Background: How LL-37 delivers double-stranded RNA (dsRNA) to activate TLR3 signaling is poorly understood.
Results: LL-37 was found to traffic to endosomes with RNA and releases the dsRNA upon endosome acidification. A peptide derived from LL-37 could antagonize LL-37·dsRNA trafficking.
Conclusion: LL-37 trafficking of dsRNA is regulated by pH.
Significance: This work establishes the mechanism of LL-37 enhancement of dsRNA-activated TLR3 signaling.
Keywords: Endosome, Innate Immunity, pH Regulation, RNA-binding Protein, Toll-like Receptor (TLR), pH
Abstract
LL-37 is a peptide secreted by human epithelial cells that can lyse bacteria, suppress signaling by Toll-like receptor 4 (TLR4), and enhance signaling to double-stranded RNA (dsRNA) by TLR3. How LL-37 interacts with dsRNA to affect signal transduction by TLR3 is not completely understood. We determined that LL-37 binds dsRNA and traffics to endosomes and releases the dsRNA in a pH-dependent manner. Using dynamic light scattering spectroscopy and cell-based FRET experiments, LL-37 was found to form higher order complexes independent of dsRNA binding. Upon acidification LL-37 will dissociate from a larger complex. In cells, LL-37 has a half-live of ∼1 h. LL-37 half-life was increased by inhibiting endosome acidification or inhibiting cathepsins, which include proteases whose activity are activated by endosome acidification. Residues in LL-37 that contact poly(I:C) and facilitate oligomerization in vitro were mapped. Peptide LL-29, which contains the oligomerization region of LL-37, inhibited LL-37 enhancement of TLR3 signal transduction. LL-29 prevented LL-37·poly(I:C) co-localization to endosomes containing TLR3. These results shed light on the requirements for LL-37 enhancement of TLR3 signaling.
Introduction
LL-37 is a 37-residue antimicrobial peptide produced by human epithelial cells by proteolytic cleavage from the C-terminal portion of the hCAP-18 protein (1). In addition to lysing bacteria, LL-37 can regulate the activities of multiple innate immune receptors (2, 3). High levels of LL-37 are also associated with autoimmune diseases such as psoriasis and asthma, suggesting that overexpression of LL-37 could be linked to diseases (3–9).
In the regulation of innate immune signaling, LL-37 has the interesting property of suppressing TLR42 signaling while enhancing TLR3 signaling (10–13). LL-37 is known to bind to agonists of multiple TLRs, including lipopolysaccharides (LPS), which can activate TLR4 signaling, and the double-stranded RNA (dsRNA) that activates TLR3. However, it is not known how LL-37 binding to the TLR agonists will differentially affect signaling.
One feature that likely contributes to these distinct outcomes in signaling is that TLR4 predominantly signals from the plasma membrane, whereas TLR3 signaling takes place in acidified endosomes (14). The antimicrobial activity and the conformation of LL-37 have been documented to be affected by pH, although the previous studies did not examine dsRNA binding or TLR3 signaling (15). We propose that should LL-37 bind to TLR agonists at neutral pH, but release them under acidic conditions, it could suppress TLR4 signaling by maintaining binding to LPS in the cytoplasm but deliver higher concentrations of dsRNA to endosomes and enhance TLR3 signaling.
In this study we document that LL-37 binding to poly(I:C) is dependent upon pH. Upon acidification of endosomes, LL-37 is released from dsRNA both in vitro and in cells. Furthermore, LL-37 is degraded in lysosomes. We also mapped the residues from LL-37 that contact dsRNA and found derivatives of LL-37 that can inhibit LL-37 enhancement of TLR3 signaling but maintain the ability to inhibit TLR4 signaling.
MATERIALS AND METHODS
Cells and Reagents
The BEAS-2B cell line was from the American Type Culture Collection and cultured in BEGM media containing supplements (11, 59). Proteasome inhibitors MG132 and lactacystin (both from Calbiochem) were dissolved in ethanol and DMSO, respectively. Cathepsin inhibitor z-FA-FMK (Santa Cruz Biotechnology) was dissolved in DMSO. Endosome acidification inhibitors ammonium chloride and chloroquine (Sigma) were dissolved in water. Bafilomycin A1 (Sigma) was dissolved in DMSO. Poly(I:C) and lipopolysaccharide (LPS) were from Invivogen. Reovirus dsRNA S4 was prepared by in vitro transcription as described in Lai et al. (11). All peptides, including ones with covalently attached fluorophores, were custom-synthesized (AnaSpec) and purified to >95% purity. Antibody to LL-37 (sc-166770) and siRNAs specific to FPRL1 (sc-40123), EGFR (sc-29301), or a nonspecific control siRNA (sc-37007) were all from Santa Cruz Biotechnology.
Fluorescence Polarization Assay
Fluorescence polarization assays used a Synergy H1 microplate reader (BioTek). All reactions were performed in phosphate buffers adjusted to the desired pH. The polarization measurements were determined as the ratio of the fluorescence intensity parallel to the excitation plane versus the fluorescence intensity perpendicular to the excitation plane. Calculations of polarization were performed using the Gene 5 software (Biotek), and the results were normalized to the starting polarization to account for possible changes in the oligomerization states of fLL-37 as a function of pH. Peptide binding to poly(I:C) used fLL-37 (0.1 μm), a version of LL-37 with an N-terminal carboxyfluorescein. Poly(I:C) was titrated added to a solution of fLL-37 to achieve a final volume of 100 μl. A complementary assay used fluorescein-labeled poly(I:C) (0.1 μm) titrated with peptides to 100-ml reactions that contained final concentrations of 10–500 nm unlabeled peptides. Interactions between LL-37 and other peptides used fLL-37 (0.1 μm) and peptides added to final concentrations of 1–1000 nm.
Förster Resonance Energy Transfer (FRET) Assays
The ability of LL-37 and poly(I:C) to interact within cells was analyzed by monitoring their ability to transfer energy, as measured by FRET assays (17). Fluorophore-labeled LL-37 and poly(I:C) were added to the cell culture media in the absence or presence of endosome acidification inhibitors and incubated for 1 h. The cells were then washed with PBS, fixed with 4% paraformaldehyde for 15 min at room temperature, and processed for microscopy as reported previously (18). Fluorescein was excited with a 488-nm laser, and emission was monitored with a Leica TCS SP5 confocal inverted-base microscope. Data analysis used the Leica LAS AF software.
Dynamic Light Scatter Spectroscopy
The hydrodynamic radii of LL-37 and other peptides was monitored by a Zetasizer Nano-S instrument (Malvern Instruments). All measurements were taken with 1 μm peptide dissolved in phosphate buffer adjusted to the desired pH at 22 °C.
Quantification of IL-6
IL-6 production was quantified by ELISA using the OptEIATM kit (BD Biosciences). A typical assay used 2 × 104 BEAS-2B cells/well grown for 24 h in flat-bottom 96-well plates. Poly(I:C) was added to a final concentration of 0.13 μg/ml. Antimicrobial peptides were added to the cell culture medium to a final concentration of 3 μm unless specified otherwise.
RNA Silencing Assays
BEAS-2B cells were seeded at 2 × 106 cells per 6-well plate for 6 h before transfection with 30 nm a mixture of three siRNAs. Transfections used Lipofectamine 2000 (Life Technologies). The level of target mRNA was analyzed using quantitative real-time-PCR and normalized to the levels of GAPDH mRNA. The sequences of the primers will be made available upon request. Poly(I:C) was added 48 h after siRNA transfection, and IL-6 levels in the culture media were collected 24 h later.
Confocal Microscopy
Cells were grown on poly-l-lysine-coated coverslips to 60% confluency. After a 1-h incubation with fluorophore-labeled peptides in the absence or presence of poly(I:C), the cells were washed with PBS and fixed with 4% paraformaldehyde for 15 min at room temperature. The cells were washed with PBS and mounted on glass slides with anti-fade mounting medium and DAPI (Life Technologies), then dried overnight in the dark. Micrographs were acquired with a Leica TCS SP5 confocal inverted-base microscope with a 63× oil objective. Images were analyzed by Leica LAS AF and Image J software. Colocalization of fluorophores was quantified using the Image J plug-in tool JACoP (16).
Statistical Analysis
All data shown are the means and ranges for 1 S.E. for a minimal of three independent samples. Data sets were compared using Student's t test calculated with GraphPad Prism 5 software.
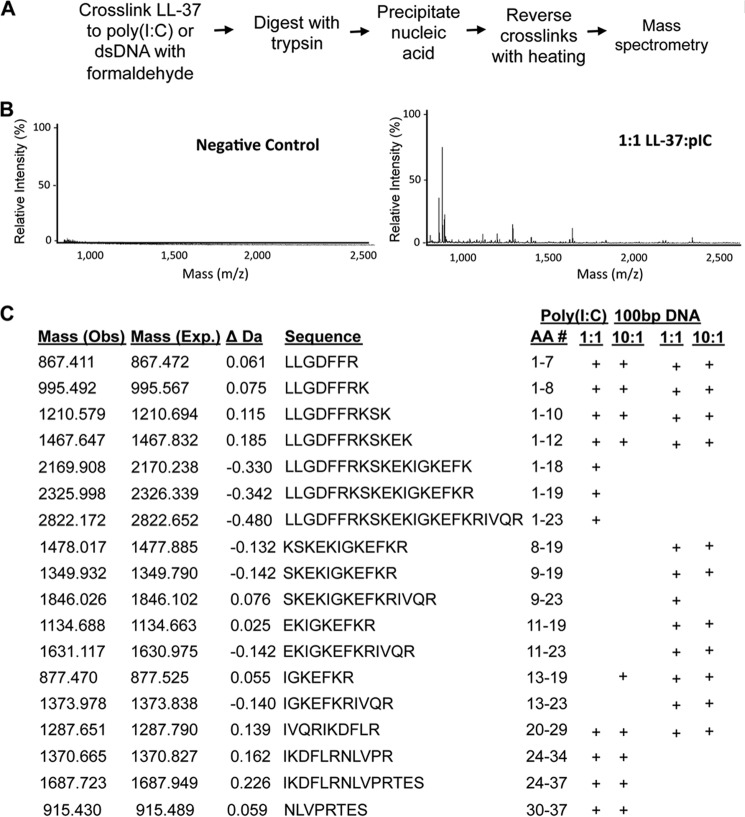
Reversible Cross-linking, Peptide Fingerprinting
LL37 was incubated with poly(I:C) at the molar ratios indicated, then cross-linked with the addition of 0.1% formaldehyde. After 15 min, formaldehyde was quenched by the addition of 2 m glycine, and LL37 was digested with 1:20 (w/w) ratio of trypsin overnight. Poly(I:C) was selectively precipitated with lithium chloride along with the covalently cross-linked peptides from LL37 as previously described (19). Protein-RNA cross-links were reversed by heating the sample for 1 h at 72 °C. The eluted peptides were analyzed using a Bruker Autoflex II MALDI-TOF mass spectrometer (Agilent Technologies).
Preparation of Necrotic DNA
BEAS-2B cells were grown to saturation for 1 week. All cells were harvested by centrifugation and subjected to 10 cycles of freezing and thawing. The cell lysates were concentrated by centrifugation at 10,000 × g for 15 min at 4 °C and adjusted to the buffers for the DNA extraction column and eluted as per the manufacturer's instructions (Qiagen). The DNA concentration was determined by spectrophotometry.
RESULTS
LL-37 Binding to Ligands in Vitro Is Dependent on pH
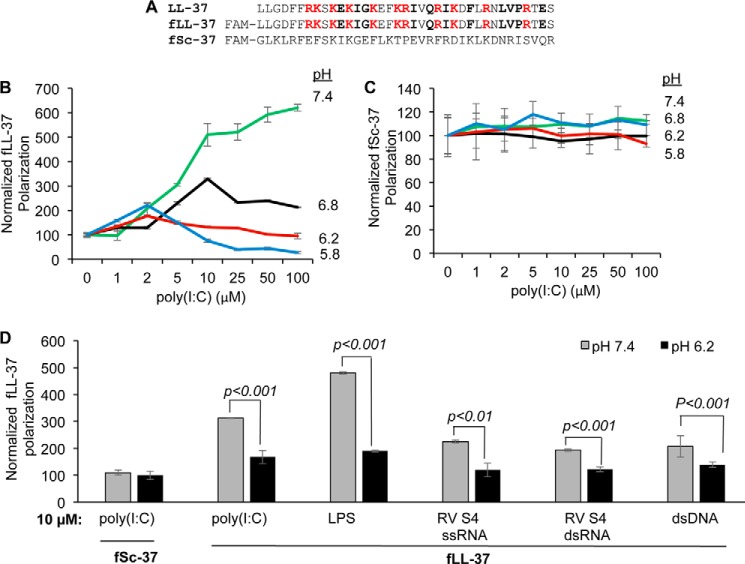
LL-37 accompanies dsRNA to endosomes containing TLR3 (18). However, it is unknown whether LL-37 will release the dsRNA in endosomes or participate in the signaling-competent TLR3·dsRNA complex. We seek to determine whether LL-37 binding to dsRNA can be affected by pH. A fluorescent polarization assay was performed with fLL-37, which has a 5′ carboxyfluorescein coupled to the N terminus of LL-37 (Fig. 1A; Refs. 11 and 18). fLL-37 was previously demonstrated to enhance TLR3 signaling by dsRNA and to suppress TLR4 signaling by LPS (18). In a pH 7.4 phosphate buffer, the fluorescence polarization of fLL-37 increased in a concentration-dependent manner upon the addition of poly(I:C), indicating that the two molecules can interact (Fig. 1B). LL37 binding to poly(I:C) significantly decreased when the pH of the buffer was decreased. fSC37, which has a scrambled order of the residues present in LL-37, did not interact with poly(I:C) titrated into the solution at any of the pH values tested (Fig. 1C). These results suggest that LL-37 can bind poly(I:C) in a pH-dependent manner.
FIGURE 1.
pH-dependent LL-37 binding to ligands. A, sequence of human antimicrobial peptide LL-37 and the scrambled peptide Sc-37. FAM denotes a carboxyfluorescein attached to the N termini of the two peptides. B, fluorescent polarization assay for LL-37 binding to poly(I:C) in buffers at different pH values. 6-Carboxyfluorescein-labeled LL-37, fLL-37, was at 0.1 μm. Poly(I:C) was titrated into the solution to form a final volume of fluorescence polarization assay with fSc-37 (C). fSc-37 was present in the assay at 0.1 μm and titrated with increasing concentrations of poly(I:C). D, binding LL-37 (0.1 μm) to different ligands at pH 7.4 and pH 6.2. fSc-37 was used as control. The values represent the increase in fluorescence polarization of fLL-37 with 10 μm concentrations of ligand at either pH 7.4 and 6.2 relative to the reaction lacking ligand. All data are the means of at least three independent experiments. p values were determined using Student's t test.
We determined whether fLL-37 binding to heteropolymeric dsRNA was dependent on the solution pH. fLL-37 binding to reovirus S4 dsRNA occurred in a concentration-dependent manner at pH 7.4, but binding was markedly decreased at pH 6.4 (data not shown). To allow comparison of the binding, the normalized fluorescence polarization at 10 μm reovirus dsRNA are shown in Fig. 1D. Other polyanionic molecules, including LPS, the sense-strand of the S4 RNA, and also dsDNA were also preferentially bound by fLL-37 at neutral pH values (Fig. 1D). These results show that LL-37 binds anionic polymers in a pH-dependent manner in vitro, although the difference in the relative degree of binding differed for the polyanionic polymers (Fig. 1D). For the remainder of this study, we will use poly(I:C) as a dsRNA ligand for LL-37.
Endosome Acidification Accompanies the Dissociation of the LL-37·Poly(I:C) Complex
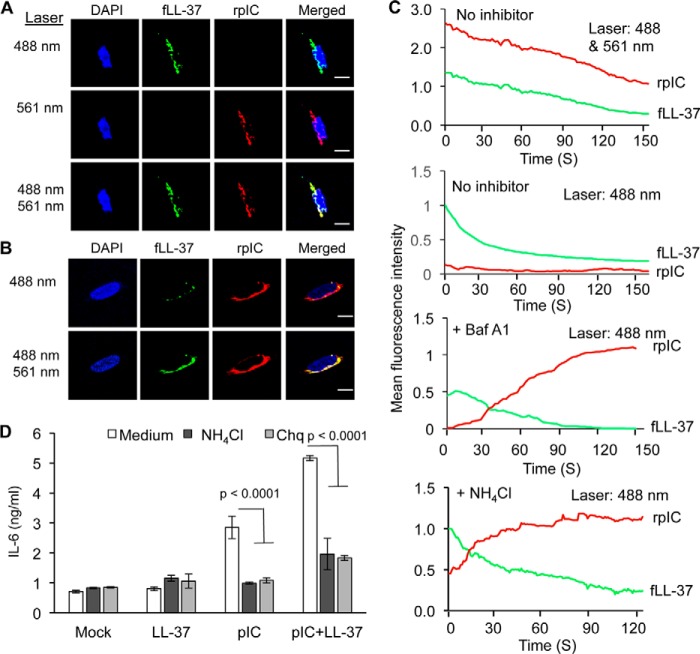
The localization of LL-37 complexed with dsRNA to endosomes and to enhance TLR3 activity have been previously characterized (18). We sought to determine whether the LL-37·poly(I:C) complex will dissociate in acidified endosomes of BEAS-2B cells, a human lung epithelial cell line that expresses endogenous FPLR-1, a receptor for the LL-37·dsRNA complex and TLR3 (11). Rhodamine-labeled poly(I:C) (rpIC) and fLL37 form a FRET pair and rpIC was previously shown to activate TLR3 signaling (11). Both fLL-37 and rpIC localized to endosomes (Fig. 2A). However, excitation of the fluorescein-labeled LL-37 did not result in energy transfer and fluorescence of the rhodamine on rpIC (Fig. 2A and 2C). This result suggests that LL-37 and poly(I:C) were no longer in physical contact in endosomes. However, cells treated with either chloroquine or ammonium chloride, or bafilomycin A1 to inhibit endosome acidification before the addition of fLL-37 and rpIC exhibited Förster resonance energy transfer (Fig. 2B, 2C and data not shown). These results suggest that endosome acidification is associated with the dissociation of the LL-37·poly(I:C) complex in cells. Inhibition of endosome acidification also reduced IL-6 production, consistent with endosome acidification being required for the activation of signal transduction by TLR3 (Fig. 2D; Refs. 20 and 21).
FIGURE 2.
LL-37 and poly(I:C) interaction in BEAS-2B cells is affected by endosome acidification. A, micrographs of the intracellular locations of fLL-37 and rpIC 60 min after their addition to BEAS-2B cells. fLL-37 is colored green, and rpIC is red. Co-localization of the two fluorophores is shown in yellow. The wavelength used to excite the fluorophores is shown to the left of the micrographs. Note that the signal for rhodamine-labeled poly(I:C) was not observed with excitation at 488 nm (upper row, third panel from the left) due to the lack of FRET from fluorescein to rhodamine. B, micrographs of the intracellular locations of fLL-37 and rpIC in BEAS-2B cells. fLL-37 and rpIC were added to the medium of the cells and the cells that were pretreated with the endosome acidification inhibitor chloroquine, ammonium chloride, or bafilomycin A1. Excitation of fluorescein was at 488 nm, and excitation of rhodamine was at 561 nm. Note that excitation of fluorescein fluorescence in fLL37 at 488 nm resulted in fluorescence from rpIC (third panel from the left, top row), indicating that FRET occurred between fLL-37 and rpIC. C, real-time monitoring of the fluorescence of the fLL-37 and rpIC in BEAS-2B cells treated or not treated with endosome inhibitors. The wavelength(s) for excitation is shown in the upper right corner for each graph. Top panel, spectra from BEAS-2B cells treated to excite fluorescence of both fLL-37 and rpIC. These results demonstrate that both molecules are present in cells. Second panel, spectra from BEAS-2B cells treated to excite fluorescence of fLL37. Note that only minimal excitation the fluorescence of rpIC was observed, likely due to the dissociation of fLL37 and rpIC. Third panel, spectra from BEAS-2B cells pretreated with bafilomycin A1 (Baf A1, 2.5 μm final concentration) to inhibit endosome acidification and then subjected to excitation of the fluorescence of fLL37. Note that energy was transferred to induce rhodamine fluorescence, suggesting that the fLL37 and rpIC complexes were physically close in space due to the lack of dissociation. Bottom panel, spectra from BEAS-2B cells pretreated with the endosome acidification inhibitor ammonium chloride (5 μm final concentration) followed by excitation of the fluorescence of fLL37. All spectra in this panel were consistent in at least three independent measurements. D, IL-6 ELISA results in the presence of endosomal acidification inhibitor, poly(I:C) and poly(I:C)/LL-37. Ammonium chloride and chloroquine (Chq) were used at final concentrations of 5 μm and 2 μg/ml, respectively. All the data represented are the mean of experiments performed in triplicate, and p values were calculated by using Student's t test.
LL-37 Oligomerization in vitro Is affected by pH
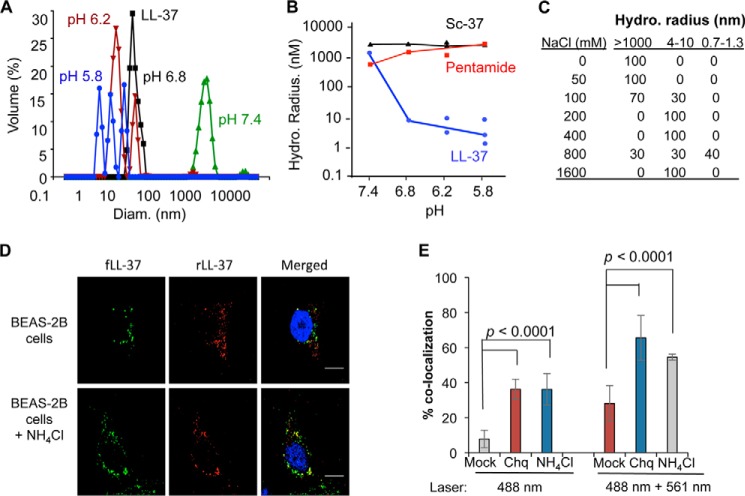
LL-37 forms higher order oligomeric complexes (22, 23). We used dynamic light scatter spectroscopy to determine whether pH will affect the oligomerization state of LL-37 in solution. At pH 7.4 the average hydrodynamic radius of LL-37 was ∼1 μm. However, a decrease in the buffer pH resulted in LL-37 dissociating to <10 nm (Fig. 3A). Sc-37 also formed a higher order complex of ∼1 μm, but this complex was not affected by acidification of the buffer (Fig. 3B). Pentamide, which had the five acidic residues in LL-37 replaced with neutral-polar residues, also failed to dissociate from a higher order complex as a function of pH.
FIGURE 3.
Oligomerization of LL-37 depends upon pH. A, LL-37 forms particles that dissociate when the pH is decreased. All assays were performed with LL-37 at a concentration of 1 μm in phosphate buffer. B, changes in the sequence of LL-37 can prevent pH-dependent dissociation. Peptides Sc-37 and pentamide form particles that have decreased dissociation in acidic buffer. Pentamide has changes of the five acidic residues to neutral-polar ones. Its sequence can be found in Table 1. C, LL-37 particles will dissociate in the presence of salt. The hydrodynamic radius of ∼1 μm LL-37 was determined in a pH 7.4 phosphate buffer adjusted to contain the concentration of NaCl indicated. D, fLL37 and rLL-37 colocalize in endosomes of BEAS-2B cells. fLL-37, rLL-37, and poly(I:C) were added to BEAS-2B cells that were either mock-treated or treated with ammonium chloride and imaged 60 min later by excitation at 488 nm or at 561 nm. The micrograph images were then merged to show the co-localization of the two fluorescent peptides. E, co-localization of fLL-37 and rLL-37 in BEAS-2B cells was quantified in the absence or presence of endosomal acidification inhibitors. To detect both peptides, the cells were excited with 488- and 561-nm lasers. To determine FRET between the two peptides, the cells were excited at 488 nm to activate fluorescence of fLL-37, and the emissions for both fluorescein and rhodamine were determined. The data shown were quantified from 20 independent cells from three independently prepared samples. The p values were calculated by using Student's t test. Chq, chloroquine.
The results with pentamide suggest that ionic interactions between LL-37 peptides contribute to oligomer formation. To address this we examined whether salt concentrations will affect the hydrodynamic radii of LL-37. The reactions were performed in a pH 7.4 buffer. In solutions with up to 50 mm NaCl, LL-37 was in complexes with hydrodynamic radii of >1000 nm. At ∼100 mm NaCl, complexes of 4–10 nm were observed (Fig. 3C). With NaCl of 200 mm or higher, LL-37 only had a hydrodynamic radii of 2–5 nm. The sensitivity of higher order complexes of LL-37 to salt is consistent with ionic interactions being responsible for the association between LL-37 subunits.
LL-37 Oligomerization in Cells Is Affected by pH
To examine whether LL-37 forms higher order oligomers in cells, we used a 1:1 mixture of fLL-37 and rhodamine-labeled LL-37 (rLL-37). When the two peptides were added to the medium of BEAS-2B cells, they co-localized to endosomes within 30 min (Fig. 3D). However, relatively little FRET was observed unless cells were treated with ammonium chloride or chloroquine to inhibit endosome acidification (Fig. 3, D and E). Identical results were observed when the cells were treated with a 1:1 mixture of fLL-37 and rLL-37 added to cell culture media along with poly(I:C) (data not shown). These results suggest that the LL-37·poly(I:C) complex dissociates upon endosome acidification.
Endosome Acidification Is Associated with LL-37 Turnover
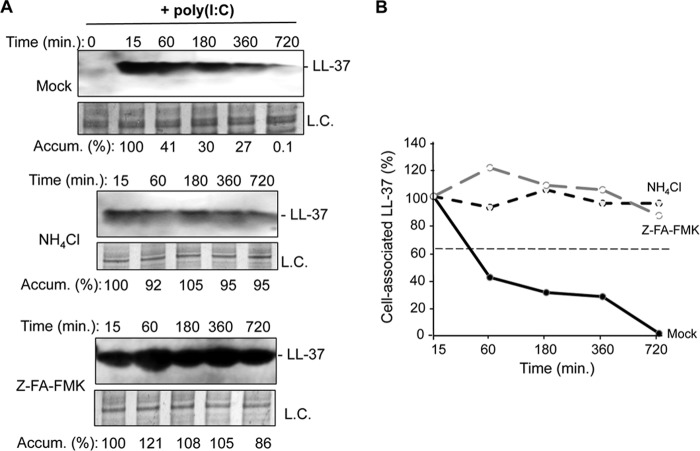
Acidification of endosomes can activate cathepsins that can cleave TLR3, -7, and -9 and increase signaling (24, 25). We examined whether LL-37 would be subject to proteolysis, possibly by cathepsins. BEAS-2B cells were treated with LL-37 in the absence or presence of poly(I:C) for 15 min, then washed with phosphate-buffered saline and incubated with fresh medium. The cells were harvested over time, lysed, and subjected to Western blot analysis. LL-37 accumulation decreased over time, with an estimated half-life of 1 h (Fig. 4, A and B). Cells treated with ammonium chloride or the cathepsin inhibitor Z-FA-FMK had half-lives in excess of 12 h (Fig. 4, A and B). These results suggest that endosome acidification can lead to proteolysis of LL-37.
FIGURE 4.
LL-37 is degraded in endosomes of BEAS-2B cells. A, Western blot assessing the accumulation of LL-37 over time after its addition along with poly(I:C) (0.13 μg/ml) to BEAS-2B cells. Upper panel, accumulation of LL-37 in untreated BEAS-2B cells. Middle panel, the accumulation of LL-37 in the presence of endosomal acidification inhibitor NH4Cl (5 μm final concentration). Bottom panel, the accumulation of LL-37 in the presence of cathepsin inhibitor Z-FA-FMK (5 μm final concentration). In all three panels LL-37 was detected by a Western blot of cell lysates electrophoresed in NuPAGE 4–12% Bis-Tris gels designed to separate small proteins. Quantification of band intensity is calculated relative to that of the loading control (L.C.). B, half-life of LL-37 as measured by band intensity of Western blots with and without endosomal acidification inhibitor or cathepsin inhibitor. A similar increase in LL-37 half-life was observed in two independent experiments.
Residues in LL-37 That Contact Poly(I:C)
We seek to determine how residues in LL-37 contact poly(I:C) using a reversible cross-linking peptide fingerprinting method (RCAP; Ref. 19). RCAP fingerprinting has been used to map the RNA-contacting regions within several protein·RNA complexes using formaldehyde, a bifunctional cross-linking agent that cross-links primary amines that are within 2 Å of each other (26).
Cross-linked LL-37·poly(I:C) were extensively digested with trypsin to cleave C-terminal of lysines and arginines (Fig. 5A). Poly(I:C) and peptide fragments cross-linked to poly(I:C) were selectively precipitated with lithium chloride. The cross-linked peptides co-purified with poly(I:C) were reversed and subjected to mass spectrometric analysis. Control reactions performed without formaldehyde did not identify peptides, indicating that the washing conditions are stringent (Fig. 5B). Several peptides derived from LL-37 were observed with the LL-37·poly(I:C) cross-linked complex at both a 1:1 and a 10:1 molar ratio of LL-37 to poly(I:C) fractionated to have an average length of 500 bp. All peptides matched to within 0.5 daltons of the expected LL-37 tryptic fragments (Fig. 5C). Both the N- and C-terminal portions of LL-37 were found to be in contact with poly(I:C), but a higher concentration of peptide resulted in a decreased cross-linking to residues 18, 19, and 23 of LL-37 (Fig. 5C). Overlapping peptides were observed, suggesting that some lysine and arginine residues were only partially cross-linked to poly(I:C), as some sites remained accessible to trypsin cleavage (Fig. 5C).
FIGURE 5.
Mapping LL-37 residues that contact poly(I:C) and double-stranded DNA. A, schematic of the RCAP fingerprinting protocol. B, sample mass spectra from the RCAP fingerprinting assay detecting peptides from LL-37 that can be cross-linked to poly(I:C). The negative control was performed with an assay containing all of the components but not treated with formaldehyde. C, summary of the peptides from LL-37 found to contact poly(I:C) of ∼500 bp and a DNA molecule of 100 bp. The peptides identified by mass spectrometry within an accuracy of 0.5 Da of the theoretical masses are denoted with a + symbol.
The RCAP fingerprinting assay was also used to map LL-37 interaction with a 100-bp DNA. RCAP fingerprinting has been validated for protein-DNA interaction (27). Again, the background was minimal, and peptides from LL-37 from the cross-linked samples were easily identified. At a 1:1 molar ration of LL-37 to DNA the N-terminal region of LL-37 also contacted the DNA. In addition, up to residues 29 of LL-37 were observed to be cross-linked to DNA (Fig. 5C) An interesting difference from the cross-link to poly(I:C) is that the middle region of LL-37 appears to be less in contact with DNA when a 10:1 molar ratio of LL-37 and DNA was analyzed. It is possible that stiffness of dsDNA could prevent it from properly adjusting the contacts with LL-37.
Residues in LL-37 Required for Function
To better define how residues in LL-37 contribute to function, several variants derived from LL-37 were tested for 1) repression of TLR4 signaling by LPS, 2) enhancement of TLR3 signaling by poly(I:C), and 3) binding to LL-37. Signaling by TLR3 and TLR4 was monitored using ELISA to detect IL-6 secreted into the medium of BEAS-2B cells.
N- and C-terminal truncations in LL-37 retained the ability to suppress TLR4 signaling (Table 1). Pentamide, which had all acidic residues in LL-37 replaced with neutral-polar residues, was the only one that failed to suppress TLR4 signaling. These results suggest that multiple regions of LL-37 can interact with LPS to suppress TLR4 signaling.
TABLE 1.
Summary of the activities of LL-37 and variants
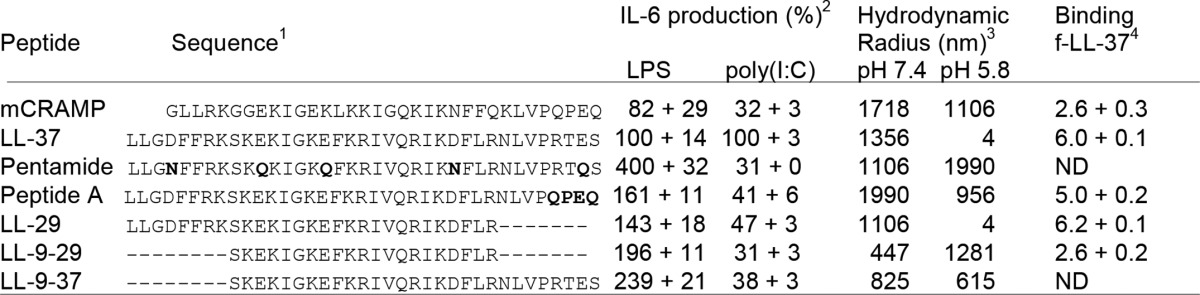
1 Amino acid residues changed from LL-27 are in bold. The residues deleted from LL-37 are shown with a dash.
2 IL-6 production in BEAS-2B cells induced with 1.0 μg/ml LPS or 130 ng/ml poly(I:C).
3 Dynamic light spectroscopy results were determined with 1.0 μm concentrations of the peptide in phosphate buffer adjusted to pH 7.4 or 5.8.
4 Fold change in the fluorescence anisotropy values of f-LL-37 between 1.0 nm and 1000 nm concentrations of added peptide.
In contrast, all LL-37 truncations negatively affected TLR3 signaling. Peptides LL9–29, pentamide, and mCRAMP failed to enhance TLR3 signaling. Removal of the N-terminal eight residues, the C-terminal eight residues of LL-37, or the replacement of the C-terminal four residues with those from mCRAMP resulted in peptides that retained partial activity for enhancing TLR3 signaling. These results suggest that both terminal regions of LL-37 are required for wild-type level of interaction with poly(I:C) to activate TLR3 signaling (Fig. 5, Table 1).
We tested whether the LL-37 variants dissociate in a pH-dependent manner using dynamic light scatter spectroscopy. LL-29, which lacks the C-terminal eight residues, dissociated into nanometer-sized particles (Table 1). Peptide A, with only the C-terminal four residues of LL-37 replaced with mCRAMP residues, was also reduced for the ability to dissociate under acidic pH conditions (Table 1). Therefore, although the terminal eight residues of LL-37 are not required for oligomer dissociation, their replacement may allow the peptides to form alternative complexes that do not respond to pH. Peptide LL9–29, that lacked both the C- and N-terminal regions, also failed to dissociate (Table 1).
Because LL-29 retains the ability to change its oligomerization state in a pH-dependent manner, we examined whether it can bind wild-type LL-37. In a fluorescence polarization assay, LL-29 interacted with fLL-37 to the same degree as LL-37 (Table 1). Peptide A retained partial interaction with fLL-37 in this assay, whereas LL9–29, which lacks the N-terminal eight residues of LL-37, did not (Table 1). The N-terminal 29 residues of LL-37 thus retain the ability to interact with LL-37 as well as to contact nucleic acids.
LL-29 Can Inhibit LL-37-enhanced TLR3 Signal Transduction
We examined whether LL-29 could affect the enhancement of TLR3 signaling by WT LL-37·poly(I:C). An increasing amount of LL-29 along with a constant amount of LL-37·poly(I:C) was used to assess IL-6 production in BEAS-2B cells. The presence of LL-29 reduced the enhancement of TLR3 signaling by LL-37 in a concentration-dependent manner (Fig. 6). At a 4 m excess of LL-29 relative to LL-37, IL-6 levels were comparable with those observed in the absence of LL-37 (Fig. 6). That is, LL-29 abrogated the ability of LL-37 to enhance TLR3 signaling. Peptide A also antagonized LL-37 enhancement of TLR3 signaling but to a lesser degree than LL-29 (Table 1).
FIGURE 6.
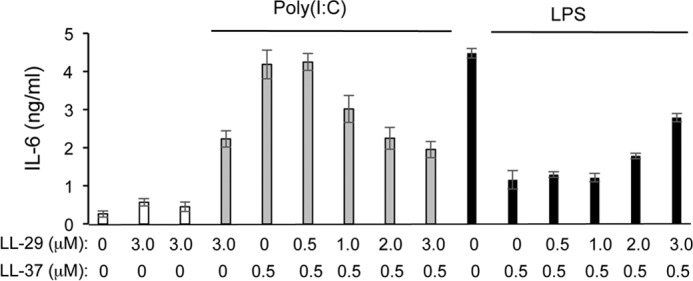
Activities of truncated LL-37 peptides on LPS- and poly(I:C)-induced signaling. IL-6 production induced by LPS or poly(I:C) in BEAS-2B cells after the addition of LL-37 in the presence or absence of LL-29. The final concentrations of the peptides are shown below the graph. Where added, poly(I:C) was to 130 ng/ml and LPS was to 1 μg/ml. All samples were performed in at least triplicates, and the results are representative of three independent experiments.
Interestingly, LL-29 added along with LL-37 to BEAS-2B cells retained the ability to suppress TLR4 signaling in response to LPS (Fig. 6). A 2 m excess of LL-29 relative to LL-37 did not reduce TLR4 signaling (Fig. 6A). At a 4 m excess of LL-29 to LL-37, IL-6 production in response to LPS was still at 64% that of the control without either of the two peptides. These results suggest that LL-29 can preferentially antagonize the LL-37 enhancement of TLR3 signaling while retaining the ability to suppress TLR4 signaling.
LL-29 Can Affect LL-37·Poly(I:C) Trafficking
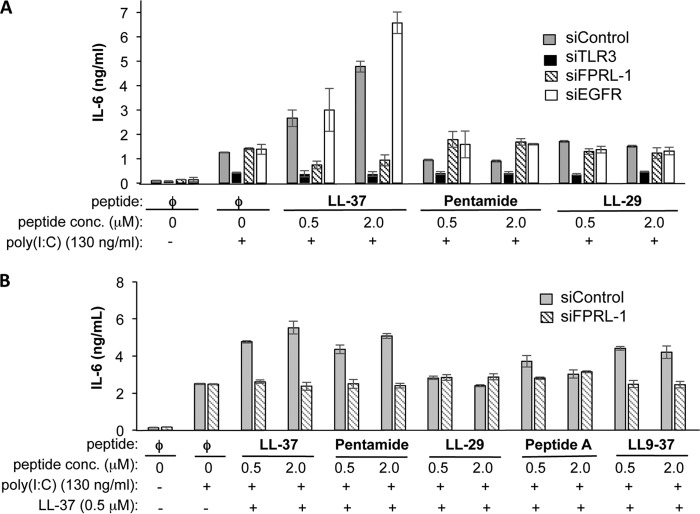
LL-37 enhancement of TLR3 signaling required trafficking of LL-37·poly(I:C) to endosomes via the formyl peptide receptor-like 1 (FPRL-1) receptor (18). Therefore, we examined whether LL-29 used the FPRL-1 receptor to enter cells. siRNA was used to knock down the expression of either FPRL-1, EGFR, or TLR3. Quantitative RT-PCR analysis showed that the FPRL-1 message was reduced to ∼20% of the control siRNA-treated samples (data not shown). Co-localization of fLL-37 and rpIC was reduced to <⅓ of the control. Cells knocked down for TLR3 expression had reduced IL-6 production in response to poly(I:C) both in the absence and presence of LL-37, demonstrating that IL-6 production requires TLR3 (Fig. 7A). Knockdowns performed with siRNAs to a nonspecific control or to EGFR had no effect on the enhancement of IL-6 production. Pentamide, LL9–37, or LL-29 added to cells along with poly(I:C) also did not enhance IL-6 production with knockdown of either FPRL-1 or EGFR (Fig. 7A). These results suggest that multiple activities of LL-37 are needed to engage the FPRL-1 receptor, including LL-37 oligomerization and binding to poly(I:C).
FIGURE 7.
LL-29 does not enhance TLR3 signaling through the FPRL-1 receptor. A, enhancement of poly(I:C)-induced TLR3 signaling by LL-37, LL-29, and pentamide. TLR signaling was assessed by measuring IL-6 secreted by BEAS-2B cells into the cell culture media 24 h after induction with poly(I:C). The cells were knocked down with TLR3, FPRL-1, EGFR, or control siRNAs 48 h before the addition of the poly(I:C) or poly(I:C) and peptides. B, effect of truncated peptides in poly(I:C)-mediated IL-6 production in the presence of LL-37. The effects of the peptides were determined in BEAS-2B cells knocked down for either a nonspecific control or for FPRL-1.
Will the addition of LL-29 affect the use of the FPRL-1 receptor by the LL-37·poly(I:C) complex? BEAS-2B cells knocked down for FPRL-1 were treated with LL-37 along with additional concentrations of LL-37, pentamide, LL-29, Peptide A, or LL9–37 (Fig. 7B). Signaling by TLR3 was determined by quantifying the IL-6 secreted in the cell culture media. Knockdown of the FPRL-1 decreased LL-37, whereas a control siRNA had no effect on WT LL-37·poly(I:C) enhancement of IL-6 production. Pentamide or LL9–37 added along with LL-37 had no effect on IL-6 production. Interestingly, the addition of Peptide A and LL-29 along with LL-37 resulted in a reduction in IL-6 levels whether the cells were treated with a siRNA to FPRL-1 or a nonspecific control. These results suggest that LL-29 and Peptide A can antagonize LL-37 enhancement of TLR3 signaling in response to poly(I:C).
LL-29 Can Affect LL-37·poly(I:C) Co-localization with TLR3 in BEAS-2B Cells
LL-29 does not use FPRL-1 to affect TLR3 signaling but can bind LL-37 in vitro (Fig. 7A and Table 1). We wondered whether it could prevent the trafficking of LL-37·poly(I:C) to endosomes that harbor TLR3. Confocal microscopy was used to examine the effect of LL-29 on the colocalization of LL-37·poly(I:C) with TLR3 in BEAS-2B cells. As expected, fLL-37 was found to colocalize with TLR3, whereas Sc-37 did not (Fig. 8). However, the addition of LL-29 reduced the co-localization between fLL-37 and TLR3 in endosomes containing TLR3 (Fig. 8).
FIGURE 8.
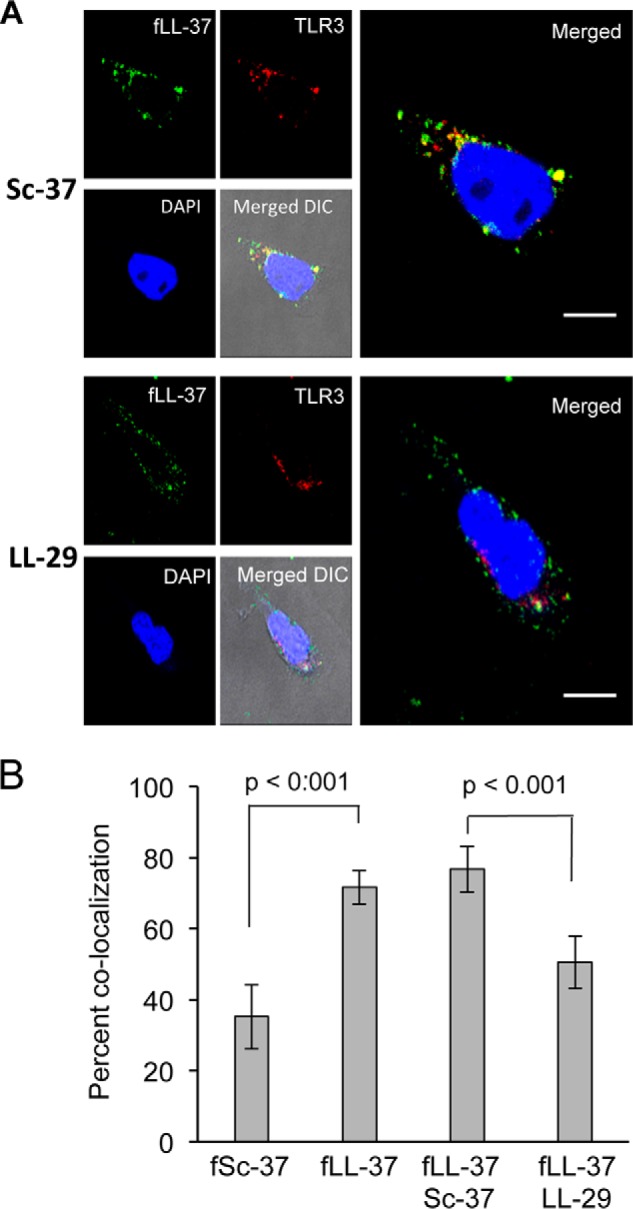
LL-29 can decrease the co-localization of fLL-37 and TLR3 in BEAS-2B cells. A, fluorescent micrographs of the locations of LL-29 or Sc-37 added to cells along with LL-37 and poly(I:C). LL-37 was added to a final concentration of 1.0 μm and poly(I:C) to a final concentration of 130 ng/ml. TLR3 was detected with goat anti-TLR3 antibody and a Alexa-488-labeled anti-goat secondary antibody. B, quantification of the percent co-localization of LL-37 and TLR3. fSc-37 was used as a control. Quantitative co-localization data are the mean values for 20 cells assessed from three independently prepared samples. The p values were calculated by using Student's t test.
DISCUSSION
LL-37 can suppress the production of proinflammatory cytokines in response to the bacterial lipopolysaccharides. Intriguingly, LL-37 can also enhance the production of proinflammatory cytokines by dsRNAs that activate TLR3. In this work we determined that the pH could affect LL-37 interaction with dsRNA both in solution and in cells. LL-37 binds to poly(I:C) at neutral pH values in vitro and releases it upon endsome acidification. Within cells, dsRNA release was determined by a loss of the Förster resonance energy transfer between fluorescently labeled LL-37 and poly(I:C). Importantly, cells inhibited for endosome acidification retained the interaction between LL-37 poly(I:C). LL-37 also exists in solution as a higher order complex that can dissociate upon acidification of the solution. Johansson et al. (15) reported that the oligomeric state of LL-37 and a neutral pH are needed for its function in suppressing the proinflammatory response to LPS. Our results show that the pH of the environment and LL-37 oligomerization state are also important factors for the LL-37 enhancement of signal transduction by TLR3. A schematic for the effects of LL-37 on dsRNA signaling by TLR3 is presented in Fig. 9.
FIGURE 9.
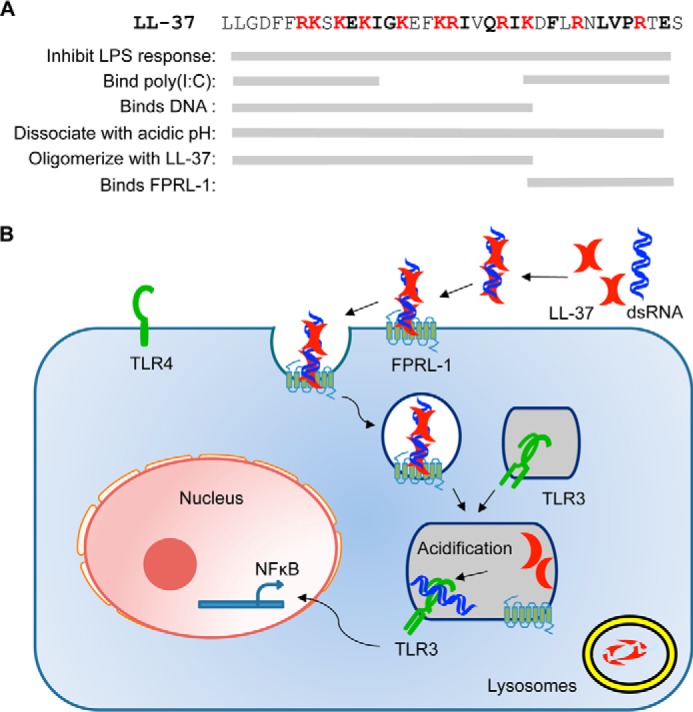
A model for LL-37 enhancement of dsRNA-induced TLR3 signaling. A, sequence of LL-37 and summary of residues involved in LPS binding, poly(I:C) binding, pH-dependent dissociation, and oligomerization. We note that the region in LL-37 delineated for a particular activity represents a minimal requirement interpreted from results in this work; it is possible and likely that other regions within LL-37 will contribute to an activity. B, schematic of LL-37-mediated poly(I:C) signaling. LL-37 binds to dsRNA and is internalized to endosomes through the FPRL-1 receptor. After maturation, the acidic environment of endosomes dissociates dsRNA and LL-37. The dsRNA is then recognized by TLR3 for signaling, whereas LL-37 is transferred to lysosomes and degraded.
The pH-dependent binding and release of dsRNA by LL-37 explains the differential effects of LL-37 on TLR3 and TLR4 signaling. Due to interactions between LL-37 and LPS taking place in the extracellular media or in the cell cytoplasm where the pH is near neutral, LL-37 does not readily release LPS. Thus, LL-37 sequesters LPS to prevent TLR4 signaling. In contrast, LL-37 bound to dsRNA traffics to endosomes where acidification can take place due to the action of proton pumps (28). The acidification of the endosome induces LL-37 to release of dsRNA where it acts as agonist for TLR3 signaling.
It is important to note that dsRNA can traffic to endosomes independent of LL-37 through scavenger receptors (29). Indeed, TLR3 can signal in response to poly(I:C) even in the absence of LL-37 (30). In the presence of LL-37, TLR3 signaling may be enhanced due to the increased concentrations of the dsRNA in endosomes where TLR3 is resident.
Thus, pH likely coordinates a number of consequential events for signaling by TLR3. Endosome acidification can also activate cathepsins, proteases to enhance the stability of signaling-competent TLR3 (31, 32) and trigger the degradation of LL-37 (Fig. 4). Notably, inhibition of endosome acidification, cathepsins B, L, and/or S (33), with the inhibitor Z-FA-FMK increased the half-life of LL-37 from 1 h to an excess of 12 h (Fig. 4B).
Despite its small size, LL-37 has numerous activities: binding to RNA, DNA, and LPS, dissociation with pH, and trafficking to endosomes via the FPRL-1 receptor. Our characterization of LL-37 and variants contributes to how LL-37 residues participate in these activities. For RNA binding, the overall positive charge of LL-37 (11 positive residues of 37) is not sufficient; both mCRAMP (9 positively charged residues out of 34) and pentamide (11 positively charged residues of 37) failed to bind dsRNA in our fluorescent polarization assay and failed to enhance TLR3 signaling (Table 1). Results from mapping studies suggest that the residues at the terminal regions of LL-37 preferentially contact both dsRNA and DNA (Fig. 5B). It is also likely that significant conformational changes in both LL-37 and the nucleic acid will take place upon complex formation, as the contacts changed with a higher ratio of LL-37 to poly(I:C). These conformational changes could be affected by the stiffness of the nucleic acid.
The truncated peptides derived from LL-37 were also informative for the interaction between LL-37 subunits. LL-29 can bind to LL-37 to the same extent as LL-37, indicating that the C-terminal eight residues of LL-37 are not required for the interaction between LL-37 peptides. We propose that the central ∼20 residues of LL-37 likely anchor the interaction between LL-37 subunits. The N-terminal residues, at least some subunits of which contact dsRNA, likely contribute to subunit interaction as a deletion of the N-terminal eight residues significantly reduced interaction with LL-37 (Table 1).
The interaction between LL-37 and FPRL-1 has more complex requirements. FPRL-1 traffics the LL-37 molecules that are complexed to dsRNA (18). Therefore, LL-37 derivatives that are defective for dsRNA binding will not endocytose using the FPRL-1 receptor. Interestingly, cells treated with LL-29 along with LL-37 and poly(I:C) were reduced for the enhancement of dsRNA signaling from the FPRL-1 receptor (Fig. 6). Furthermore, LL-37 co-localization with TLR3 in endosomes was decreased by the presence of LL-29 (Fig. 7). Given that LL-29 can interact with LL-37, it is likely that a heterocomplex of LL-37 and LL-29 prevents trafficking of the LL-37·dsRNA from use of the FPRL-1 receptor. The decrease in dsRNA concentration in endosomes likely abrogates the LL-37 enhancement of signal transduction by TLR3.
The ability of LL-29 to antagonize the activity of LL-37 could be developed to generate agents that can reduce the elevated immune responses associated with high levels of LL-37. Elevated levels of LL-37 are associated with the inflammatory response to self-DNA by TLR9 (34) and to viral dsRNAs that are the agonists for TLR3 (10, 11). Truncated parathyroid peptides are currently in use to promote bone morphogenesis with reduced ability to promote bone resorption (35). Notably, LL-29 can antagonize LL-37 activation of TLR3 signaling while retaining the ability to suppress TLR4 signaling (Table 1). It is, therefore, possible that antagonists of LL-37 derived from LL-29 can remain competent as antimicrobial peptides that can suppress the inflammatory response associated with bacterial infections.
Acknowledgments
We thank members of the Kao laboratory for numerous helpful discussions. We thank L. Kao for editing the manuscript and Jim Powers and the Indiana University Light Microscopy Center for use of the confocal microscopy facility that is supported by Indiana METACyt.
Footnotes
- TLR
- Toll-like receptor
- dsRNA
- double-stranded RNA
- FPRL-1
- formyl peptide receptor-like 1
- rpIC
- rhodamine-labeled poly(I:C)
- Z-FA-FMK
- benzyloxycarbonyl-Phe-Ala-fluoromethyl ketone
- EGFR
- EGF receptor
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- RCAP
- reversible cross-linking peptide.
REFERENCES
- 1. Sørensen O. E., Follin P., Johnsen A. H., Calafat J., Tjabringa G. S., Hiemstra P. S., Borregaard N. (2001) Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3. Blood 97, 3951–3959 [DOI] [PubMed] [Google Scholar]
- 2. Doss M., White M. R., Tecle T., Hartshorn K. L. (2010) Human defensins and LL-37 in mucosal immunity. J. Leukoc. Biol. 87, 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lande R., Ganguly D., Facchinetti V., Frasca L., Conrad C., Gregorio J., Meller S., Chamilos G., Sebasigari R., Riccieri V., Bassett R., Amuro H., Fukuhara S., Ito T., Liu Y. J., Gilliet M. (2011) Neutrophils activate plasmacytoid dendritic cells by releasing self-DNA-peptide complexes in systemic lupus erythematosus. Sci. Transl. Med. 3, 73ra19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim S. K., Park S., Lee E. S. (2010) Toll-like receptors and antimicrobial peptides expressions of psoriasis: correlation with serum vitamin D level. J. Korean Med. Sci. 25, 1506–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gambichler T., Kobus S., Kobus A., Tigges C., Scola N., Altmeyer P., Kreuter A., Bechara F. G., Skrygan M. (2011) Expression of antimicrobial peptides and proteins in etanercept-treated psoriasis patients. Regul. Pept. 167, 163–166 [DOI] [PubMed] [Google Scholar]
- 6. Goleva E., Searing D. A., Jackson L. P., Richers B. N., Leung D. Y. (2012) Steroid requirements and immune associations with vitamin D are stronger in children than adults with asthma. J. Allergy Clin. Immunol. 129, 1243–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sun J., Dahlén B., Agerberth B., Haeggström J. Z. (2013) The antimicrobial peptide LL-37 induces synthesis and release of cysteinyl leukotrienes from human eosinophils: implications for asthma. Allergy 68, 304–311 [DOI] [PubMed] [Google Scholar]
- 8. Rohde G., Message S. D., Haas J. J., Kebadze T., Parker H., Laza-Stanca V., Khaitov M. R., Kon O. M., Stanciu L. A., Mallia P., Edwards M. R., Johnston S. L. (2014) CxC-chemokines and antimicrobial peptides in rhinovirus-induced experimental asthma exacerbations. Clin. Exp. Allergy 44, 930–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dombrowski Y., Schauber J. (2012) Cathelicidin LL-37: a defense molecule with a potential role in psoriasis pathogenesis. Exp. Dermatol. 21, 327–330 [DOI] [PubMed] [Google Scholar]
- 10. Lai Y., Adhikarakunnathu S., Bhardwaj K., Ranjith-Kumar C. T., Wen Y., Jordan J. L., Wu L. H., Dragnea B., San Mateo L., Kao C. C. (2011) LL37 and cationic peptides enhance TLR3 signaling by viral double-stranded RNAs. PLoS ONE 6, e26632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lai Y., Yi G., Chen A., Bhardwaj K., Tragesser B. J., Rodrigo A., Valverde, Zlotnick A., Mukhopadhyay S., Ranjith-Kumar C. T., Kao C. C. (2011) Viral double-strand RNA-binding proteins can enhance innate immune signaling by toll-like Receptor 3. PLoS ONE 6, e25837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li G., Domenico J., Jia Y., Lucas J. J., Gelfand E. W. (2009) NF-κB-dependent induction of cathelicidin-related antimicrobial peptide in murine mast cells by lipopolysaccharide. Int. Arch. Allergy Immunol. 150, 122–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wan M., van der Does A. M., Tang X., Lindbom L., Agerberth B., Haeggström J. Z. (2014) Antimicrobial peptide LL-37 promotes bacterial phagocytosis by human macrophages. J. Leukoc. Biol. 95, 971–981 [DOI] [PubMed] [Google Scholar]
- 14. Phelps M. A., Foraker A. B., Gao W., Dalton J. T., Swaan P. W. (2004) A novel rhodamine-riboflavin conjugate probe exhibits distinct fluorescence resonance energy transfer that enables riboflavin trafficking and subcellular localization studies. Mol. Pharm. 1, 257–266 [DOI] [PubMed] [Google Scholar]
- 15. Johansson J., Gudmundsson G. H., Rottenberg M. E., Berndt K. D., Agerberth B. (1998) Conformation-dependent antibacterial activity of the naturally occurring human peptide LL-37. J. Biol. Chem. 273, 3718–3724 [DOI] [PubMed] [Google Scholar]
- 16. Bolte S., Cordelières F. P. (2006) A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 [DOI] [PubMed] [Google Scholar]
- 17. Sekar R. B., Periasamy A. (2003) Fluorescence resonance energy transfer (FRET) microscopy imaging of live cell protein localizations. J. Cell Biol. 160, 629–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Singh D., Qi R., Jordan J. L., San Mateo L., Kao C. C. (2013) The human antimicrobial peptide LL-37, but not the mouse ortholog, mCRAMP, can stimulate signaling by poly(I:C) through a FPRL1-dependent pathway. J. Biol. Chem. 288, 8258–8268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vaughan R., Fan B., You J. S., Kao C. C. (2012) Identification and functional characterization of the nascent RNA contacting residues of the hepatitis C virus RNA-dependent RNA polymerase. RNA 18, 1541–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ranjith-Kumar C. T., Duffy K. E., Jordan J. L., Eaton-Bassiri A., Vaughan R., Hoose S. A., Lamb R. J., Sarisky R. T., Kao C. C. (2008) Single-stranded oligonucleotides can inhibit cytokine production induced by human toll-like receptor 3. Mol. Cell. Biol. 28, 4507–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leonard J. N., Ghirlando R., Askins J., Bell J. K., Margulies D. H., Davies D. R., Segal D. M. (2008) The TLR3 signaling complex forms by cooperative receptor dimerization. Proc. Natl. Acad. Sci. U.S.A. 105, 258–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Oren Z., Lerman J. C., Gudmundsson G. H., Agerberth B., Shai Y. (1999) Structure and organization of the human antimicrobial peptide LL-37 in phospholipid membranes: relevance to the molecular basis for its non-cell-selective activity. Biochem. J. 341, 501–513 [PMC free article] [PubMed] [Google Scholar]
- 23. Xhindoli D., Pacor S., Guida F., Antcheva N., Tossi A. (2014) Native oligomerization determines the mode of action and biological activities of human cathelicidin LL-37. Biochem. J. 457, 263–275 [DOI] [PubMed] [Google Scholar]
- 24. Ewald S. E., Engel A., Lee J., Wang M., Bogyo M., Barton G. M. (2011) Nucleic acid recognition by Toll-like receptors is coupled to stepwise processing by cathepsins and asparagine endopeptidase. J. Exp. Med. 208, 643–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sepulveda F. E., Maschalidi S., Colisson R., Heslop L., Ghirelli C., Sakka E., Lennon-Duménil A. M., Amigorena S., Cabanie L., Manoury B. (2009) Critical role for asparagine endopeptidase in endocytic Toll-like receptor signaling in dendritic cells. Immunity 31, 737–748 [DOI] [PubMed] [Google Scholar]
- 26. Lu K., Ye W., Zhou L., Collins L. B., Chen X., Gold A., Ball L. M., Swenberg J. A. (2010) Structural characterization of formaldehyde-induced cross-links between amino acids and deoxynucleosides and their oligomers. J. Am. Chem. Soc. 132, 3388–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pérez-Vargas J., Vaughan R. C., Houser C., Hastie K. M., Kao C. C., Nemerow G. R. (2014) Isolation and characterization of the DNA and protein binding activities of adenovirus core protein V. J. Virol. 88, 9287–9296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Forgac M. (2007) Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 8, 917–929 [DOI] [PubMed] [Google Scholar]
- 29. Limmon G. V., Arredouani M., McCann K. L., Corn Minor R. A., Kobzik L., Imani F. (2008) Scavenger receptor class-A is a novel cell surface receptor for double-stranded RNA. FASEB J. 22, 159–167 [DOI] [PubMed] [Google Scholar]
- 30. Takiguchi T., Morizane S., Yamamoto T., Kajita A., Ikeda K., Iwatsuki K. (2014) Cathelicidin antimicrobial peptide LL-37 augments IFN-β expression and anti-viral activity induced by double-stranded RNA in keratinocytes. Br. J. Dermatol. 10.1111/bjd.12942 [DOI] [PubMed] [Google Scholar]
- 31. de Bouteiller O., Merck E., Hasan U. A., Hubac S., Benguigui B., Trinchieri G., Bates E. E., Caux C. (2005) Recognition of double-stranded RNA by human Toll-like receptor 3 and downstream receptor signaling requires multimerization and an acidic pH. J. Biol. Chem. 280, 38133–38145 [DOI] [PubMed] [Google Scholar]
- 32. Kuznik A., Bencina M., Svajger U., Jeras M., Rozman B., Jerala R. (2011) Mechanism of endosomal TLR inhibition by antimalarial drugs and imidazoquinolines. J. Immunol. 186, 4794–4804 [DOI] [PubMed] [Google Scholar]
- 33. Garcia-Cattaneo A., Gobert F. X., Müller M., Toscano F., Flores M., Lescure A., Del Nery E., Benaroch P. (2012) Cleavage of Toll-like receptor 3 by cathepsins B and H is essential for signaling. Proc. Natl. Acad. Sci. U.S.A. 109, 9053–9058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lande R., Gregorio J., Facchinetti V., Chatterjee B., Wang Y. H., Homey B., Cao W., Wang Y. H., Su B., Nestle F. O., Zal T., Mellman I., Schröder J. M., Liu Y. J., Gilliet M. (2007) Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature 449, 564–569 [DOI] [PubMed] [Google Scholar]
- 35. Martin T. J. (2014) Bone biology and anabolic therapies for bone: current status and future prospects. J. Bone Metab. 21, 8–20 [DOI] [PMC free article] [PubMed] [Google Scholar]