Background: ATP-bound RecA cannot catalyze DNA recombination without crucial accessory factors.
Results: DprA facilitates RecA assembly onto SsbA- or SsbB-coated single-stranded DNA, but only DprA and SsbA enable RecA·ATP to mediate strand exchange.
Conclusion: RecA·ATP transition to an active state requires DprA and SsbA.
Significance: DprA and SsbA may serve as a two-component mediator during chromosomal transformation.
Keywords: DNA-binding Protein, DNA Recombination, DNA Transformation, DNA-Protein Interaction, Protein-DNA Interaction
Abstract
Bacillus subtilis competence-induced RecA, SsbA, SsbB, and DprA are required to internalize and to recombine single-stranded (ss) DNA with homologous resident duplex. RecA, in the ATP·Mg2+-bound form (RecA·ATP), can nucleate and form filament onto ssDNA but is inactive to catalyze DNA recombination. We report that SsbA or SsbB bound to ssDNA blocks the RecA filament formation and fails to activate recombination. DprA facilitates RecA filamentation; however, the filaments cannot engage in DNA recombination. When ssDNA was preincubated with SsbA, but not SsbB, DprA was able to activate DNA strand exchange dependent on RecA·ATP. This work demonstrates that RecA·ATP, in concert with SsbA and DprA, catalyzes DNA strand exchange, and SsbB is an accessory factor in the reaction. In contrast, RecA·dATP efficiently catalyzes strand exchange even in the absence of single-stranded binding proteins or DprA, and addition of the accessory factors marginally improved it. We proposed that the RecA-bound nucleotide (ATP and to a lesser extent dATP) might dictate the requirement for accessory factors.
Introduction
Horizontal gene transfer, or the rapid acquisition of genes, enables bacteria to gain genetic diversity that cannot be obtained through mutational processes alone (1). Virus-mediated transduction and natural transformation are the main routes of horizontal gene transfer of chromosomal DNA segments among bacteria (reviewed in Refs. 2–4). Natural transformation is a bacterium-encoded powerful mechanism for the acquisition of new genetic traits to adapt changing environmental conditions, to evade vaccines by switching their capsular serotype, and to acquire antibiotic resistance and virulence genes of both episomal and chromosomal origin, which are becoming the major threats to modern medicine (2–4). Genetic transformation has been documented in ∼85 species, with Bacillus subtilis, Streptococcus pneumoniae, Haemophilus influenzae, and Neisseria gonorrhoeae being the best characterized systems (2–4). The existence of this bacterium-programmed mechanism of genetic recombination suggests that natural transformation offered evolutionary advantages, a situation akin to what is believed for sexual recombination in eukaryotes (1, 3, 4).
During B. subtilis competence development, DNA replication and cell division are halted, the expression of RecA, DprA, SsbB, and SsbA, among many other genes, are induced, and the competence uptake machinery is built at one of the cell poles. The cytosolic RecA, DprA, and SsbB proteins, which physically interact with one another in vivo (5), transiently localize to the cell pole and co-localize with the uptake machinery (reviewed in Refs. 2–4). The localization of SsbA, which is an essential factor, remains elusive. The uptake machinery actively processes exogenous dsDNA, and it takes up and internalizes this DNA in its single-stranded form in a nonpolar fashion (2–4). This is consistent with the observations that end-processing functions (e.g. AddAB (counterpart of Escherichia coli RecBCD, RecBCDEco) and RecJ) play no role in the initial stages of chromosomal transformation and that DprA is the main RecA mediator (3, 4).
RecA, at the entry pole, with the help of accessory proteins, polymerizes on the internalized ssDNA as soon as it leaves the entry channel. Then RecA recombines the internalized ssDNA with a resident genome via homologous recombination (4). The accessory proteins that act before RecA nucleation are subdivided into two classes: SSB4 proteins (SsbA and SsbB in the vast majority of naturally transformable bacteria), which can limit or inhibit RecA activity, thereby suppressing unwanted RecA·ssDNA nucleoprotein filament (NPF) formation, and two different classes of mediators (RecO and DprA) (6, 7). This is consistent with genetic analysis showing that: (i) SsbA is an essential protein; (ii) the absence of SsbB, RecO, or DprA results in ∼5-, ∼3-, and ∼100-fold reduction in chromosomal transformation, respectively; and (iii) the absence of both RecO and DprA leads to a 103-fold reduction of these process (6, 7), suggesting that both RecO and DprA play a crucial role in natural chromosomal transformation. (Note that, unless stated otherwise, the indicated genes and products are of B. subtilis origin.5)
Previously, it has been postulated that RecA loading onto the internalized ssDNA is a multistep processes. First, the ssDNA is coated mostly either by DprA (8), by SsbB (9), or by SsbA as soon as it leaves the entry channel of the uptake apparatus (4). Second, SsbA loads RecO and SsbB loads DprA onto ssDNA (5, 10). Finally, the interaction of RecO or DprA with SsbA- or SsbB-coated ssDNA, respectively, recruits RecA onto the incoming ssDNA (6, 7). In vitro, however, RecA from Firmicutes competent bacteria (i.e. B. subtilis, S. pneumoniae) show a higher activity with dATP as the nucleotide co-factor. RecA·dATP·Mg2+ (referred to here as RecA·dATP) or RecASpn·dATP catalyzes DNA strand exchange even in the absence of accessory factors (11–14). The RecA·dATP transitional states resemble those described previously for RecAEco, designated as O, Ac, Ao, and P states (Fig. 1) (15–18). In short, RecA in the O state is largely inactive, but RecA·dATP induces a high affinity DNA binding state, which leads to cooperative polymerization of RecA·dATP onto SSB-coated ssDNA (Fig. 1) (6, 7). This RecA·dATP NPF facilitates the transition to the A state. In the presence of dsDNA, RecA·dATP is converted to the P state (Fig. 1), which is active to catalyze DNA strand exchange even in the absence of DprA and RecO (6, 7). RecA·dATP NPFs catalyze DNA pairing and DNA strand exchange with high efficiency, and the addition of DprA, SsbA, or SsbB marginally stimulates RecA-mediated DNA strand exchange reaction (<2-fold) when compared with RecA alone (6, 7). However, dATP might not be the primary in vivo nucleotide co-factor used by RecA. The ATP pool is 100–500-fold higher than that of dATP (Refs. 19 and 20 and our unpublished results), but the relative affinity of RecA for ATP or dATP is similar (21), suggesting that the catalytic site of RecA would contain ATP primarily. This premise creates a paradox: RecA·ATP can nucleate onto ssDNA, but it cannot catalyze DNA strand exchange (11, 12, 14). Furthermore, RecA·ATP neither nucleates nor polymerizes onto SsbA-coated ssDNA (14).
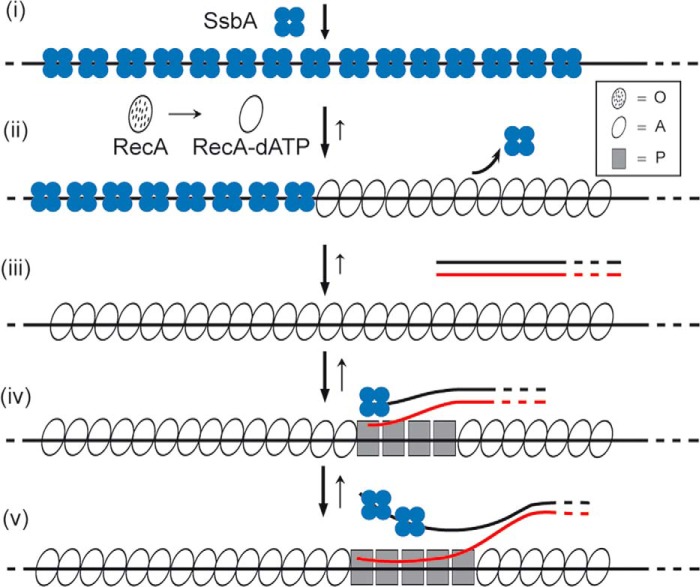
FIGURE 1.
Polymerization on DNA, pairing, and strand exchange mediated by RecA in the presence of dATP. The template for RecA·dATP·Mg2+ assembly in vivo is SsbB- or SsbA-coated ssDNA (step i). Based primarily on biochemical analyses, there are at least three significant functional stages of RecA. In the first state, RecA cannot bind ssDNA (apo RecA, dotted ovals, O state), but upon binding to dATP·Mg2+, it forms the active state denoted as RecA-dATP (empty oval, A state) that can nucleate on the SsbB- or SsbA-coated ssDNA (step ii). RecA-dATP upon nucleation undergoes a second functional transition, which allows efficient displacement of SsbA and polymerization on the ssDNA (step iii). RecA-dATP by binding to dsDNA acquires a third structural transition state (filled squares, P state) that can catalyze DNA pairing (step iv) and DNA strand exchange (step v). The overall reaction (denoted by an arrow) is reversible, and the length of the arrow denotes the preferred direction of the reaction.
To gain insights into the molecular basis of DNA strand pairing and exchange mediated by RecA·ATP, the conditions that help overcome inhibition by either of the SSB proteins were investigated. We have shown that RecA·ATP cannot compete with sub- or saturating concentrations of SSB proteins. RecA·ATP nucleates and polymerizes on the DprA·ssDNA complex more efficiently than on protein-free ssDNA (see Fig. 3). These RecA NPFs, which are active for DNA pairing, are unable to catalyze DNA strand exchange. We show that RecA·ATP polymerizes on the DprA·ssDNA·SsbA complex with similar efficiency than on the DprA·ssDNA·SsbB complex, but only the former NPFs are proficient in the catalysis of DNA pairing and strand exchange. We propose that RecA·ATP, in concert with DprA and SsbA, should undergo different transition states to ensure that genetic recombination occurs when all partners are available.
FIGURE 3.
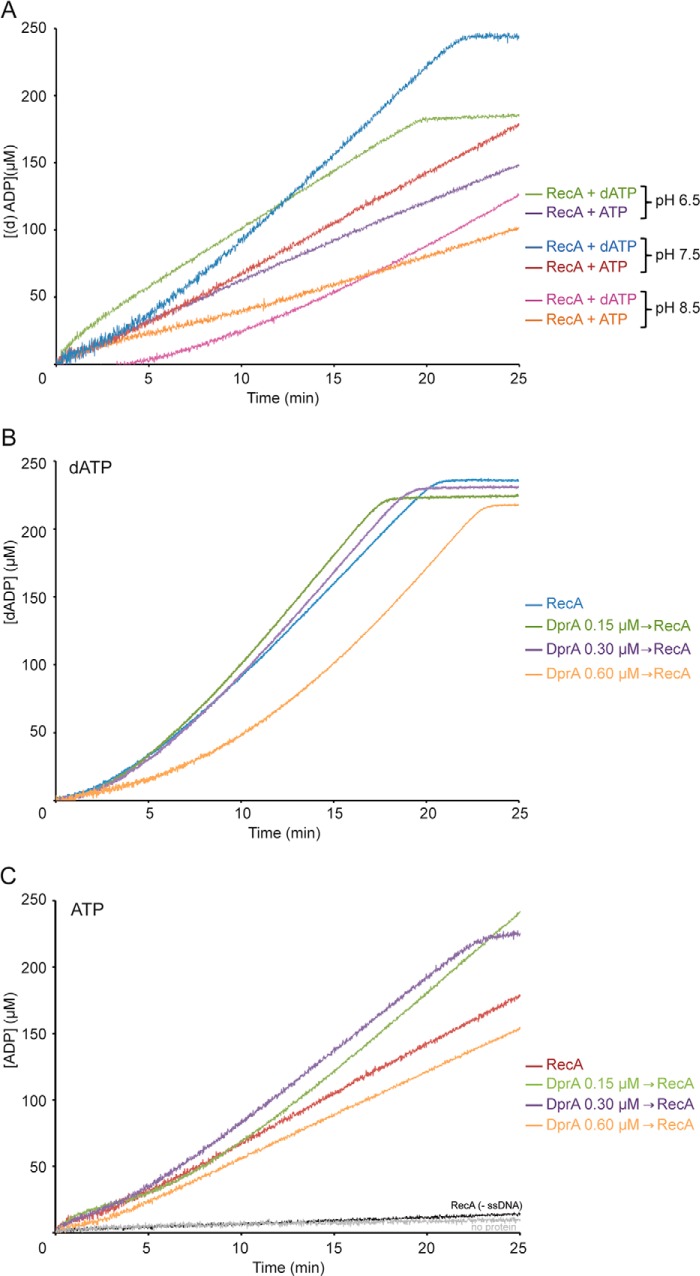
RecA preferentially hydrolyzes dATP. A, circular 3,199-nt ssDNA (10 μm in nt) was incubated with RecA (0.8 μm) in buffer B, A, or C (pH 6.5, 7.5, or 8.5) containing 5 mm ATP or dATP. Then the ssDNA-dependent (d)ATPase activity was measured for 25 min. B and C, circular ssDNA was preincubated with DprA (0.15, 0.3, or 0.6 μm) for 5 min, and RecA was then added in buffer A containing 5 mm dATP (B) or ATP (C) at 37 °C. The ssDNA-dependent dATPase or ATPase ([d]ATPase) activity was measured for 25 min. The reaction was carried out under standard conditions, and the amount of ATP or dATP hydrolyzed was calculated as described under “Experimental Procedures.” All reactions were repeated three or more times with similar results.
EXPERIMENTAL PROCEDURES
Enzymes, Reagents, DNA, and Protein Purification
All chemicals used in this study were of analytical grade. Isopropyl β-d-thiogalactopyranoside was from Calbiochem, and polyethyleneimine, DTT, ATP, and dATP were from Sigma. DNA restriction enzymes were supplied by Fermentas. DEAE, Q- and SP-Sepharose, Sephadex G-100, and Superose 12 were from GE Healthcare, and phosphocellulose was from Whatman. Plasmid pGEM3Zf(+) and pGEM3Zf(+) ssDNA (Promega) were purified and used for RecA-mediated DNA strand exchange studies as described (14).
E. coli BL21(DE3)[pLysS] cells bearing pCB722 ssbA, pCB777 ssbB, or pCB888 dprA under the control of a phage T7 promoter were used to overexpress SsbA, SsbB, and DprA proteins, respectively, as described (7, 14, 22). B. subtilis BG214 cells bearing pBT61, containing recA under the control of its own promoter, were used to overexpress RecA (23). SsbA (18.7 kDa), SsbB (12.4 kDa), DprA (32.7 kDa), and RecA (38.0 kDa) proteins were purified as described (7, 14, 23). All proteins were purified to 98% homogeneity (Fig. 2). The molar extinction coefficients for SsbA, SsbB, RecA, and DprA were calculated as 11,400, 13,000, 15,200, and 45,000 m−1 cm−1, respectively, at 280 nm, as previously described (23). The protein concentrations were determined using the aforementioned molar extinction coefficients. RecA and DprA are expressed as moles of monomers and SsbA and SsbB as tetramers. In the text, the protein concentrations are expressed in their ratios with ssDNA, which was expressed as moles of nt, whereas in the figure legends the molar concentrations of proteins and ssDNA are presented.
FIGURE 2.
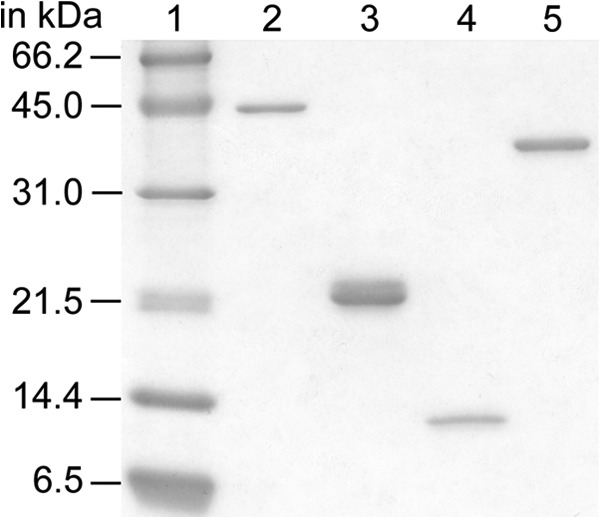
Purified B. subtilis DNA recombination proteins. Purified RecA (38.0 kDa, lane 2), SsbA (18.7 kDa, lane 3), SsbB (12.4 kDa, lane 4), and DprA (32.7 kDa, lane 5) are shown. Approximately 400 ng of each purified protein was fractionated on a 17% SDS-PAGE and stained with Coomassie Brilliant Blue. The molecular masses (in kDa) of the control proteins (lane 1) are indicated.
RecA (d)ATP Hydrolysis Assays
The ssDNA-dependent dATP or ATP (denoted as (d)ATP) hydrolysis activity of RecA protein was performed as described (6, 24). RecA-mediated (d)ATP hydrolysis was not affected within the first 180 min of incubation, even upon binding a silicon nanowire transistor (25). Absorbance measurements were taken with a Shimadzu CPS-240A dual beam spectrophotometer equipped with a temperature controller (6). Rates of ssDNA-dependent RecA-mediated (d)ATP hydrolysis and the lag times were measured in Buffer B, A, or C (50 mm Tris-HCl, pH 6.5, 7.5, or 8.5, respectively), 1 mm DTT, 80 mm NaCl, 10 mm MgOAc, 50 μg/ml BSA, 5% glycerol) containing 5 mm (d)ATP for varying times at 37 °C in a 50-μl reaction volume. The orders of addition of 3,197-nt pGEM3Zf(+) ssDNA (10 μm), the purified proteins, and their concentrations are indicated in the text. A (d)ATP regeneration system (0.5 mm phosphoenolpyruvate, 10 units/ml pyruvate kinase) and a coupling system (0.25 mm NADH, 10 units/ml lactate dehydrogenase) were also included (6).
To determine the lag time of RecA nucleation, we have measured the steady state rate of RecA-mediated (d)ATP hydrolysis and the time observed in achieving this rate. As previously reported (24, 26), the lag time, which represents the delay in reaction progress relative to a theoretically reaction curve lacking it, was derived from the time intercept of a linear regression line fit to the steady state portion of data in (d)ATP hydrolysis assays.
The amount of hydrolyzed (d)ADP was calculated as described previously (6). In the absence of RecA, neither DprA (1 DprA/11 nt) nor SsbA (1 SsbA/33 nt) or SsbB (1 SsbB/33 nt) exhibited (d)ATP hydrolysis activity when compared with the mock reaction (data not shown), indicating that the hydrolysis of (d)ATP observed in the assays can be solely attributed to the presence of RecA.
RecA-mediated (d)ATP-dependent DNA Strand Exchange
Under physiological Mg2+ concentrations (< 2 mm), RecA protein is inactive for recombination activities (14), and the SSB proteins occlude 35-nt (SSB35 binding mode). However, under optimal RecA conditions (10 mm Mg2+), the SSB proteins occlude 65-nt, with the ssDNA wrapping around all four subunits of the tetramer (SSB65) (27, 28). In this study, the experiments were performed under optimal RecA conditions (10 mm MgOAc); hence the SSB proteins were expected to be in the SSB65 binding mode (see above), a DprA dimer should bind 30–40 nt (7), and a RecA monomer should bind 3 nt (29).
The 3,197-bp KpnI-cleaved pGEM3Zf(+) dsDNA (20 μm) and homologous circular 3,197-nt ssDNA (10 μm) were incubated with the indicated concentrations of protein or protein combinations in Buffer A containing 2 mm (d)ATP for 60 min at 37 °C in a final volume of 20 μl. A (d)ATP regeneration system (8 units/ml creatine phosphokinase and 8 mm phosphocreatine) was included. The samples were deproteinized as described (22, 30) and separated by 0.8% agarose gel electrophoresis with ethidium bromide staining. The signal was quantified using a Gel Doc (Bio-Rad) system as described (10).
RESULTS
Examination of Nucleotide Hydrolysis Conditions by RecA
RecA hydrolyzes dATP with higher efficiency than ATP when bound to ssDNA (10, 11, 14, 21). To investigate whether changes in the reaction condition can overcome the barrier of the RecA ATPase, the rate of nucleotide hydrolysis was measured at different pH. RecA-mediated dATP hydrolysis was also measured as a control.
RecA-mediated (d)ATP hydrolysis was used as an indirect inference of nucleation and polymerization of RecA on the ssDNA, although ssDNA-dependent RecA-mediated nucleotide hydrolysis cannot reliably distinguish between nucleation and filament growth. Hence, we deduced the time of nucleation by extrapolation as described under “Experimental Procedures.” The intracellular pH is maintained in the range of 7.4–7.8, and RecA has an isoelectric point of ∼5.0, which suggests that the protein will have a net negative charge at physiological pH.
RecA-mediated dATP hydrolysis was slow and preceding establishment of the maximal hydrolysis rate at pH 7.5 (Fig. 3A). We have measured the steady state rate of RecA-mediated dATP hydrolysis and the lag time observed in achieving this rate. Under the conditions used, RecA hydrolyzed dATP at near the previously observed kcat of 18.2 ± 0.4 min−1 (1 RecA monomer/12 nt) (Fig. 3A and Table 1) (6, 7, 10, 21) with a delay to reach it of 4 ± 0.5 min (Fig. 3A and Table 1) (6, 7, 10). We interpreted the data to mean that the rate of RecA nucleation and subsequent filament formation on protein-free ssDNA was biphasic. Nucleation took ∼4 min, and the rate of dATP hydrolysis represents the equilibrium state of end-dependent filament assembly and disassembly, taken as filament growth (18, 31, 32). The loading of RecAEco onto ssDNA is also slow, typically proceeding with a lag of 15 min or more (24).
TABLE 1.
Rates of ssDNA-dependent ATP hydrolysis and lag time measurements
N.D., not detected; —, not done in the previous work.
| Proteinsa | Condition | Lag time | kcat |
|---|---|---|---|
| min | min−1 | ||
| RecA (1 RecA/12 nt) | dATP | 4 ± 0.5 | 18.2 ± 0.4 |
| ATP | <1 | 9.0 ± 0.3 | |
| RecA + DprA (1 DprA/66 nt) | dATP | 4 ± 0.4 | 17.4 ± 0.2 |
| ATP | 4 ± 0.3 | 14.2 ± 0.3 | |
| RecA + DprA (1 DprA/33 nt) | dATP | 4 ± 0.4 | 17.3 ± 0.3 |
| ATP | 2 ± 0.4 | 17.7 ± 0.5 | |
| RecA + DprA (1 DprA/16 nt) | dATP | 8 ± 0.6 | 17.1 ± 0.5 |
| ATP | 3 ± 0.4 | 7.9 ± 0.5 | |
| RecA + SsbA (1 SsbA/125 nt) | dATPb | — | — |
| ATP | <1 | 4.1 ± 0.2 | |
| RecA + SsbA (1 SsbA/90 nt) | dATPb | — | — |
| ATP | <1 | 2.0 ± 0.2 | |
| RecA + SsbA (1 SsbA/33 nt) | dATPb | >9 | 13.1 ± 0.4 |
| ATP | N.D. | 0.8–1.5 | |
| RecA + SsbB (1 SsbB/125 nt) | dATPb | — | — |
| ATP | 4 ± 0.3 | 5.7 ± 0.3 | |
| RecA + SsbB (1 SsbB/90 nt) | dATPb | — | — |
| ATP | 6.5 ± 0.2 | 2.9 ± 0.5 | |
| RecA + SsbB (1 SsbB/33 nt) | dATPb | ∼8 | 17.6 ± 0.5 |
| ATP | N.D. | 1.0–2.0 | |
| RecA + SsbA + DprA | dATPb | ∼5 ± 1 | 25.0 ± 0.5 |
| (1 SsbA or DprA/33 nt) | ATP | 3 ± 0.3 | 18.1 ± 0.2 |
| RecA + SsbB + DprA (1 SsbB or DprA/33 nt) | dATPb | ∼1 | 25–30 |
| ATP | 3 ± 0.3 | 18.6 ± 0.3 | |
| RecA (pH 6.5)c (1 RecA/12 nt) | dATP | <1 | 11.2 ± 0.2 |
| ATP | <1 | 7.5 ± 0.2 | |
| RecA (pH 8.5) (1 RecA/12 nt)d | dATP | 7.5 ± 0.2 | 8.0 ± 0.2 |
| ATP | <1 | 5.0 ± 0.3 | |
| dATP 5:0 | 3.5 | 18.5 | |
| RecA (1 RecA/12 nt)e | dATP:ATP 4:1 | 1.7 | 15.9 |
| dATP:ATP 1:1 | 1.9 | 14.3 | |
| dATP:ATP 1:4 | 1.0 | 12.0 | |
| ATP 0:5 | <1 | 8.9 |
a Rates of RecA-mediated (d)ATP hydrolysis and the nucleation lag times were measured at pH 7.5 as indicated under “Experimental Procedures.” The steady state kinetic parameters for RecA (1 RecA/12 nt) were derived from the data presented in Figs. 3–5 and 8.
b The data of RecA-mediated dATP hydrolysis are from (7).
c RecA-mediated (d)ATP hydrolysis and nucleation lag time at pH 6.5.
d RecA-mediated (d)ATP hydrolysis and nucleation lag time at pH 8.5.
e RecA-mediated (d)ATP hydrolysis and nucleation lag time at different dATP:ATP ratios.
At pH 6.5, RecA nucleation onto ssDNA did not show a lag phase, but the final rate of RecA-mediated dATP hydrolysis was significantly decreased (1.6-fold) when compared with pH 7.5 (Fig. 3A and Table 1). At pH 8.5, RecA nucleation in the presence of dATP was significantly delayed when compared with pH 7.5 (1.7-fold), and the rate of RecA-mediated dATP hydrolysis was further reduced (2.2-fold) (Fig. 3A and Table 1).
RecA-mediated ATP hydrolysis increased without an apparent lag and at nearly the previously observed kcat of 9.0 ± 0.3 min−1 (1 RecA monomer/12 nt) (Fig. 3A and Table 1) at pH 7.5 (14). The final rate of RecA-mediated ATP hydrolysis marginally decreased at pH 6.5 (1.2-fold) and decreased 1.8-fold at pH 8.5 (Fig. 3A and Table 1). Therefore, the optimal rate of ATP or dATP hydrolysis occurs at physiological pH.
Because the RecA filaments formed on ssDNA in the presence of dATP or ATP (23) were indistinguishable from those formed by RecAEco (reviewed in Refs. 18 and 32), we have assumed that the slow rate of RecA·ATP polymerization on the ssDNA, when compared with RecA·dATP, is either limited by secondary structures or RecA is partially inactive to form NPFs. If the latter hypothesis is correct, a mediator should be needed to recruit RecA·ATP to ssDNA.
DprA Stimulates RecA·ATP Assembly on ssDNA
To investigate whether the DprA mediator can facilitate assembly of the RecA NPFs, we measured ssDNA-dependent ATP hydrolysis by RecA. Titration of DprA was carried out to define optimal reaction conditions in the possible loading of RecA·ATP onto protein-free ssDNA.
In the presence of dATP, addition of low DprA concentrations (1 DprA/66 nt) did not significantly facilitate RecA nucleation onto ssDNA when compared with RecA alone (Fig. 3B and Table 1). The steady state rate of RecA-mediated dATP hydrolysis, in the presence of DprA, was comparable with RecA alone (Fig. 3B and Table 1), suggesting that efficient nucleation and polymerization of RecA·dATP is not significantly improved by addition of the DprA mediator. Similar results were previously described (7).
When ATP was provided in place of dATP as the nucleotide co-factor, biphasic curves of ATP hydrolysis were observed with the addition of sub- and stoichiometric DprA concentrations (1 DprA/66 to 33 nt) (Fig. 3C). At a low ratio (1 DprA/66 nt), a slow nucleation preceding establishment of the maximal hydrolysis rate was observed (Fig. 3C and Table 1). At stoichiometric concentrations with ssDNA, DprA (1 DprA/33 nt) reduced the lag phase of RecA nucleation and significantly increased the final rate of ATP hydrolysis (Fig. 3C and Table 1). In other words, in the presence of stoichiometric DprA concentrations, RecA·ATP nucleates and polymerizes on the ssDNA with similar efficiency as RecA·dATP in the absence of any accessory factor (Fig. 3, B and C, and Table 1). Because RecA and DprA physically interact (5, 7), it is likely that such interaction contributes to the loading of RecA·ATP onto ssDNA, and RecA might undergo a functional transition after interacting with ATP and DprA. Alternatively, DprA, which preferentially binds ssDNA, might stimulate RecA-mediated (d)ATP hydrolysis by removing DNA secondary structure. We consider this hypothesis unlikely because DprA forms one of few discrete blobs per ssDNA molecule (7).
Independent of the nucleotide co-factor used, when the DprA concentration approached parity with RecA (1 DprA/16 nt and 1 RecA/12 nt), the lag time was significantly increased, but the final rate of RecA (d)ATP hydrolysis did not significantly change when compared with RecA alone (Fig. 3, A and B, and Table 1). As previously proposed for RecA·dATP (7), DprA upon interacting with RecA (5, 7) might form a transient inactive binary complex that sequestered RecA from ssDNA.
SsbA or SsbB Inhibits RecA-mediated ATP Hydrolysis
When ssDNA was preincubated with stoichiometric or substoichiometric amounts of SsbB or SsbA (1 SSB tetramer/33 or 66 nt), the final rate of RecA-mediated ATP hydrolysis was ∼6- and ∼8-fold lower, respectively, than observed for RecA alone (Fig. 4, A and B, and Table 1).
FIGURE 4.
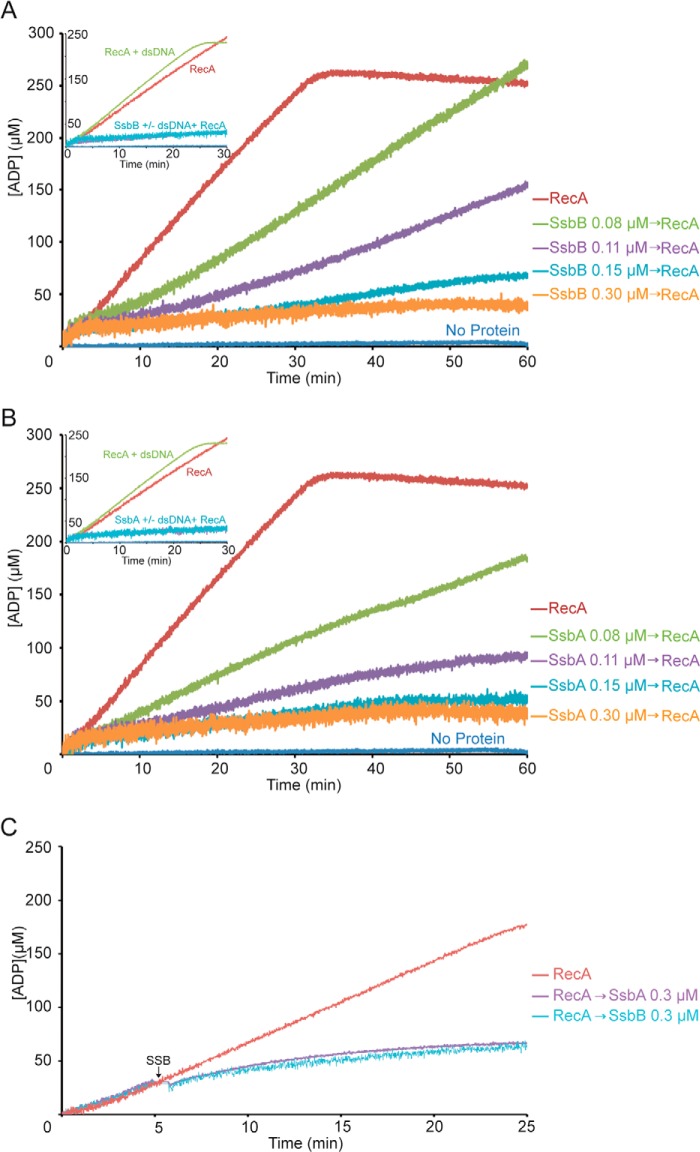
Stoichiometric amounts of SsbA or SsbB inhibit RecA-mediated ATP hydrolysis. A and B, circular ssDNA (10 μm in nt) was preincubated with increasing SsbB (A) or SsbA (B) concentrations (0.08, 0.11, 0.15, and 0.3 μm) for 5 min at 37 °C in buffer A containing 5 mm ATP. Then RecA (0.8 μm) was added, and the ATPase activity was measured for 60 min. Insets, circular ssDNA (10 μm in nt) was preincubated with stoichiometric amounts of SsbB or SsbA (0.3 μm) for 5 min at 37 °C in buffer A containing 5 mm ATP. Then RecA (0.8 μm) and dsDNA (20 μm in nt) was added, and the ATPase activity was measured for 25 min. C, effect of SsbA or SsbB on RecA filament extension in the presence of ATP. Circular ssDNA (10 μm in nt) was incubated with RecA (0.8 μm) at 37 °C in buffer A containing 5 mm ATP, and the ATPase activity was measured (time 0). Five min later, stoichiometric amounts of SsbB or SsbA (0.3 μm) were added, and the ssDNA-dependent ATPase activity was further measured for 20 min. All reactions were repeated three or more times with similar results. The amount of nucleotide hydrolyzed was calculated as described in the legend to Fig. 3.
To test whether RecA can compete with the SSB protein for ssDNA binding, the rate of RecA-mediated ATP hydrolysis was measured in the presence of limiting SsbB or SsbA concentrations. When ssDNA was preincubated with limiting amounts of SSB (1 SsbB/SsbA 90 to 125 nt), the rate of RecA-mediated ATP hydrolysis significantly decreased (Fig. 4, A and B, and Table 1). It is likely that: (i) RecA·ATP cannot compete with SsbB or SsbA (1 SSB for every 2.6 RecA molecules) for nucleation onto ssDNA, and (ii) RecA·ATP can partially displace limiting SsbB (1 SSB for every 10 or 7 RecA molecules) at longer incubation times. Alternatively, RecA·ATP needs to be activated, for example, by interacting with dsDNA. To test this, a stoichiometric amount of SsbA or SsbB was preincubated with ssDNA, and then the rate of RecA-mediated hydrolysis of ATP in the presence of dsDNA was measured. The presence of dsDNA did not alter the rate of RecA-mediated ATP hydrolysis (Fig. 4, A and B, insets).
To confirm whether nucleated RecA can displace SsbB or SsbA from the ssDNA, RecA was preincubated with ssDNA, ATP hydrolysis was measured, and then 5 min later SsbA or SsbB (1 SSB/33 nt) was added (Fig. 4C). RecA hydrolyzed ATP in the presence of protein-free ssDNA at the expected rate (Figs. 3 and 4, A and B) during the first 5 min. However, RecA-mediated hydrolysis of ATP plunged 6–9 min after SSB addition (Fig. 4C), suggesting that RecA cannot displace SsbA or SsbB.
DprA Overcomes the Inhibition Exerted by SSB Proteins on RecA-mediated ATP Hydrolysis
To learn whether DprA can overcome the inhibition exerted by SsbA or SsbB on RecA-mediated ATP hydrolysis, SsbA or SsbB was preincubated with ssDNA, DprA was added, and ssDNA-dependent ATP hydrolysis by RecA was measured. RecA·dATP nucleation and filament growth onto protein-free ssDNA was run as a control (Fig. 5, A and B).
FIGURE 5.
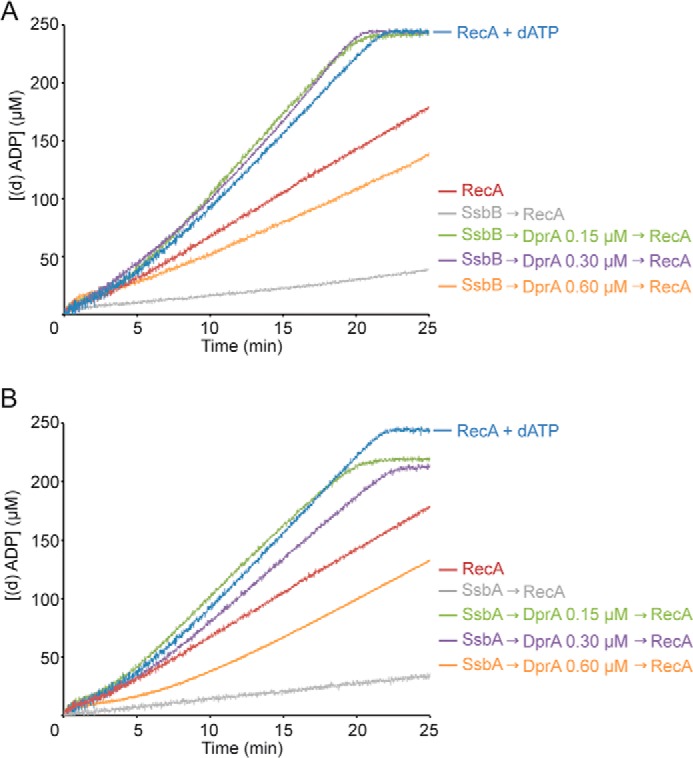
DprA overcomes the SsbA or SsbB inhibition for RecA-mediated ATP hydrolysis. A, circular ssDNA (10 μm in nt) was preincubated with SsbB (0.3 μm) for 5 min, and DprA (0.15, 0.3, or 0.6 μm) was added for 5 min at 37 °C in buffer A containing 5 mm ATP or dATP. Then RecA (0.8 μm) was added, and the ATPase activity was measured for 25 min. The activity of RecA in the presence of dATP was measured as control. B, circular ssDNA was preincubated with SsbA (0.3 μm) for 5 min, and DprA (0.15, 0.3, or 0.6 μm) was added for 5 min at 37 °C in buffer A containing 5 mm ATP or dATP. Then RecA (0.8 μm) was added, and the ATPase activity was measured for 25 min. The activity of RecA in the presence of dATP was measured as control. All reactions were repeated three or more times with similar results. The amount of nucleotide hydrolyzed was calculated as described in the legend to Fig. 3.
DprA overcame the SSB kinetic blockage for RecA nucleation and filament formation (Fig. 5, A and B, and Table 1). In the presence of sub- or stoichiometric amounts of DprA (1 DprA/66 to 33 nt), RecA nucleation and subsequent filament formation onto SsbB- or SsbA-coated ssDNA was biphasic, with a slow nucleation step preceding establishment of the maximal hydrolysis rate of comparable with that of RecA·dATP (Fig. 5, A and B, and Table 1). At a SSB:DprA ratios of 1, RecA nucleation and subsequent filament formation onto SsbB- or SsbA-coated ssDNA was comparable with levels of RecA·dATP in the absence of any accessory factor (Fig. 5, A and B, and Table 1). Similar results were observed at SSB:DprA ratios of 2:1 (Table 1). At a higher DprA concentration (1 DprA/16 nt), the interaction between DprA and RecA might lead to inactive binary complex (see above).
SsbA- or SsbB-bound ssDNA Cannot Reverse the Inability of RecA·ATP to Catalyze DNA Recombination
RecA from naturally transforming bacteria (e.g. RecA, RecASpn, or RecADra) cannot catalyze DNA strand exchange in the absence of accessory proteins (11–14, 33). A DNA strand exchange assay that uses circular ssDNA and linear duplex DNA was employed to investigate whether SsbA or SsbB helps overcome the inability of RecA·ATP to catalyze DNA recombination.
The 3,197-bp linear dsDNA (lds, 20 μm in nt) and homologous circular 3,197-nt ssDNA (css, 10 μm in nt) were incubated with RecA in buffer containing 2 mm (d)ATP for 60 min at 37 °C. RecA initiates DNA recombination by pairing the lds with the complementary css, leading to the formation of a joint molecule (jm) between the free end of lds and circular css, followed by extensive DNA strand exchange to generate the nicked circular duplex (nc) and the linear ssDNA (lss) products (Fig. 6A). In this reaction, unless the displaced lss product is sequestered by SSB, it can be further engaged in a reverse reaction with the nc product that yields css or lds as either products or a network of interlinked intermediates (reviewed in Ref. 34). This explains why css and lds are always present when reactions are analyzed.
FIGURE 6.
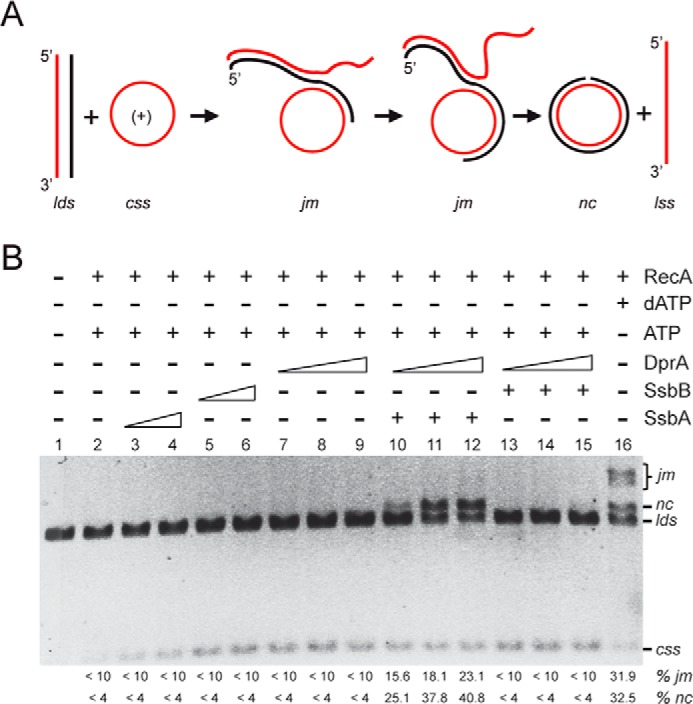
SsbA and DprA contribute to RecA-promoted DNA strand exchange in the presence of ATP. A, scheme of the three-strand exchange reaction between circular ssDNA (css, in red) and the linear duplex (lds, in black and red) substrate and the expected intermediates (jm) and final products (nc and lss) by RecA-mediated DNA strand exchange. B, circular ssDNA (10 μm in nt) and homologous linear dsDNA (20 μm in nt) were preincubated with increasing concentrations of SsbA, SsbB (0.15 and 0.3 μm), or DprA (0.025, 0.05, and 0.1 μm) for 10 min at 37 °C in buffer A containing 2 mm ATP or dATP, and then DprA was added to the preformed SsbA·ssDNA or SsbB·ssDNA (SSB 0.3 μm) complexes and incubated for 10 min at 37 °C. Finally, RecA (0.8 μm) was added, and the reaction was incubated for 60 min at 37 °C. The products of the reactions were deproteinized, separated, and quantified as described under “Experimental Procedures.” The positions of the bands corresponding to css, cds, lds, nc, and jm are indicated. + and − denote the presence and absence, respectively, of the indicated protein. The amount of recombination intermediates (jm) and products (nc) are indicated as percentages and are the average values obtained from more than three independent experiments (the results given stand within a 5% standard error).
To follow the strand exchange process, the disappearance of the lds and the appearance of jm intermediates and nc products were monitored by agarose gel electrophoresis and ethidium bromide staining upon deproteinization of the reaction (Fig. 6B). In the presence of dATP and a subsaturating amount of RecA (1 RecA/12 nt), ∼64% of the lds substrate was converted into either jm (∼32%) or the final product nc (∼32%) in 60 min (Fig. 6B, lane 16). The addition of SsbA or SsbB slightly stimulated the reaction (6, 7, 10).
When ATP was used instead, RecA was not able to catalyze much DNA strand exchange. RecA·ATP rendered low levels of DNA pairing intermediates (5–9% of lds being converted onto jm, denoted as <10%) and only a trace of the final nc product (<4%) (Fig. 6B, lane 2). An increase in the RecA concentration (1 RecA/6 to 3 nt) did not significantly increase the yield of jm or nc (Ref. 14 and our unpublished results). We surmised that recombination occurs under these circumstances, but the loading of RecA·ATP onto the displaced ssDNA strand prompts the invasion of the nascent DNA joint to result in the reversal of the recombination reaction (Fig. 6A). To test this hypothesis and also to gain insights about the inhibition of ATP hydrolysis by SsbA and SsbB, the ssDNA was preincubated with either SSB protein. When SsbA or SsbB (1 SSB/66 to 33 nt) was preincubated with ssDNA, RecA exhibited low levels of ATP-dependent DNA pairing, and only a trace level of the final nc product was produced (Fig. 6B, lanes 3–6). It is likely that the low yield of nc products is due to the low efficiency of RecA·ATP NPF formation (Fig. 4) rather than to the presence of DNA secondary structures.
DprA Cannot Overcome the Inability of RecA·ATP to Catalyze DNA Strand Exchange
RecA·ATP can nucleate and polymerize on the DprA·ssDNA complex more efficiently than on protein-free ssDNA (Fig. 3B). Then it was asked whether DprA helps to overcome the inability of RecA·ATP to catalyze the three-stranded DNA exchange reaction (Fig. 6B). The addition of DprA (1 DprA/400 to 100 nt) increased neither the efficiency of DNA pairing nor DNA strand exchange when compared with RecA·ATP alone (Fig. 6B, lanes 2 and 7–9).
DprA and SsbA Facilitate RecA-mediated DNA Strand Exchange in the Presence of ATP
In preceding sections, by measuring the efficiencies of ATP hydrolysis and DNA strand exchange, we were able to deduce that: (i) RecA·ATP can nucleate and polymerize on DprA·ssDNA, DprA·ssDNA·SsbA or DprA·ssDNA·SsbB complexes with a similar efficiency as RecA·dATP on protein-free ssDNA (Figs. 3 and 5), and (ii) RecA·ATP cannot efficiently catalyze DNA strand exchange in the presence of SsbA, SsbB or DprA (Fig. 6B, lanes 3–9). The requirement of SsbA or SsbB on RecA-mediated recombination in the presence of DprA was then tested.
When the ssDNA was preincubated with SsbA (1 SsbA/33 nt) and two different amounts of DprA (1 DprA/100 nt), ∼23 and ∼40% of the lds substrate was converted into the jm and nc product, respectively (Fig. 6B, lane 12). With a lower DprA amount (1 DprA/400 nt), ∼15 and ∼25% of the lds substrate was converted into the jm and nc product, respectively (Fig. 6B, lanes 10), and intermediate effect was observed in the presence of 1 DprA/200-nt (Fig. 6B, lanes 11).
When SsbB was used instead of SsbA, however, DprA, at different concentrations, did not enable RecA to catalyze DNA strand exchange (Fig. 6B, lanes 13–15). The addition of various DprA concentrations only enabled RecA to catalyze ATP-dependent DNA pairing up to the levels of RecA alone (Fig. 6B, lanes 5–9 and 13–15). This outcome was unexpected because it was reported that: (i) in vivo SsbB interacts with both DprA and RecA (5), and (ii) RecA·dATP can nucleate on the DprA·ssDNA·SsbB complexes more efficiently than on ssDNA·SsbB complexes (7). We hypothesized that the RecA·ATP cannot be converted to a P-like “active” state in the presence of SsbB and DprA (Fig. 1), and a functional interaction with SsbA and DprA is necessary and sufficient for RecA·ATP activation. If this hypothesis is correct, then limiting SsbA concentrations should overcome the inability of RecA to catalyze DNA strand exchange in the presence of SsbB and DprA. This idea was tested in experiments described in the next section.
Limiting SsbA Facilitates RecA-mediated DNA Strand Exchange in the Presence of SsbB and DprA
To test whether limiting SsbA reverses the inactive state of the RecA·ATP NPF formed in the presence of DprA and SsbB, DNA strand exchange assays were performed. Different amounts of SsbA (1 SsbA/200, 100, 50, and 40 nt) and SsbB (1 SSbA/40, 50, 60, and 200 nt) were mixed so that the final stoichiometry of SSB:ssDNA remained constant, and the SSB mixtures were preincubated with ssDNA, followed by addition of a fixed amount of DprA (1 DprA/100 nt) to the preformed SsbA·ssDNA·SsbB complexes. Finally, RecA (1 RecA/10 nt) was added, and ATP-dependent RecA-mediated DNA strand exchange was analyzed (Fig. 7).
FIGURE 7.
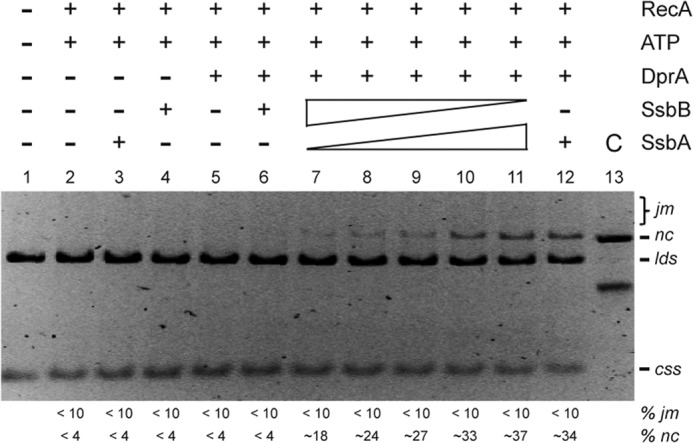
Limiting SsbA in concert with SsbB and DprA contributes to RecA-promoted DNA strand exchange in the presence of ATP. Circular ssDNA (css; 10 μm in nt) and homologous linear dsDNA (lds; 20 μm in nt) were preincubated with increasing concentrations of SsbA (0.05, 0.1, 0.15, 0.2, and 0.25 mm) and then with decreasing SsbB (0.25, 0.2, 0.15, 0.1, and 0.05 μm) for 10 min at 37 °C in buffer A containing 2 mm ATP. Then DprA (0.1 μm) was added to the preformed SsbA·ssDNA, SsbB·ssDNA, or the SsbA·ssDNA·SsbB complex (SSB 0.3 μm) and incubated for 10 min at 37 °C. Finally, RecA (0.8 μm) was added, and the reaction was incubated for 60 min at 37 °C. The separation of the products and the symbols are those described in the legend to Fig. 6. The amounts of recombination intermediates (jm) and products (nc) are indicated as percentages and are the average values obtained from more than three independent experiments (the results given stand within a 5% standard error).
In the presence of limiting SsbA concentrations (1 SsbA/200 nt), DprA, and SsbB, the accumulation of the final nc product (∼18%) was significantly increased when compared with the absence of SsbA or when SsbB was omitted (<4% nc) (Fig. 7, compare lane 5 with lane 7). Moreover, the rate and extent of DNA strand exchange increased with the SsbA concentration in the reaction (Fig. 7, lanes 7–11).
Limiting dATP Concentrations Cannot Facilitate RecA·ATP-mediated DNA Strand Exchange in the Absence of Accessory Factors
RecA·dATP efficiently polymerizes onto ssDNA, and in the presence of homologous duplex DNA, it catalyzes DNA strand exchange even in the absence of accessory factors (Figs. 6, lane 16, and 7, lane 2). Conversely, RecA·ATP polymerizes onto ssDNA with lower efficiently than RecA·dATP (Table 1 and Fig. 8A), and it cannot catalyze DNA strand exchange in the absence of DprA and SsbA (Figs. 6, lanes 10–12, and 8B, lane 5). We believe that RecA bound to dATP or ATP is found in the active (Fig. 1) or inactive states for DNA strand exchange, respectively. Because the affinity of RecA for both dATP and ATP is similar, but the dATP pool in the cytosol is 100–500-fold lower than that of ATP, the nucleotide bound to RecA molecules in NPFs will be mostly ATP, but a few RecA molecules will be bound to dATP. We hypothesize that those few RecA·dATP molecules activate RecA·ATP NPFs and facilitate DNA strand exchange. To address this hypothesis, we tested RecA filamentation onto ssDNA and DNA strand exchange (Fig. 8, A and B). The final rate of RecA (d)ATP hydrolysis increased when the dATP:ATP ratio increased (Table 1 and Fig. 8A).
FIGURE 8.
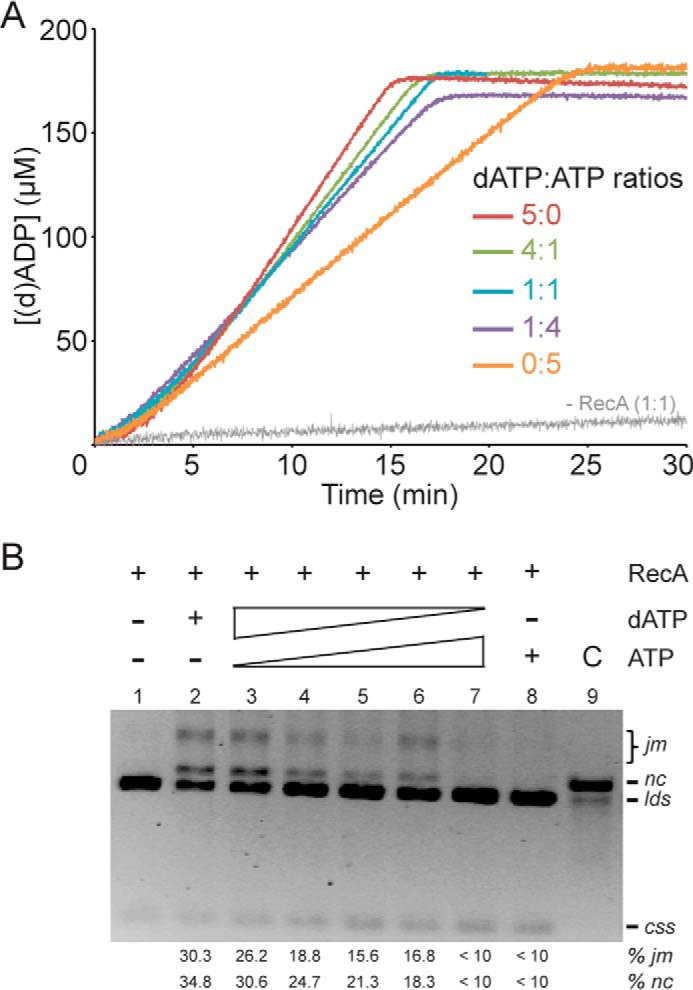
Limiting dATP concentrations facilitate RecA·ATP NPF formation and DNA strand exchange. A, circular 3,199-nt ssDNA (10 μm in nt) was incubated with RecA (0.8 μm) in buffer A containing a total of 5 mm of dATP/ATP at different ratios (5:0, 4:1, 1:1, 1:4, or 0:5) at 37 °C, and the ssDNA-dependent (d)ATPase activity was measured for 30 min. The amount of nucleotide hydrolyzed was calculated as described in the legend to Fig. 3. B, circular ssDNA (10 μm in nt) and homologous linear dsDNA (20 μm in nt) were incubated with RecA (0.8 μm) for 60 min at 37 °C in buffer A containing a total of 5 mm of dATP/ATP at different ratios (5:0, 4:1, 3:2, 1:1, 2:3, 1:4, or 0:5). The separation of the products and the symbols are those described in the legend to Fig. 6. The amount of recombination intermediates (jm) and products (nc) are indicated as percentages and are the average values obtained from more than three independent experiments (the results given stand within a 5% standard error).
RecA·ATP NPFs catalyze DNA pairing with low efficiency, but it cannot catalyze DNA strand exchange (Figs. 6B, lane 16, and 8B, lane 8). This effect, however, was reversed by increasing the dATP:ATP ratios (Fig. 8B, lanes 2–6). These results suggest that: (i) mixed RecA-dATP/RecA·ATP filaments might be formed; (ii) RecA catalyzes DNA strand exchange at equimolar dATP:ATP ratios; and (iii) limiting dATP concentrations (1:4 dATP:ATP ratios) are not sufficient to activate RecA·ATP to catalyze DNA strand exchange in the absence of accessory factors.
DISCUSSION
Here we have reported a quantitative description of the effect of ATP on RecA nucleation and filament extension on ssDNA and also the RecA-mediated DNA strand exchange in the presence of different accessory factors induced during natural competence. This is the process by which environmental DNA, internalized as ssDNA, recombines with a resident genome via homologous recombination. The RecA·ATP activities measured are germane for understanding natural chromosomal transformation where the internalized linear ssDNA recombines with the recipient homologous duplex in the presence of SsbA and DprA.
RecA·dATP Catalyzes DNA Strand Exchange in the Absence of Accessory Factors
In vitro, RecA preferentially hydrolyzes dATP when compared with ATP and catalyzes DNA strand exchange in the absence of the accessory factors (Fig. 6B, lane 16) (6, 7, 10). Conversely, RecA in the ATP-bound form cannot catalyze DNA strand exchange (Fig. 6B, lane 2) (11, 14, 21). However, RecA shows a similar Km for ATP and dATP (21), and the in vivo ATP pool is 100–500-fold higher than that of dATP both in exponential or post-exponential cells growing under defined medium (Ref. 20 and our unpublished results). A similar ATP:dATP ratio has been reported for different organisms ranging from E. coli to humans (19, 35).
Why does RecA prefer dATP in vitro? This apparent paradox can be explained if we assume that unscheduled RecA-mediated chromosomal transformation is an undesired reaction, and during competence induction there are two RecA subpopulations. In the hypothetical minor subpopulation, which might operate under “extreme” conditions (e.g. absence of accessory factors), the small fraction of RecA bound to dATP (or to both) enters into an “activated state” that is sufficient to catalyze DNA strand exchange even in the absence of DprA and SsbA (Fig. 8B, lanes 2–6). RecA·dATP on its own or in concert with RecA·ATP should promote assembly of “continuous” NPFs on protein-free ssDNA because of its high affinity for ssDNA and reduced sensitivity to DNA secondary structures and to facilitate DNA recombination under conditions in which the ATP pool is significantly reduced (Fig. 8). This is consistent with the observation that chromosomal transformation is reduced ∼103-fold in the absence of both mediators (RecO and DprA), but it is reduced >104-fold in the absence of RecA (6, 7). It is likely that in the ΔrecO ΔdprA background, dATP might have a small but significant contribution to chromosomal transformation provided that the ATP pool is significantly reduced. Conversely, in the major subpopulation, RecA bound to ATP is in an auto-inhibited state.
RecA·ATP Needs Activation to Catalyze DNA Recombination
The RecA·ATP NPFs catalyze DNA pairing, but these intermediates might dissociate, halted or reversed before extensive DNA strand exchange can occur. Similarly, other RecA proteins from naturally transforming bacteria (e.g. RecASpn or RecADra) in the ATP-bound form are unable to catalyze DNA strand exchange (12, 13, 33).
Previously, it was hypothesized that bacterial RecA is sensitive to DNA secondary structures in the presence of ATP and the presence of SSB overcomes the barrier or favor the forward recombination reaction (33). Indeed, the inability of RecADra-mediated DNA strand exchange can be overcome either by decreasing the pH of the reaction or even by adding a heterologous SSB protein (e.g. SSBEco) after RecADra·ATP (33). These proposed mechanisms do not seem to be universally conserved, because decreasing the pH of the reaction has little effect on RecA-mediated ATP hydrolysis (Fig. 3A) or on the ability of RecA·ATP to catalyze DNA strand exchange with ssDNA coated with SsbA or SsbB (Fig. 6).
DprA Cannot Activate RecA·ATP to Catalyze DNA Strand Exchange
DprA is an ubiquitous protein and has been shown to play an important role in all naturally competent species in which it has been studied and in the mobilization of gene transfer agents (7, 36–39). Gene transfer agents are phage-like particles that apparently mobilize random DNA fragments of α-proteobacteria (e.g. Rhodobacter capsulatous and Rhodopseudomonas palustris) rather than virus genes (1). The structure of R. palustris DprA (DprARpa) (Protein Data Bank code 3MAJ) and DprASpn (Protein Data Bank code 3UQZ) and the C-terminally truncated structure of H. pylori DprA (DprAHpy) complexed with ssDNA (Protein Data Bank code 4LJK) were solved. The DprARpa structure contains three domains: the sterile alpha motif, which is often involved in protein·protein interaction, the central Rossman fold domain, and the DD3 or DML1-like C-terminal domain. DprAHpy lacks the sterile alpha motif (40), whereas DprA and DprASpn lack the DD3 or DML1-like domain (41).
DprA, which preferentially binds ssDNA (7, 37, 40) and physically interacts with SsbB and RecA in vivo (5) and with RecA in vitro (7, 8, 41, 42), has two activities: to mediate annealing of two complementary strands coated by SsbB and to a lesser extent by SsbA or naked ssDNA and to recruit RecA·dATP onto SsbB- or SsbA-coated ssDNA (7). The first activity is required for the annealing of the incoming oligomeric plasmid ssDNA to reconstitute a dsDNA circular plasmid molecule (plasmid transformation) (7). The second activity is intrinsic to directing RecA loading onto SsbB- or SsbA-coated ssDNA (Fig. 4). We have shown that DprA, which forms discrete blobs on ssDNA (7), significantly enhances RecA·ATP NPF formation in the presence of SsbB (Fig. 5), but these RecA NPFs render low yield of RecA-mediated recombination (Fig. 6). It is likely that the inability of RecA to catalyze DNA strand exchange is not at the level of NPF formation. Conversely, DprASpn is able to facilitate RecAEco·ATP-mediated DNA strand exchange in the presence or absence of SSBEco (8). The relevance of this finding, with heterologous proteins, to the mechanism of DNA strand exchange remains to be confirmed.
DprA and SsbA Activate RecA·ATP to Catalyze DNA Strand Exchange
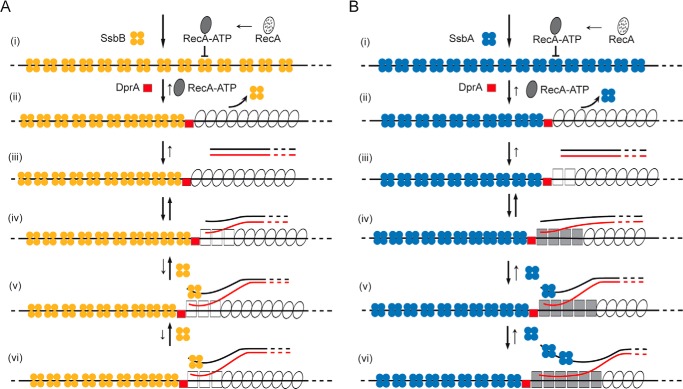
The molecular basis of RecA·ATP activation, the regulatory mechanisms, and their controls have hitherto remained elusive for RecA from naturally transforming bacteria. We propose that RecA-mediated homology-dependent integration of the internalized ssDNA into the genome during natural transformation is a multistep reaction entailing at least four different functional transitions of the RecA protein (Fig. 9). First, RecA bound to ATP induces the ssDNA binding state (first transition state). RecA·ATP leads to ssDNA binding and subsequent polymerization onto protein-free ssDNA, but these RecA NPFs are unable to catalyze DNA strand exchange (Figs. 6 and 8). Second, as soon as the internalized ssDNA leaves the entry channel, SsbB, which co-localizes with the DNA uptake machinery, should coat the ssDNA (Fig. 9A). However, cytosolic SsbA, which shows ∼20-fold higher affinity for ssDNA and forms a more stable complex with ssDNA than does SsbB (6), should effectively compete with SsbB for binding to ssDNA (Fig. 9B). Third, a stoichiometric amount of SsbA or SsbB bound to ssDNA blocks RecA·ATP nucleation and filament extension on the DNA (Fig. 4), and SsbA or SsbB fails to activate RecA-mediated ATP-dependent DNA strand exchange (Fig. 6). Fourth, unlike RecAEco that can displace SSBEco from circular ssDNA (18, 31, 32), RecA nucleation onto SsbA- or SsbB-coated ssDNA or RecA·ATP polymerization onto ssDNA after SsbA or SsbB addition is blocked (Fig. 4). Fifth, DprA, which interacts with RecA both in vivo and in vitro (5, 7), facilitates RecA·ATP filament extension on protein-free ssDNA to levels comparable with RecA·dATP (Fig. 3C). This suggests that DprA might promote a third RecA functional transition state (Fig. 9, ii). Sixth, DprA, which forms discrete complexes on ssDNA (7), is necessary but not sufficient to activate RecA·ATP to promote DNA strand exchange (Fig. 6B, lanes 7–9). Seventh, DprA overcomes the negative effect exerted by stoichiometric SsbB concentrations on RecA nucleation and filament formation, but these RecA NPFs cannot catalyze DNA strand exchange when DprA and SsbB were added before RecA (Fig. 6B). It is likely that elimination of discontinuous RecA NPF formation by DprA and the removal of secondary structure by SsbB are necessary but not sufficient to promote the RecA functional transition upon interaction with dsDNA (Fig. 9A, iii–vi). Eighth, DprA and SsbA facilitate the fourth functional transition state of RecA·ATP. The presence of DprA facilitates RecA·ATP nucleation onto SsbA-coated ssDNA and efficiently stimulates RecA-mediated DNA strand exchange when SsbA was added before RecA (Fig. 6), suggesting that DprA and SsbA facilitate the fourth functional transition state of RecA·ATP to make it proficient to catalyze genetic recombination (Fig. 9B, iv–vi). Ninth, in the presence of SsbB and DprA, the transition to an active state only takes place if limiting concentrations of SsbA are present in the reaction (Fig. 7). It is likely that: (i) DprA and SsbA are crucial to enable RecA to catalyze ATP-dependent DNA strand exchange; (ii) SsbA and SsbB are not mutually interexchangeable to activate RecA-mediated DNA strand exchange; and (iii) SsbB might work as an auxiliary protein to protect the internalized ssDNA. Alternatively, SsbA acting in conjunction with DprA overcomes the inhibition posed by SsbB. Because SsbB interacts with RecA and DprA in vivo (5), we favor the hypothesis that DprA and SsbA work as a two-component mediator during chromosomal transformation. Finally, the uncoupling of SsbB protection for the internalized ssDNA upon exit of the entry channel and RecA-mediated DNA strand exchange in concert with DprA and SsbA could help to ensure more than one transformation events in the same competent cell. This uncoupling will also contribute to genetic plasticity of natural competent cell (9). It is likely that the activation of RecA·ATP ensures that genetic recombination occurs only when sufficient SsbA is available to bind to the displaced strand and RecA and DprA to interact with ssDNA and dsDNA, thus providing a novel mechanism of regulation.
FIGURE 9.
RecA polymerization and RecA-mediated pairing and strand exchange in the presence of different accessory proteins and ATP. The template for RecA·ATP·Mg2+ assembly in vivo is SsbB- (A) or SsbA-coated ssSDNA (B). Based primarily on biochemical analyses, we propose at least four different transitional states of the RecA protein. In the first step, RecA (dotted oval), upon binding ATP·Mg2+ (denoted as RecA·ATP), goes through its first structural transition (filled oval), but it cannot nucleate on the SsbB- or SsbA-coated ssDNA (step i). In the second step, the interaction of DprA with RecA involves a second transition stage (empty ovals) (step ii). DprA partially dislodges SsbB (A) or SsbA (B) and RecA efficiently nucleates and polymerizes on the SsbA·ssDNA·DprA or SsbB·ssDNA·DprA complexes (steps ii and iii). RecA·ATP, by binding to dsDNA, acquires a third transition state (empty squares) that can catalyze DNA pairing upon interacting with a homologous duplex (step iv). DprA and SsbA (B) promote a fourth functional transition of RecA (filled squares) when the complementary duplex is introduced to the filament and activates it to catalyze the forward strand exchange reaction (steps v and vi). DprA and SsbB (A) cannot facilitate RecA fourth transition stage, and it only promotes DNA pairing (steps v and vi). Limiting SsbA, however, can stimulate the fourth transition state in the presence of SsbB and DprA. The overall reaction (denoted by an arrow) is reversible, and the length of the arrow denotes the preferred direction of the reaction.
Acknowledgments
We thank S. Ayora and Patrick Sung for comments on the manuscript.
This work was supported in part by the Ministerio de Economía y Competitividad, Dirección General de Investigación FU2012-39879-C02-01 and by Comunidad de Madrid Grant S2009MAT-1507 (to J. C. A.).
The nomenclature used to denote the origin of proteins from other bacteria is based on the bacterial genus and species (e.g. E. coli RecA is referred to as RecAEco). SSB is used to describe this type of protein in generic terms, and SSB with a subscript (e.g. SSBEco) in specific terms. RecA in the rATP·Mg2+- or dATP·Mg2+-bound form is denoted as RecA·ATP or RecA·dATP.
- SSB
- single-stranded binding
- css
- circular ssDNA
- jm
- joint molecules
- kb
- kilo bp
- lds
- linear dsDNA
- lss
- linear ssDNA
- nc
- nicked circular
- NPF
- nucleoprotein filament
- nt
- nucleotide.
REFERENCES
- 1. Takeuchi N., Kaneko K., Koonin E. V. (2014) Horizontal gene transfer can rescue prokaryotes from Muller's ratchet: benefit of DNA from dead cells and population subdivision. G3 (Bethesda) 4, 325–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen I., Christie P. J., Dubnau D. (2005) The ins and outs of DNA transfer in bacteria. Science 310, 1456–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Claverys J. P., Martin B., Polard P. (2009) The genetic transformation machinery: composition, localization, and mechanism. FEMS Microbiol. Rev. 33, 643–656 [DOI] [PubMed] [Google Scholar]
- 4. Kidane D., Ayora S., Sweasy J. B., Graumann P. L., Alonso J. C. (2012) The cell pole: the site of cross talk between the DNA uptake and genetic recombination machinery. Crit. Rev. Biochem. Mol. Biol. 47, 531–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kramer N., Hahn J., Dubnau D. (2007) Multiple interactions among the competence proteins of Bacillus subtilis. Mol. Microbiol. 65, 454–464 [DOI] [PubMed] [Google Scholar]
- 6. Yadav T., Carrasco B., Myers A. R., George N. P., Keck J. L., Alonso J. C. (2012) Genetic recombination in Bacillus subtilis: a division of labor between two single-strand DNA-binding proteins. Nucleic Acids Res. 40, 5546–5559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yadav T., Carrasco B., Hejna J., Suzuki Y., Takeyasu K., Alonso J. C. (2013) Bacillus subtilis DprA recruits RecA onto single-stranded DNA and mediates annealing of complementary strands coated by SsbB and SsbA. J. Biol. Chem. 288, 22437–22450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mortier-Barrière I., Velten M., Dupaigne P., Mirouze N., Piétrement O., McGovern S., Fichant G., Martin B., Noirot P., Le Cam E., Polard P., Claverys J. P. (2007) A key presynaptic role in transformation for a widespread bacterial protein: DprA conveys incoming ssDNA to RecA. Cell 130, 824–836 [DOI] [PubMed] [Google Scholar]
- 9. Attaiech L., Olivier A., Mortier-Barrière I., Soulet A. L., Granadel C., Martin B., Polard P., Claverys J. P. (2011) Role of the single-stranded DNA-binding protein SsbB in pneumococcal transformation: maintenance of a reservoir for genetic plasticity. PLoS Genet. 7, e1002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Manfredi C., Carrasco B., Ayora S., Alonso J. C. (2008) Bacillus subtilis RecO nucleates RecA onto SsbA-coated single-stranded DNA. J. Biol. Chem. 283, 24837–24847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lovett C. M., Jr., Roberts J. W. (1985) Purification of a RecA protein analogue from Bacillus subtilis. J. Biol. Chem. 260, 3305–3313 [PubMed] [Google Scholar]
- 12. Steffen S. E., Katz F. S., Bryant F. R. (2002) Complete inhibition of Streptococcus pneumoniae RecA protein-catalyzed ATP hydrolysis by single-stranded DNA-binding protein (SSB protein): implications for the mechanism of SSB protein-stimulated DNA strand exchange. J. Biol. Chem. 277, 14493–14500 [DOI] [PubMed] [Google Scholar]
- 13. Grove D. E., Anne G., Hedayati M. A., Bryant F. R. (2012) Stimulation of the Streptococcus pneumoniae RecA protein-promoted three-strand exchange reaction by the competence-specific SsbB protein. Biochem. Biophys. Res. Comm. 424, 40–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carrasco B., Manfredi C., Ayora S., Alonso J. C. (2008) Bacillus subtilis SsbA and dATP regulate RecA nucleation onto single-stranded DNA. DNA Repair (Amst.) 7, 990–996 [DOI] [PubMed] [Google Scholar]
- 15. Bazemore L. R., Takahashi M., Radding C. M. (1997) Kinetic analysis of pairing and strand exchange catalyzed by RecA. Detection by fluorescence energy transfer. J. Biol. Chem. 272, 14672–14682 [DOI] [PubMed] [Google Scholar]
- 16. Yancey-Wrona J. E., Camerini-Otero R. D. (1995) The search for DNA homology does not limit stable homologous pairing promoted by RecA protein. Curr. Biol. 5, 1149–1158 [DOI] [PubMed] [Google Scholar]
- 17. Voloshin O. N., Camerini-Otero R. D. (2004) Synaptic complex revisited; a homologous recombinase flips and switches bases. Mol. Cell 15, 846–847 [DOI] [PubMed] [Google Scholar]
- 18. Cox M. M. (2007) Motoring along with the bacterial RecA protein. Nat. Rev. Mol. Cell Biol. 8, 127–138 [DOI] [PubMed] [Google Scholar]
- 19. Bennett B. D., Kimball E. H., Gao M., Osterhout R., Van Dien S. J., Rabinowitz J. D. (2009) Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lioy V. S., Machon C., Tabone M., Gonzalez-Pastor J. E., Daugelavicius R., Ayora S., Alonso J. C. (2012) The ζ toxin induces a set of protective responses and dormancy. PLoS One 7, e30282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Steffen S. E., Bryant F. R. (1999) Reevaluation of the nucleotide co-factor specificity of the RecA protein from Bacillus subtilis. J. Biol. Chem. 274, 25990–25994 [DOI] [PubMed] [Google Scholar]
- 22. Ayora S., Missich R., Mesa P., Lurz R., Yang S., Egelman E. H., Alonso J. C. (2002) Homologous-pairing activity of the Bacillus subtilis bacteriophage SPP1 replication protein G35P. J. Biol. Chem. 277, 35969–35979 [DOI] [PubMed] [Google Scholar]
- 23. Carrasco B., Ayora S., Lurz R., Alonso J. C. (2005) Bacillus subtilis RecU Holliday-junction resolvase modulates RecA activities. Nucleic Acids Res. 33, 3942–3952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hobbs M. D., Sakai A., Cox M. M. (2007) SSB protein limits RecOR binding onto single-stranded DNA. J. Biol. Chem. 282, 11058–11067 [DOI] [PubMed] [Google Scholar]
- 25. Chiesa M., Cardenas P. P., Otón F., Martinez J., Mas-Torrent M., Garcia F., Alonso J. C., Rovira C., Garcia R. (2012) Detection of the early stage of recombinational DNA repair by silicon nanowire transistors. Nano Lett. 12, 1275–1281 [DOI] [PubMed] [Google Scholar]
- 26. Vlašić I., Mertens R., Seco E. M., Carrasco B., Ayora S., Reitz G., Commichau F. M., Alonso J. C., Moeller R. (2014) Bacillus subtilis RecA and its accessory factors, RecF, RecO, RecR and RecX, are required for spore resistance to DNA double-strand break. Nucleic Acids Res. 42, 2295–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shereda R. D., Kozlov A. G., Lohman T. M., Cox M. M., Keck J. L. (2008) SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 43, 289–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ha T., Kozlov A. G., Lohman T. M. (2012) Single-molecule views of protein movement on single-stranded DNA. Annu. Rev. Biophys. 41, 295–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen Z., Yang H., Pavletich N. P. (2008) Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature 453, 489–494 [DOI] [PubMed] [Google Scholar]
- 30. Ayora S., Piruat J. I., Luna R., Reiss B., Russo V. E., Aguilera A., Alonso J. C. (2002) Characterization of two highly similar Rad51 homologs of Physcomitrella patens. J. Mol. Biol. 316, 35–49 [DOI] [PubMed] [Google Scholar]
- 31. Radding C. M. (1991) Helical interactions in homologous pairing and strand exchange driven by RecA protein. J. Biol. Chem. 266, 5355–5358 [PubMed] [Google Scholar]
- 32. Kowalczykowski S. C., Eggleston A. K. (1994) Homologous pairing and DNA strand-exchange proteins. Annu. Rev. Biochem. 63, 991–1043 [DOI] [PubMed] [Google Scholar]
- 33. Ngo K. V., Molzberger E. T., Chitteni-Pattu S., Cox M. M. (2013) Regulation of Deinococcus radiodurans RecA protein function via modulation of active and inactive nucleoprotein filament states. J. Biol. Chem. 288, 21351–21366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kowalczykowski S. C., Dixon D. A., Eggleston A. K., Lauder S. D., Rehrauer W. M. (1994) Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 58, 401–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Traut T. W. (1994) Physiological concentrations of purines and pyrimidines. Mol. Cell Biochem. 140, 1–22 [DOI] [PubMed] [Google Scholar]
- 36. Bergé M., Mortier-Barrière I., Martin B., Claverys J. P. (2003) Transformation of Streptococcus pneumoniae relies on DprA- and RecA-dependent protection of incoming DNA single strands. Mol. Microbiol. 50, 527–536 [DOI] [PubMed] [Google Scholar]
- 37. Brimacombe C. A., Ding H., Beatty J. T. (2014) Rhodobacter capsulatus DprA is essential for RecA-mediated gene transfer agent (RcGTA) recipient capability regulated by quorum-sensing and the CtrA response regulator. Mol. Microbiol. 92, 1260–1278 [DOI] [PubMed] [Google Scholar]
- 38. Dwivedi G. R., Sharma E., Rao D. N. (2013) Helicobacter pylori DprA alleviates restriction barrier for incoming DNA. Nucleic Acids Res. 41, 3274–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sinha S., Mell J. C., Redfield R. J. (2012) Seventeen Sxy-dependent cyclic AMP receptor protein site-regulated genes are needed for natural transformation in Haemophilus influenzae. J. Bacteriol. 194, 5245–5254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang W., Ding J., Zhang Y., Hu Y., Wang D. C. (2014) Structural insights into the unique single-stranded DNA-binding mode of Helicobacter pylori DprA. Nucleic Acids Res. 42, 3478–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Quevillon-Cheruel S., Campo N., Mirouze N., Mortier-Barrière I., Brooks M. A., Boudes M., Durand D., Soulet A. L., Lisboa J., Noirot P., Martin B., van Tilbeurgh H., Noirot-Gros M. F., Claverys J. P., Polard P. (2012) Structure-function analysis of pneumococcal DprA protein reveals that dimerization is crucial for loading RecA recombinase onto DNA during transformation. Proc. Natl. Acad. Sci. U.S.A. 109, E2466–E2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lisboa J., Andreani J., Sanchez D., Boudes M., Collinet B., Liger D., van Tilbeurgh H., Guérois R., Quevillon-Cheruel S. (2014) Molecular determinants of the DprA-RecA interaction for nucleation on ssDNA. Nucleic Acids Res. 42, 7395–7408 [DOI] [PMC free article] [PubMed] [Google Scholar]