Background: Methoprene-tolerant (Met) and Germ-cell expressed belonging to the bHLH-PAS family have been identified as juvenile hormone (JH) receptors in Drosophila.
Results: Physical interaction with Hsp83 facilitates nuclear import of Met and JH action.
Conclusion: Hsp83 modulates JH signaling through mediating the nuclear localization of Met.
Significance: Our study helps in understanding the complicated molecular mechanisms of JH signaling.
Keywords: Basic Helix-loop-helix Transcription Factor (bHLH), Drosophila, Heat Shock Protein 90 (Hsp90), Hormone Receptor, Insect, bHLH Transcription Factor, Cytoplasm-Nucleus Shuttling, Insect Hormones, Molecular Chaperone;
Abstract
Juvenile hormone (JH) receptors, methoprene-tolerant (Met) and Germ-cell expressed (Gce), transduce JH signals to induce Kr-h1 expression in Drosophila. Dual luciferase assay identified a 120-bp JH response region (JHRR) in the Kr-h1α promoter. Both in vitro and in vivo experiments revealed that Met and Gce transduce JH signals to induce Kr-h1 expression through the JHRR. DNA affinity purification identified chaperone protein Hsp83 as one of the proteins bound to the JHRR in the presence of JH. Interestingly, Hsp83 physically interacts with PAS-B and basic helix-loop-helix domains of Met, and JH induces Met-Hsp83 interaction. As determined by immunohistochemistry, Met is mainly distributed in the cytoplasm of fat body cells of the larval when the JH titer is low and JH induces Met nuclear import. Hsp83 was accumulated in the cytoplasm area adjunct to the nucleus in the presence of JH and Met/Gce. Loss-of-function of Hsp83 attenuated JH binding and JH-induced nuclear import of Met, resulting in a decrease in the JHRR-driven reporter activity leading to reduction of Kr-h1 expression. These data show that Hsp83 facilitates the JH-induced nuclear import of Met that induces Kr-h1 expression through the JHRR.
Introduction
Juvenile hormones (JH)2 are a family of sesquiterpenoid compounds synthesized primarily by the corpus allatum (CA) in insects. JH and ecdysteroids (20-hydroxyecdysone (20E) is the most active form) coordinately control insect molting and metamorphosis. One major function of JH is to inhibit some actions of 20E during the larval stages and therefore, JH is usually referred to as the “status quo” hormone (1). In the fruit fly, Drosophila melanogaster, a dipteran insect, the status quo action of JH is subtle but functionally important. As demonstrated in a JH-deficient animal from which the CA is genetically ablated, JH prevents 20E-triggered apoptosis of the larval fat body (2) and precocious differentiation of the optic lobe of adult brain (3) during the larval-pupal metamorphosis in Drosophila.
Significant progress has been made on the identification of the JH receptor and JH signal transduction pathway during the last two decades. Based on a genetic screen to isolate mutants that are resistant to a JH agonist, methoprene, in Drosophila, methoprene-tolerant (Met), a transcription factor belonging to the bHLH-PAS family, was suggested to be involved in JH action (4, 5). Met forms homodimers or heterodimers with its paralog, Germ-cell expressed (Gce), and JH reduces this dimerization (6). Although both Met and gce null mutants are viable, the Met gce double mutant dies during the larval-pupal transition (7), resembling what is seen in the JH-deficient animal (2, 3). Functionally, Met and Gce mediate JH action to prevent 20E-triggered metamorphic events and thus maintaining the status quo action (2, 3, 7). In the red flour beetle, Tribolium castaneum, reduction of Met expression by RNAi results in precocious metamorphosis (8, 9). Importantly, both Drosophila and Tribolium Met bind to JH at physiological concentrations in vitro (10, 11), suggesting strongly that Met is a bona fide JH receptor.
Krüppel homolog 1 (Kr-h1), encoding a zinc finger transcription factor, was first identified as a JH response gene in Drosophila (12). Kr-h1 consists of three isoforms: Kr-h1α, Kr-h1β, and Kr-h1γ (13). Both Kr-h1α and Kr-h1γ are JH responsive, whereas the mRNA of Kr-h1α is much more abundant than that of Kr-h1γ (12, 13). Later studies showed that Met and Gce transduce JH signals to induce Kr-h1 expression (7). In Tribolium, reduction of Kr-h1 expression by RNAi results in precocious metamorphosis and Kr-h1 works downstream of Met (14, 15). Studies in the mosquito, Aedes aegypti (16), and the silkworm, Bombyx mori (17), further confirm the general concept that Met mediates JH signals to induce Kr-h1 expression in insects.
Besides Met and Kr-h1, other proteins are also involved in JH signaling. The p160/SRC/NCoA-like molecule (known as Taiman in Drosophila, FISC in Aedes, and SRC in Tribolium and Bombyx), which belong to bHLH-PAS family transcriptional regulators, directly interact with Met and is required for JH signal transduction (15, 17–19). Cycle, which is also a bHLH-PAS transcriptional regulator, forms a heterodimer with Met to mediate circadian expression of JH-induced genes in Aedes (16). JH response elements (JHRE) in the Aedes AaET promoter and Bombyx, Aedes, and Tribolium Kr-h1 promoters have been identified as a C box sequence (CACGCG, an E-box-like motif) or a canonical E box sequence (CACGTG), which are essential for JH-induced gene expression through Met (16–19). A recent study in Aedes revealed that a consensus 9-mer Met-binding motif, CACG(C/T)G(A/G)(T/A)G, is conserved in the promoter regions of many JH response genes (20).
To better understand the JH signal transduction pathway in Drosophila, we identified a JHRR (which contains three E-box-like motifs) in the Kr-h1 promoter and used it to purify nuclear proteins bound to the JHRR. The chaperone protein, heat shock protein 83 (Hsp83), was identified as one of the proteins bound to JHRR in the presence of JH. We further illustrate that, in the presence of JH, Hsp83 interacts with Met and facilitates its nuclear import. Met then participates in induction of Kr-h1 expression through the JHRR.
EXPERIMENTAL PROCEDURES
Fly Strains and Genetics
Flies were raised at 25 °C using standard cornmeal/molasses/yeast medium. Aug21-GAL4, UAS-grim, Lsp2-GAL4, and Met27gce2.5k were reported previously (2, 7). The Hsp83 mutant, Hsp8308445, and UAS-Hsp83-RNAi (stock numbers 32996 and 33947) were obtained from the Bloomington Drosophila Stock Center. The UAS-Met-V5 construct was generated using the pUAST vector, whereas the JHRR-LacZ construct was generated using the pCaSpeR vector. Transgenic flies were produced by P-element-mediated germline transformation. Hsp8308445::Lsp2-GAL4, Hsp8308445::UAS-Met-V5, Lsp2-GAL4::UAS-Met-V5, and Lsp2-GAL4::JHRR-LacZ were obtained by genetic recombination. For the GAL4/UAS experiments, Lsp2-GAL4 or the flies recombined with Lsp2-GAL4 was crossed with the UAS lines. For the Flp-out experiments, hsFlpase; Act>CD2>GAL4; UAS-GFP was crossed with UAS-Met-V5 or UAS-Met-V5::UAS-Hsp83-RNAi to generate mosaic clones. Heat shock was performed at 6 h after egg laying (AEL) for 15 min.
Synchronization of Larval Development
The synchronization was performed at egg laying for studies at 96 h AEL (equal to the end of day 1 of the third instar or the onset of day 2 of the third instar) and at the initiation of wandering for studies at 3 h after the initiation of wandering (AIW) (21). The JH titer (22, 23), JH biosynthesis by the CA (24), and jhamt mRNA levels (25) are low at 96 h AEL and high at 3 h AIW, respectively.
β-Galactosidase Activity Assay and Immunohistochemistry
The larval fat body tissues were collected from different Drosophila lines at 96 h AEL and 3 h AIW. To test the expression of JHRR-LacZ, β-galactosidase activity was measured using the Animal Tissue β-galactosidase Activity Detection Kit (GENMED, China) according to the manufacturer's instructions.
The cellular localization of Met-V5 and Hsp83 was measured by immunohistochemistry as described previously (7, 21, 26, 27). Fat body tissues were incubated with the V5 mouse monoclonal antibody (R960–25, Invitrogen) or Hsp83 antibody (number 4874S, Cell Signaling), followed with Alexa Fluor 546 goat anti-mouse IgG (A11003, Invitrogen) and Alexa Fluor 488 goat anti-rabbit IgG (A11008, Invitrogen). Fluorescence signals were observed using an Olympus Fluoview FV1000 confocal microscope under the same conditions throughout the studies.
Cell Culture
Drosophila Kc cells were routinely maintained in Schneider's Drosophila medium (Lonza, Switzerland) supplemented with 5% fetal bovine serum (HyClone). When needed, 1 μm methoprene (stock concentration is 100 μg/μl, dissolved in DMSO) (Dr. Ehrenstorfer GmbH, Germany), 10 μg/ml of cycloheximide (Sigma), and/or 1 μg/ml of geldanamycin (GA) (Cell Signaling) were added to Kc cells. In these experiments, DMSO was used as a control. Transient transfection assays were carried out for 48 h using Effectene (Qiagen). The final DNA concentration was 0.2 μg/ml, and the DNA:Effectene ratio used was 1:25 (26–29). Using the T7 RiboMAX Express RNAi System (Promega), dsRNAs of Met, gce, Hsp83, and EGFP (as a control) were synthesized. Reduction of gene expression by RNAi in Kc cells was performed by transfecting dsRNAs using Effectene at a final concentration of 2 μg/ml of dsRNA (26–29). The transfected cells were cultured for 48 h.
Culture of Fat Body Tissues
The larval fat body tissues were collected from different Drosophila lines at 96 h AEL and 3 h AIW and cultured in Schneider's Drosophila medium supplemented with 5% fetal bovine serum. When needed, 1 μm methoprene and/or 1 μg/ml of GA were added to the medium.
Quantitative Real-time PCR and Western Blotting
Quantitative real-time PCR (qPCR) and Western blotting were performed as described previously (2, 26–29). The primary antibodies used for Western blotting were V5 mouse monoclonal antibody (R960–25, Invitrogen), c-Myc antibody (9E10) (sc-40, Santa Cruz Biotechnology), and monocloned ANTI-FLAG M2 antibody produced in mouse (F1804, Sigma).
Dual Luciferase Assay
For identification of the Kr-h1 JHRR, a 2-kb region of the Kr-h1α promoter upstream of the transcription start site was cloned into SacI and EcoRI sites of pGL3 basic vector containing the hsp70 minimal promoter. The deletions and mutations of promoter regions were also constructed into pGL3. After transient transfection in Kc cells with the reporter pGL3 vector and the reference pRL vector, the dual luciferase assays were performed as previously described (26–29).
Chromatin Immunoprecipitation Assay
Kc cells were transfected with the Met-V5 expression plasmid. At 48 h after transfection, the cells were exposed to 1 μm methoprene (or DMSO as a control) for 10 min. Then the cells were fixed and subjected to a chromatin immunoprecipitation assay (ChIP) using the PierceTM agarose ChIP Kit (Thermo) and the V5 mouse monoclonal antibody (R960-25, Invitrogen). Mock immunoprecipitations with preimmune serum were used for negative controls. The precipitated DNA and input were analyzed by qPCR to detect binding between Met-V5 and JHRR using primers 5′-CCACACCATGATGAAGCGCA-3′ and 5′-CTGGCAGAGTTCGCGTGGC-3′. In addition, the binding between Met-V5 and a 5′-UTR of Kr-h1α was also performed as a control by qPCR using primers 5′-GAATACGACATAACAGCC-3′ and 5′-CGATTTCCGTGAATATGTTCT-3′.
Affinity Purification and Identification of Nuclear Proteins Bound to JHRR
The nuclear proteins isolated from methoprene-treated Kc cells were used for affinity purification of nuclear proteins bound to the JHRR. Cells in 10 × 100-mm tissue culture dishes were treated with 1 μm methoprene (or DMSO as a control) for 10 min, and the cell pellets were collected in 50-ml tubes after centrifugation for 5 min at 1,000 × g. As published previously (29, 30), the nuclear proteins were extracted according to the manufacturer's protocols. Total protein content in the extracts was quantified by a BCA protein assay kit (Beyotime) and the extracts were stored at −80 °C until use.
JHRR fragments were synthesized by PCR amplification and phosphorylated using T4 polynucleotide kinase, followed by ligation with T4 DNA ligase to produce the polymerized JHRRs (31). Polymerized JHRRs were then coupled to commercial CNBr-activated Sepharose 4B resin (GE Healthcare) according to the manufacturer's instructions. The resulting polymerized JHRR affinity column was then used to isolate nuclear proteins bound to the JHRR as described previously (30). The eluted JHRR-binding proteins were digested with trypsin and subjected to HPLC-MS/MS analysis (Shanghai Applied Protein Technology, China) (2, 28).
Immunoprecipitation
Met-V5 (its mutated constructs and Flag-gce) and Myc-Hsp83 were cloned into pAc5.1 (Invitrogen) to generate the expression constructs. The constructs were transfected into Kc cells, at 48 h after transfection, the cells were harvested and lysed in ice-cold NP-40 lysis buffer (Beyotime, China). The procedure for immunoprecipitation has been previously described in detail (26, 28). The immunoprecipitates were separated by SDS-PAGE and analyzed by Western blotting.
Ligand Binding Assay
Dextran-coated charcoal assay was performed as described previously (10, 11). Kc cells were transfected with Met or EGFP overexpression plasmid. Prior to harvesting, cells were treated with GA for 5 h. The total proteins were extracted and incubated with 3H-JH III (20 Ci/mmol, PerkinElmer Life Sciences; final concentration = 100 nm) for 90 min at 22 °C. This was followed by the addition of 5% charcoal-dextran solution, gentle mixing for 2 min, and centrifugation for 1 min at 3,000 × g. The supernatant was collected into a scintillation vial and incubated overnight at 4 °C. Radioactivity was measured using a liquid scintillation counter (LS-6000IC, Beckman).
Statistics
Data were analyzed using the Student's t test and analysis of variance (ANOVA), t test: *, p < 0.05; **, p < 0.01. ANOVA analysis is shown as the bars labeled with different lowercase letters as significantly different (p < 0.05). The mean ± S.D. from at least three independent experiments is shown.
RESULTS
Identification of Response Regions to Methoprene in Kr-h1 Promoter
Kr-h1 was first identified as a JH response gene in Drosophila (12). We here found that methoprene induces Kr-h1 expression in a rapid and reversible manner in Drosophila Kc cells (Fig. 1A). The mRNA level of Kr-h1 increased about 5-fold within 5 min after methoprene treatment, gradually increased up to 8-fold by half an hour, and the induction was steadily decreased after 1 h (Fig. 1A). Moreover, the rapid induction of Kr-h1 expression by methoprene was not blocked by a protein synthesis inhibitor, cycloheximide, suggesting that Kr-h1 is a JH primary-response gene (Fig. 1B).
FIGURE 1.
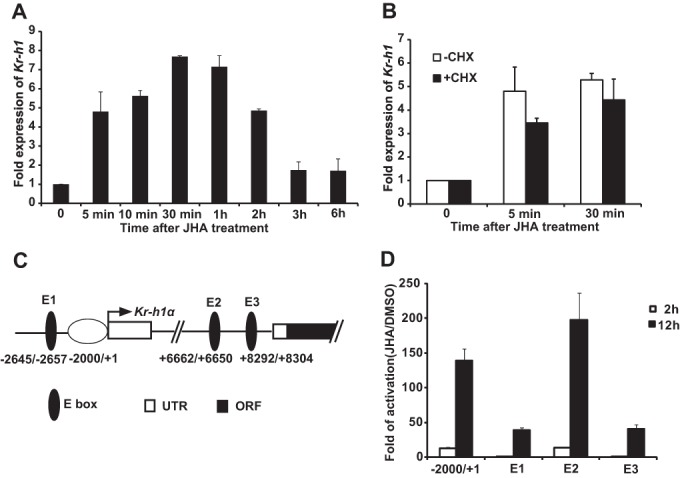
JH induces Kr-h1 expression through four JH response regions. A and B, for detecting the mRNA level of Kr-h1, Kc cells were pretreated with 10 μg/ml of cycloheximide (CHX, a protein synthesis inhibitor) for 2 h and treated with 1 μm methoprene (JHA) for the indicated time, and the relative Kr-h1 mRNA levels were measured by quantitative real-time PCR. A, time course of JH-induced Kr-h1 expression. B, Kr-h1 is a JH primary-response gene (t test showing no statistical difference). C, schematic representation of the four JHRRs in the promoter region and the first intron of Kr-h1α. Arrow represents the transcription start site (+1), and numbers indicate the distance from the transcription start site. D, Kc cells were co-transfected with the pGL3 basic plasmids containing the indicated promoter regions of Kr-h1, the hsp70 basal promoter regulating expression of firefly luciferase (Fluc), and a reference reporter plasmid carrying Rellina luciferase (Rluc). After 48 h of transfection, the cells were treated with 1 μm methoprene (JHA) for 2 or 12 h, and then the dual luciferase assays were performed. E1–E3, approximate 120-bp regions containing the canonical E-boxes in the middle.
To identify response regions to methoprene in the Kr-h1 promoter, we employed the dual luciferase assay system and Kc cells. A 2-kb Kr-h1α promoter region (−2000 to +1 bp upstream of the transcriptional start site) was cloned into pGL3 vector. This promoter region supported ∼10- and 140-fold increase in the luciferase activity within 2 and 12 h after methoprene treatment, respectively. Three other regions (E1, E2, and E3, about 120 bp in each region) containing canonical E-boxes of possible JHREs (17), which are present in the promoter region beyond 2 kb from the transcriptional start site and the first intron of Kr-h1α (Fig. 1C), were also cloned into pGL3. Among these three regions, E2 showed response to methoprene treatment similar to the 2-kb promoter region, whereas E1 and E3 showed lower response (Fig. 1D). Although the 2-kb promoter region and E2 showed the highest response to methoprene, it is likely that all four regions are involved in JH-induced Kr-h1 expression.
Identification of a JHRR in 2-kb Promoter Region of Kr-h1
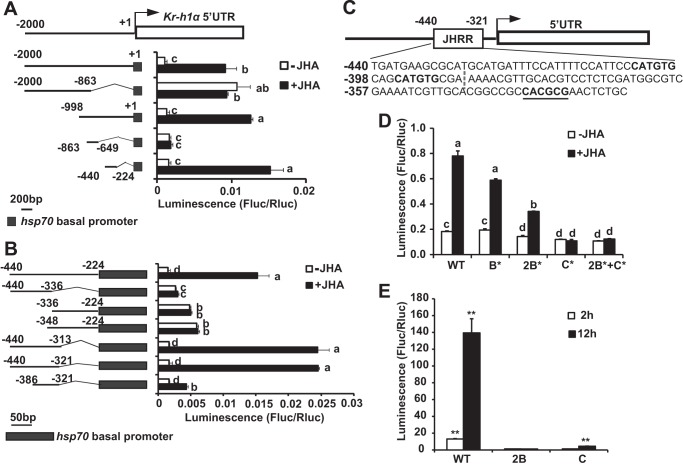
The 2-kb promoter region of Kr-h1α was chosen for further studies because it showed high response to methoprene treatment and is located right before the transcriptional start site. Subsequent reporter assays with a series of deletion constructs (−2000 to −863, −998 to +1, −863 to −649, and −440 to −224) showed that the region crucial for response to methoprene is located between −440 and −224 (Fig. 2A). Further reporter analyses with shorter deletion constructs (−440 to −336, −336 to −224, −348 to −224, −440 to −313, −440 to −321, and −386 to −321) identified a 120-bp region (−440 to −321) that is responsible for the majority of response to methoprene and is referred to as the JHRR (Fig. 2B).
FIGURE 2.
Identification of a JHRR in Kr-h1 promoter. A and B, Kc cells were co-transfected with the pGL3 basic plasmids containing truncations of 2-kb (A) or 216-bp (B) promoter regions of Kr-h1α and the hsp70 basal promoter regulating expression of firefly luciferase (Fluc) and a reference reporter plasmid carrying Rellina luciferase (Rluc). At 48 h after transfection, the cells were treated with 1 μm methoprene (JHA) for 2 h, and then dual luciferase assays were performed. Arrow represents the transcriptional start site (+1), numbers indicate the distance from the transcriptional start site, and shaded box represent the hsp70 basal promoter. A, the luciferase reporter activity supported by the −2000 to 1 of Kr-h1α promoter region truncations are shown; B, the luciferase reporter activity supported by −440 to −224 of the Kr-h1α promoter region truncations are shown. C, schematic representation of the location of JHRR (−440 to −321), which contains three E-box-like motifs. The nucleotide sequences are shown below the gene structure. Bold plus underlined nucleotides are C box, bold nucleotides are B boxes, and the dash separates the JHRR into two regions: −440 to −387 and −386 to −321. D, elucidation of the individual contribution of each E-box-like motif in the JHRR using the dual luciferase assays. WT, wild type JHRR; 1B*, mutation to the first B box of JHRR; 2B*, mutations to both B boxes of JHRR; C*, mutation to C box of JHRR; 2B*+C*, mutations to the two B boxes and C box of JHRR. E, Kc cells were co-transfected with pGL3 basic plasmids containing the WT JHRR, two copies of the two B boxes, or four copies of the C box (2B or C), the hsp70 basal promoter regulating expression of firefly luciferase (Fluc) and a reference reporter plasmid carrying Rellina luciferase (Rluc). After 48 h of transfection, the cells were treated with 1 μm JHA for 2 or 12 h, and dual luciferase assays were performed.
We noticed that the −386 to −321 region, which showed a much lower response to methoprene than the JHRR, contains a single C box (CACGCG), whereas the JHRR contains two additional B boxes (CATGTG) (Fig. 2, B and C). bHLH-PAS transcription factors are known to bind E-box-like motifs (CANNTG), including both C box and B box (32). To elucidate the individual contribution of each E-box-like motif in the JHRR, point mutations were introduced. Mutations to both B boxes resulted in significant reduction in the JH-induced luciferase activity, whereas a single mutation to the first B box caused no obvious reduction. However, mutation to the C box or all three E-box-like motifs completely abolished the luciferase activity (Fig. 2D). Moreover, the pGL3 reporter constructs carrying two copies of the two B boxes or four copies of the C box showed no response to methoprene within 2 h. The former construct still showed no induction of luciferase activity after 12 h of methoprene treatment, whereas the later construct showed 4.5-fold induction (Fig. 2E). In conclusion, all three E-box-like motifs in the JHRR are required for full response, but their relative contributions require further studies.
Met and Gce Transduce JH Signals to Induce Kr-h1 Expression through JHRR
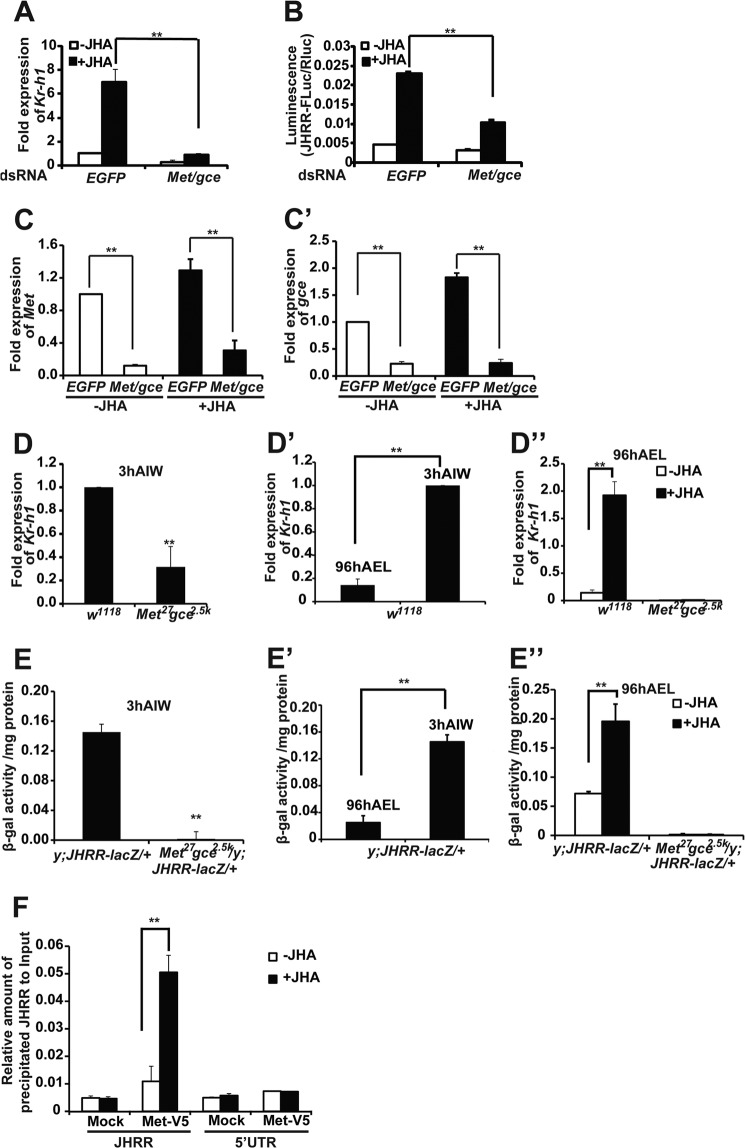
It has been documented that Met and Gce transduce JH signals to induce Kr-h1 expression in Drosophila (7). In Kc cells, the JH-induced Kr-h1 expression (Fig. 3A) and JHRR-driven luciferase activity (Fig. 3B) were significantly decreased when the mRNA levels of Met and gce were simultaneously reduced by RNAi (Fig. 3, C and C′). To avoid possible off-targeting effects of RNAi (33), multiple sets of Met and gce dsRNAs were generated and similar results were obtained (data not shown).
FIGURE 3.
Met and Gce are required for JH-induced Kr-h1 expression through JHRR. A and B, the JH-induced Kr-h1 expression and JHRR-driven luciferase activity were measured after Met and gce were simultaneously reduced by RNAi in Kc cells. EGFP RNAi was used as the control. A, for detecting the mRNA level of Kr-h1, Kc cells were treated with 1 μm methoprene (JHA) (DMSO as the control) for 30 min, and the relative Kr-h1 mRNA levels were measured by qPCR. B, for detecting the JHRR-driven luciferase activity, Kc cells were treated with 1 μm JHA (DMSO as the control) for 2 h, and then dual luciferase assays were performed. C and C′, RNAi efficiency of Met (C) and gce (C′) in Kc cells. D–D″) The mRNA levels of Kr-h1 in the fat body isolated from w1118 and Met27gce2.5k larvae at the indicated stage. 3 h AIW the JH titer is high. At 96 h AEL the JH titer is low. In D″, the isolated fat body tissues were treated with 1 μm methoprene (JHA) (DMSO as the control) for 30 min. E–E″, the JHRR-driven β-galactosidase (β-gal) activities were measured in the fat body isolated from the +/y; JHRR-LacZ and Met27gce2.5k/y; JHRR-LacZ/+ larvae at indicated stage. In E″, the isolated fat body tissues were treated with 1 μm methoprene (JHA) (DMSO as the control) for 2 h. F, ChIP. Kc cells were transfected with Met-V5 expression plasmid for 48 h followed methoprene (or DMSO as a control) treatment for 10 min. ChIP was performed using the PierceTM Agarose ChIP Kit (Thermo) and the V5 antibody to detect the binding between Met-V5 and the JHRR DNA or DNA in a 5′-UTR of Kr-h1α (in the 1st exon, lacking E-box-like motifs). The precipitated DNA and input were analyzed by qPCR. EGFP, enhanced green fluorescent protein.
Similar to the previous report that JH fails to induce Kr-h1 expression in the Met gce double mutant, Met27gce2.5k, during larval molts (7), Kr-h1 expression was significantly reduced in the fat body at 3 h AIW (Fig. 3D). In comparison with 3-h AIW when the JH titer is high, Kr-h1 expression was much lower in the wild type w1118 larvae at 96 h AEL when the JH titer is low (Fig. 3D′). Moreover, at 96 h AEL, Kr-h1 expression was significantly enhanced by the addition of methoprene to the cultured fat body tissues isolated from the w1118 larvae (Fig. 3D″, left panel) but not from the Met27gce2.5k larvae (Fig. 3D″, right panel).
We further produced JHRR-LacZ transgenic flies, and put JHRR-LacZ in the Met gce double mutant background to generate Met27gce2.5k/y; JHRR-LacZ/+. At 3 h AIW, the JHRR-driven β-galactosidase activity was very high in the fat body of JHRR-LacZ larvae, but became nearly undetectable in Met27gce2.5k/y; JHRR-LacZ/+ (Fig. 3E). In comparison to the 3-h AIW, the JHRR-driven β-galactosidase activity was lower in the JHRR-LacZ larvae at 96 h AEL (Fig. 3E′). Moreover, at 96 h AEL, the JHRR-driven β-galactosidase activity was significantly increased by addition of methoprene to fat body tissues isolated from JHRR-LacZ larvae (Fig. 3E″, left panel) but not from the Met27gce2.5k/y; JHRR-LacZ/+ larvae (Fig. 3E″, right panel). Taken together, these in vitro and in vivo data show that Met and Gce transduce JH signals to induce Kr-h1 expression through the JHRR.
Furthermore, we performed ChIP to examine whether Met directly binds to the JHRR, which contains three E-box-like motifs, for inducing Kr-h1 expression. The binding of Met-V5 to DNA was detected using the V5 antibody and cross-linked chromatin isolated from Kc cells, which were transfected with the Met-V5 expression plasmid. As measured by qPCR, V5 antibody weakly precipitated JHRR DNA, and the addition of methoprene increased the amount of precipitated JHRR DNA by 5-fold (Fig. 3F, left panel). By contrast, V5 antibody did not precipitate DNA from the 5′-UTR region of Kr-h1α, which lies in the first exon of Kr-h1α and contains no E-box-like motifs, and methoprene did not increase the precipitation (Fig. 3F, right panel). The ChIP-qPCR experimental data conclusively showed that Met directly binds to the JHRR for inducing Kr-h1 expression.
Identification of Hsp83 as One of the Proteins Bound to JHRR
To identify the proteins binding to the JHRR, nuclear proteins isolated from methoprene- or DMSO-treated Kc cells were purified using the polymerized JHRR affinity column and subjected to HPLC-MS/MS analysis. The chaperone protein Hsp83 (homolog of the mammalian Hsp90) was identified as a JHRR-bound nuclear protein present only in purified nuclear proteins isolated from methoprene-treated Kc cells but not from the DMSO-treated cells (Fig. 4). The other JHRR-bound nuclear proteins identified in either control or methoprene-treated Kc cells are listed in Tables 1 and 2.
FIGURE 4.
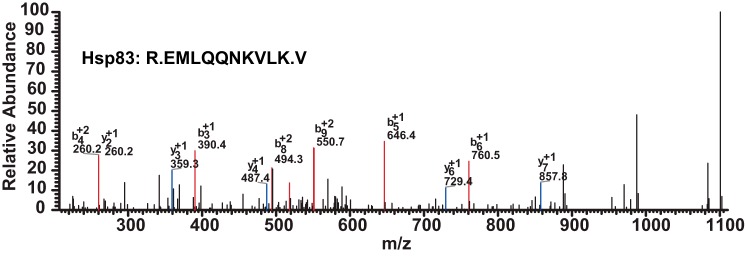
MS identification of Hsp83 as a nuclear protein bound to JHRR. Nuclear proteins isolated from methoprene- or DMSO-treated Kc cells were purified through the polymerized JHRR affinity column and subjected to HPLC-MS/MS analysis. The R.EMLQQNKVLK.V peptide fragment of Hsp83 was detected in the purified nuclear proteins isolated from methoprene-treated Kc cells but not from the DMSO-treated control Kc cells. PepCount, 3; UniquePepCount, 1; MW, 81865.49; PI, 4.91; MH+, 1247.4903; Diff(MH+), 0.2713; Charge, 2; Rank, 1.
TABLE 1.
The JHRR-bound nuclear proteins identified in methoprene-treated Kc cells using HPLC-MS/MS
| Reference | Pep count | Unique Pep count | Cover percent | Mr | PI | Sequence | MH+ | Diff (MH+) | Charge |
|---|---|---|---|---|---|---|---|---|---|
| HSP83 | 3 | 1 | 1.39 | 81865 | 4.91 | R.EM*LQQNKVLK.V | 1247.4903 | 0.2713 | 2 |
| Nup358 | 1 | 1 | 0.44 | 296385 | 5.76 | R.LNSNQLITEGNR.A | 1359.471 | 1.014 | 2 |
| Lon | 7 | 3 | 4.27 | 112893 | 6.89 | K.IENPLVLIDEVDK.I | 1497.7154 | 0.8384 | 2 |
| K.IVEVSNPIIMDLLR.R | 1612.9594 | −0.0616 | 2 | ||||||
| K.VVPEAIMTVIDEELTK.L | 1788.0962 | 1.3302 | 2 | ||||||
| HMGZ | 1 | 1 | 12.61 | 12656 | 9.28 | K.EYEANGGTDSGAPK.K | 1396.3986 | −0.8754 | 2 |
| dbe | 1 | 1 | 3.67 | 37849 | 10.22 | R.VLQDDIGCDIIK.I | 1389.5703 | 0.3233 | 2 |
| CG32701 | 2 | 1 | 4.97 | 33030 | 4.74 | K.GAEEFVIETVEASFR.Y | 1684.828 | 0.534 | 2 |
| CG2199 | 1 | 1 | 1.60 | 83801 | 8.92 | K.VVDEIDDQVNNK.K | 1388.4627 | 0.0837 | 2 |
| CG1600 | 1 | 1 | 4.61 | 47029 | 5.75 | R.SMHCLSKGGVVLISDEVAEK.L | 2160.4714 | −1.5266 | 2 |
| Snr1 | 1 | 1 | 4.59 | 42151 | 5.58 | R.TYAFSEAPLSTIDVPFR.N | 1915.1348 | −0.1432 | 2 |
| Row | 1 | 1 | 2.16 | 67032 | 9.52 | K.NIVQAAATDNTAK.H | 1317.4309 | −0.0921 | 2 |
| EIF-4a | 2 | 1 | 2.99 | 45766 | 5.43 | R.DVIAQAQSGTGK.T | 1175.2744 | 0.9414 | 2 |
| bel | 1 | 1 | 1.88 | 85099 | 7.18 | R.SGDCPILVATAVAAR.G | 1501.7039 | −0.2061 | 2 |
| CG9143 | 2 | 1 | 2.21 | 91575 | 8.85 | K.IVDITSSQQTAQTLTESR.L | 1979.1356 | −0.4984 | 2 |
| 14-3-3 | 3 | 2 | 11.92 | 29570 | 4.71 | K.AAFDDAIAELDTLSEESYK.D | 2089.1989 | 0.2789 | 2 |
| R.DICSDILNVLEK.H | 1419.5964 | −0.5866 | 2 | ||||||
| ballchen | 1 | 1 | 2.90% | 68467 | 9.79 | R.IGPSIGVGGFGEIYAACK.V | 1797.0381 | 0.6281 | 2 |
| Sle | 9 | 3 | 3.43 | 15980 | 5.14 | K.ELEFVDLDDDEPVFPSEAPK.Y | 2292.4372 | −0.0058 | 2 |
| K.LSEAAQLLNVSEEK.S | 1531.6902 | −0.8658 | 2 | ||||||
| R.TNAGYVTVVDEPPTK.V | 1591.7441 | 1.3241 | 2 | ||||||
| CG5787 | 6 | 3 | 4.21 | 100257 | 9.75 | K.NDAYGIGQR.R | 994.0435 | 0.2655 | 2 |
| R.GGGNYSNNSNSGGNNSYPR.S | 1916.86 | 0.434 | 2 | ||||||
| R.GGPSSNFGQANR.G | 1192.2236 | −0.8514 | 2 | ||||||
| CG31910 | 1 | 1 | 5.73 | 31301 | 6.12 | K.KHYGIEAEVLGLLVK.L | 1669.9891 | −0.7329 | 2 |
| CG4069 | 1 | 1 | 3.14 | 56351 | 6.27 | K.LGEANIADIIQLLEAK.E | 1711.9812 | 1.5982 | 2 |
| CG11943 | 7 | 4 | 3.59 | 235147 | 6.2 | K.EITGLVTNYAESLVDGSGILGR.L | 2265.506 | −0.422 | 2 |
| K.FISLASDLLPQTLFK.A | 1694.0074 | −0.7916 | 2 | ||||||
| K.SIADNLLQQR.A | 1158.2902 | 0.8982 | 2 | ||||||
| R.DCEASAEAGANSR.A | 1338.3141 | 0.6181 | 2 |
TABLE 2.
The JHRR-bound nuclear proteins identified in DMSO-treated Kc cells using HPLC MS/MS
| Reference | Pep count | Unique pepcount | Cover percent | Mr | PI | Sequence | MH+ | Diff (MH+) | Charge |
|---|---|---|---|---|---|---|---|---|---|
| Tim17 a1 | 1 | 1 | 4.95% | 24158.97 | 8.77 | R.EPNLLQDIPVK.S | 1266.4686 | −1.7814 | 2 |
| Maggie | 1 | 1 | 8.11% | 16152.05 | 4.52 | R.NAVGAVSSATVK.S | 1104.2393 | 0.3603 | 2 |
| CG5873 | 1 | 1 | 2.21% | 62098.11 | 5.92 | K.EVMEKFGLVLQK.D | 1421.7304 | −1.3596 | 2 |
| CG11670 | 1 | 1 | 2.86% | 51380.58 | 9.24 | R.ILHPVMQEVAKAR.R | 1492.815 | −0.509 | 2 |
| Lamin B receptor | 1 | 1 | 2.51% | 80235.07 | 9.83 | R.SSVGPLTGSGSGSSLPIK.A | 1631.8098 | 0.4828 | 2 |
| α-Est7 | 1 | 1 | 1.92% | 65435.93 | 6.09 | K.EKLDSWSAQIR.D | 1333.4751 | −0.5879 | 2 |
| L(2)35Df | 1 | 1 | 0.95% | 118925.62 | 6.32 | K.SATELSGPLR.S | 1031.1452 | 0.9902 | 2 |
| Moira | 2 | 1 | 1.26% | 129025.02 | 5.44 | K.TSDNSNTQEFSSSAK.E | 1603.5839 | 0.5489 | 2 |
| X16 | 1 | 1 | 7.75% | 27867.42 | 11.56 | R.SGGGGGGGGGGGGGGGLGGR.D | 1345.3213 | −0.9317 | 2 |
| Brahma | 2 | 1 | 0.80% | 184643.99 | 7.11 | K.PVGLDPITLLQER.E | 1451.6931 | 0.8301 | 2 |
| Ost48 | 1 | 1 | 4.01% | 49980.65 | 5.37 | R.LAQFVDDGGNVLVAGSEK.S | 1819.9934 | −0.3046 | 2 |
| Nopp140 | 11 | 4 | 6.71% | 70602.36 | 8.88 | K.FSGGDQDEATPNK.K | 1366.3725 | 1.1105 | 2 |
| K.IADAIVLEYLQSK.D | 1463.7006 | 0.2986 | 2 | ||||||
| K.LSEILQFYQTK.S | 1370.5753 | 1.8633 | 2 | ||||||
| R.TEDVVVDSR.V | 1020.076 | −0.168 | 2 | ||||||
| Black pearl | 1 | 1 | 10.64% | 15766.68 | 9.59 | K.YIAQIIVLGAQAVGR.A | 1572.8767 | −0.3743 | 2 |
| CG12592 | 4 | 2 | 2.03% | 159801.18 | 5.14 | K.LSEAAQLLNVSEEK.S | 1531.6902 | −1.2028 | 2 |
| R.TNAGYVTVVDEPPTK.V | 1591.7441 | −0.1329 | 2 | ||||||
| CG11920 | 1 | 1 | 4.03% | 34682.95 | 9.57 | R.LIFSNDPGIINK.D | 1331.5423 | −0.2397 | 2 |
Hsp83 Is Required for JH-induced Kr-h1 Expression
The mammalian Hsp90 is critical for regulating transcriptional activity by interacting with ligand-activated transcription factors, including some nuclear receptors such as glucocorticoid receptor (GR) (34, 35) and a bHLH-PAS transcription factor, the aryl hydrocarbon receptor (AhR) (36, 37). In Drosophila, Hsp83 is necessary for activating the 20E nuclear receptor complex, EcR-USP, that mediates 20E signal transduction (38). To this end, we investigated whether Hsp83 is involved in the JH-induced Kr-h1 expression.
First, we tested this hypothesis in Kc cells. The JH-induced Kr-h1 expression (Fig. 5A) and JHRR-driven luciferase activity (Fig. 5B) were significantly decreased when the cells were pre-treated with GA, a potent and highly specific inhibitor of Hsp90/Hsp83, which binds to the ADP/ATP binding pocket located in the N terminus of Hsp90/Hsp83 for inactivating its ATPase activity (39), or after the mRNA level of Hsp83 was reduced by RNAi.
FIGURE 5.
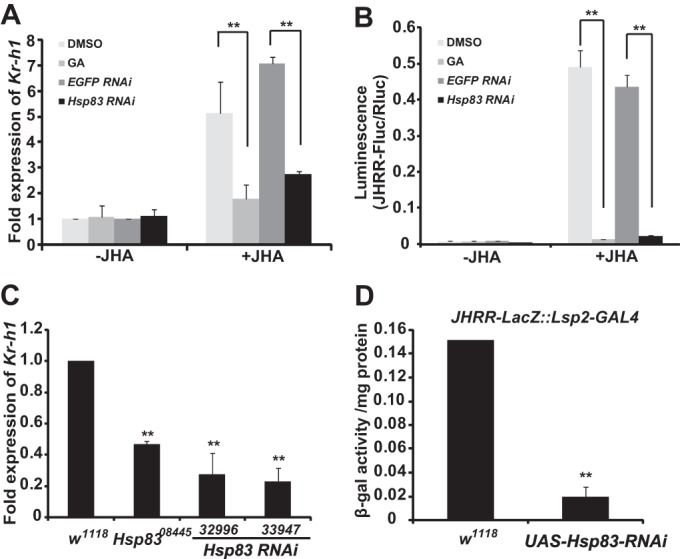
Hsp83 is required for JH-induced Kr-h1 expression through JHRR. A and B, the JH-induced Kr-h1 expression (A) and JHRR-driven luciferase activity (B) were measured after Kc cells were pretreated with 1 μg/ml of GA (DMSO as the control) for 2 h or after Hsp83 was reduced by RNAi (EGFP RNAi as the control). C, the mRNA levels of Kr-h1 in the fat body isolated from w1118, Hsp8308445 (an Hsp83 weak mutant), and two Hsp83 RNAi (32996 and 33947; Lsp2-GAL4>UAS-Hsp83-RNAi) larvae at 3 h after the initiation of wandering when the JH titer is high. D, the JHRR-driven β-gal activities were measured in the fat body isolated from the JHRR-LacZ::Lsp2-GAL4/+ and JHRR-LacZ::Lsp2-GAL4/UAS-Hsp83-RNAi larvae at 3 h AIW. EGFP, enhanced green fluorescent protein.
Next, we tested this hypothesis in the larval fat body at 3 h AIW. In comparison with the w1118 larvae, the Kr-h1 mRNA level of an Hsp83 weak mutant, Hsp8308445, was reduced to about 50%. To avoid any non-tissue autonomous effects, the binary GAL4/UAS system was used to reduce Hsp83 expression specifically in the larval fat body by RNAi. Kr-h1 expression was reduced to ∼25% in two lines of the Lsp2-GAL4>UAS-Hsp83-RNAi larvae (32996 and 33947) (Fig. 5C). Moreover, the JHRR-driven β-galactosidase activity in JHRR-LacZ::Lsp2-GAL4/UAS-Hsp83-RNAi larvae was decreased to about 15% of that in the control larvae, JHRR-LacZ::Lsp2-GAL4/+ (Fig. 5D). These data demonstrate that Hsp83 is required for JH-induced Kr-h1 expression through the JHRR and suggest that Hsp83 might influence the action of Met in JH signal transduction.
Hsp83 Interacts with Met through Its PAS-B and bHLH Domains
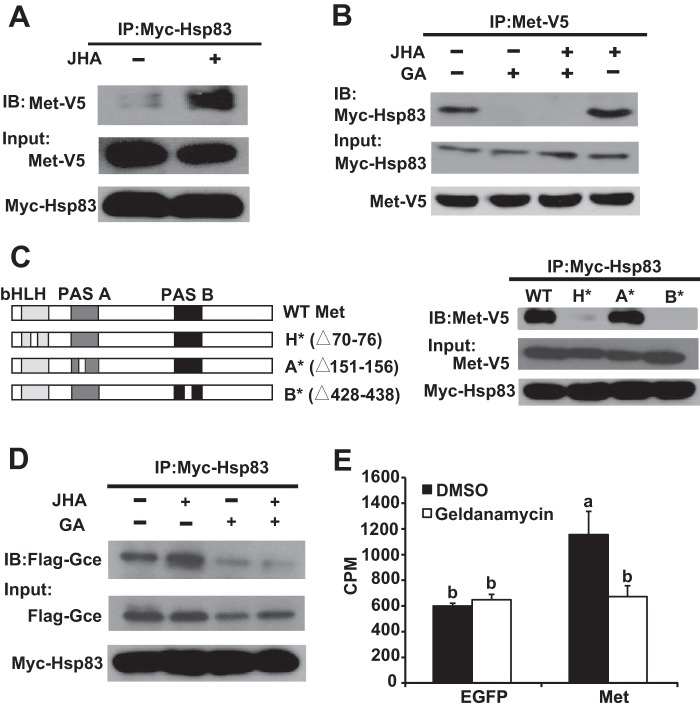
It is well known that mammalian Hsp90 physically interacts with AhR to facilitate its transcriptional activity (36, 37). Thus, we investigated whether Hsp83 physically interacts with Met after Myc-Hsp83 and Met-V5 were co-overexpressed in Kc cells. Co-immunoprecipitation experiments revealed that Met associated with Hsp83 and JH was able to induce physical interaction between Met and Hsp83 (Fig. 6, A and B). Importantly, GA significantly decreased the basal and JH-induced Met-Hsp83 interaction (Fig. 6B). We then individually mutated the bHLH, PAS-A, and PAS-B domains of Met. Mutation to the PAS-B domain of Met abolished the Met-Hsp83 interaction, mutation to the bHLH domain significantly decreased the interaction, but mutation to the PAS-A domain had no obvious effect (Fig. 6C), indicating that the PAS-B and bHLH domains of Met are essential for the Met-Hsp83 interaction. In addition, Gce also associated with Hsp83, and GA decreased the basal and JH-induced Gce-Hsp83 interaction (Fig. 6D).
FIGURE 6.
Hsp83 physically interacts with Met/Gce and facilitates Met to bind to JH. A, B, and D, Kc cells were co-transfected with the pAC5.1 expression constructs of Myc-Hsp83 and Met-V5 or Flag-Gce for 48 h, pretreated with 1 μg/ml of GA for 2 h and treated with 1 μm methoprene (JHA) or DMSO for 10 min as indicated, and then cell lysates were subjected to immunoprecipitation (IP) with an anti-Myc (A and D) and an anti-V5 (B) antibody, respectively. The interacting proteins were detected on immunoblot (IB). Input panels represent 10% of the initial material. C, left panel: the bHLH, PAS-A, and PAS-B domains of Met (H*, A*, and B*) were mutated (partially deleted) individually. Right panel: besides the WT (wild type), the mutated Met-V5 constructs were used, the other conditions are the same as in A. E, GA decreased the JH binding activity of Met. Kc cells were transfected with the pAC5.1 expression constructs of EGFP or Met-V5 for 48 h, pretreated with 1 μg/ml of GA for 2 h, the JH-binding activity of total proteins isolated from Kc cells was measured. EGFP, enhanced green fluorescent protein.
Hsp83 Is Required for JH Binding to Met
The JH binding activity of total proteins extracted from the Met-overexpressed Kc cells was signfiicatnly higher than proteins isolated from control Kc cells overexpressing EGFP. Importantly, GA treatment had no apparent effect on the JH binding activity of total proteins extracted from EGFP-overexpressed cells, but decreased that of the Met-overexpressed cells to the control level (Fig. 6E), suggesting that Hsp83 facilitates JH binding to Met.
Hsp83 Facilitates JH-induced Met Nuclear Import
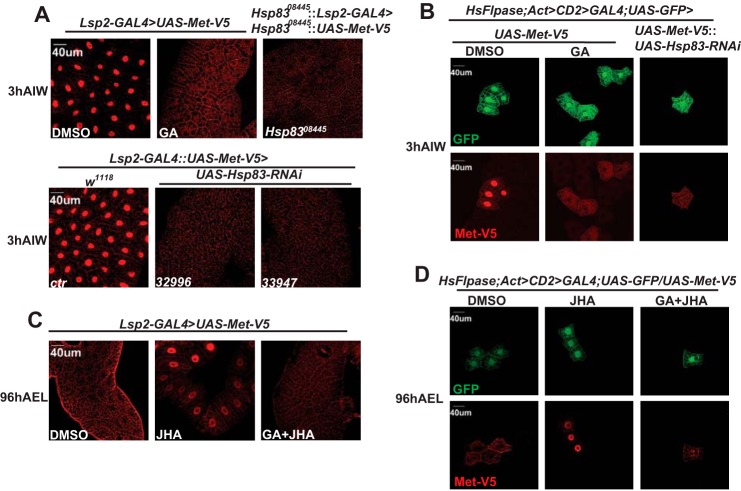
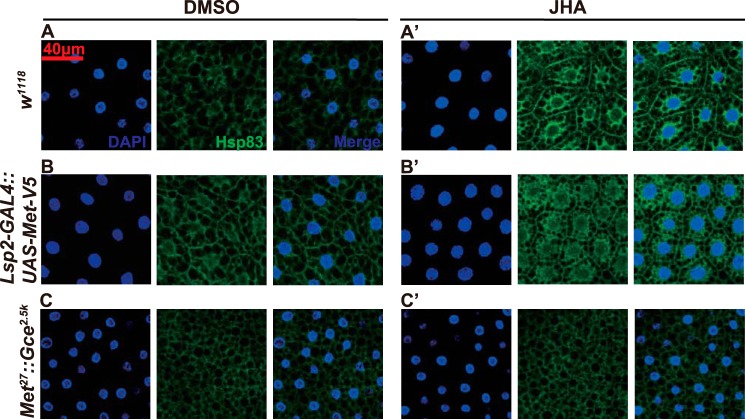
The mammalian Hsp90 regulates subcellular localization of the ligand-activated GR (40, 41) and AhR (36). It has been reported that Met is predominantly localized in the nuclei of Drosophila-cultured cells (10, 42) and tissues (43). As detected by immunohistochemistry using a V5 antibody, Met-V5 was predominantly localized in the nuclei of fat body cells isolated from Lsp2-GAL4>UAS-Met-V5 (Fig. 7A, first image in the upper panel) and Lsp2-GAL4::UAS-Met-V5 (Fig. 7A, first image in the lower panel) larvae at 3 h AIW. Interestingly, the nuclear localization of Met-V5 was inhibited when fat body tissues were exposed to GA (Fig. 7A, second image in the upper panel). To confirm the GA effects, we examined whether mutation to or RNAi of Hsp83 affects subcellular localization of Met-V5 at 3 h AIW. Consistent with the GA effects, Met-V5 became mainly distributed in the cytoplasm of the fat body cells isolated from the Hsp8308445::UAS-Met-V5/Hsp8308445::Lsp2-GAL4 larvae (Fig. 7A, third image in the upper panel) and two lines of the Lsp2-GAL4::UAS-Met-V5/UAS-Hsp83-RNAi larvae (Fig. 7A, second and third images in the lower panel). To avoid any non-cell autonomous effects of Hsp83, we further used the Flp-out technique to detect the cytoplasm-nucleus shuttling of Met-V5 in clones of Met-V5-overexpressed fat body cells, which were marked with GFP, at 3 h AIW. Again, Met-V5 was predominantly localized in the nuclei of cloned Met-V5-overexpressed fat body cells isolated from the hsFlpase; Act>CD2>GAL4; UAS-GFP/UAS-Met-V5 larvae (Fig. 7B, first images in both panels), and GA significantly inhibited the nuclear import of Met-V5 resulting in predominant localization of Met-V5 in the cytoplasm (Fig. 7B, second images in both panels). Moreover, Met-V5 was mainly distributed in the cytoplasm of the cloned fat body cells isolated from the hsFlpase; Act>CD2>GAL4; UAS-GFP/UAS-Met-V5::UAS-Hsp83-RNAi larvae (Fig. 7B, third images in both panels). These data demonstrate that Hsp83 is required for Met nuclear import.
FIGURE 7.
Hsp83 facilitates JH-induced Met nuclear import. Immunohistochemistry with the anti-V5 antibody reveals the subcellular localization of Met-V5 in fat body cells isolated from the indicated genotypes and different treatments. GA, in vitro treatment with 1 μg/ml of geldanamycin for 2 h; JHA, in vitro treatment with 1 μm methoprene for 2 h; GA+JHA, GA for 2 h and JHA for 2 h; DMSO, the control treatment. A, Met-V5 was detected at 3 h AIW when the JH titer is high. B, heat shock was performed at 6 h AEL for 15 min, and Met-V5 was detected in clones of Met-V5-overexpressed fat body cells, which were marked with GFP, at 3 h AIW. C, Met-V5 was detected at 96 h AEL when the JH titer is low. D, heat shock was performed at 6 h AEL for 15 min, and Met-V5 was detected in cell clones at 96 h AEL.
We also detected the subcellular localization of Met-V5 in Lsp2-GAL4>UAS-Met-V5 larvae at 96 h AEL. To our great surprise, Met-V5 was mainly distributed in the cytoplasm of fat body cells at this stage. Moreover, when fat body tissues were exposed to methoprene, Met-V5 shifted from the cytoplasm to the nucleus, and the methoprene-induced nuclear import of Met-V5 was dramatically inhibited by GA (Fig. 7C). Consistently, Met-V5 was mainly distributed in the cytoplasm of cloned Met-V5-overexpressed fat body cells in the hsFlpase; Act>CD2>GAL4; UAS-GFP/UAS-Met-V5 larvae at 96 h AEL, methoprene induced the nuclear import of Met-V5, and GA blocked the methoprene-induced nuclear import of Met-V5 (Fig. 7D). The combined genetic data demonstrate that Hsp83 facilitates the JH-induced nuclear import of Met in the larval fat body.
Hsp83 Is Accumulated in the Cytoplasm Area Adjunct to the Nucleus in the Presence of JH and Met/Gce
Because JH induces Met nuclear import, we investigated whether JH also induces Hsp83 nuclear import at 96 h AEL. As detected by immunohistochemistry, Hsp83 was almost evenly distributed in the fat body cells isolated from w1118 larvae (Fig. 8A). When the fat body tissues were exposed to methoprene, Hsp83 was accumulated in the cytoplasm area adjunct to the nucleus (Fig. 8A′). Importantly, Hsp83 was slightly accumulated in the cytoplasm area adjunct to the nucleus in fat body cells isolated from the Lsp2-GAL4>UAS-Met-V5 larvae (Fig. 8B), whereas methoprene further increased this accumulation (Fig. 8B′). Moreover, it appears that localization of Hsp83 was evenly distributed in the fat body cells isolated from the Met27gce2.5k larvae (Fig. 8C), and methoprene had no effect on its subcellular localization (Fig. 8C′). These data show that Hsp83 is accumulated in the cytoplasm area adjunct to the nucleus in the presence of JH and Met/Gce.
FIGURE 8.
Hsp83 is accumulated in the cytoplasm area adjunct to the nucleus in the presence of JH and Met/Gce. Immunohistochemistry with the anti-Hsp83 antibody reveals the subcellular localization of Hsp83 in fat body cells isolated from the indicated genotypes (A and A′, the w1118 larvae; B and B′, the Lsp2-GAL4>UAS-Met-V5 larvae; C and C′, the Met27gce2.5k larvae) at 96 h after egg laying, without (left panels, DMSO as control) or with (right panels) in vitro treatment with 1 μm methoprene (JHA) for 2 h.
DISCUSSION
Mammalian Hsp90 forms a molecular chaperone complex with other co-chaperone proteins, including Hsp70, Hop, p23, and a tetratricopeptide repeat-domain protein to regulate the correct maturation and activation of the “client proteins” (44). Ligand-activated transcription factors (e.g. GR and AhR) usually undergo a constant and dynamic cytoplasm-nucleus shuttling, which is regulated by the Hsp90 chaperone complex (35, 36, 40, 41, 44, 45). In this study Hsp83 was identified as a nuclear protein bound to the JHRR in the presence of JH (Fig. 4). Importantly, we have shown that Hsp83 facilitates JH binding and JH-induced nuclear import of Met, which then binds to the JHRR and induces Kr-h1 expression (Figs. 3–7). Surprisingly, Met, Gce, Taiman, and Cycle were not detected in either nuclear protein isolated from control and methoprene-treated Kc cells in affinity purification experiments. This is probably due to their relatively low abundance in nuclear proteins. As shown by cross-linking studies for several nuclear receptors, the stoichiometry of the receptor to Hsp90 is 1:2 (34), so the amount of Hsp83 bound to JHRR should be relatively more abundant than Met/Gce and/or Taiman/Cycle in the nuclear proteins.
Previous studies have shown that the PAS-B domain of Met accounts for Met-Met and Met-Gce dimerization (6) as well as JH binding (11), and that its PAS-A and PAS-B domains are indispensable for Met-FISC dimerization (18). Apparently, bHLH, PAS-A, and PAS-B domains of Met have overlappng binding sites, which also occur in AhR (37). In the absence of JH, Met/Gce forms dimers, and JH reduces Met/Gce dimerization (6) and induces the Met-Hsp83 interaction (Fig. 6, A–C). Moreover, GA attenuated the JH-binding activity of total proteins isolated from Met-overexpressed Kc cells (Fig. 6E), suggesting that the JH-induced Met-Hsp83 interaction is important for Met to form a proper conformation for binding to JH.
It has been long considered that in Drosophila, Met localizes in the nuclei no matter whether JH is present or not (10, 42). Inconsistent with those reports, we observed that Met-V5 is predominantly localized in the nuclei of fat body cells at 3 h AIW when the JH titer is high. In contrast, Met-V5 is mainly distributed in the cytoplasm of fat body cells at 96 h AEL when the JH titer is low, and moreover, JH induces Met-V5 nuclear import (Fig. 7). It was reported that the JH-binding activity of proteins in the cytoplasm isolated from fat body cells of the Met mutants was much lower than that of wild type larvae (46), suggesting that Met may also present in the cytoplasm, at least in some circumstances. A constant and dynamic cytoplasm-nucleus shuttling of Met and the JH-induced Met nuclear import are supported by the identification of nuclear import and export signals of Met, which are predominantly localized in PAS-A and PAS-B domains, respectively (42). In conclusion, similar to the other ligand-activated transcription factors, nuclear import of the JH receptor Met is aided by Hsp83 in Drosophila (Fig. 7). Moreover, Hsp83 is accumulated in the cytoplasm area adjunct to the nucleus in the presence of JH and Met/Gce (Fig. 8). This increase is likely due to an increase in import of Met/Gce, JH, and Hsp83 complex. A recent report on Helicoverpa armigera (Lepidoptera) also suggests that both 20E and JH stimulate nuclear import of Hsp90/Hsp83 in cultured cells (47). Nup358, a nuclear pore protein containing a tetratricopeptide repeat domain, was also identified as a JHRR-bound nuclear protein from methoprene-treated Kc cells (Table 1). Our preliminary data imply Hsp83 physically interacts with the tetratricopeptide repeat domain of Nup358, suggesting that this protein may be involved in Hsp83-facilicated JH-induced Met nuclear import and thus JH signaling (data not shown). These observations suggest that whereas Hsp83 is involved in JH-induced Met nuclear localization, it is not sufficient to complete the process and other components are necessary.
Hsp90/Hsp83 does not contain a nuclear localization signal. In mammalian cells, nuclear import of the GR-Hsp90 complex through the nuclear pore complex is mediated by its interaction with Nup62, the integral nuclear pore glycoprotein, and Importin, the classical pathway of nuclear import. By interacting with the Hsp90 chaperone complex, motor proteins are involved in passing the GR-Hsp90 complex through the nuclear pore (40, 41, 44, 45). Whether the above molecular mechanism applies to the JH-induced Met nuclear import remains unknown and requires further studies.
Based on the data presented in this paper and previous studies, we propose a model that Hsp83 facilitates Met nuclear import to modulate JH signaling in Drosophila (Fig. 9). Prior to binding to JH in the cytoplasm, Met remains in an inactive conformation and only weakly associates with Hsp83, and JH induces the physical interaction between the PAS-B and bHLH domains of Met and Hsp83. In turn, the JH-induced Met-Hsp83 interaction permits a proper conformational change in Met and an increase in binding to JH. Thus, JH induces nuclear import of the Met-Hsp83 complex, possibly along with other transcription co-factors (e.g. Taiman or Cycle). Upon entering the nucleus, the JH-bound Met directly binds to the JHRR and induces the expression of JH response genes such as Kr-h1.
FIGURE 9.
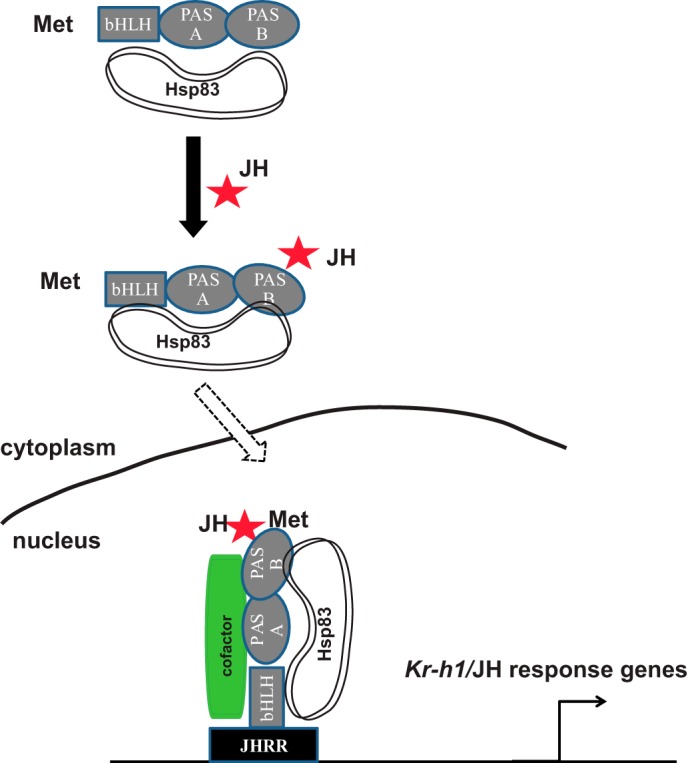
A model showing that Hsp83 interacts with Met to regulate JH signaling. See ”Discussion“ for details of the model. Met contains bHLH, PAS-A, and PAS-B domains; red star, JH; Hsp83 interacts with the bHLH and PAS-B domains of Met; cofactor, Taiman/FISC/SRC or Cycle. The line represents nuclear membrane separating cytoplasm from nucleus.
It is well documented that the mammalian Hsp90 chaperone complex is critical for regulating transcriptional activity of several ligand-activated transcription factors (i.e. GR and AhR). However, this is the first time to show that Hsp90/Hsp83 plays an essential role in the regulation of a bHLH-PAS transcription factor in invertebrates. Interestingly, nuclear import of both AhR and Met is regulated by the Hsp90 Hsp83 molecular chaperone complex, suggesting this mechanism is evolutionarily conserved for the ligand-activated bHLH-PAS transcription factors. This study adds to our knowledge on the comprehensive molecular mechanism of JH signaling and the general roles of bHLH-PAS transcription factors.
This work was supported in part by the National Science Foundation of China Grant 31330072 and the 973 program Grant 2012CB114605 (to S. L.).
- JH
- juvenile hormones
- CA
- corpus allatum
- 20E
- 20-hydroxyecdysone
- Met
- methoprene-tolerant
- JHRR
- JH response region
- AEL
- after egg laying
- AIW
- after initiation of wandering
- GA
- geldanamycin
- qPCR
- quantitative real-time PCR
- DMSO
- dimethyl sulfoxide
- bHLH
- basic helix-loop-helix
- GR
- glucocorticoid receptor
- AhR
- aryl hydrocarbon receptor
- Gce
- Germ-cell expressed
- ChIP
- chromatin immunoprecipitation
- Kr-h1
- Krüppel homolog 1.
REFERENCES
- 1. Jindra M., Palli S. R., Riddiford L. M. (2013) The juvenile hormone signaling pathway in insect development. Annu. Rev. Entomol. 58, 181–204 [DOI] [PubMed] [Google Scholar]
- 2. Liu Y., Sheng Z., Liu H., Wen D., He Q., Wang S., Shao W., Jiang R. J., An S., Sun Y., Bendena W. G., Wang J., Gilbert L. I., Wilson T. G., Song Q., Li S. (2009) Juvenile hormone counteracts the bHLH-PAS transcription factors MET and GCE to prevent caspase-dependent programmed cell death in Drosophila. Development 136, 2015–2025 [DOI] [PubMed] [Google Scholar]
- 3. Riddiford L. M., Truman J. W., Mirth C. K., Shen Y. C. (2010) A role for juvenile hormone in the prepupal development of Drosophila melanogaster. Development 137, 1117–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wilson T. G., Fabian J. (1986) A Drosophila melanogaster mutant resistant to a chemical analog of juvenile hormone. Dev. Biol. 118, 190–201 [DOI] [PubMed] [Google Scholar]
- 5. Ashok M., Turner C., Wilson T. G. (1998) Insect juvenile hormone resistance gene homology with the bHLH-PAS family of transcriptional regulators. Proc. Natl. Acad. Sci. U.S.A. 95, 2761–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Godlewski J., Wang S., Wilson T. G. (2006) Interaction of bHLH-PAS proteins involved in juvenile hormone reception in Drosophila. Biochem. Biophys. Res. Commun. 342, 1305–1311 [DOI] [PubMed] [Google Scholar]
- 7. Abdou M. A., He Q., Wen D., Zyaan O., Wang J., Xu J., Baumann A. A., Joseph J., Wilson T. G., Li S., Wang J. (2011) Drosophila Met and Gce are partially redundant in transducing juvenile hormone action. Insect Biochem. Mol. Biol. 41, 938–945 [DOI] [PubMed] [Google Scholar]
- 8. Konopova B., Jindra M. (2007) Juvenile hormone resistance gene methoprene-tolerant controls entry into metamorphosis in the beetle Tribolium castaneum. Proc. Natl. Acad. Sci. U.S.A. 104, 10488–10493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Parthasarathy R., Tan A., Palli S. R. (2008) bHLH-PAS family transcription factor methoprene-tolerant plays a key role in JH action in preventing the premature development of adult structures during larval-pupal metamorphosis. Mech. Dev. 125, 601–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miura K., Oda M., Makita S., Chinzei Y. (2005) Characterization of the Drosophila methoprene-tolerant gene product: juvenile hormone binding and ligand-dependent gene regulation. FEBS J. 272, 1169–1178 [DOI] [PubMed] [Google Scholar]
- 11. Charles J. P., Iwema T., Epa V. C., Takaki K., Rynes J., Jindra M. (2011) Ligand-binding properties of a juvenile hormone receptor, methoprene-tolerant. Proc. Natl. Acad. Sci. U.S.A. 108, 21128–21133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Minakuchi C., Zhou X., Riddiford L. M. (2008) Krüppel homolog 1 (Kr-h1) mediates juvenile hormone action during metamorphosis of Drosophila melanogaster. Mech. Dev. 125, 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pecasse F., Beck Y., Ruiz C., Richards G. (2000) Kruppel-homolog, a stage-specific modulator of the prepupal ecdysone response, is essential for Drosophila metamorphosis. Dev. Biol. 221, 53–67 [DOI] [PubMed] [Google Scholar]
- 14. Minakuchi C., Namiki T., Shinoda T. (2009) Krüppel homolog 1, an early juvenile hormone-response gene downstream of methoprene-tolerant, mediates its anti-metamorphic action in the red flour beetle Tribolium castaneum. Dev. Biol. 325, 341–350 [DOI] [PubMed] [Google Scholar]
- 15. Zhang Z., Xu J., Sheng Z., Sui Y., Palli S. R. (2011) Steroid receptor co-activator is required for juvenile hormone signal transduction through a bHLH-PAS transcription factor, methoprene tolerant. J. Biol. Chem. 286, 8437–8447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shin S. W., Zou Z., Saha T. T., Raikhel A. S. (2012) bHLH-PAS heterodimer of methoprene-tolerant and cycle mediates circadian expression of juvenile hormone-induced mosquito genes. Proc. Natl. Acad. Sci. U.S.A. 109, 16576–16581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kayukawa T., Tateishi K., Shinoda T. (2013) Establishment of a versatile cell line for juvenile hormone signaling analysis in Tribolium castaneum. Sci. Rep. 3, 1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li M., Mead E. A., Zhu J. (2011) Heterodimer of two bHLH-PAS proteins mediates juvenile hormone-induced gene expression. Proc. Natl. Acad. Sci. U.S.A. 108, 638–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kayukawa T., Minakuchi C., Namiki T., Togawa T., Yoshiyama M., Kamimura M., Mita K., Imanishi S., Kiuchi M., Ishikawa Y., Shinoda T. (2012) Transcriptional regulation of juvenile hormone-mediated induction of Kruppel homolog 1, a repressor of insect metamorphosis. Proc. Natl. Acad. Sci. U.S.A. 109, 11729–11734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zou Z., Saha T. T., Roy S., Shin S. W., Backman T. W., Girke T., White K. P., Raikhel A. S. (2013) Juvenile hormone and its receptor, methoprene-tolerant, control the dynamics of mosquito gene expression. Proc. Natl. Acad. Sci. U.S.A. 110, E2173–E2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu H., Jia Q., Tettamanti G., Li S. (2013) Balancing crosstalk between 20-hydroxyecdysone-induced autophagy and caspase activity in the fat body during Drosophila larval-prepupal transition. Insect Biochem. Mol. Biol. 43, 1068–1078 [DOI] [PubMed] [Google Scholar]
- 22. Sliter T. J., Sedlak B. J., Baker F. C., Schooley D. A. (1987) Juvenile hormone in Drosophila melanogaster: identficiation and titer determination during development. Insect Biochem. Mol. Biol. 17, 161–165 [Google Scholar]
- 23. Jones D., Jones G., Teal P. E. (2013) Sesquiterpene action, and morphogenetic signaling through the ortholog of retinoid X receptor, in higher Diptera. Gen. Comp. Endocrinol. 194, 326–335 [DOI] [PubMed] [Google Scholar]
- 24. Richard D. S., Applebaum S. W., Gilbert L. I. (1989) Developmental regulation of juvenile hormone biosynthesis by the ring gland of Drosophila. J. Comp. Physiol. B 159, 383–387 [DOI] [PubMed] [Google Scholar]
- 25. Niwa R., Niimi T., Honda N., Yoshiyama M., Itoyama K., Kataoka H., Shinoda T. (2008) Juvenile hormone acid O-methyltransferase in Drosophila melanogaster. Insect Biochem. Mol. Biol. 38, 714–720 [DOI] [PubMed] [Google Scholar]
- 26. Guo E., He Q., Liu S., Tian L., Sheng Z., Peng Q., Guan J., Shi M., Li K., Gilbert L. I., Wang J., Cao Y., Li S. (2012) MET is required for the maximal action of 20-hydroxyecdysone during Bombyx metamorphosis. PLoS ONE 7, e53256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hossain M. S., Liu Y., Zhou S., Li K., Tian L., Li S. (2013) 20-Hydroxyecdysone-induced transcriptional activity of FoxO upregulates brummer and acid lipase-1 and promotes lipolysis in Bombyx fat body. Insect Biochem. Mol. Biol. 43, 829–838 [DOI] [PubMed] [Google Scholar]
- 28. Wang S., Wang J., Sun Y., Song Q., Li S. (2012) PKC-mediated USP phosphorylation at Ser35 modulates 20-hydroxyecdysone signaling in Drosophila. J. Proteome Res. 11, 6187–6196 [DOI] [PubMed] [Google Scholar]
- 29. Tian L., Ma L., Guo E., Deng X., Ma S., Xia Q., Cao Y., Li S. (2013) 20-Hydroxyecdysone upregulates Atg genes to induce autophagy in the Bombyx fat body. Autophagy 9, 1172–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li Y., Zhang Z., Robinson G. E., Palli S. R. (2007) Identification and characterization of a juvenile hormone response element and its binding proteins. J. Biol. Chem. 282, 37605–37617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kadonaga J. T., Tjian R. (1986) Affinity purification of sequence-specific DNA binding proteins. Proc. Natl. Acad. Sci. U.S.A. 83, 5889–5893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kewley R. J., Whitelaw M. L., Chapman-Smith A. (2004) The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int. J. Biochem. Cell Biol. 36, 189–204 [DOI] [PubMed] [Google Scholar]
- 33. Baumann A., Barry J., Wang S., Fujiwara Y., Wilson T. G. (2010) Paralogous genes involved in juvenile hormone action in Drosophila melanogaster. Genetics 185, 1327–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pratt W. B., Toft D. O. (1997) Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr. Rev. 18, 306–360 [DOI] [PubMed] [Google Scholar]
- 35. Cano L. Q., Lavery D. N., Bevan C. L. (2013) Mini-review: foldosome regulation of androgen receptor action in prostate cancer. Mol. Cell. Endocrinol. 369, 52–62 [DOI] [PubMed] [Google Scholar]
- 36. Kazlauskas A., Sundström S., Poellinger L., Pongratz I. (2001) The hsp90 chaperone complex regulates intracellular localization of the dioxin receptor. Mol. Cell. Biol. 21, 2594–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Soshilov A., Denison M. S. (2011) Ligand displaces heat shock protein 90 from overlapping binding sites within the aryl hydrocarbon receptor ligand-binding domain. J. Biol. Chem. 286, 35275–35282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Arbeitman M. N., Hogness D. S. (2000) Molecular chaperones activate the Drosophila ecdysone receptor, an RXR heterodimer. Cell 101, 67–77 [DOI] [PubMed] [Google Scholar]
- 39. Bedin M., Gaben A. M., Saucier C., Mester J. (2004) Geldanamycin, an inhibitor of the chaperone activity of HSP90, induces MAPK-independent cell cycle arrest. Int. J. Cancer 109, 643–652 [DOI] [PubMed] [Google Scholar]
- 40. Echeverría P. C., Mazaira G., Erlejman A., Gomez-Sanchez C., Piwien Pilipuk G., Galigniana M. D. (2009) Nuclear import of the glucocorticoid receptor-hsp90 complex through the nuclear pore complex is mediated by its interaction with Nup62 and importin. Mol. Cell Biol. 29, 4788–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Galigniana M. D., Echeverría P. C., Erlejman A. G., Piwien-Pilipuk G. (2010) Role of molecular chaperones and TPR-domain proteins in the cytoplasmic transport of steroid receptors and their passage through the nuclear pore. Nucleus 1, 299–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Greb-Markiewicz B., Orłowski M., Dobrucki J., Ożyhar A. (2011) Sequences that direct subcellular traffic of the Drosophila methoprene-tolerant protein (MET) are located predominantly in the PAS domains. Mol. Cell. Endocrinol. 345, 16–26 [DOI] [PubMed] [Google Scholar]
- 43. Pursley S., Ashok M., Wilson T. G. (2000) Intracellular localization and tissue specificity of the Methoprene-tolerant (Met) gene product in Drosophila melanogaster. Insect Biochem. Mol. Biol. 30, 839–845 [DOI] [PubMed] [Google Scholar]
- 44. Jackson S. E. (2013) Hsp90: structure and function. Top. Curr. Chem. 328, 155–240 [DOI] [PubMed] [Google Scholar]
- 45. Freitas N., Cunha C. (2009) Mechanisms and signals for the nuclear import of proteins. Curr. Genomics 10, 550–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shemshedini L., Wilson T. G. (1990) Resistance to juvenile-hormone and an insect growth-regulator in Drosophila is associated with an altered cytosolic juvenile hormone-binding protein. Proc. Natl. Acad. Sci. U.S.A. 87, 2072–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu W., Zhang F. X., Cai M. J., Zhao W. L., Li X. R., Wang J. X., Zhao X. F. (2013) The hormone-dependent function of Hsp90 in the crosstalk between 20-hydroxyecdysone and juvenile hormone signaling pathways in insects is determined by differential phosphorylation and protein interactions. Biochim. Biophys. Acta 1830, 5184–5192 [DOI] [PubMed] [Google Scholar]