEB J. 17, 94–96 [DOI] [PubMed] [Google Scholar]
- 39. Beraza N., Lüdde T., Assmus U., Roskams T., Vander Borght S., Trautwein C. (2007) Hepatocyte-specific IKKγ/NEMO expression determines the degree of liver injury. Gastroenterology 132, 2504–2517 [DOI] [PubMed] [Google Scholar]
- 40. Luedde T., Beraza N., Kotsikoris V., van Loo G., Nenci A., De Vos R., Roskams T., Trautwein C., Pasparakis M. (2007) Deletion of NEMO/IKKγ in liver parenchymal cells causes steatohepatitis and hepatocellular carcinoma. Cancer Cell 11, 119–132 [DOI] [PubMed] [Google Scholar]
- 41. Beraza N., Ofner-Ziegenfuss L., Ehedego H., Boekschoten M., Bischoff S. C., Mueller M., Trauner M., Trautwein C. (2011) Nor-ursodeoxycholic acid reverses hepatocyte-specific nemo-dependent steatohepatitis. Gut 60, 387–396 [DOI] [PubMed] [Google Scholar]
- 42. Higuchi H., Grambihler A., Canbay A., Bronk S. F., Gores G. J. (2004) Bile acids up-regulate death receptor 5/TRAIL-receptor 2 expression via a c-Jun N-terminal kinase-dependent pathway involving Sp1. J. Biol. Chem. 279, 51–60 [DOI] [PubMed] [Google Scholar]
- 43. Mundt B., Wirth T., Zender L., Waltemathe M., Trautwein C., Manns M. P., Kühnel F., Kubicka S. (2005) Tumour necrosis factor related apoptosis inducing ligand (TRAIL) induces hepatic steatosis in viral hepatitis and after alcohol intake. Gut 54, 1590–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.