Background: Proline is an important amino acid for stress resistance in different organisms.
Results: Depletion of proline biosynthesis disrupts redox homeostasis and increases sensitivity to endoplasmic reticulum (ER) stress in yeast.
Conclusion: Proline biosynthesis is critical for maintaining the intracellular redox environment and the UPR during ER stress.
Significance: Proline metabolism is shown to have an important role in ER stress tolerance that was previously unknown.
Keywords: Amino Acid, Biosynthesis, Endoplasmic Reticulum Stress (ER Stress), Metabolism, Redox, Glutathione, Proline
Abstract
The amino acid proline is uniquely involved in cellular processes that underlie stress response in a variety of organisms. Proline is known to minimize protein aggregation, but a detailed study of how proline impacts cell survival during accumulation of misfolded proteins in the endoplasmic reticulum (ER) has not been performed. To address this we examined in Saccharomyces cerevisiae the effect of knocking out the PRO1, PRO2, and PRO3 genes responsible for proline biosynthesis. The null mutants pro1, pro2, and pro3 were shown to have increased sensitivity to ER stress relative to wild-type cells, which could be restored by proline or the corresponding genetic complementation. Of these mutants, pro3 was the most sensitive to tunicamycin and was rescued by anaerobic growth conditions or reduced thiol reagents. The pro3 mutant cells have higher intracellular reactive oxygen species, total glutathione, and a NADP+/NADPH ratio than wild-type cells under limiting proline conditions. Depletion of proline biosynthesis also inhibits the unfolded protein response (UPR) indicating proline protection involves the UPR. To more broadly test the role of proline in ER stress, increased proline biosynthesis was shown to partially rescue the ER stress sensitivity of a hog1 null mutant in which the high osmolality pathway is disrupted.
Introduction
Proline metabolism has been linked to stress tolerance in a broad range of organisms. In addition to being a proteogenic amino acid, proline functions as an osmolyte, oxidative stress protectant, chemical chaperone, and as a source of nitrogen and energy under nutrient limiting conditions. Proline is synthesized from glutamate in three enzymatic steps as shown in Fig. 1A. Glutamate is first converted to glutamate-γ-semialdehyde (GSA)2 via the coordinated activities of glutamate kinase and γ-glutamyl phosphate reductase. In yeast Saccharomyces cerevisiae, glutamate kinase (Pro1p) and γ-glutamyl phosphate reductase (Pro2p) are separate enzymes, whereas in plants and humans glutamate kinase and γ-glutamyl phosphate reductase are combined into the bifunctional enzyme Δ1-pyrroline-5-carboxylate synthetase. GSA spontaneously cyclizes to Δ1-pyrroline-5-carboxylate (P5C), which is reduced to proline by P5C reductase (Pro3p). The proline biosynthesis pathway is subject to proline feedback inhibition with proline-inhibiting glutamate kinase (Pro1p) (1). P5C can also be generated from ornithine by ornithine-δ-aminotransferase (Car2p). Thus, genetic deletion of PRO1 and PRO2 does not completely disrupt proline biosynthesis in yeast. In eukaryotes proline is degraded back to glutamate in the mitochondrion by proline dehydrogenase (Put1p) and P5C dehydrogenase (Put2p).
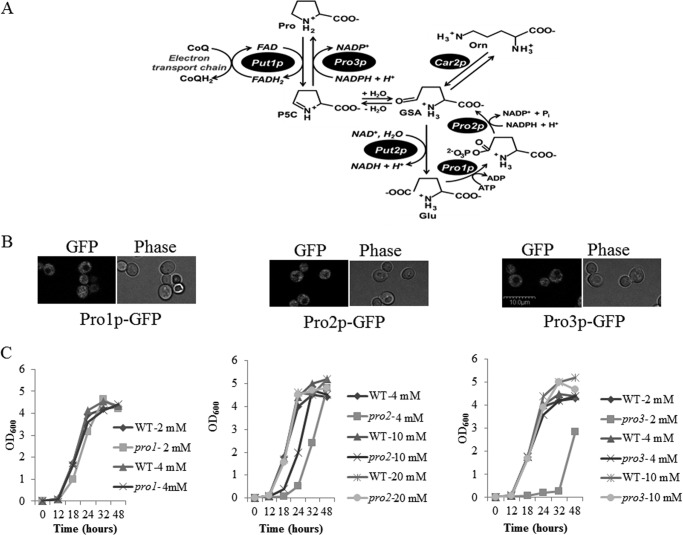
FIGURE 1.
Proline metabolic pathway and characterization of proline biosynthesis enzyme localization and null mutants. A, proline metabolic pathway in S. cerevisiae. Proline is synthesized from glutamate in the cytosol starting with Pro1p (glutamate kinase) and Pro2p (γ-glutamyl phosphate reductase). The intermediate, GSA, spontaneously cyclizes to P5C, which is then reduced to proline by Pro3p (P5C reductase). Alternatively, GSA is generated from ornithine (Orn) via Car2p. Proline is oxidized to glutamate by Put1p (proline dehydrogenase) and Put2p (P5C dehydrogenase) in the mitochondrion. Put1p couples proline oxidation to reduction of ubiquinone (CoQ) in the electron transport chain. B, fluorescence visualization by confocal microscopy of Pro1p-, Pro2p-, and Pro3p-GFP fusion proteins in strain A44 (PRO1-GFP:KanMX6), A46 (PRO2-GFP:KanMX6), and A48 (PRO3-GFP:KanMX6), respectively. All GFP fusion proteins are distributed throughout the cells, indicating cytosolic localization. C, growth rates of the BY4741 WT strain and null mutants pro1 (left panel), pro2 (middle panel), and pro3 (right panel) in SD medium supplemented with different concentrations of proline (2, 4, 10, and 20 mm) as indicated. Cells were diluted to A600 of 0.01 and inoculated into 3 ml of medium and grown at 30 °C with shaking (220 rpm) in 15 ml of cell culture tubes. The growth rate was monitored at A600 at the indicated time points. The scale bar indicates 10 μm.
Understanding the mechanisms by which proline metabolism influences cellular responses to stress is complicated by its multifaceted roles in the cell. In mammalian cells the breakdown of proline has been shown to generate mitochondrial reactive oxygen species (ROS), which ultimately induce cell signaling cascades that promote cell apoptosis or survival (2–5) and in Caenorhabditis elegans increase lifespan (6). Proline biosynthesis has been shown to be up-regulated in plants under different stress conditions with proline accumulation generally associated with increased stress tolerance (2, 7, 8). In humans inborn errors in human P5C reductase isozyme 1 (PYCR1) (9, 10) and P5C synthetase (P5CS) (11, 12) are associated with deficiencies in connective tissue and progeroid features. Fibroblasts from patients with impaired PYCR1 activity show altered mitochondrial morphology and increased apoptosis upon oxidative stress (10). Increased expression of PYCR1 is found in different human cancer cells (4, 13–16) and is thought to enhance cell proliferation (4, 13). In yeast S. cerevisiae, when a Pro1p variant that is insensitive to feedback inhibition by proline was overexpressed in a PUT1 deletion strain (put1), proline levels were increased, and the yeast strain was more resistant to various stressors such as freezing, desiccation, hydrogen peroxide, and ethanol (17). Accordingly, a pro1 null mutant strain was found to be sensitive to freeze-thaw stress (18).
In this study we examine how proline biosynthesis influences endoplasmic reticulum (ER) stress response in yeast S. cerevisiae. The ER is an important organelle for folding and assembling secretory and membrane proteins. The process for protein folding is highly regulated with disruption of protein assembly causing accumulation of misfolded proteins and ER stress. ER stress has been linked to several human diseases such as type II diabetes, neurodegeneration, heart disease, and cancer (19). ER stress is often accompanied by oxidative stress and eventual apoptosis, which is one of the causes of ER stress-induced cell death. In responding to ER stress, intracellular signaling pathways such as the unfolded protein response (UPR) are evoked to help cells adapt (20). In yeast, there are several pathways involved in ER stress response including the much studied UPR pathway comprising the protein kinase Ire1p and its downstream transcription factor Hac1p, the ER-associated protein degradation pathway (21), and the calcium signal pathway (22, 23). Recently it has been reported that the high osmolarity glycerol (HOG) mitogen-activated protein kinase (MAPK) pathway also has an important role in ER stress survival (24, 25), suggesting glycerol is an important ER stress protectant.
Although there have been several studies of proline protection against various types of environmental stress, very little is known about the role of proline in ER stress response. Previously, a pro1 mutant was identified by screening a null-mutant library of S. cerevisiae for sensitivity to dithiothreitol (DTT), a reducing agent that causes ER stress (26). In addition, proline has been shown to inhibit protein aggregation in vitro and in vivo (27–29). Here, we test the effect of genetic limitation of proline on ER stress tolerance in the S. cerevisiae null mutants pro1, pro2, and pro3. The proline biosynthesis mutants are shown to be more sensitive to ER stress-inducing agents than the wild-type strain. Insights into the mechanism by which genetic depletion of proline biosynthesis affects ER stress tolerance were gained by investigating intracellular redox markers and the UPR. The protective role of proline in ER stress was also examined more broadly by exploring whether proline biosynthesis can rescue the ER stress phenotype of ire1 and hog1 null mutants.
EXPERIMENTAL PROCEDURES
Chemicals, Yeast Strains, and Culture Conditions
All chemicals, culture media, and buffers were purchased from Fisher and Sigma unless otherwise indicated. The S. cerevisiae strains used in this study are provided in Table 1. Gene knock-out mutant strains and strains for expression of GFP C-terminal fusion proteins were constructed using PCR-mediated gene knock-out or C-terminal tagging techniques (30). The gene knock-out strain was constructed by replacing the corresponding gene ORF with the HIS3MX6 cassette, and the mutant for expression of GFP C-terminal fusion protein was constructed by fusing the GFP(S65T)-kanMX6 cassette to the corresponding C terminus of the gene. Each mutant was confirmed by genotyping using the corresponding primers provided in Table 2. Non-proline auxotrophs were maintained in YPD medium (2% (w/v) d-glucose, 2% (w/v) bactopeptone, 1% (w/v) yeast extract) or SD medium (0.17% (w/v) yeast nitrogen base without amino acids, 0.5% (w/v) ammonium sulfate, 2% (w/v) d-glucose) supplemented unless otherwise indicated with leucine (100 mg/liter), methionine (20 mg/liter), histidine (20 mg/liter), and uracil (20 mg/liter). Proline auxotrophs were maintained in SD medium supplemented with varying concentrations of proline (10–20 mm). Growth profiles of the different strains were followed by measuring the optical density at 600 nm (A600) with log phase considered to be at A600 of 0.6–1.0 and stationary phase at A600 ≥ 3.0. Cells were also cultured anaerobically using a deoxygenated jar with an oxygen scrubbing catalyst (BD Biosciences) and an anaerobic indicator (BD Biosciences). Cells were grown anaerobically on SD medium containing 20 mg/liter ergosterol and 0.05% Tween 80 at 30 °C for 3 days.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Open Biosystems |
| A143 | As BY4741, but pro1::His3 | This study |
| A18 | As BY4741, but pro2::KanMX | Open Biosystems |
| A98 | As BY4741, but pro3::His3 | This study |
| A2 | As BY4741, but put1::KanMX | Open Biosystems |
| A64 | As BY4741, but ire1::KanMX | Open Biosystems |
| A65 | As BY4741, but hog1::KanMX | Open Biosystems |
| A145 | As BY4741, but pro2::KanMX pro1::His3 | This study |
| A142 | As BY4741, but pro2::KanMX pro3::His3 | This study |
| A140 | As BY4741, but pro2::KanMX put1::His3 | This study |
| A116 | As BY4741, but put1::KanMX pro3::His3 | This study |
| A44 | As BY4741, but PRO1-GFP:KanMX6 | This study |
| A46 | As BY4741, but PRO2-GFP:KanMX6 | This study |
| A48 | As BY4741, but PRO3-GFP:KanMX6 | This study |
TABLE 2.
Primers used in this study
F, forward primer; R, reverse primer.
| Primer ID | Sequence (5′ to 3′) | Application |
|---|---|---|
| DB-96-F | tataggatccatgaaggatgctaatgagag | PRO1-GFP genotyping |
| DB-102-F | gcgaatatgtcgctcatagagaaaatttggcattcccacctcgtcggatccccgggttaattaa | GFP tagging for PRO1 |
| DB-103-R | taagaaatatcattaagggaatcctctataactttttagttcgttgaattcgagctcgtttaaac | GFP tagging for PRO1, PRO1 knock out |
| DB-116-F | ggtcattttatcagctactgtaatatagtcataagaacgcgaaacggatccccgggttaattaa | PRO1 knock out |
| DB-174-F | ttgagcaccagttaaagtatgaaca | PRO1 knockout genotyping |
| DB-98-F | tataggatccatgtccagttcacaacaaat | PRO2 CDNA synthesis, PRO2-GFP genotyping |
| DB-99-R | tcaactcgagtcaagcgtaatctggaacatcgtatgggtataatgtcacagtctttatat | PRO2 cDNA synthesis |
| DB-120-F | caaagcttttgttcacaaggatttagatataaagactgtgacattacggatccccgggttaattaa | GFP tagging for PRO2 |
| DB-121-R | gtatacacgtccttcatacatggaacttagctttatcctaaaaatttgaattcgagctcgtttaaac | GFP tagging for PRO2 |
| DB-100-F | tataggatccatgacttacacattggcaatt | PRO3 cDNA synthesis, PRO3-GFP genotyping |
| DB-101-R | tcaactcgagtcaagcgtaatctggaacatcgtatgggtatttcttcttttggcctaatt | PRO3 cDNA synthesis |
| DB-122-F | gaagaggcagcccgtgttgcgtcacaattaggccaaaagaagaaacggatccccgggttaattaa | GFP tagging for PRO3 |
| DB-105-R | tatacggctacaaagcgaacatacatacatttcacgcaaagcaatgaattcgagctcgtttaaac | GFP tagging for PRO3, PRO3 knock out |
| DB-104-F | aatagaagtaacacaacaccaatagcaaacaaaccaactacagatcggatccccgggttaattaa | PRO3 knock out |
| DB-175-F | ataaagtcgcatacgtacaaaagga | PRO3 knockout genotyping |
| DB-173-R | gtcgattacagcaaatggtc | GFP fusion mutant and gene knockout mutant genotyping |
| DB-158-F | agaaggcctccaagggtatt | KAR2 mRNA levels |
| DB-159-R | taagacaccagcttgaacgg | KAR2 mRNA levels |
| DB-176-F | caaggcatgatgcagatttc | SLT2 mRNA levels |
| DB-177-R | ctagaagcgtgccgttatca | SLT2 mRNA levels |
| DB-182-F | ccttaaacgaaggcgaagag | GSC2 mRNA levels |
| DB-183-R | gggacgtctacgaccattct | GSC2 mRNA levels |
| DB-156-F | ccaagatgttcgttggaatg | GSH1 mRNA levels |
| DB-157-R | cctccaaatccgttcttgtt | GSH1 mRNA levels |
| DB-140-F | atttggccggtagagatttg | ACT1 mRNA levels |
| DB-141-R | tccaaggcgacgtaacatag | ACT1 mRNA levels |
| DB-223-F | ttggcagacccactctg | HAC1i mRNA levels |
| DB-225-R | ctgcgcttctggattacg | HAC1i mRNA levels |
Overexpression of PRO1(D154N), PRO2, and PRO3 in Yeast Cells
The cDNA of PRO1, PRO2, and PRO3 were synthesized from RNA extracted from wild-type BY4741 strain and amplified by PCR using corresponding primers (see Table 2 for a list of primers; a HA tag was introduced by the reversed primer). The resulting PCR products were verified by sequencing and cloned into p413TEF (PRO1, PRO2) or p416GPD (PRO3) vectors at BamH1 and Xho1 sites. The PRO1(D154N) construct was made by site-directed mutagenesis (Stratagene site-directed mutagenesis kit) of p413TEF-PRO1. All the constructs were confirmed by DNA sequencing (Eurofins MWG Operon) and transformed into their respective yeast mutant strain via the LiAc-based assay. Expression of each protein was indicated by complementation of the corresponding proline auxotroph strain.
Spot Assays
Cells were grown overnight to stationary phase in SD medium alone except for the pro1, pro2, and pro3 mutant strains, which were grown in SD medium supplemented with 10 mm proline. Cells were then pelleted and resuspended in deionized water to an A600 of 2.0 and were serially diluted (1:10) with deionized water. Cells were spotted on the indicated culture plates using a replica plater and grown 2–3 days at 30 °C.
Determination of Intracellular Superoxide, Glutathione, and Proline
Superoxide anion radicals were estimated with dihydroethidium (Invitrogen) using a flow cytometer, BD Biosciences FACSCanto II, with a blue laser (488 nm) at PE channel (585/42, 556LP). Cells were incubated with SD medium supplemented with 10 μm dihydroethidium at 30 °C for 15 min, washed once with PBS, and resuspended in 500 μl of PBS and analyzed by fluorescence detection using flow cytometry.
Intracellular free glutathione was extracted as described previously (31). Briefly, cells were grown in SD minimal medium with or without proline to log phase before the indicated treatments. Yeast cells were washed twice with PBS and resuspended in ice-cold HCl and 1.3% (w/v) 5-sulfosalicylic acid. Cells were broken with acid-washed glass beads using 3 cycles of vortexing (1 min) and incubation on ice (1 min). The broken cells were then incubated on ice for an additional 15 min. Cell debris and proteins were pelleted by centrifugation at 15,000 × g for 15 min at 4 °C. The supernatant was used for measuring free glutathione. The reduced and oxidized forms of glutathione were determined as described previously (32).
Intracellular free proline was measured as described using the ninhydrin assay (33). Briefly, about 5 ml of log phase grown cells were spun down, washed with 0.9% NaCl, and resuspended in 0.5 ml of distilled water. The cells were transferred to a boiling water bath, and intracellular free amino acids were extracted by boiling for 10 min followed by centrifugation at 15,000 × g for 5 min. The concentration of proline was determined in the resulting extract.
Enzyme Assays
Glutathione reductase (GLR) activity was measured as described (34) but with a slight modification. Cells were broken with acid-washed glass beads in lysis buffer (0.1 m potassium phosphate buffer, pH 7.5, 1 mm EDTA) as described. Cell debris was then pelleted by centrifugation at 15,000 × g for 15 min at 4 °C. The GLR activity was measured in the supernatant, and the total protein concentration was measured with the Bradford assay. One unit of GLR activity is the amount of enzyme that oxidizes 1 μmol of NADPH/min at 25 °C (pH 7.5).
Determination of Intracellular NADP(H)
NADP(H) was extracted with base and acid for NADPH and NADP+ measurements, respectively, as described but with a slight modification (35). Briefly, about 10 absorbance units of yeast cells were resuspended either in 0.1 m NaOH or 0.1 m HCl. Cells were broken with glass beads as described above for glutathione. After centrifuging the broken cells at 15,000 × g for 10 min at 4 °C, the resulting supernatant was heated at 60 °C for 10 min. The extract was then neutralized with an equal volume of 0.1 m HCl or 0.1 m NaOH followed by NADP(H) measurement via a spectrophotometric enzymatic cycling assay as described (36).
Real-time PCR
Total RNA was extracted from yeast cells using the RNeasy mini kit (Qiagen). Cells were grown to log phase before the indicated treatments. The extracted RNA was treated with RNase-free DNase (Fermentas) before cDNA synthesis with M-MulV reverse transcriptase and oligo(dT) primers (Fermentas). mRNA levels were quantified by real-time PCR using the IQ-SYBR-supero mix (Bio-Rad) and MyIQ real-time PCR detection system (Bio-Rad). Relative mRNA levels were calculated using the 2−ΔΔCT method and the actin gene ACT1 as the internal control. PCR products were also analyzed by 2% agarose gel electrophoresis to confirm product size and specificity. Spliced HAC1 (HAC1i) mRNA was quantified by real-time PCR using the reverse primer designed to span the exon-exon junction (Table 2) (37). The specificity of the HAC1i PCR product was confirmed by control assays in ire1 mutant cells, which lack HAC1 splicing due to the absence of Ire1, the nuclease responsible for cleaving HAC1 mRNA during ER stress (38).
Cell Viability Assays by Propidium Iodide (PI) Staining
Yeast cells were treated as indicated, spun down, and resuspended in 1 ml of PBS containing 3 μg/ml PI for 10 min followed by flow cytometry analysis of the PI-stained cells. Cell death rate is the percentage of PI-positive cells per 10,000 cells counted.
RESULTS
Localization of the Proline Biosynthesis Enzymes and Growth Profiles of Null Mutants
The intracellular localization of Pro1p, Pro2p, and Pro3p was determined by making GFP fusion proteins driven by the native promoter of the corresponding gene. All three enzymes were previously reported to be in the cytosol, but the evidence was indirect such as complementation of the pro1 mutant with the proB gene from Escherichia coli (39) and isolation of Pro3p from the cytosolic fraction (40). In agreement with previous findings, Pro1p, Pro2p, and Pro3p were expressed exclusively in the cytosol (Fig. 1B). Next, the growth profiles of the pro1, pro2, and pro3 null mutants were characterized in SD medium supplemented with increasing amounts of proline. Without proline, the pro1 and pro2 mutants exhibited growth, whereas pro3 did not grow (Fig. 2A, top panel). These results are consistent with arginine catabolism supplying P5C for PRO3 in pro1 and pro2 cells and a strict requirement for proline in pro3 cells (41). The growth profiles of the pro1, pro2, and pro3 mutants were restored to that of the parent wild-type strain at 4, 20, and 4 mm proline, respectively (Fig. 1C). Thus, the pro2 mutant requires the highest proline concentration for growth in SD medium. A similar growth dependence on proline was observed on SD plates (Fig. 2A, top panel).
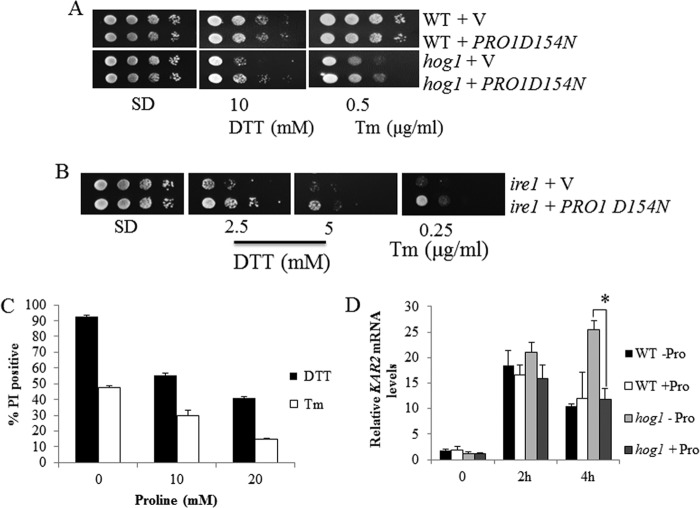
FIGURE 2.
Proline biosynthesis mutants are sensitive to ER stress. A, growth of BY4741 WT cells and null mutants put1, pro1, pro2, and pro3 on SD plates with increasing proline concentrations (0–20 mm) in the absence and presence of Tm (0.5 μg/ml) or DTT (10 mm). B, growth of WT cells and null mutants pro2 and pro3 harboring empty vector (V) or construct for expression of recombinant PRO2 or PRO3. C, growth of wild-type cells on SD medium with histidine, leucine, and uracil concentrations reduced to of normal concentrations. Cells were spotted on SD plates in the absence and presence of Tm (0.5 μg/ml) or DTT (10 mm).
Proline Biosynthesis Mutants Are Sensitive to ER Stress
The ER stress sensitivities of the pro1, pro2, and pro3 mutants and the proline catabolic put1 mutant were tested by growing each strain to stationary phase and spotting on SD plates containing different concentrations of proline (0, 2, 4, 10, and 20 mm) and DTT or tunicamycin (Tm). DTT and Tm are chemicals used commonly for inducing ER stress (42). Both chemicals induce ER stress but do so via different mechanisms. DTT induces ER stress by reducing thiols and disrupting protein disulfide bond formation (43, 44), whereas Tm blocks N-linked glycosylation leading to accumulation of misfolded proteins (35, 45).
All of the proline biosynthesis mutants were more sensitive to Tm and DTT than the wild-type strain at 2 and 4 mm proline (Fig. 2A). The strongest Tm stress phenotype was observed for the pro3 mutant, whereas pro2 cells appeared most sensitive to DTT. The pro1 mutant was least sensitive to DTT/Tm relative to pro2 and pro3. Increasing proline to 10 mm rescued the Tm sensitivity of all the mutants. With DTT, 10 mm proline protected the pro1 and pro3 mutants, whereas 20 mm proline was needed to fully rescue the pro2 mutant. In contrast to the proline biosynthesis mutants, disruption of proline catabolism in the put1 null mutant had only a minimal effect on sensitivity to Tm and DTT (Fig. 2A).
The results shown in Fig. 2A indicate that the pro2 mutant has the highest sensitivity to DTT, whereas pro3 is most sensitive to Tm. To confirm that the sensitivity of these mutants was due to the loss of functional PRO2 and PRO3 genes, the pro2 and pro3 mutants were transformed with recombinant PRO2 and PRO3, respectively. Overexpression of PRO2 and PRO3 in the corresponding mutants rescued growth in SD medium without proline and increased resistance to DTT and Tm (Fig. 2B). These results show that the ER stress-sensitive phenotype of pro2 and pro3 is due to the knock-out of PRO2 and PRO3, respectively, and suggest that proline biosynthesis helps yeast cells survive ER stress under limiting proline conditions.
One mechanism by which cells adapt to ER stress is to down-regulate global protein synthesis, which in yeast occurs during amino acid starvation (46). How limiting other amino acids influences ER stress tolerance was examined in the BY4741 wild-type strain, which is an auxotroph for histidine, leucine, and uracil. These supplements were simply reduced to of the normal amount in SD medium (2 mg/liter histidine, 10 mg/liter leucine, and 2 mg/liter uracil). Fig. 2C shows that in contrast to proline, wild-type cells grown under limiting conditions for histidine, leucine, and uracil do not show increased sensitivity to DTT and Tm. In fact, the Tm stress resistance of wild-type cells appears to be slightly improved by limiting histidine and leucine, consistent with lower protein synthesis benefiting cells during ER stress. Thus, the ER stress sensitivity of the proline biosynthesis mutants appears to contrast that of general amino acid limitation.
N-Acetyl-l-cysteine and Anaerobic Conditions Rescue the pro3 Mutant
Because Tm has been shown previously to increase oxidative stress in yeast (35), we examined if N-acetyl-l-cysteine (NAC) and reduced glutathione (GSH) could mitigate the Tm sensitivity of pro3. Fig. 3A shows that both NAC and GSH rescue the Tm sensitive-phenotype of pro3, whereas methionine has no effect. The effects of NAC and GSH are similar to proline. Other chemicals with known antioxidant properties such as butylated hydroxyanisole (47) and diphenyleneiodonium (an NADPH-oxidase inhibitor) (48) did not rescue the Tm sensitivity of pro3 (Fig. 3A). These results suggest that GSH is important for repairing ER-associated oxidative stress such as helping to resolve non-native disulfide bonds formed as a result of oxidative stress or enzyme-catalyzed oxidation (49).
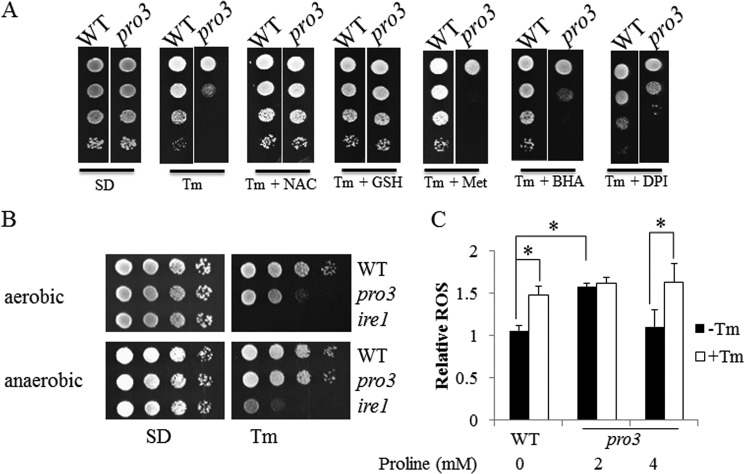
FIGURE 3.
ER stress protection of pro3 by reducing agents and anaerobic conditions. A, spot assays of WT and pro3 cells grown on SD plates with 4 mm proline alone (SD) or with 4 mm proline in the presence of Tm (0.5 μg/ml), Tm and N-acetyl-l-cysteine (Tm + NAC), Tm and GSH (Tm + GSH), Tm and methionine (Tm + Met), Tm and butylated hydroxyanisole (Tm + BHA), and Tm and diphenyleneiodonium (Tm + DPI). B, spot assays of WT, pro3, and ire1 cells on SD medium with 4 mm proline alone (SD) or in the presence of Tm (0.5 μg/ml) under aerobic (top panel) and anaerobic conditions (bottom panel) at 30 °C for 3 days. C, relative superoxide levels in WT (no proline) and pro3 (2 and 4 mm proline) cells in SD medium treated with (+Tm, 1 μg/ml) or without (−Tm) for 4 h. Superoxide was measured by ROS probe dihydroethidium bromide using flow cytometry and normalized to the lowest level. Data are the mean ± S.D. from three independent experiments (*, p < 0.05).
Additional evidence for oxidative stress contributing to the ER sensitivity of pro3 was obtained by treating the pro3 mutant with Tm under anaerobic conditions. Fig. 3B shows that pro3 is less sensitive to Tm under anaerobic conditions. Wild-type and ire1 mutant cells showed no significant change to Tm treatment when switched to anaerobic growth (Fig. 3B).
ROS levels in wild-type and pro3 were then measured using dihydroethidium (50). Under non-stressed conditions, the ROS levels in pro3 (2 mm proline) were 1.5-fold higher than wild type (Fig. 3C). Increasing proline in the SD medium to 4 mm lowered ROS levels in pro3 to that of wild type. Upon treating cells with Tm, ROS levels increased by 1.5-fold in wild-type and pro3 (4 mm proline). At 2 mm proline, ROS levels did not further increase in pro3 cells with Tm treatment (Fig. 3C).
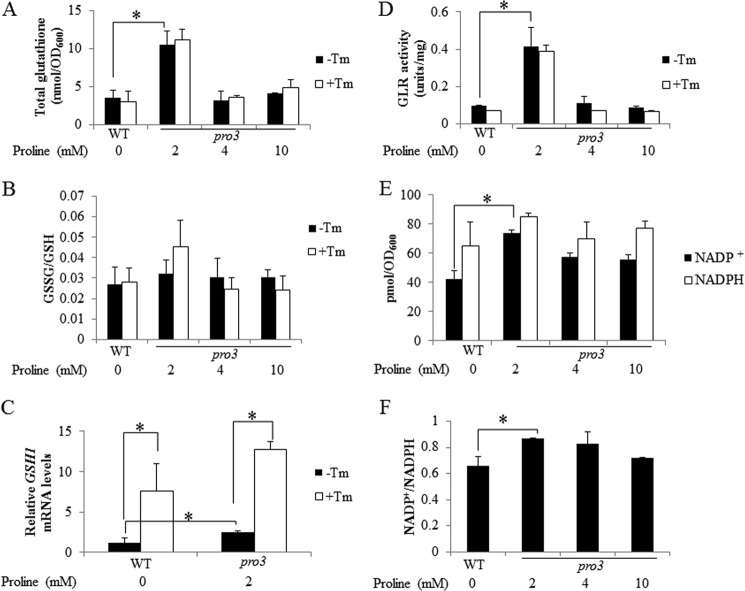
Because GSH reversed the ER stress sensitivity of the pro3 mutant, we assessed GSH status in pro3. GSH levels were measured in wild-type and pro3 cells grown to log phase with different proline concentrations (2, 4, and 10 mm). Total GSH was 3-fold higher in pro3 with 2 mm proline relative to wild type (Fig. 4A). At higher concentrations of proline (4 and 10 mm) GSH levels were similar to wild type (Fig. 4A). In all conditions Tm did not change total GSH levels in wild type and pro3 (Fig. 4A). The ratio of oxidized to reduced glutathione (GSSG/GSH) was shown to be similar between wild type (WT) and pro3 with no significant change upon treatment with Tm (Fig. 4B). We also measured the mRNA levels of γ-glutamylcysteine synthetase 1 (GSH1), the rate-limiting enzyme of GSH biosynthesis. Under growth conditions with 2 mm proline, the GSH1 transcript levels were ∼2-fold higher in pro3 relative to wild-type cells (Fig. 4C), indicating that increased GSH1 expression may be the cause of higher glutathione in pro3. We also observed that after Tm treatment, GSH1 increased by 3- and 5-fold in wild type and pro3, respectively (Fig. 4C), indicating that GSH is required for cells recovering from ER stress. Interestingly, GSH1 levels remained nearly 2-fold higher in pro3 relative to wild type after Tm treatment. GSH metabolism was explored further by measuring the activity of GLR, which catalyzes the NADPH-dependent reduction of GSSG to 2GSH. GLR activity was observed to be 4-fold higher in pro3 at 2 mm proline than in wild-type cells (Fig. 4D). At higher concentrations of proline, GLR activity in pro3 was similar to wild-type cells. No change in GLR activity was observed with Tm treatment in wild type or pro3. We last measured NADP+ and NADPH and found significantly higher NADP+ and a NADP+/NADPH ratio in pro3 at 2 mm proline relative to wild type (Fig. 4, E and F). At higher proline concentrations (4 and 10 mm proline) no significant difference in NADP+ and NADPH was observed between wild-type and the pro3 mutant.
FIGURE 4.
Proline limitation results in redox imbalance in pro3. WT cells and pro3 cells were grown to log phase in SD medium with varying concentrations of proline (0–10 mm) as indicated and treated with (+Tm, 1 μg/ml) or without Tm (−Tm) for 2 h. The following parameters were then determined. A, total intracellular glutathione. B, ratio of oxidized (GSSG) to reduced glutathione (GSH). C, relative GSH1 mRNA levels (4 h of Tm treatment) determined by real-time PCR with mRNA levels normalized to the internal control ACT1 and the lowest expression set at 1. D, GLR activity. The concentrations of NADP+ and NADPH (E) and the corresponding ratio of NADP+/NADPH (F) were determined in WT and pro3 cells grown in SD medium as above but without Tm treatment. Data are the mean ± S.D. from three independent experiments (*, p < 0.05).
Oxidative Stress and the UPR
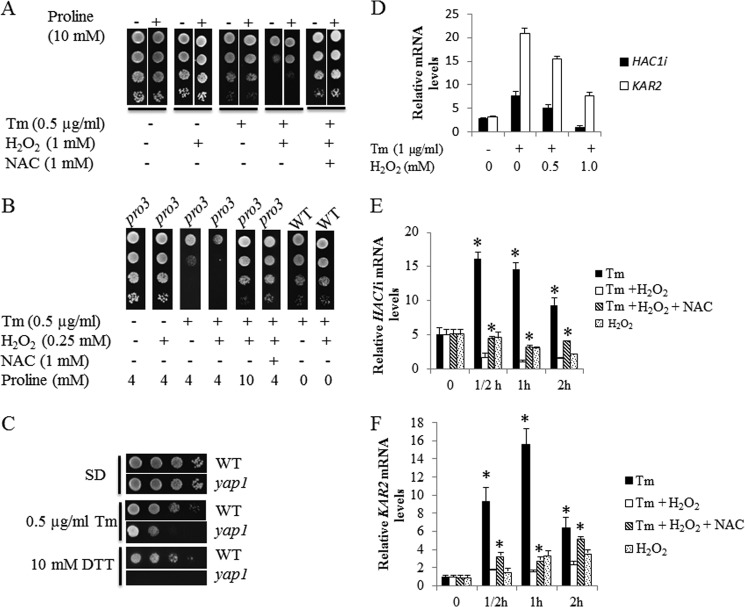
To test whether higher ROS levels as observed in the pro3 mutant (Fig. 3C) exacerbate Tm sensitivity, wild-type cells were treated with Tm in the presence of hydrogen peroxide (H2O2). The addition of H2O2 (1 mm) during Tm treatment significantly decreased cell survival, indicating a compounding effect of oxidative stress on ER stress (Fig. 5A). H2O2 alone did not diminish cell survival (Fig. 5A). The addition of NAC dramatically rescued cells from Tm and H2O2 toxicity. Interestingly, the addition of 10 mm proline also partially repressed the combined toxicity of Tm and H2O2 in wild-type cells (Fig. 5A). The sensitivity of the pro3 mutant strain to Tm was also enhanced by H2O2 but at a lower concentration (0.25 mm H2O2) (Fig. 5B). As in wild-type cells, NAC prevented cell death induced by Tm and H2O2 treatment (Fig. 5B), consistent with proline limitation, resulting in higher intracellular oxidative stress. Increasing the proline concentration from 4 to 10 mm also rescued the pro3 cells from Tm and H2O2 toxicity (Fig. 5B). Note that at 0.25 mm, H2O2 does not increase the Tm sensitivity of wild-type cells (Fig. 5B). The role of cellular antioxidants in ER stress defense was further evaluated by testing the sensitivity of the yap1 mutant to Tm or DTT. Fig. 5C shows yap1 cells exhibit significantly increased sensitivity to Tm and DTT relative to wild-type cells.
FIGURE 5.
Oxidative stress increases Tm toxicity and inhibits the UPR. A–C, spot assays of wild-type (A), wild-type and pro3 (B), and wild-type and yap1 cells (C) on SD plates with (+) or without (−) the indicated supplements. D–F, real-time PCR quantification of spliced HAC1 (HAC1i) or KAR2 mRNA in BY4741 wild-type cells grown to log phase in SD medium treated with the following. D, without (-) or with (+) Tm (1 μg/ml) in the presence of increasing H2O2 (0–1 mm) for 2 h; E and F) Tm (1 μg/ml) alone (Tm), Tm and 1 mm H2O2 (Tm + H2O2), Tm, 1 mm H2O2 and 1 mm NAC (Tm + H2O2 + NAC), and 1 mm H2O2 alone (H2O2) for the indicated hours (h). mRNA levels were normalized to the internal control ACT1 with the lowest expression level set at 1. *, p < 0.05 compared with Tm + H2O2 at the corresponding time point. Data are the mean ± S.D. from three independent experiments.
In yeast and mammalian cells, a functional UPR is essential for cell survival under ER stress (21). To test whether oxidative stress impacts the UPR, HAC1 mRNA splicing (37) was examined via real-time PCR after Tm treatment with and without H2O2. As expected, HAC1i (spliced HAC1) levels increased (∼3-fold) in wild-type cells after Tm treatment (Fig. 5D), indicating induction of the UPR. In the presence of H2O2 (1 mm), Tm induction of HAC1i mRNA splicing was significantly reduced. A dose-dependent inhibition of the UPR was also observed by following the ER stress-inducible UPR-target gene KAR2 (38). KAR2 mRNA levels were reduced by nearly 4-fold with H2O2 relative to Tm treatment alone (Fig. 5D).
The effect of H2O2 on the time course (0.5–2 h) of the UPR was also examined. At each time point H2O2 significantly blocked the UPR as determined by HAC1i (Fig. 5E) and KAR2 (Fig. 5F) mRNA levels. The addition of NAC partially rescued the UPR with 2–3-fold higher levels of HAC1i and KAR2 (Fig. 5, E and F) at each time point relative to cells treated with Tm and H2O2. Treatment of cells with H2O2 alone resulted in 2-fold lower HAC1i mRNA levels (Fig. 5E) by 2 h and a ∼3-fold increase in KAR2 mRNA (Fig. 5F) by 1 h.
Proline Limitation Inhibits the UPR
To test if proline limitation impacts the UPR, HAC1i and KAR2 mRNA levels were examined in pro3 mutant cells grown in SD medium supplemented with increased concentrations of proline. With 2 mm proline, pro3 cells exhibited a significantly lower induction of HAC1i (Fig. 6A) and KAR2 (Fig. 6B) mRNA by Tm relative to wild-type cells. Increasing the proline concentration (4 and 10 mm) rescued the Tm-dependent UPR as observed by higher HAC1i and KAR2 mRNA levels (Fig. 6, A and B). NAC partially rescued HAC1i mRNA levels in pro3 cells at 2 mm proline (Fig. 6C) similar to that observed with wild-type cells treated simultaneously with Tm and H2O2 (Fig. 5E). For comparison, other conditions of nutrient starvation (1/10 of normal concentration in SD medium) such as leucine and uracil were tested for effects on the UPR. Imposing leucine or uracil starvation on wild-type cells results in a significant drop in HAC1i mRNA to nearly undetectable levels, similar to that observed in the ire1 mutant strain (Fig. 6A). Also, no increase in HAC1i mRNA was observed with Tm stress (data not shown). However, limiting leucine or uracil did not increase the toxicity of Tm or DTT (Fig. 2C). Therefore, the dampened UPR under leucine or uracil starvation is most likely due to generally lower protein synthesis caused by amino acid starvation and, thus, lower misfolded protein loading in the ER. In contrast, proline limitation does not appear to decrease overall misfolded protein loading, as HAC1i mRNA levels are similar between wild-type and pro3 cells before Tm treatment. This implies that proline limitation results in higher misfolded protein levels relative to leucine or uracil, suggesting that proline-depleted cells are under ER stress before Tm treatment.
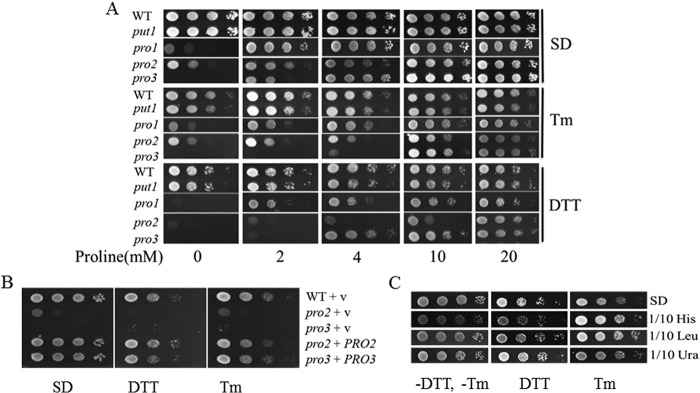
FIGURE 6.
Proline limitation diminishes the UPR response induced by Tm. A and B, relative spliced HAC1 (HAC1i) mRNA or KAR2 mRNA levels of log phase cells of BY4741 WT, ire1, and pro3 null mutant grown in SD medium with increasing concentrations of proline treated without (−Tm) or with (+Tm) Tm (1 μg/ml) for 2 h. C, relative HAC1i mRNA levels of pro3 grown in SD medium with 2 mm proline treated with Tm alone (Tm) and Tm and 1 mm NAC (Tm + NAC) for 2 h. mRNA levels were normalized to the internal control ACT1 with the lowest expression level set at 1. *, p < 0.05 compared with WT or pro3 with the addition of 10 mm proline treated with Tm (A and B) or as indicated (C). Data are the mean ± S.D. from three independent experiments.
Because ER stress activates the calcium signaling pathway as well as the UPR (22, 23), we tested the possibility of impaired calcium import in pro3. Exogenous proline was previously reported to enhance calcium import in plants (51), indicating proline levels may influence calcium signaling. Calcium import, however, was observed not to be affected in the pro3 mutant as determined by monitoring expression levels of SLT2 (MPK1), a protein kinase that is critical for calcium homeostasis and is up-regulated by Tm (22) and GSC2, a gene known to be up-regulated by increased calcium levels (52) (data not shown).
Proline Biosynthesis Gene Expression in Response to Tm Treatment
To determine whether ER stress influences the expression of proline biosynthesis genes, we examined protein expression levels of Pro1p, Pro2p, and Pro3p after Tm treatment. The fluorescence intensity of Pro1p-GFP, Pro2p-GFP, and Pro3p-GFP fusion proteins with expression of each gene under control of the corresponding native promoter was measured using flow cytometry. No significant change in the fluorescence of the GFP fusion proteins was observed after 2 and 4 h of Tm treatment (data not shown). Consistent with no change in proline biosynthesis gene expression, no significant change in proline content was observed after Tm treatment (data not shown).
Intermediates in the Proline Metabolic Pathway Do Not Contribute to ER Stress Sensitivity
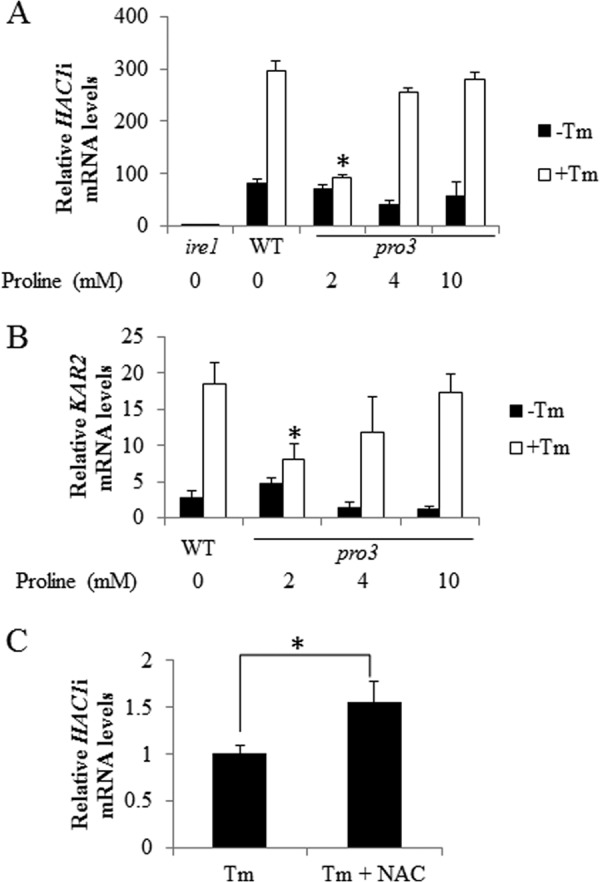
The ER stress sensitivity of the pro2 and pro3 mutants may also be impacted by intermediate metabolites in the proline biosynthesis pathway. In the first step Pro1p converts glutamate to γ-glutamyl phosphate, which is unstable and undergoes cyclization to form 5-oxoproline, which may increase in pro2 (53). The second intermediate of proline biosynthesis is P5C/GSA, which could increase in the pro3 mutant and is thought to be toxic to yeast and induce apoptotic cell death (2). Because proline is a feedback inhibitor of Pro1p (54), supplementing the pro2 and pro3 strains with proline may reduce buildup of these potentially harmful intermediates. To test the effects of the intermediates γ-glutamyl phosphate and P5C/GSA, double mutants pro1/pro2 and pro2/pro3 were tested for sensitivity to DTT and Tm, respectively. Fig. 7 shows that the sensitivity of the double mutants pro1/pro2 and pro2/pro3 to DTT and Tm, respectively, is similar to that observed with the single pro2 and pro3 mutants. In addition, the stress phenotype of the double mutants was rescued by proline. Thus, the intermediates of proline biosynthesis do not appear to contribute significantly to the observed ER stress phenotypes of pro2 and pro3.
FIGURE 7.
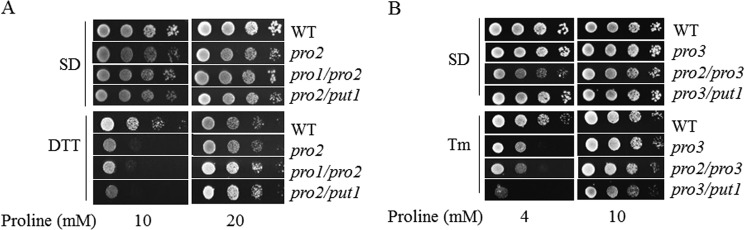
Intermediates of the proline biosynthesis pathway do not appear to contribute to ER stress. A, spot assays of WT, pro2, pro1/pro2, and pro2/put1 cells on SD medium with different concentrations of proline as indicated with (DTT) and without (SD) DTT (10 mm). B, spot assays of WT, pro3, pro2/pro3, and pro3/put1 cells on SD medium with different concentrations of proline as indicated with (Tm) and without (SD) Tm (0.5 μg/ml).
It is also possible that the breakdown of proline by Put1p would accentuate problems with inadequate proline levels in the pro2 and pro3 mutant strains. However, knocking out the PUT1 gene in the double mutants pro2/put1 and pro3/put1 did not rescue the sensitivity of the pro2 and pro3 mutants to DTT and Tm, respectively (Fig. 7). Instead, the double mutant pro3/put1 appeared to be more sensitive to Tm but was still rescued by proline.
Proline Protects hog1 and ire1 Null Mutants against ER Stress
Finally, we tested whether proline can rescue ER stress-sensitive mutants apart from the proline metabolic pathway. The effect of proline on the ER stress-sensitive mutants hog1 and ire1 was examined by overexpressing the mutant Pro1p(D154N) in these mutants. Pro1p is feedback-inhibited by proline, thereby limiting accumulation of proline in the cell. Previously, it was shown that Pro1p(D154N) is less sensitive to feedback inhibition by proline with overexpression of Pro1p(D154N), resulting in higher intracellular proline levels (17). We used the same strategy here to increase intracellular proline levels in hog1 and ire1 cells. Cells overexpressing the Pro1p(D154N) mutant were determined to have 3-fold higher intracellular proline levels than control cells with the empty vector. Interestingly, overexpression of Pro1p(D154N) in hog1 and ire1 increased survival of these mutants under DTT or Tm stress (Fig. 8A and B). The effect of Pro1p(D154N) was most evident in the hog1 mutant, as rescue of ire1 was only observed at lower concentrations of DTT and Tm (Fig. 8).
FIGURE 8.
Proline protects hog1 and ire1 null mutants against ER stress. A and B, spot assays of yeast WT, ire1, and hog1 cells containing an empty vector (V) or the vector expressing the PRO1 mutant (PRO1-D154N) on SD plates alone (SD) or with DTT and Tm. C, cell death rate of hog1 cells assayed by PI staining. Mutant cells were grown in SD medium to log phase and were then treated with 2 mm DTT or 1 μg/ml Tm in the presence of varying concentrations of proline (0, 10, and 20 mm) for 16 h. Dead cells were then stained with PI. D, KAR2 mRNA levels were analyzed by real-time PCR in WT and hog1 cells grown in SD medium to log phase without (−Pro) or with (+Pro) 20 mm proline followed by treatment with Tm (1 μg/ml) for 0, 2, and 4 h. The mRNA levels were normalized to the internal control ACT1 with the lowest expression set at 1 (*, p < 0.05). Data are the mean ± S.D. from three independent experiments.
Proline protection of hog1 was further tested by determining the death rate of the hog1 mutant cells after the addition of 2 mm DTT or 1 μg/ml Tm in liquid SD medium containing 0, 10, or 20 mm proline for 16 h. The death rate of hog1 mutant cells treated with DTT or Tm in SD medium was reduced by 60 and 30%, respectively, when supplemented with proline (Fig. 8C). To gain molecular insight into proline protection of the hog1 mutant, we examined the expression of the ER stress marker KAR2 in hog1 and wild-type cells under Tm stress with and without proline (Fig. 8D). In cells without proline, KAR2 levels remained elevated in the hog1 mutant much longer than in wild-type cells, showing that the hog1 mutant recovers more slowly from Tm stress, which is consistent with a previous report (25). In cells supplemented with proline, however, the KAR2 expression profile in the hog1 mutant is similar to that of wild type, indicating that proline helps hog1 recover in the late phase of ER stress response (Fig. 8D).
DISCUSSION
In this study we found that genetic depletion of individual steps in proline biosynthesis results in distinct growth and ER stress phenotypes. The pro3 mutant has a strict requirement for proline (41) and showed high sensitivity to ER stress-inducing agents. Proline limitation in yeast S. cerevisiae, as demonstrated in the pro3 mutant, leads to ROS accumulation, higher NADP+ and total GSH levels, and inhibition of the UPR. Tm sensitivity of the pro3 mutant was rescued by NAC or GSH treatment and by anaerobic growth conditions, indicating proline limitation negatively impacts antioxidant defense systems and the UPR during ER stress recovery.
Amino acid limitation effectively reduces ER protein loading and is thought to benefit cells during ER stress (55–57). Yeast mutants deficient for ribosomal large subunits or with less cytosolic mRNA were found to be more resistant to Tm, which may be due to lower protein synthesis overall (58, 59). In agreement with this notion, we found that limiting leucine, uracil, and histidine slightly improved tolerance to Tm. In contrast, we found that genetic depletion of proline biosynthesis decreased ER stress tolerance. This implies that proline may have a unique role in ER stress protection. The expression levels of Pro1p, Pro2p, and Pro3p do not change significantly after DTT or Tm treatment. In a yeast gene microarray experiment, only 28 genes were induced by DTT, although 195 DTT stress-sensitive null mutants were identified (26). Thus, a large number of genes that are critical for DTT stress survival are not necessarily induced by DTT (26). During ER stress, proline levels did not change. However, we did detect 4-fold higher PUT1 mRNA levels after Tm treatment (data not shown), indicating increased proline degradation. Thus, an important role of the proline biosynthesis pathway may be to maintain intracellular proline levels during ER stress.
Protein misfolding is often connected with oxidative stress and depletion of GSH (60, 61). Glutathione is critical for mitigating ROS generated by the ER during oxidative protein folding (49, 61–64). In mammalian cells, glutathione biosynthesis and assimilation are up-regulated during ER stress via the antioxidant transcription factor Nrf2 (65) and the eIF2α signaling pathway (55, 66). Disruption of these signaling pathways leads to increased ROS- and ER stress-induced cell death, which can be rescued by NAC or GSH (55, 66). Previously in yeast, ROS was detected in wild-type cells treated with Tm (35) and a yeast ER-associated protein degradation mutant in which misfolded protein was overexpressed (67). GSH supplementation was shown to mitigate ROS accumulation and the ER stress sensitivity of the null ER-associated protein degradation mutant (67). Yeast null mutants of superoxide dismutase (SOD1) and YAP1 have also been shown to be sensitive to Tm treatment (35). Consistent with these earlier results, we also show here that ROS levels and glutathione synthesis increase in wild-type and pro3 cells after Tm treatment. Thus, our data suggest a role for ROS defense in yeast during ER stress.
Characterization of the pro3 mutant suggests an underlying mechanism by which proline limitation increases ER stress sensitivity involves disruption of redox homeostasis. At 2 mm proline, a redox imbalance in pro3 cells is evidenced by increased intracellular ROS levels, an elevated NADP+/NADPH ratio (68), and higher expression and activity of GSH1 and GLR, respectively, which are regulated by Yap1p in response to oxidative stress (69–71). Under proline limitation, we also observed higher total glutathione levels, which may be caused by the increased ROS stimulated GSH synthesis, which has been observed in other yeast mutants defective in oxidative stress defense (72, 73). Because the Tm sensitivity of the pro3 mutant is reversed by anaerobic growth conditions and by treatment with NAC or GSH, the redox imbalance in pro3 cells likely heightens ER stress sensitivity. Exactly how proline limitation leads to a more oxidative environment in pro3 cells is not clear. In addition to nutrient starvation (74–76), ER stress or protein misfolding may contribute to the oxidative stress observed in pro3 cells. Increasing the proline concentration to 4 mm restored the redox status of the pro3 mutant to near that of wild-type cells and greatly diminished DTT sensitivity. A higher concentration of proline (10 mm), however, was needed to fully repress the Tm sensitivity of pro3.
Oxidative and ER stress coexist in many pathologic states and may trigger one another, but exactly how they interact is not clear (77). Our data show that oxidative stress exacerbates ER stress sensitivity via inhibition of the UPR. The UPR is essential for yeast cells to survive under ER stress resulting in increased chaperone synthesis, inhibition of protein synthesis to reduce ER protein loading, and increased antioxidant defenses (21, 61). We observed that the UPR response to Tm is rapidly inhibited by H2O2 in wild-type cells and by proline limitation in pro3 cells (2 mm proline), demonstrating oxidative stress profoundly impacts ER stress tolerance. Previous studies in mammalian cells showed that exogenous oxidants such as peroxides and ROS generators can trigger several aspects of the UPR (77). Interestingly in yeast, H2O2 alone does not induce HAC1 splicing; rather, it inhibits HAC1 splicing induced by Tm (Fig. 5, D and E). NAC partially restored the UPR upon Tm in H2O2-treated wild-type cells and in pro3 cells under proline limitation. Proline fully rescues the UPR in the pro3 mutant, which correlates with an improved intracellular redox environment at higher proline concentrations (4 and 10 mm). Thus, one of the mechanisms by which NAC and proline decrease Tm toxicity involves protection of the UPR, likely by removing oxidative stress. Besides the UPR, diminished oxidative stress would also benefit ER stress tolerance by down-regulation of cellular death processes such as apoptosis. Proline has been shown previously to function as an anti-apoptotic factor in yeast (78).
Reports are sparse about how the UPR is inhibited by H2O2. In a study of the mutant sod1, NADP+ levels increased whereas NAPDH levels remained constant during ER stress. It was proposed that increases in superoxide interrupt the overall NADP(H) flux leading to a higher NADP+/NADPH ratio and thereby reducing the UPR during ER stress (35). Similarly, we observed a higher NADP+/NADPH ratio due to increased NADP+ in pro3 cells under proline limiting conditions. Thus, in an analogous manner to that of sod1, increased ROS caused by proline limitation may interfere with NADP(H) flux leading to inhibition of the UPR. The increased NADP+ found in pro3 cells at 2 mm proline may result from an imbalance of higher GLR activity and diminished flux of the pentose phosphate pathway during proline limitation (68). Proline metabolism was shown previously to be coupled with the pentose phosphate pathway (79), indicating a potential mechanism by which proline impacts intracellular redox homeostasis (2).
In contrast to ROS and NADP+, the higher GSH levels detected in pro3 cells are less likely to inhibit the UPR. It was previously reported that disruption of TRR1 results in a 3-fold increase in GSH levels, which was proposed to contribute to more misfolded protein and induce the UPR (73). Overexpression of GSH1 also led to a 30% increase in UPR induction (63). Thus, it is unlikely that higher glutathione levels lead to a reduced UPR in pro3 cells. However, the higher GSH levels would be expected to exacerbate DTT toxicity as seen for pro3 and pro2 cells. Depletion of thioredoxin in the double mutant trx1/trx2 resulted in a 3-fold increase in total GSH and increased sensitivity to DTT (73), indicating that elevated GSH increases sensitivity to reductive stress in the ER (63). Besides interruption of redox homeostasis, another mechanism by which proline depletion inhibits the UPR may occur via decreased synthesis or destabilization of a protein that is critical to the UPR.
Overall, the results from this study indicate that proline biosynthesis and proline have fundamental roles in ER stress protection. We propose that the major mechanism by which proline biosynthesis protects against ER stress is 2-fold involving maintenance of intracellular redox homeostasis and the UPR. However, other potential roles of proline in ER stress should not be ignored as evidenced by partial rescue of the ER stress-sensitive hog1 and ire1 mutant strains with overexpression of the Pro1p(D154N) mutant. The ability of proline to protect hog1 cells against ER stress indicates that proline can also function as an osmolyte substitute for glycerol in hog1. As an established antioxidant and osmolyte, proline may serve as an important stabilizer of redox balance and protein folding during ER stress that benefits overall redox homeostasis, the UPR, and protein folding.
Understanding which ROS defense systems are most critical for combating ER stress will be an important avenue of research in the future. Although the yap1 mutant is sensitive to ER stress, we have observed that other mutant strains such as sod2 and cta1 are only slightly sensitive to Tm-induced ER stress despite being highly sensitive to superoxide and H2O2, respectively (data not shown). These results indicate that defense against ER stress-induced oxidative stress may rely on specific ROS defense systems. This notion is also supported by our data showing that cells under proline limitation are more sensitive to ER stress than oxidative stress alone (see Fig. 5B). Thus, proline may have a more important role in combating ROS that originates from ER stress rather than oxidative stress in general. Determining how the redox state of the ER is impacted by proline limitation may provide novel insights into the mechanism of proline protection.
Acknowledgments
We thank Terri Fangman and Dr. You Zhou from the University of Nebraska Microscopy Core Facility for expert advice and technical assistance with cell imaging.
This work was supported, in whole or in part, by National Institutes of Health Grants GM079393 and P30GM103335. This work was also supported by the University of Nebraska Agricultural Research Division.
- GSA
- glutamate-γ-semialdehyde
- ER
- endoplasmic reticulum
- GLR
- glutathione reductase
- GSH1
- γ-glutamylcysteine synthetase 1
- IRE1
- inositol-requiring protein
- NAC
- N-acetyl-l-cysteine
- PI
- propidium iodide
- P5C
- Δ1-pyrroline-5-carboxylate
- ROS
- reactive oxygen species
- Tm
- tunicamycin
- UPR
- unfolded protein response
- NADP+
- nicotinamide adenine dinucleotide phosphate
- NADPH
- reduced form of NADP+.
REFERENCES
- 1. Pérez-Arellano I., Carmona-Álvarez F., Gallego J., Cervera J. (2010) Molecular mechanisms modulating glutamate kinase activity. Identification of the proline feedback inhibitor binding site. J. Mol. Biol. 404, 890–901 [DOI] [PubMed] [Google Scholar]
- 2. Liang X., Zhang L., Natarajan S. K., Becker D. F. (2013) Proline mechanisms of stress survival. Antioxid. Redox Signal. 19, 998–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Krishnan N., Dickman M. B., Becker D. F. (2008) Proline modulates the intracellular redox environment and protects mammalian cells against oxidative stress. Free Radic. Biol. Med. 44, 671–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Natarajan S. K., Zhu W., Liang X., Zhang L., Demers A. J., Zimmerman M. C., Simpson M. A., Becker D. F. (2012) Proline dehydrogenase is essential for proline protection against hydrogen peroxide-induced cell death. Free Radic. Biol. Med. 53, 1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu W., Le A., Hancock C., Lane A. N., Dang C. V., Fan T. W., Phang J. M. (2012) Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc. Natl. Acad. Sci. U.S.A. 109, 8983–8988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zarse K., Schmeisser S., Groth M., Priebe S., Beuster G., Kuhlow D., Guthke R., Platzer M., Kahn C. R., Ristow M. (2012) Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial l-proline catabolism to induce a transient ROS signal. Cell Metab. 15, 451–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Szabados L., Savouré A. (2010) Proline: a multifunctional amino acid. Trends Plant Sci. 15, 89–97 [DOI] [PubMed] [Google Scholar]
- 8. Székely G., Abrahám E., Cséplo A., Rigó G., Zsigmond L., Csiszár J., Ayaydin F., Strizhov N., Jásik J., Schmelzer E., Koncz C., Szabados L. (2008) Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 53, 11–28 [DOI] [PubMed] [Google Scholar]
- 9. Yildirim Y., Tolun A., Tuysuz B. (2011) The phenotype caused by PYCR1 mutations corresponds to geroderma osteodysplasticum rather than autosomal recessive cutis laxa type 2. Am. J. Med. Genet. A 155, 134–140 [DOI] [PubMed] [Google Scholar]
- 10. Reversade B., Escande-Beillard N., Dimopoulou A., Fischer B., Chng S. C., Li Y., Shboul M., Tham P. Y., Kayserili H., Al-Gazali L., Shahwan M., Brancati F., Lee H., O'Connor B. D., Schmidt-von Kegler M., Merriman B., Nelson S. F., Masri A., Alkazaleh F., Guerra D., Ferrari P., Nanda A., Rajab A., Markie D., Gray M., Nelson J., Grix A., Sommer A., Savarirayan R., Janecke A. R., Steichen E., Sillence D., Hausser I., Budde B., Nürnberg G., Nürnberg P., Seemann P., Kunkel D., Zambruno G., Dallapiccola B., Schuelke M., Robertson S., Hamamy H., Wollnik B., Van Maldergem L., Mundlos S., Kornak U. (2009) Mutations in PYCR1 cause cutis laxa with progeroid features. Nat. Genet. 41, 1016–1021 [DOI] [PubMed] [Google Scholar]
- 11. Baumgartner M. R., Hu C. A., Almashanu S., Steel G., Obie C., Aral B., Rabier D., Kamoun P., Saudubray J. M., Valle D. (2000) Hyperammonemia with reduced ornithine, citrulline, arginine and proline: a new inborn error caused by a mutation in the gene encoding delta(1)-pyrroline-5-carboxylate synthase. Hum. Mol. Genet. 9, 2853–2858 [DOI] [PubMed] [Google Scholar]
- 12. Bicknell L. S., Pitt J., Aftimos S., Ramadas R., Maw M. A., Robertson S. P. (2008) A missense mutation in ALDH18A1, encoding Δ1-pyrroline-5-carboxylate synthase (P5CS), causes an autosomal recessive neurocutaneous syndrome. Eur. J. Hum. Genet. 16, 1176–1186 [DOI] [PubMed] [Google Scholar]
- 13. Ernst T., Hergenhahn M., Kenzelmann M., Cohen C. D., Bonrouhi M., Weninger A., Klären R., Gröne E. F., Wiesel M., Güdemann C., Küster J., Schott W., Staehler G., Kretzler M., Hollstein M., Gröne H. J. (2002) Decrease and gain of gene expression are equally discriminatory markers for prostate carcinoma: a gene expression analysis on total and microdissected prostate tissue. Am. J. Pathol. 160, 2169–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jariwala U., Prescott J., Jia L., Barski A., Pregizer S., Cogan J. P., Arasheben A., Tilley W. D., Scher H. I., Gerald W. L., Buchanan G., Coetzee G. A., Frenkel B. (2007) Identification of novel androgen receptor target genes in prostate cancer. Mol. Cancer 6, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Okabe H., Satoh S., Kato T., Kitahara O., Yanagawa R., Yamaoka Y., Tsunoda T., Furukawa Y., Nakamura Y. (2001) Genome-wide analysis of gene expression in human hepatocellular carcinomas using cDNA microarray: identification of genes involved in viral carcinogenesis and tumor progression. Cancer Res. 61, 2129–2137 [PubMed] [Google Scholar]
- 16. Nilsson R., Jain M., Madhusudhan N., Sheppard N. G., Strittmatter L., Kampf C., Huang J., Asplund A., Mootha V. K. (2014) Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat. Commun. 5, 3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Takagi H. (2008) Proline as a stress protectant in yeast: physiological functions, metabolic regulations, and biotechnological applications. Appl. Microbiol. Biotechnol. 81, 211–223 [DOI] [PubMed] [Google Scholar]
- 18. Ando A., Nakamura T., Murata Y., Takagi H., Shima J. (2007) Identification and classification of genes required for tolerance to freeze-thaw stress revealed by genome-wide screening of Saccharomyces cerevisiae deletion strains. FEMS Yeast Res. 7, 244–253 [DOI] [PubMed] [Google Scholar]
- 19. Lin J. H., Walter P., Yen T. S. (2008) Endoplasmic reticulum stress in disease pathogenesis. Annu. Rev. Pathol. 3, 399–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tabas I., Ron D. (2011) Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat. Cell Biol. 13, 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Patil C., Walter P. (2001) Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 13, 349–355 [DOI] [PubMed] [Google Scholar]
- 22. Bonilla M., Cunningham K. W. (2003) Mitogen-activated protein kinase stimulation of Ca2+ signaling is required for survival of endoplasmic reticulum stress in yeast. Mol. Biol. Cell 14, 4296–4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bonilla M., Nastase K. K., Cunningham K. W. (2002) Essential role of calcineurin in response to endoplasmic reticulum stress. EMBO J. 21, 2343–2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Torres-Quiroz F., García-Marqués S., Coria R., Randez-Gil F., Prieto J. A. (2010) The activity of yeast Hog1 MAPK is required during endoplasmic reticulum stress induced by tunicamycin exposure. J. Biol. Chem. 285, 20088–20096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bicknell A. A., Tourtellotte J., Niwa M. (2010) Late phase of the endoplasmic reticulum stress response pathway is regulated by Hog1 MAP kinase. J. Biol. Chem. 285, 17545–17555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rand J. D., Grant C. M. (2006) The thioredoxin system protects ribosomes against stress-induced aggregation. Mol. Biol. Cell 17, 387–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Samuel D., Kumar T. K., Ganesh G., Jayaraman G., Yang P. W., Chang M. M., Trivedi V. D., Wang S. L., Hwang K. C., Chang D. K., Yu C. (2000) Proline inhibits aggregation during protein refolding. Protein Sci. 9, 344–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kumar T. K., Samuel D., Jayaraman G., Srimathi T., Yu C. (1998) The role of proline in the prevention of aggregation during protein folding in vitro. Biochem. Mol. Biol. Int. 46, 509–517 [DOI] [PubMed] [Google Scholar]
- 29. Ignatova Z., Gierasch L. M. (2006) Inhibition of protein aggregation in vitro and in vivo by a natural osmoprotectant. Proc. Natl. Acad. Sci. U.S.A. 103, 13357–13361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 31. Grant C. M., Perrone G., Dawes I. W. (1998) Glutathione and catalase provide overlapping defenses for protection against hydrogen peroxide in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 253, 893–898 [DOI] [PubMed] [Google Scholar]
- 32. Vandeputte C., Guizon I., Genestie-Denis I., Vannier B., Lorenzon G. (1994) A microtiter plate assay for total glutathione and glutathione disulfide contents in cultured/isolated cells: performance study of a new miniaturized protocol. Cell Biol. Toxicol. 10, 415–421 [DOI] [PubMed] [Google Scholar]
- 33. Chen C., Wanduragala S., Becker D. F., Dickman M. B. (2006) Tomato QM-like protein protects Saccharomyces cerevisiae cells against oxidative stress by regulating intracellular proline levels. Appl. Environ. Microbiol. 72, 4001–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carlberg I., Mannervik B. (1985) Glutathione reductase. Methods Enzymol. 113, 484–490 [DOI] [PubMed] [Google Scholar]
- 35. Tan S. X., Teo M., Lam Y. T., Dawes I. W., Perrone G. G. (2009) Cu,Zn superoxide dismutase and NADP(H) homeostasis are required for tolerance of endoplasmic reticulum stress in Saccharomyces cerevisiae. Mol. Biol. Cell 20, 1493–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zerez C. R., Lee S. J., Tanaka K. R. (1987) Spectrophotometric determination of oxidized and reduced pyridine nucleotides in erythrocytes using a single extraction procedure. Anal. Biochem. 164, 367–373 [DOI] [PubMed] [Google Scholar]
- 37. Cox J. S., Walter P. (1996) A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87, 391–404 [DOI] [PubMed] [Google Scholar]
- 38. Cox J. S., Shamu C. E., Walter P. (1993) Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73, 1197–1206 [DOI] [PubMed] [Google Scholar]
- 39. Tomenchok D. M., Brandriss M. C. (1987) Gene-enzyme relationships in the proline biosynthetic pathway of Saccharomyces cerevisiae. J. Bacteriol. 169, 5364–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brandriss M. C., Magasanik B. (1981) Subcellular compartmentation in control of converging pathways for proline and arginine metabolism in Saccharomyces cerevisiae. J. Bacteriol. 145, 1359–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brandriss M. C. (1979) Isolation and preliminary characterization of Saccharomyces cerevisiae proline auxotrophs. J. Bacteriol. 138, 816–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Balasubramanian M. N., Butterworth E. A., Kilberg M. S. (2013) Asparagine synthetase: regulation by cell stress and involvement in tumor biology. Am. J. Physiol. Endocrinol. Metab. 304, E789–E799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jämsä E., Simonen M., Makarow M. (1994) Selective retention of secretory proteins in the yeast endoplasmic reticulum by treatment of cells with a reducing agent. Yeast 10, 355–370 [DOI] [PubMed] [Google Scholar]
- 44. Pollard M. G., Travers K. J., Weissman J. S. (1998) Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol. Cell 1, 171–182 [DOI] [PubMed] [Google Scholar]
- 45. Barnes G., Hansen W. J., Holcomb C. L., Rine J. (1984) Asparagine-linked glycosylation in Saccharomyces cerevisiae: genetic analysis of an early step. Mol. Cell. Biol. 4, 2381–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hinnebusch A. G. (2005) Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59, 407–450 [DOI] [PubMed] [Google Scholar]
- 47. Han J., Back S. H., Hur J., Lin Y. H., Gildersleeve R., Shan J., Yuan C. L., Krokowski D., Wang S., Hatzoglou M., Kilberg M. S., Sartor M. A., Kaufman R. J. (2013) ER stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 15, 481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen C., Dickman M. B. (2004) Dominant active Rac and dominant negative Rac revert the dominant active Ras phenotype in Colletotrichum trifolii by distinct signalling pathways. Mol. Microbiol. 51, 1493–1507 [DOI] [PubMed] [Google Scholar]
- 49. Chakravarthi S., Jessop C. E., Bulleid N. J. (2006) The role of glutathione in disulphide bond formation and endoplasmic-reticulum-generated oxidative stress. EMBO Rep. 7, 271–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhao H., Kalivendi S., Zhang H., Joseph J., Nithipatikom K., Vásquez-Vivar J., Kalyanaraman B. (2003) Superoxide reacts with hydroethidine but forms a fluorescent product that is distinctly different from ethidium: potential implications in intracellular fluorescence detection of superoxide. Free Radic. Biol. Med. 34, 1359–1368 [DOI] [PubMed] [Google Scholar]
- 51. Chen J., Zhang Y., Wang C., Lü W., Jin J. B., Hua X. (2011) Proline induces calcium-mediated oxidative burst and salicylic acid signaling. Amino Acids 40, 1473–1484 [DOI] [PubMed] [Google Scholar]
- 52. Stathopoulos A. M., Cyert M. S. (1997) Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 11, 3432–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Marco-Marín C., Gil-Ortiz F., Pérez-Arellano I., Cervera J., Fita I., Rubio V. (2007) A novel two-domain architecture within the amino acid kinase enzyme family revealed by the crystal structure of Escherichia coli glutamate 5-kinase. J. Mol. Biol. 367, 1431–1446 [DOI] [PubMed] [Google Scholar]
- 54. Sekine T., Kawaguchi A., Hamano Y., Takagi H. (2007) Desensitization of feedback inhibition of the Saccharomyces cerevisiae γ-glutamyl kinase enhances proline accumulation and freezing tolerance. Appl. Environ. Microbiol. 73, 4011–4019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Harding H. P., Zhang Y., Zeng H., Novoa I., Lu P. D., Calfon M., Sadri N., Yun C., Popko B., Paules R., Stojdl D. F., Bell J. C., Hettmann T., Leiden J. M., Ron D. (2003) An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619–633 [DOI] [PubMed] [Google Scholar]
- 56. Patil C. K., Li H., Walter P. (2004) Gcn4p and novel upstream activating sequences regulate targets of the unfolded protein response. PLoS Biol. 2, E246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jousse C., Oyadomari S., Novoa I., Lu P., Zhang Y., Harding H. P., Ron D. (2003) Inhibition of a constitutive translation initiation factor 2α phosphatase, CReP, promotes survival of stressed cells. J. Cell Biol. 163, 767–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Delaney J. R., Ahmed U., Chou A., Sim S., Carr D., Murakami C. J., Schleit J., Sutphin G. L., An E. H., Castanza A., Fletcher M., Higgins S., Jelic M., Klum S., Muller B., Peng Z. J., Rai D., Ros V., Singh M., Wende H. V., Kennedy B. K., Kaeberlein M. (2013) Stress profiling of longevity mutants identifies Afg3 as a mitochondrial determinant of cytoplasmic mRNA translation and aging. Aging Cell 12, 156–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Steffen K. K., McCormick M. A., Pham K. M., MacKay V. L., Delaney J. R., Murakami C. J., Kaeberlein M., Kennedy B. K. (2012) Ribosome deficiency protects against ER stress in Saccharomyces cerevisiae. Genetics 191, 107–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bhandary B., Marahatta A., Kim H. R., Chae H. J. (2012) An involvement of oxidative stress in endoplasmic reticulum stress and its associated diseases. Int. J. Mol. Sci. 14, 434–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Santos C. X., Tanaka L. Y., Wosniak J., Laurindo F. R. (2009) Mechanisms and implications of reactive oxygen species generation during the unfolded protein response: roles of endoplasmic reticulum oxidoreductases, mitochondrial electron transport, and NADPH oxidase. Antioxid. Redox Signal. 11, 2409–2427 [DOI] [PubMed] [Google Scholar]
- 62. Jessop C. E., Bulleid N. J. (2004) Glutathione directly reduces an oxidoreductase in the endoplasmic reticulum of mammalian cells. J. Biol. Chem. 279, 55341–55347 [DOI] [PubMed] [Google Scholar]
- 63. Cuozzo J. W., Kaiser C. A. (1999) Competition between glutathione and protein thiols for disulphide-bond formation. Nat. Cell Biol. 1, 130–135 [DOI] [PubMed] [Google Scholar]
- 64. Molteni S. N., Fassio A., Ciriolo M. R., Filomeni G., Pasqualetto E., Fagioli C., Sitia R. (2004) Glutathione limits Ero1-dependent oxidation in the endoplasmic reticulum. J. Biol. Chem. 279, 32667–32673 [DOI] [PubMed] [Google Scholar]
- 65. Cullinan S. B., Zhang D., Hannink M., Arvisais E., Kaufman R. J., Diehl J. A. (2003) Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol. Cell. Biol. 23, 7198–7209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rouschop K. M., Dubois L. J., Keulers T. G., van den Beucken T., Lambin P., Bussink J., van der Kogel A. J., Koritzinsky M., Wouters B. G. (2013) PERK/eIF2α signaling protects therapy resistant hypoxic cells through induction of glutathione synthesis and protection against ROS. Proc. Natl. Acad. Sci. U.S.A. 110, 4622–4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Haynes C. M., Titus E. A., Cooper A. A. (2004) Degradation of misfolded proteins prevents ER-derived oxidative stress and cell death. Mol. Cell 15, 767–776 [DOI] [PubMed] [Google Scholar]
- 68. Ying W. (2008) NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxid. Redox Signal. 10, 179–206 [DOI] [PubMed] [Google Scholar]
- 69. Wu A. L., Moye-Rowley W. S. (1994) GSH1, which encodes γ-glutamylcysteine synthetase, is a target gene for yAP-1 transcriptional regulation. Mol. Cell. Biol. 14, 5832–5839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Stephen D. W., Rivers S. L., Jamieson D. J. (1995) The role of the YAP1 and YAP2 genes in the regulation of the adaptive oxidative stress responses of Saccharomyces cerevisiae. Mol. Microbiol. 16, 415–423 [DOI] [PubMed] [Google Scholar]
- 71. Grant C. M., Collinson L. P., Roe J. H., Dawes I. W. (1996) Yeast glutathione reductase is required for protection against oxidative stress and is a target gene for yAP-1 transcriptional regulation. Mol. Microbiol. 21, 171–179 [DOI] [PubMed] [Google Scholar]
- 72. Drakulic T., Temple M. D., Guido R., Jarolim S., Breitenbach M., Attfield P. V., Dawes I. W. (2005) Involvement of oxidative stress response genes in redox homeostasis, the level of reactive oxygen species, and ageing in Saccharomyces cerevisiae. FEMS Yeast Res. 5, 1215–1228 [DOI] [PubMed] [Google Scholar]
- 73. Trotter E. W., Grant C. M. (2002) Thioredoxins are required for protection against a reductive stress in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 46, 869–878 [DOI] [PubMed] [Google Scholar]
- 74. Scherz-Shouval R., Elazar Z. (2007) ROS, mitochondria and the regulation of autophagy. Trends Cell Biol. 17, 422–427 [DOI] [PubMed] [Google Scholar]
- 75. Scherz-Shouval R., Shvets E., Fass E., Shorer H., Gil L., Elazar Z. (2007) Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 26, 1749–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Chen Y., Azad M. B., Gibson S. B. (2009) Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ. 16, 1040–1052 [DOI] [PubMed] [Google Scholar]
- 77. Cao S. S., Kaufman R. J. (2014) Endoplasmic reticulum stress and oxidative stress in cell fate decision and human disease. Antioxid. Redox Signal. 21, 396–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chen C., Dickman M. B. (2005) Proline suppresses apoptosis in the fungal pathogen Colletotrichum trifolii. Proc. Natl. Acad. Sci. U.S.A. 102, 3459–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hagedorn C. H., Phang J. M. (1986) Catalytic transfer of hydride ions from NADPH to oxygen by the interconversions of proline and Δ1-pyrroline-5-carboxylate. Arch. Biochem. Biophys. 248, 166–174 [DOI] [PubMed] [Google Scholar]