Background: Glycosaminoglycan (GAG) lyases have been widely isolated from terrestrial but not marine bacteria.
Results: A novel GAG lyase (HCLase) was identified for the first time from a marine bacterium.
Conclusion: The HCLase has very low homology to the characterized conventional GAG lyases and possesses very unique biochemical characteristics.
Significance: HCLase will be useful for chondroitin sulfate/hyaluronan-related research and applications.
Keywords: Chondroitin Sulfate, Glycosaminoglycan, Hyaluronan, Hyaluronidase, Proteoglycan, Chondroitinase, Eliminase, Lyase, Marine Bacterium
Abstract
Lyases cleave glycosaminoglycans (GAGs) in an eliminative mechanism and are important tools for the structural analysis and oligosaccharide preparation of GAGs. Various GAG lyases have been identified from terrestrial but not marine organisms even though marine animals are rich in GAGs with unique structures and functions. Herein we isolated a novel GAG lyase for the first time from the marine bacterium Vibrio sp. FC509 and then recombinantly expressed and characterized it. It showed strong lyase activity toward hyaluronan (HA) and chondroitin sulfate (CS) and was designated as HA and CS lyase (HCLase). It exhibited the highest activities to both substrates at pH 8.0 and 0.5 m NaCl at 30 °C. Its activity toward HA was less sensitive to pH than its CS lyase activity. As with most other marine enzymes, HCLase is a halophilic enzyme and very stable at temperatures from 0 to 40 °C for up to 24 h, but its activity is independent of divalent metal ions. The specific activity of HCLase against HA and CS reached a markedly high level of hundreds of thousands units/mg of protein under optimum conditions. The HCLase-resistant tetrasaccharide Δ4,5HexUAα1-3GalNAc(6-O-sulfate)β1-4GlcUA(2-O-sulfate)β1-3GalNAc(6-O-sulfate) was isolated from CS-D, the structure of which indicated that HCLase could not cleave the galactosaminidic linkage bound to 2-O-sulfated d-glucuronic acid (GlcUA) in CS chains. Site-directed mutagenesis indicated that HCLase may work via a catalytic mechanism in which Tyr-His acts as the Brønsted base and acid. Thus, the identification of HCLase provides a useful tool for HA- and CS-related research and applications.
Introduction
Chondroitin sulfate (CS),3 synthesized as the glycosaminoglycan (GAG) side chains of proteoglycans (1), is widely expressed on cell surfaces and in extracellular matrices and participates in various biological events, including development of the central nervous system (2, 3), wound repair (4, 5), viral attachment (6–8), growth factor signaling (9, 10), morphogenesis (11), and cytokinesis (12–14). The various functions of CS have been attributed to their structural diversity. CS chains are composed of repeating disaccharide units of GlcUA-GalNAc in which GlcUA and GalNAc represent d-glucuronic acid and N-acetyl-d-galactosamine, respectively. In biosynthesis, after or while CS chains are polymerized by various enzyme complexes, each of which is formed by heterologous combinations of two of the six chondroitin synthase family members (15–17), CS chains are further modified by differential sulfation by specific sulfotransferases at C-2 of GlcUA/l-iduronic acid and/or C-4 and/or C-6 of GalNAc to yield prominent structural diversity (18). Furthermore, some GlcUA residues are epimerized into l-iduronic acid (IdoUA) by the action of glucuronyl C-5 epimerase, and the chain that contains repeating disaccharide units of -IdoUA-GalNAc- has been designated as dermatan sulfate (DS) (19, 20). Therefore, CS and DS chains are often detected as co-polymeric structures (CS-DS) and are more likely to be periodically distributed in cell/tissue-specific manners (14, 21). The introduction of these unique structural domains into CS-DS chains in respective tissues results in different responses to various CS- or DS-binding proteins such as heparin-binding growth factors and cytokines (22).
Prominent structural heterogeneity has hampered detailed structural and functional analyses of CS and DS chains. Spectroscopic techniques such as nuclear magnetic resonance (NMR) and mass spectrometry (MS) are widely used to structurally determine CS/DS oligosaccharides (23). Furthermore, some CS/DS-degrading enzymes, particularly bacterial CS/DS lyases with specific activities, are being used in the structural and functional analyses of CS/DS chains in conjunction with spectroscopic techniques (23–25). CS/DS lyases depolymerize CS/DS via a β-elimination reaction, which cleaves the galactosaminidic linkages to uronic acid residues to yield an unsaturated 4,5-bond between C-4 and C-5 on the uronic acid residue at the site of cleavage (26). Several CS/DS lyases are now commercially available, including chondroitinase (CSase) ABC from Proteus vulgaris (27), which digests both CS and DS as well as hyaluronan (HA). CSase ACI from Flavobacterium heparinum (27) and CSase ACII from Arthrobacter aurescens (28) both specifically cleave CS, whereas CSase B from F. heparinum only cleaves the galactosaminidic linkages attached to the IdoUA residues of DS as well as CS-DS hetero- and polymers. CS/DS lyases are not only useful tools for investigating the structure-function relationship and the preparation of bioactive oligosaccharides of CS/DS but are also potential therapeutic agents for the treatment of injuries to the central nervous system (29). Therefore, screening and identifying novel CS/DS lyases with high activity and specificity are important for both academic research and applications.
The ocean is regarded as the origin of life on Earth and hosts most life forms. The unique marine environment as a powerful selective force prompts marine organisms to generate specific and potent bioactive molecules to adapt to complicated marine environments. Marine animals are immense sources of GAGs with unique structures and bioactivities. Three of the five commercially available CS/DS subtypes, namely CS-C, CS-D, and CS-E, are derived from shark cartilage, shark fins, and squid cartilage, respectively. In addition, we and others have purified various novel CS, DS, and CS-DS preparations from sea animals (30–34). The abundant presence of these unique structures in marine environments indicates the existence of CS/DS-degrading microbes. Therefore, marine microbes may be an ideal source for identifying novel CS/DS lyases with extraordinary properties. However, to the best of our knowledge, no CS/DS lyase from marine bacteria has yet been characterized in detail.
In the present study, a marine Vibrio strain with high GAG-degrading capacity was isolated using CS-C from shark cartilage as the sole carbon source. The bacterial genome was sequenced to identify the genes of GAG-degrading enzymes. A putative GAG lyase gene (HA and CS lyase (hclase)) was expressed in Escherichia coli, and the recombinant HCLase protein was purified to analyze sequence properties, substrate spectra, and enzymatic characteristics. An HCLase-resistant CS tetrasaccharide was isolated from CS-D and sequenced by enzymatic digestion followed by HPLC analysis. The key amino acid residues at the active site of HCLase were identified by site-directed mutagenesis. Our results demonstrated that HCLase exhibited very high activity and specificity for the digestion of CS and HA and will be a useful tool for basic research on and applications of CS and HA.
EXPERIMENTAL PROCEDURES
Materials
The strains and plasmids used in this study are listed in Table 1. SDS, proteinase K, PrimeSTARTM HS DNA polymerases, restriction endonuclease, and other genetic engineering enzymes were purchased from Takara Inc. (Dalian, China). Standard unsaturated disaccharides, chondroitin, CS-A from whale cartilage, CS-C and CS-D from shark cartilage, CS-E from squid cartilage, and DS from porcine skin were purchased from Seikagaku Corp. (Tokyo, Japan). 2-Aminobenzamide (2-AB), cyanoborohydride (NaBH3CN), CSase ABC (EC 4.2.2.20), heparin, heparan sulfate, alginate, and xanthan were obtained by Sigma. All other chemicals and reagents were of the highest quality available. CS or HA tetra-, hexa-, and octasaccharides were prepared by the digestion of CS-A or HA, respectively, using CSase ABC followed by gel filtration chromatography on a SuperdexTM Peptide 10/300 GL column as described previously (24).
TABLE 1.
Bacterial strains, plasmids, and primers used for sequencing in the present study
Restriction enzyme sites are underlined. Apr, ampicillin-resistant; Kanr, kanamycin-resistant.
| Strains and plasmids | Description | Source |
|---|---|---|
| Strains | ||
| Vibrio sp. FC509 | A chondroitin sulfate-degrading marine bacterium that secretes multiple GAG lyases (patented as CGMCC 8913) | This study |
| E. coli BL21(DE3) | F−, ompT, hsdSB (rB−, mB−), dcm, gal,λ (DE3), pLysS, Cmr | Novagen |
| E. coli Top10 | F− mcrA Δ (mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 deoR recA1 araD139Δ (araA-leu)7697 galU galK rpsL endA1 nupG | Invitrogen |
| Plasmids | ||
| pBAD/gIII A | Expression vector; Apr | Invitrogen |
| pCold TF | Expression vector; Apr | Takara |
| pET22b | Expression vector; Apr | Novagen |
| pET30a | Expression vector; Kanr | Novagen |
| pET22b-HCLase | pET22b carrying an amplified NcoI-XhoI fragment encoding the recombinant protein of HCLase fused with a His6 tag at the C terminus | This study |
| Sequencing primers | ||
| HCLase-F | 5′-GCCATGGATATGCGATGACCACCAGTTCACTG-3′ | |
| HCLase-R | 5′-GCTCGAGTCGCACTGAAAATTGATAACTTTGTCC-3′ | |
| HCLase-W492A-F | 5′-CCCGTGGTCGATAACCATCGACTCGC-3′ | |
| HCLase-W492A-R | 5′-TGCGTAATCGGTGTAGTGGTCTTGCTGGC-3′ | |
| HCLase-H285A-F | 5′-GACATTGCTTATAACGGCACTTATGGC-3′ | |
| HCLase-H285A-R | 5′-GCCTGCCTGAATAAAGGAGCCATCTTGG-3′ | |
| HCLase-Y290A-F | 5′-AACCGGCACTTATGGCAACGTGCTACTGGG-3′ | |
| HCLase-Y290A-R | 5′-TGCAGCAATGTCGCCATGCTGAATAAAGGAG-3′ | |
| HCLase-Y294A-F | 5′-GGCAACGTGCTACTGGGTGGGCTTGGC-3′ | |
| HCLase-Y294A-R | 5′-TGCAGTGCCGTTATAAGCAATGTCGCATGCTG-3′ | |
Identification of Marine CS-degrading Bacteria
Coastal sediments were collected from Jiaozhou Bay (N36°3′39″–N36°3′43″, E120°18′31″–E120°19′15″) nearby Qingdao City in Shandong Province, China. Basal medium (BM) composed of 3.0% (w/v) NaCl, 0.3% (w/v) KH2PO4, 0.7% (w/v) K2HPO4·3H2O, 0.2% (w/v) (NH4)2SO4, 0.01% (w/v) MgSO4, 0.01% (w/v) FeSO4·7H2O, 0.05% (w/v) chondroitin sulfate from shark cartilage (CS-C), and 1.5% (w/v) agar (pH 7.2) was used to isolate CS-degrading bacteria from the sediments. After being incubated at 30 °C for 72 h, colonies on the BM were randomly selected and transferred to fresh plates for further purification. To assay the polysaccharide-degrading abilities of the isolates, BM broth without agar was supplemented with various polysaccharides (alginate, chondroitin sulfates C, dermatan sulfate, heparin, heparin sulfate, hyaluronate, and xanthan) as the sole carbon source at a final concentration of 0.5% (w/v). Bacterial growth was evaluated by measuring absorbance at 600 nm (A600).
The genomic DNA of individual CS-degrading bacteria was prepared using SDS and a proteinase K treatment. PCR amplification of the 16 S rRNA gene sequence was performed using the bacterial universal primer pair 27f (5′-GAGTTTGATCCTGGCTCAG-3′) and 1492r (5′-AAGGAGGTGATCCAGCC-3′) (35). Gel-recovered PCR products were cloned into the pMD19-T vector (Takara Inc.) for sequencing. The sequence was analyzed against the GenBankTM database using the on-line BLAST program to search for the most similar sequences. A phylogenetic tree was generated using the neighbor joining method of Nei and co-workers (36) with MEGA version 5.05.
Cloning of GAG Lyase Genes
The draft genome of Vibrio sp. FC509 was sequenced at Meiji Biotech Inc. (Shanghai, China) using Roche Applied Science 454 GsFLX, Illumina GAIIx technology. The sequence of Vibrio sp. FC509 was annotated at Oak Ridge National Laboratory using their genome annotation pipeline. This included the application of a number of annotation programs beginning with open reading frame (ORF) prediction using Prodigal (37) followed by manual annotation using the JGI GenePRIMP pipeline (38). Automated protein function prediction was then performed using a number of databases, including protein domains (Pfam), UniProt (39), TIGRFAMs (40), KEGG (41), InterPro (42), and COG (43); metabolic reconstruction analysis using PRIAM (44); signal peptide prediction using SignalP (45); tRNA prediction using tRNAscan-SE (46); and rRNA prediction using RNAmmer (47).
Sequence Analyses of Genes and Proteins of Chondroitin Lyases
Promoter motifs of the 5′-flanking DNA region upstream of the ORF were identified using Primer Premier version 5.0 (PREMIER Biosoft International, Palo Alto, CA) and the Promoter 2.0 Prediction Server. The GC content (G+C%) of the ORF was calculated using Bio-Edit version 7.0.5.3.
A similarity search of the protein sequence was performed using the BLASTp algorithm online. Secretion signal peptides and their types were identified using the SignalP 4.0 server and LipoP 1.0 server, respectively. The molecular mass of the protein was estimated using the peptide mass tool on the ExPASy server of the Swiss Institute of Bioinformatics. Sequence alignment and phylogenetic analysis were performed using MEGA version 5.05. Protein modules and domains were identified using the Simple Modular Architecture Research Tool, Pfam database, and Carbohydrate-Active Enzyme database.
Heterologous Expression of the HCLase Gene
To express HCLase in E. coli strains, the full-length gene of HCLase was amplified using primer pairs (as listed in Table 1) and high fidelity PrimeSTAR HS DNA polymerases (Takara Inc.). Primer pairs with restriction enzyme sites (underlined) were designed according to the inserting site sequences of the expression plasmids, including pBAD/gIII A (Invitrogen), pET-22b(+), pET-30a(+) (Novagen), and pCold TF (Takara Inc.). Gel-recovered PCR products were cloned into these expression vectors. The expression plasmid (pBA-HCLase) constructed from pBAD/gIII A was transformed into E. coli Top10 cells, whereas another expression vector from pET-22b(+) (pE22b-rHCLase), pET-30a(+) (pE30a-rHCLase), or pCold TF (pCTF-rHCLase) was transformed into E. coli BL21(DE3) cells. The integrity of the nucleotide sequences of all constructed plasmids was confirmed via DNA sequencing.
E. coli cells harboring an expression vector were initially cultured in LB broth. When cell density reached an A600 of 0.8–1.0, the broth was supplemented with the inducer (l-arabinose or isopropyl 1-thio-β-d-galactopyranoside) at a final concentration of 0.01 mm to start the expression of targeting proteins. After a continual cultivation for an additional 24 h at 16 °C, cells were harvested by centrifugation at 6,000 × g for 15 min, washed twice using ice-cold buffer A (50 mm Tris-HCl, 150 mm NaCl (pH 8.0)), resuspended in buffer A, and disrupted by sonication (50 repetitions, 5 s) in an ice-cold environment. After centrifugation at 15,000 × g for 30 min, the supernatant was collected for further purification of soluble targeting proteins.
Purification of Recombinant Protein rHCLase
To purify the rHCLase protein, the supernatant containing the soluble native enzyme was loaded onto a column packed with nickel-SepharoseTM 6 Fast Flow (GE Healthcare), then the column was washed with buffer A containing 50 mm imidazole to remove impurities, and rHCLase was finally eluted from the Ni-NTA column using a gradient concentration of imidazole, ranging from 50 to 250 mm. The purity of rHCLase was analyzed using SDS-PAGE. SDS-PAGE was performed using 13.2% polyacrylamide gels according to Sambrook et al. (48). Coomassie Brilliant Blue R-250 was used to stain proteins in gels. Protein concentrations were determined by the Folin-Lowry method (49).
Assay of rHCLase Activity toward Various Polysaccharide Substrates
To determine the substrates of rHCLase, various polysaccharides (e.g. alginate, CS-A, CS-C, CS-D, CS-E, DS, heparin, heparin sulfate, hyaluronan, and xanthan) were individually dissolved in deionized water to prepare stock solutions (10 mg/ml). Each stock solution (10 μl) was mixed with 20 μl of 250 mm NaH2PO4-Na2HPO4 buffer (pH 7.0), 60 μl of water, and 10 μl of the appropriately diluted enzyme and then incubated at 37 °C for 12 h. Enzyme-treated polysaccharide samples were heated in boiling water for 10 min and then cooled in ice-cold water for 10 min. After being centrifuged at 15,000 × g for 15 min, the supernatant was collected and analyzed by the absorbance at 232 nm (27) and gel filtration HPLC.
Biochemical Characterization of the Recombinant Protein rHCLase
To determine the optimal pH for rHCLase activity, HA and CS-C (1 mg/ml) were digested, respectively, with rHCLase (2 ng) in buffers with different pH values, including a final concentration of 50 mm NaAc-HAc buffer (pH 5.0–6.0), 50 mm NaH2PO4-Na2HPO4 buffer (pH 6.0–8.0), and 50 mm Tris-HCl buffer (pH 7.0–10.0) in a total volume of 100 μl, at 30 °C for 10 min. After the optimum pH was determined, the effects of temperature on rHCLase activity were tested in 50 mm NaH2PO4-Na2HPO4 (pH 8.0) at temperatures from 0 to 90 °C for 10 min. The effects of metal ions/chelating reagent (5 mm) and concentrations of NaCl (0–1 m) on the HA- and CS-degrading activities of rHCLase were investigated at the optimum pH and temperature described above. To determine the thermostability of rHCLase, the enzyme in 50 mm NaH2PO4-Na2HPO4 buffer (pH 8.0) containing 0.5 m NaCl was preincubated for 0–24 h at a temperature from 0 to 90 °C, and the residual HA/CS-degrading activity was determined at 30 °C. All reactions were performed in triplicate, and after each treatment, the activity of enzyme was estimated by measuring the absorbance at 232 nm (27).
Optimum Assay Conditions of rHCLase
The activities of rHCLase were measured according to the method provided by Yamagata et al. (27). Briefly, rHCLase (2 ng) was added to 1 mg/ml GAGs in 50 mm NaH2PO4-Na2HPO4, 500 mm NaCl buffer (pH 8.0) in a total volume of 1 ml. The reaction mixture was incubated at 30 °C. At various time intervals (up to 10 min), aliquots of 100 μl were withdrawn in duplicate, boiled for 10 min, and then cooled in ice-cold water for 10 min. After being centrifuged at 15,000 × g for 15 min, the supernatant was collected, diluted five times, and analyzed by absorbance at 232 nm. One unit of enzyme was defined as the amount of enzyme that produced 1 μmol of unsaturated carbon bonds/min.
Gel Filtration Chromatography
Samples digested with rHCLase were analyzed by gel filtration chromatography on a Superdex Peptide 10/300 GL column. The mobile phase was 0.20 m NH4HCO3 at a flow rate of 0.4 ml/min, and the eluted fractions were monitored at 232 nm using a UV detector. Online monitoring and data analysis (e.g. molar ration determination) were performed using the software LCsolution version 1.25.
Digestion Pattern of Polysaccharides by rHCLase
To determine the degradation pattern of rHCLase, the digests of HA and CS (1 mg/ml) by rHCLase (1 unit/ml) were traced at 30 °C. Aliquots of the degradation products (10 μg) were removed for time course experiments to analyze gel filtration patterns by monitoring at 232 nm.
To further determine the molecular weights and structural characteristics of the oligosaccharide products, 1 ml of various polysaccharides (1 mg/ml) was digested using rHCLase (1 unit/ml) at 30 °C for 10 min. The reaction mixture was heated in boiling water for 10 min and subsequently cooled to 4 °C. After being centrifuged at 15,000 × g for 30 min, the supernatant was loaded onto a pre-equilibrated Superdex Peptide 10/300 GL column. Fractionated oligosaccharide samples were collected for pure monomers by online monitoring at 232 nm and freeze-dried repeatedly to remove NH4HCO3 for further identification.
The major disaccharide fractions purified from HA and CS-A were further identified by electrospray ionization MS on an ion trap TOF hybrid mass spectrometer (LCMS-IT-TOF, Shimadzu, Japan). Electrospray ionization MS analysis was set in the negative ion mode and with the following parameters: source voltage at 3.6 kV, nebulizer nitrogen gas flow rate at 1.5 liter/min, heat block and curved desolvation line temperature at 200 °C, and detector voltage at 1.8 kV. The mass acquisition range was set at 200–600.
Isolation and Sequence of an rHCLase-resistant Tetrasaccharide from CS-D
To prepare rHCLase-resistant tetrasaccharides, 200 μl of CS-D (1 mg/ml) was exhaustively digested using rHCLase (100 units/ml) at 30 °C for 72 h. The reaction mixture was heated in boiling water for 10 min and subsequently cooled to 4 °C. After being centrifuged at 15,000 × g for 30 min, the supernatant was loaded onto a pre-equilibrated Superdex Peptide 10/300 GL column. The tetrasaccharide fraction was collected by online monitoring at 232 nm and desalted by repeating freeze-drying. The tetrasaccharide fraction was subfractionated by anion-exchange HPLC on a YMC-Pack Polyamine II column (YMC, Kyoto, Japan). The tetrasaccharide fraction was loaded on the column equilibrated with 16 mm NaH2PO4 and then eluted with a linear gradient from 16 to 460 mm NaH2PO4 over 60 min at a flow rate of 1.0 ml/min at room temperature. The eluates were monitored by measuring absorbance at 232 nm, and a major peak was collected and desalted with a Superdex Peptide column as described above.
The purified HCLase-resistant tetrasaccharide was sequenced at a low picomole level by CSase digestion in conjunction with HPLC. Briefly, to identify the composition of the disaccharide, an aliquot (5 pmol) of the tetrasaccharide fraction was digested with CSase ABC (Sigma) and labeled with 2-AB and sodium cyanoborohydride reagents as described by Bigge et al. (50). Free 2-AB was removed by extraction with chloroform. The tetrasaccharide fraction (30 pmol) used for sequencing was labeled with 2-AB and purified by paper chromatography, and an aliquot (5 pmol) of 2-AB-labeled tetrasaccharide sample was then treated with a novel exo-CSase from Vibrio sp. FC509.4 All these preparations were individually analyzed by anion-exchange HPLC on a YMC-Pack Polyamine II column eluted with a linear gradient from 16 to 460 mm NaH2PO4 over a 60-min period and monitored using a fluorescence detector.
Effects of 2-AB Labeling on the Digestion of Oligosaccharides by rHCLase
To investigate the effects of 2AB labeling on the digestion of oligosaccharides by rHCLase, pure HA/CS oligosaccharide monomers were fluorescently labeled with 2-AB and digested by rHCLase. The resulting digests (5-pmol samples) were analyzed by gel filtration on a Superdex Peptide 10/300 GL column (GE healthcare), and the samples were monitored using a fluorescence detector with excitation and emission wavelengths of 330 and 420 nm, respectively (51).
Expression and Characterization of Mutant rHCLase
To investigate the functions of the specific amino acid residues in the HA/CS-degrading activities of HCLase, the protein sequence of HCLase was directly submitted to the SWISS-MODEL automatic modeling server to search for the homologous contribution of the three-dimensional structure online (52). The modeled structure was observed using SPD-Viewer version 3.7 (53).
The computational model showed that the Trp-492, His-285, Tyr-290, and Tyr-294 residues may affect the interaction between the enzyme and substrate. To characterize the role of these residues, we used site-directed mutagenesis of the pET22-rHCLase plasmid by swapping these residues with alanine residues using rapid PCR amplification with high fidelity DNA polymerase of PrimeSTAR Max Premix (Takara Inc.) to form a series of gene mutation products. Corresponding primer pairs are listed in Table 1.
Amplified gene products were individually phosphorylated at the 5′-end, circulated, and transformed into E. coli BL21(DE3) cells, and protein expression was then induced. GAG-degrading activities were measured as described above.
RESULTS
Identification of Chondroitin Sulfate-degrading Bacteria
Using the BM screening plates, 276 bacterial strains were isolated from nine coastal sediment samples. The results obtained from an examination of bacterial growth and consumption of polysaccharides indicated that 15 isolates could degrade more than three types of polysaccharides by utilizing them as the sole carbon sources (Table 2). BLASTn analysis of the 16 S rRNA genes showed that all 15 polysaccharide-degrading bacterial strains belonged to the Vibrio genus.
TABLE 2.
Growth of various bacterial strains in medium containing a sole carbon source
| Strains | Agarose | HA | DS | CS-C | Heparin | Heparin sulfate | Alginate | Xanthan |
|---|---|---|---|---|---|---|---|---|
| FC202 | + | +++ | +++ | |||||
| FC207 | +++ | +++ | + | |||||
| FC208 | +++ | +++ | + | |||||
| FC301 | ++ | |||||||
| FC303 | +++ | + | +++ | |||||
| FC305 | +++ | ++ | +++ | + | ++ | |||
| FC306 | + | +++ | +++ | |||||
| FC307 | +++ | ++ | +++ | + | ++ | |||
| FC309 | +++ | +++ | + | |||||
| FC310 | ++ | + | + | |||||
| FC503 | +++ | + | +++ | |||||
| FC504 | ++ | + | + | |||||
| FC509 | +++ | +++ | +++ | + | + | ++ | ||
| FC510 | ++ | ++ | + | |||||
| FC511 | +++ | +++ | + |
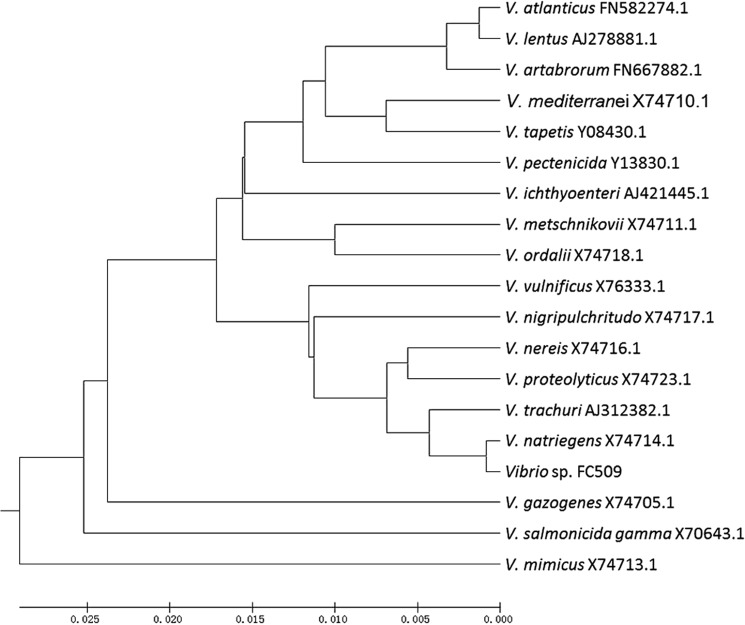
The Vibrio sp. FC509 strain, one of the 15 polysaccharide-degrading marine bacteria, could degrade and utilize alginate, DS, CS-C, heparin, and HA for its growth. The growth of FC509 was enhanced when either CS-C or HA was used as the sole carbon source with the highest cell density to give an A600 ranging from 2.8 to 3.0. Thus, these results showed that Vibrio sp. FC509 is a marine bacterium that can efficiently degrade multiple GAGs. The results of further phylogenetic analyses indicated that the Vibrio sp. FC509 strain was the most homologous to Vibrio natriegens (Fig. 1).
FIGURE 1.
Phylogenetic analysis of CS-degrading bacteria based on the sequences of the 16 S rRNA gene. The phylogenetic analysis was performed using the neighbor joining method in MEGA 5.1.
Information on the HCLase Gene and Protein Sequences
The draft genome sequence of Vibrio sp. FC509 was predicted to be ∼5.2 Mb in size. The genome obtained encoded at least 4500 ORFs with a length of longer than 100 amino acids. One putative GAG-degrading gene (GenBank TM accession number KJ885185), named hclase, was 2436 bp in length and had a GC content of 50.5%. The hclase gene encoded a protein (HCLase) consisting of 811 amino acid residues.
The molecular mass of the putative HCLase protein was 90.2 kDa. The isoelectric point (pI) was 7.23. A BLASTp search showed that HCLase shared sequence identities of higher than 30% to putative GAGs lyases from bacteria such as Bacillus, Photobacterium, Aliivibrio, Vibrio, and Yersinia strains. However, of the proteins elucidated, HCLase shared the highest sequence identity (34%) to a hyaluronidase purified from Bacillus sp. A50 (54) followed by a xanthan lyase (32%) from Bacillus sp. GL1 (55). Carbohydrate-Active Enzyme database and Simple Modular Architecture Research Tool analyses showed that the HCLase protein contained an N-terminal signal peptide and GAG lyase module. SignalP 3.0 and LipoP 1.0 analyses indicated that the type I signal peptide contained 22 amino acid residues. A BLASTp search showed that the GAG lyase module contained five active site regions and four candidate catalytic site residues (Trp-492, His-285, Tyr-290, and Tyr-294) that were conserved within the elucidated GAG lyases.
Heterologous Expression of HCLase in E. coli
The full-length sequence of the HCLase ORF was amplified directly from the genomic DNA of Vibrio sp. FC509. The PCR product was recovered and cloned into the pET-22b(+) vector following a T7 promoter. In this HCLase expression vector (pE22b-rHCLase), a pelB leader peptide was added at the N terminus, and a His6 tag was added at the C terminus. SDS-PAGE analysis indicated that E. coli BL21(DE3) cells harboring the pE22b-rHCLase plasmid could form soluble products (∼20 mg/liter) with a correct molecular mass of 90 kDa. Expression of the HCLase gene using the pBA-HCLase vector in E. coli Top10 cells or pCTF-HCLase in E. coli BL21(DE3) cells also succeeded in producing soluble fusion proteins (∼50–100 mg/liter).
The crude enzyme was extracted from the cultures of pE22b-rHCLase-harboring host cells by sonication and centrifugation. The recombinant enzyme (rHCLase) was further purified by Ni-NTA affinity chromatography. As shown in Fig. 2, SDS-PAGE showed that the rHCLase protein could be eluted from the Ni-NTA column using a gradient of imidazole concentrations ranging from 50 to 250 mm. Purified rHCLase had a purity of >97%, recovery of ∼80%, and initial protein concentration of 20 μg/ml.
FIGURE 2.
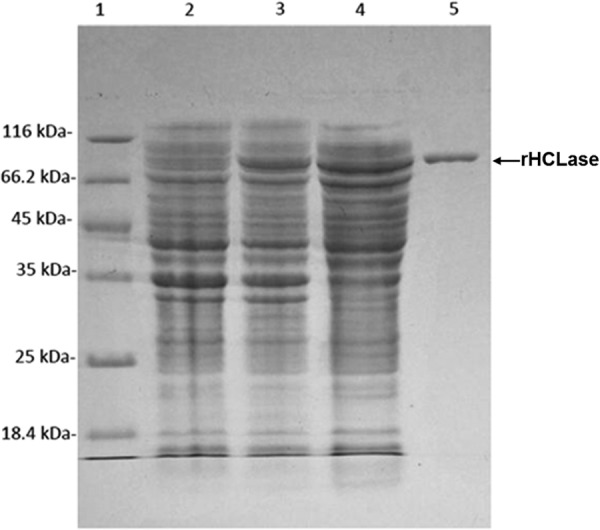
Purification of recombinant rHCLase from E. coli by Ni2+ chelation chromatography. Enzyme purity following each fractionation step was assessed by SDS-PAGE using 13.2% polyacrylamide gels followed by staining with Coomassie Brilliant Blue. Lane 1, unstained protein molecular weight marker SM 0431 (Thermo); lane 2, uninduced cell lysate; lane 3, induced cell lysate; lane 4, supernatant fluid of the induced cell lysate; lane 5, purified recombinant rHCLase. Molecular weight markers and their corresponding masses are also indicated.
Enzymatic Characteristics of rHCLase
Purified rHCLase was able to degrade HA, CS-A, CS-C, CS-D, and CS-E to produce oligosaccharides with an absorbance at 232 nm, which is characteristic of typical unsaturated oligosaccharides. However, rHCLase could hardly digest DS and had no effect on heparan sulfate or heparin. These results showed that the HCLase protein was a GAG lyase with high specificity to HA and CS.
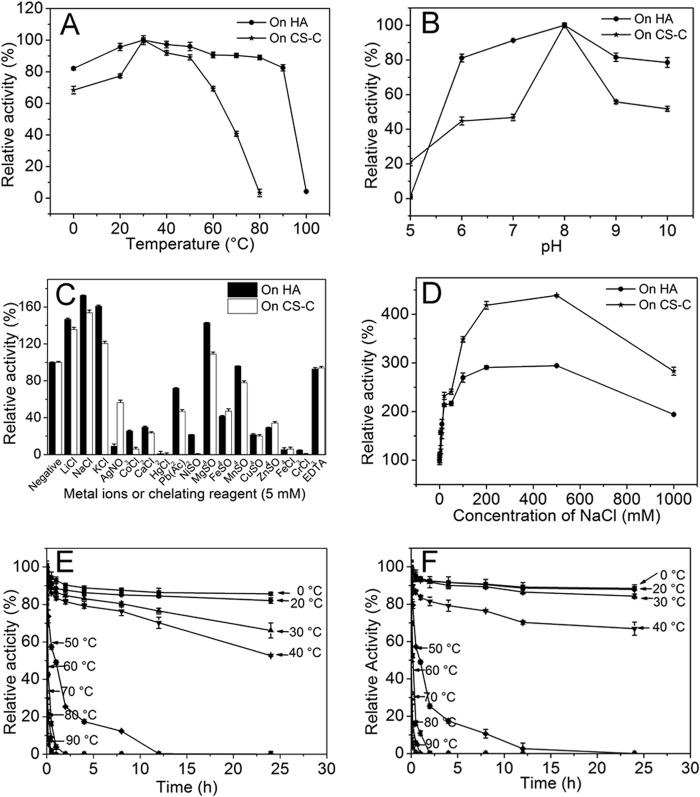
The rHCLase showed the optimum temperature at 30 °C when either HA or CS was used as the substrate (Fig. 3A). A thermostability assay further showed that the HA/CS-degrading activity of rHCLase was very stable at temperatures from 0 to 40 °C and retained more than 50% activity even if it was preincubated at a corresponding temperature for 24 h (Fig. 3, E and F). The optimal pH determined at 30 °C in 50 mm NaH2PO4-Na2HPO4 buffer was 8.0, and the hyaluronidase activity of rHCLase was retained at at least 80% of the highest activity obtained at pH 5–9. However, in the case of its CSase activity, only slightly more than 45% of that at pH 7–8 remained (Fig. 3B). These results suggested that the CS-degrading activity of rHCLase was more sensitive to pH than its HA-degrading activity.
FIGURE 3.
Biochemical reaction conditions for recombinant rHCLase. A, effects of temperature. The enzyme activities of rHCLase were measured using HA and CS-C as substrates in the 50 mm NaH2PO4-Na2HPO4 buffer (pH 8.0) at different temperatures for 10 min. Data are shown as the percentage of the activity of that obtained at 30 °C (100%). B, effects of pH. The activities of rHCLase against HA and CS-C were measured in buffers with varying pH values from 5 to 10 at 30 °C for 10 min. Data are shown as the percentage of the activity of that obtained in the NaH2PO4-Na2HPO4 buffer (pH 8.0). C, effects of metal ions. The activities of rHCLase against HA and CS-C were measured in the NaH2PO4-Na2HPO4 buffer (pH 8.0) containing a 5 mm concentration of various metal ions at 30 °C for 10 min. Data are shown as the percentage of the activity of that obtained in the buffer without tested metal ions. D, effects of NaCl concentrations. The activities of rHCLase against HA and CS-C were measured in the NaH2PO4-Na2HPO4 buffer (pH 8.0) containing 0-1 m NaCl at 30 °C for 10 min. Data are shown as the percentage of the activity obtained in the buffer without NaCl. E and F, thermostability of rHCLase. The enzyme in 50 mm NaH2PO4-Na2HPO4 buffer (pH 8.0) was preincubated for 0–24 h at temperature from 0 to 90 °C, and the residual activity against HA (E) or CS-C (F) was estimated at 30 °C. Data are shown as the activity relative to that of untreated rHCLase. Error bars represent means of triplicates ±S.D.
The HA/CS-degrading activities of rHCLase were strongly inhibited by Ag+, Hg2+, Pb2+, Ni2+, Fe2+, Cu2+, Zn2+, Fe3+, and Cr3+ at a concentration of 5.0 mm. In contrast, basic alkali metal ions such as Li+, Na+, and K+ at 5.0 mm significantly promoted the HA/CS lyase activity of rHCLase as in the case of other halophilic lyases from marine microorganisms. The HA/CS-degrading activity of HCLase was slightly affected or even significantly inhibited by divalent metal salts (CaCl2, MgCl2, MnCl2, and CoCl2) and 5.0 mm EDTA, which indicated that divalent metal ions were not needed for the lyase activity of rHCLase (Fig. 3C).
To determine the effects of ionic strength on enzyme activities, different concentrations of sodium chloride were added to the basic reaction buffer (50 mm NaH2PO4-Na2HPO4 (pH 8.0)), and the activity of rHCLase was then measured. Fig. 3D shows that HA/CS-degrading activities markedly increased as the concentration of NaCl was raised from 0 to 0.5 m. The optimum concentration of the sodium ion was 500 mm, which was close to the concentration of salt in seawater, and when it was increased to 1 m, HA and CS lyase activities were maintained at 300 and 200%, respectively, of that obtained using a NaCl-free buffer.
Under the optimal conditions of 30 °C in 50 mm NaH2PO4-Na2HPO4 and 500 mm NaCl (pH 8.0), the specific activity of rHCLase was measured using HA, chondroitin, and various types of commercial CS variants as described under “Experimental Procedures.” The specific activities of rHCLase using HA, chondroitin, CS-A, CS-C, CS-D, and CS-E were 450,000, 330,000, 202,000, 193,000, 175,000, and 164,000 units/mg of protein, respectively (Table 3). The specific activities of HCLase were hundreds to thousands of times higher than those of the best characterized hyaluronidases and chondroitinases (56).
TABLE 3.
Activity analysis for rHCLase and its mutant proteins
| rHCLase | rHCLase-Y290A | rHCLase-W492A, -H285A, -Y294A | |
|---|---|---|---|
| units/mg | units/mg | units/mg | |
| HA | 450,000 | 8.5 | <1.0 |
| CS-A | 202,000 | 7.5 | <1.0 |
| CS-C | 193,000 | 7.9 | <1.0 |
| CS-D | 175,000 | 7.6 | <1.0 |
| CS-E | 164,000 | 6.7 | <1.0 |
| Chondroitin | 330,000 | 8.2 | <1.0 |
| DS | <1.0 | <1.0 | <1.0 |
Digestion Pattern of Polysaccharides by rHCLase
To determine the degradation pattern of rHCLase, the degradation of HA and CS-A (1 mg/ml) by rHCLase (1 unit/ml) was monitored at 30 °C. The digestion time varied from 0, 0.5, 1, to 10 min for HA (Fig. 4A), and from 0, 1, 2, to 10 min for CS-A (Fig. 4B). The digests were loaded onto a Superdex Peptide 10/300 GL column and monitored by absorbance at 232 nm. Whenever HA or CS-A was used as the substrate, rHCLase initially produced higher molecular mass oligosaccharides and then smaller oligomers with strong absorbance at 232 nm, which suggested that the HCLase protein was a GAG endolyase.
FIGURE 4.
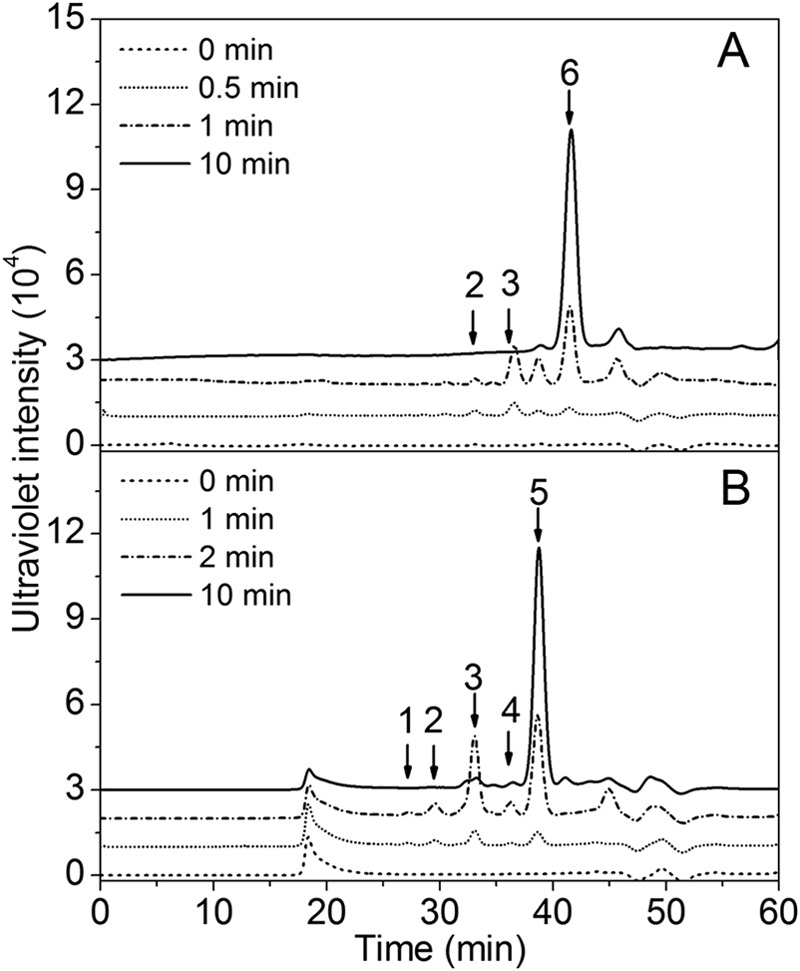
Time course experiments of rHCLase degradation of HA and CS-A. HA (A) or CS-A (B) (1 mg/ml) was treated with rHCLase (1 unit/ml), and a 10-μl aliquot was taken at different time points for gel filtration analysis as described under “Experimental Procedures.” The elution positions of the following standard oligosaccharides are indicated by arrows: 1, CS or HA octasaccharide fraction; 2, CS or HA hexasaccharide fraction; 3, CS or HA tetrasaccharide fraction; 4, disulfated CS disaccharide fraction; 5, monosulfated CS disaccharide fraction; 6, nonsulfated CS or HA disaccharide.
Furthermore, HA, CS-A, CS-C, CS-D, and CS-E were exhaustively digested with rHCLase to determine their final products. The resulting oligosaccharides were analyzed and assigned by gel filtration analysis using HA- and CS-derived authentic unsaturated oligosaccharides. As shown in Fig. 5, nonsulfated and monosulfated disaccharides were the main final products for HA (Fig. 5A) and CS-A (Fig. 5B), respectively. The digests of oversulfated CS-E caused two main disaccharide peaks that corresponded to mono- and disulfated disaccharides (Fig. 5E). Except for the major peak of monosulfated disaccharides, a minor tetrasaccharide peak was detected in the digests of both CS-C (Fig. 5C) and CS-D (Fig. 5D), indicating the existence of HCLase-resistant structures in these digests. It is well known that CS/DS from marine animals are usually rich in unique oversulfated disaccharides (30–34), and CS-C and CS-D from shark cartilage contain disulfated disaccharide GlcUA(2S)β1-3GalNAc(6S) (where 2S and 6S represent 2-O-sulfate and 6-O-sulfate, respectively) the galactosaminidic linkage bound to which may be resistant to the action of rHCLase as in the case of CSase ACI (57).
FIGURE 5.
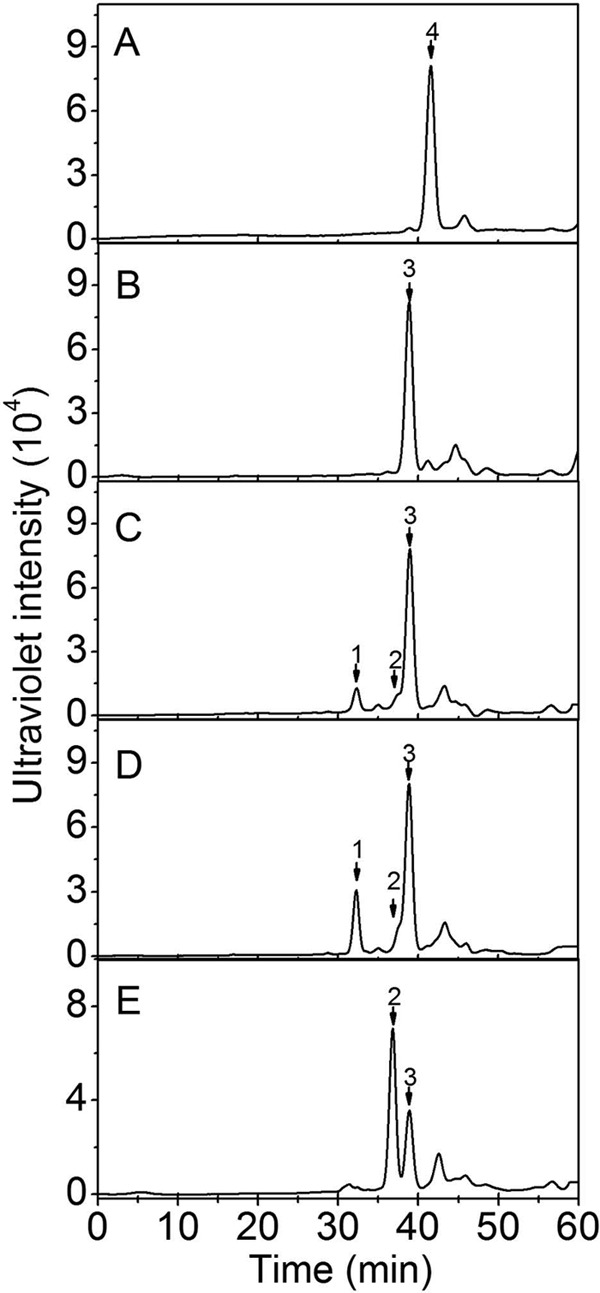
Analysis of the final products of HA and CS digested by rHCLase. Ten micrograms each of HA (A), CS-A (B), CS-C (C), CS-D (D), or CS-E (E) was exhaustively digested with rHCLase and then separated into its constituent oligosaccharides by gel filtration chromatography on a Superdex Peptide column as described under “Experimental Procedures.” The elution positions of the following standard oligosaccharides are indicated by arrows: 1, CS tetrasaccharides; 2, CS disulfated disaccharides; 3, CS monosulfated disaccharides; 4, nonsulfated CS or HA disaccharides.
To further characterize the structures of the final products of HA and CS-A, the major disaccharide fractions were collected and analyzed by electrospray ionization MS spectrometry in a negative ion mode. The major peaks of the HA and CS-A disaccharides were detected at m/z 378.1028 and 458.0587 (Fig. 6), which could be assigned by mass calculation to nonsulfated unsaturated Δ4,5HexUA-GlcNAc derived from HA and monosulfated unsaturated Δ4,5HexUA-GalNAc derived from CS-A, respectively. These results further confirmed that HCLase was a lyase that digested HA and CS through a β-elimination mechanism.
FIGURE 6.
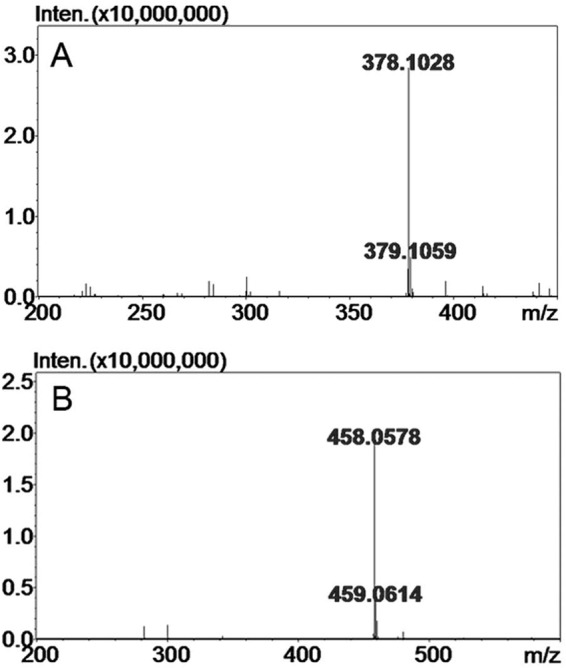
Time-of-flight mass spectra of the disaccharides of HA/CS digested by HCLase. The main final products obtained from HA (A) and CS-A (B) digested by HCLase were identified by electrospray ionization MS on an ion trap TOF hybrid mass spectrometer as described under “Experimental Procedures.” Inten., intensity.
Sequencing of the rHCLase-resistant Tetrasaccharide
To investigate the structure of the HCLase-resistant tetrasaccharide, CS-D was exhaustively digested with rHCLase, and the digest was fractionated by gel filtration as described under the “Experimental Procedures.” The fraction of HCLase-resistant tetrasaccharides was collected and further subfractionated by anion-exchange chromatography on a YMC-Pack Polyamine II column. A single tetrasaccharide peak was detected and collected for the subsequent characterization. To confirm the homogeneity of the tetrasaccharide fraction, an aliquot (5 pmol) of the purified sample was labeled with 2-AB and analyzed by anion-exchange HPLC again, and a single peak was detected again, confirming its high purity (Fig. 7A). The disaccharide analysis showed that the purified fraction was composed of HexUA1-3GalNAc(6S) and HexUA(2S)1-3GalNAc(6S) in a molar ratio of 1.0:1.0 (Fig. 7B), which suggested that the HCLase-resistant tetrasaccharide consisted of 1 mol each of saturated or unsaturated HexUA1-3GalNAc(6S) and HexUA(2S)1-3GalNAc(6S). To sequence the tetrasaccharide, the 2AB-labeled tetrasaccharide fraction (5 pmol) was digested with a new CSase from the same Vibrio sp. FC509 that is an exo-CSase and could effectively cleave 2-AB-labeled HA/CS tetrasaccharides.4 The digest was subjected to an anion-exchange HPLC assay, and a single peak was detected at the position corresponding to 2-AB-labeled Δ4,5HexUA(2S)α1-3GalNAc(6S) (Fig. 7C), indicating that the disaccharide GlcUA(2S)β1-3GalNAc(6S) was located at the reducing end of the HCLase-resistant tetrasaccharide. Based on these results, it was concluded that the resistant tetrasaccharide had the following structure: Δ4,5HexUAα1-3GalNAc(6S)β1-4GlcUA(2S)β1-3GalNAc(6S).
FIGURE 7.
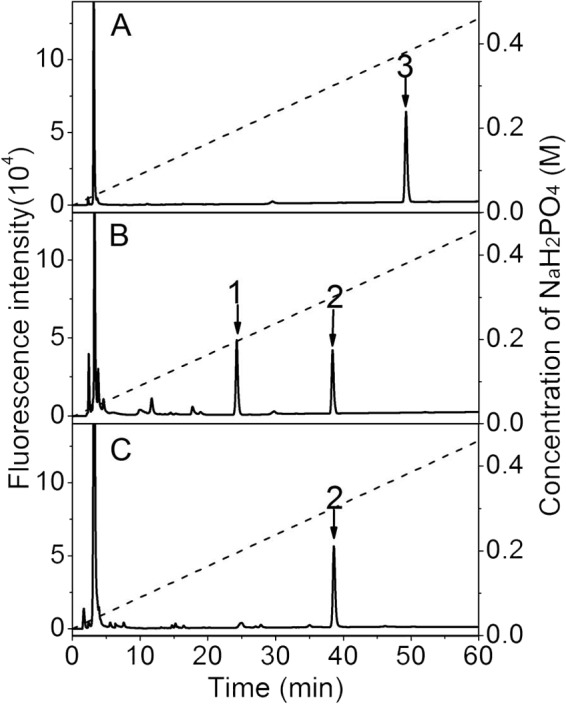
Sequencing analysis of the HCLase-resistant tetrasaccharide fraction. A, the purity of the HCLase-resistant tetrasaccharide fraction from CS-D was confirmed through 2-AB labeling followed by anion-exchange HPLC. B, the disaccharide composition of the resistant tetrasaccharide was determined by comparing the elution positions of the 2-AB-labeled unsaturated disaccharides produced by digestion using CSase ABC with those of authentic 2-AB-derivatized unsaturated CS disaccharides indicated by arrows. C, the disaccharide moiety at the reducing end of the resistant tetrasaccharide was identified by digesting the 2-AB-labeled tetrasaccharides with a novel CSase from Vibrio sp. FC509. All samples were analyzed by HPLC on a YMC-Pack Polyamine II column using a NaH2PO4 gradient (indicated by the dashed line). 1, Δ4,5HexUAα1-3GalNAc(6S); 2, Δ4,5HexUA(2S)α1-3GalNAc(6S); 3, the resistant tetrasaccharide.
Effects of 2-AB Labeling on the Digestion of Oligosaccharides by rHCLase
Fluorescent labeling, in particular using 2-AB, is often used in the disaccharide composition and structural analyses of GAGs because of its high sensitivity and selective derivation to the reducing end. However, labeling may affect the cleavage of GAG chains by GAG-degrading enzymes such as CSase ABC, which cannot cleave 2-AB-labeled tetrasaccharides (51). To investigate the effects of 2AB labeling on the digestion of oligosaccharides by rHCLase, purified HA/CS oligosaccharides (3 pmol) were individually labeled at the reducing ends using 2-AB and then treated with HCLase. As shown in Fig. 8, the 2-AB-labeled HA tetrasaccharide was not further attacked by HCLase (Fig. 8A). In contrast, the 2-AB-labeled HA hexasaccharide was digested to release a 2-AB-labeled tetrasaccharide (peak 2) and unlabeled disaccharide (Fig. 8B). The same phenomenon was observed in the case of oligosaccharides from CS-A and CS-C (data not shown), suggesting that HCLase could not degrade the tetrasaccharides at the reducing end of HA/CS chains when labeled at the reducing ends, similar to CSase ABC.
FIGURE 8.
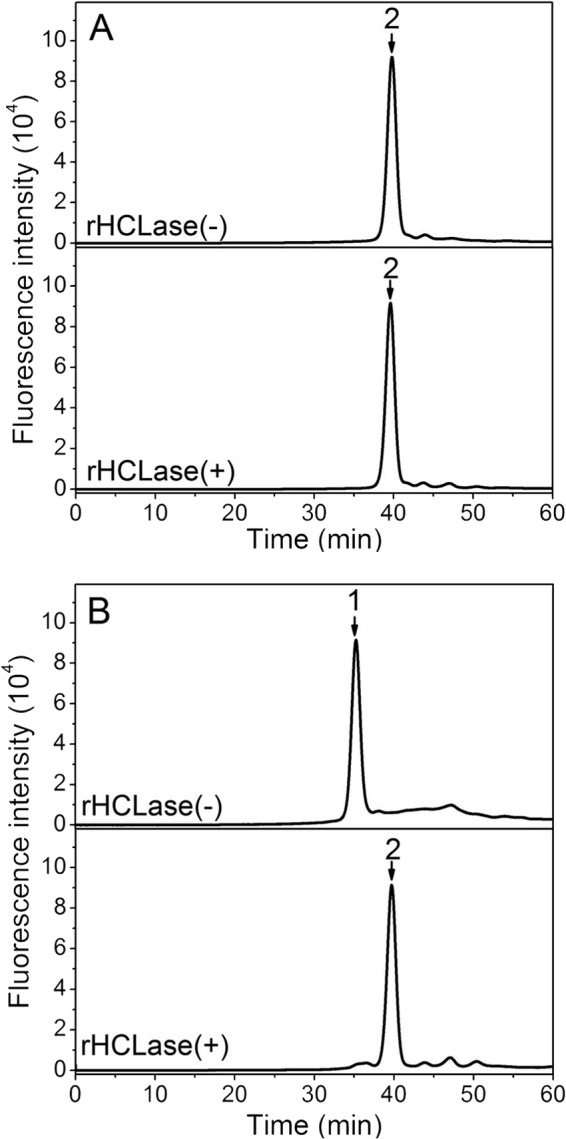
Analysis of the final products of 2-AB-labeled oligosaccharides digested by rHCLase. Five picomoles of 2-AB-labeled HA tetrasaccharides (A) or hexasaccharides (B) was exhaustively digested without (top panel) or with rHCLase (100 units/ml) (bottom panel) at 30 °C for 72 h. The resulting product was analyzed by gel filtration as described above. 1, 2-AB-labeled unsaturated HA hexasaccharide; 2, 2-AB-labeled unsaturated HA tetrasaccharide.
Study of rHCLase Site-directed Mutagenesis
To preliminarily investigate the catalytic mechanism underlying HCLase, four conserved amino acid residues (Trp-492, His-285, Tyr-290, and Tyr-294) were identified by comparing HCLase with homologous sequences. Each conserved residue was replaced by alanine (Ala) through site-directed mutagenesis. The mutant proteins were expressed using the pET22b system, and the soluble recombinant mutants rHCLase-W492A, rHCLase-H285A, rHCLase-Y290A, and rHCLase-Y294A were assayed for their degrading abilities. As shown in Fig. 9, the mutants rHCLase-W492A, rHCLase-H285A, and rHCLase-Y294A were no longer able to degrade HA, whereas the mutant rHCLase-Y290A could still digest HA into disaccharides as the major final products; however, its HA/CS-degrading activity was markedly decreased (Table 3). These results suggested that these residues may play crucial roles in the catalytic mechanism of HCLase when digesting HA and CS.
FIGURE 9.
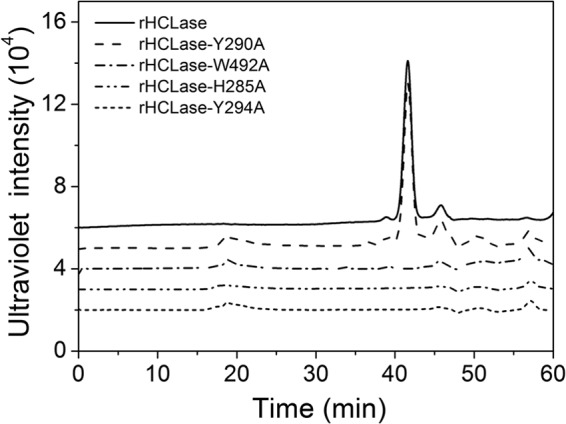
Gel filtration chromatography of HA treated with rHCLase mutant proteins. HA (10 μg) was exhaustively digested with rHCLase and its mutant proteins and then analyzed by gel filtration chromatography on a Superdex Peptide column as described under “Experimental Procedures.”
DISCUSSION
All known GAG lyases to date have been isolated from terrestrial bacteria such as P. vulgaris (27), F. heparinum (27), and A. aurescens (28). However, marine bacterium-derived GAG lyases remain unexplored even though various GAGs have been found in and isolated from marine sources for basic research and industrial purposes. In the present study, a novel HA/CS lyase, HCLase, was identified in a newly isolated marine Vibrio sp. FC509 for the first time. A sequence similarity search showed that HCLase had very low homology (≤34%) to the various GAG lyases characterized so far. HCLase specifically digested HA and CS into disaccharides as the main final products and had very unique biochemical characteristics such as very high activity and excellent thermostability. In addition, it exhibited the highest activity at approximately neutral pH. All of these characteristics strongly indicated that HCLase may belong to a new unique GAG lyase family and be very useful for investigating the structure-function and preparation of bioactive oligosaccharides from HA and various CS variants.
In most cases, HA and CS depolymerases can digest both HA and CS with different degrees of activity. Except for the hyaluronidase (EC 4.2.2.1) from Streptomyces hyalurolyticus that is specific for HA (58), both bacterial and vertebrate hyaluronidases degrade chondroitin and various kinds of CS variants, although degradation occurs more slowly than that of HA. The various CSases characterized so far have also shown a certain degree of HA-degrading activity: CSase ABC primarily degrades all types of CS and DS but has low activity against HA, CSase ACI acts on HA with 20% of the activity of that toward CS, and CSase C shows similar levels of activities toward CS-C and HA. A plausible explanation for these phenomena is that hyaluronidase originally evolved from the pre-existing CSases (59, 60). Although HCLase had the highest activity (450,000 IU/mg) against HA, the degradation rates of chondroitin and various types of CS isoforms were relatively high (73–36% of hyaluronidase activity), and these were very different from other conventional hyaluronidases, which generally show markedly lower activity toward chondroitin and CS than HA. In view of the very high activities for both HA and CS, this new enzyme has been designated as HCLase rather than hyaluronidase to avoid a misnomer.
Being different from most hyaluronidases, which are the most active at acidic pHs, the optimum pH (pH 8.0) for the activity of HCLase toward both HA and CS was approximately neutral pH, which was similar to that of the activities of the CSases ABC, ACI, and B. Although the optimum temperature and pH for HCLase to digest HA and CS were the same, HA-degrading activity was maintained at high levels over a wide range of temperatures and pH, whereas CS-degrading activity was very sensitive to changes in temperature and pH. These results suggested that substrates may affect the stability of the structure of HCLase, but the mechanism behind these observations remains to be investigated. A thermostability assay showed that both the HA- and CS-degrading activities of HCLase were very stable at a temperature from 0 to 40 °C for a significant time period (up to 24 h).
Marine bacteria have been classified as halophilic microorganisms (61) and represent the main sources for isolating halophilic enzymes. The HA- and CS-digesting activities of HCLase were both significantly stimulated by alkali metal ions, in particular Na+. Moreover, these enzyme activities acutely increased as sodium chloride concentrations were raised from 0 to 0.2 m and peaked at a concentration of 0.5 m, which is close to the concentration of the salinity of seawater. HCLase maintained relatively high activity even when the concentration of sodium chloride was increased to 1.0 m. These results suggested that HCLase may be a typical halophilic enzyme.
The specific activity of GAG lyases such as the well known hyaluronidases from S. hyalurolyticus and Streptococcus dysgalactiae as well as CSases ABC, ACI, and ACII is tens to thousands of international units per milligram of the enzyme protein. In contrast, an enzyme with very high activities for HA and CS was purified from an oral Peptostreptococcus species and characterized. The specific enzyme activity for HA reached 600,000 IU/mg protein (62). Another HA/CS lyase was more recently purified from Bacillus sp. and exhibited the greatest activity against HA (1,020,000 IU/mg), and the degradation rates of CS-A and CS-C were 39 and 13.5%, respectively, of the rate for HA (54). These findings suggested the existence of GAG lyases with very high activities. Nevertheless, these enzymes have not been cloned, expressed, or identified in detail. The recombinant expression and characterization of HCLase in the present study confirmed the existence of such “super GAG lyases” at the genetic level and suggested the possibility of producing such enzymes by genetic engineering.
Except for CSase ABC, which can cleave almost all types of natural CS variants and DS with various sulfation patterns, the ability of most enzymes to digest CS has limitations in the susceptible structures. CS-degrading hyaluronidases from either bacteria or animals are typically able to digest non- or low-sulfated CS but not highly sulfated CS. CSase ACI cannot cleave the galactosaminidic bond bound to the D unit (GlcUA(2S)β1-3GalNAc(6S)) (63). Although HCLase can digest various other types of galactosaminidic bonds to generate disaccharides as the main final products, an HCLase-resistant tetrasaccharide ΔHexUAα1-3GalNAc(6S)β1-4GlcUA(2S)β1-3GalNAc(6S) was found in the digest of CS-D, the structure of which indicated that, as in the case of CSase ACI (57), HCLase was also unable to act on the galactosaminidic linkage bound to the D unit. Moreover, we found that HCLase could not cleave 2AB-labeled HA or CS tetrasaccharides, similar to CSases ABC and ACI, whereas CSase ACII could (51).
Based on the comparison of homologous amino acid sequences, four conserved residues, Trp-492, His-285, Tyr-290, and Tyr-294, were identified in the amino acid sequence of HCLase. In site-directed mutagenesis, the substitution of these conserved residues by alanine completely abolished or strongly inhibited the activity of HCLase toward both HA and CS, which suggested key roles for these amino acids in the active sites of HCLase. Structural and biochemical data indicate that His and Tyr residues act as the acceptor and donor for protons, respectively, whereas Trp acts as the hydrophobic residue, which facilitates the interaction between the enzyme and substrate in the β-elimination mechanism of lyases. Because divalent ions are not necessary for enzyme activity, the action of HCLase may utilize the catalytic mechanism of Tyr-His but not Arg/Lys to act as the Brønsted base and acid (26). However, a structural investigation of HCLase to determine its precise mechanism of action has yet to be performed.
In conclusion, HCLase, as a marine bacterium-derived GAG lyase, possesses a series of unique features such as very high activity toward HA and CS, highest activity at neutral pH, high activity over a wide range of temperatures and pH values, and halophilicity, indicating that it will be a novel powerful enzyme for the structural analysis and oligosaccharide preparation of HA and CS variants. Hyaluronidase has been used clinically as a spreading agent for other medications as well as in the management of the adverse effects of HA soft tissue fillers and treatments for sclerodermoid lesions, lymphedema, and keloids (64). Recent studies have revealed that CS proteoglycans are potential inhibitors against axon regeneration after injuries to the central nervous system, and CSase treatments represent a promising in vivo approach to overcome inhibition by CS proteoglycans (65). HCLase, which has various excellent properties, is a potential candidate for such medical applications.
This work was supported by National High Technology Research and Development Program of China Grant 2012AA021504, Major State Basic Research Development Program of China Grant 2012CB822102, Shandong Province Science and Technology Development Plan Grant 2013GSF12106, General Financial Grant from China Postdoctoral Science Foundation Grant 2013M531588, and Specialized Research Fund for the Doctoral Program of Higher Education Grant 20130131120079 and in part by Grant-in-aid for Challenging Exploratory Research 25670018 from the Japan Society for the Promotion of Science.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) KJ885185.
W. Wang, W. Han, M. Zhao, and F. Li, unpublished data.
- CS
- chondroitin sulfate
- DS
- dermatan sulfate
- HA
- hyaluronan
- GAG
- glycosaminoglycan
- GlcUA
- d-glucuronic acid
- HexUA
- hexuronic acid
- Δ4,5HexUA
- Δ4,5-unsaturated hexuronic acid
- 2S
- 2-O-sulfate
- 6S
- 6-O-sulfate
- CSase
- chondroitinase
- 2-AB
- 2-aminobenzamide
- IdoUA
- l-iduronic acid
- BM
- basal medium
- HCLase
- HA and CS lyase
- rHCLase
- recombinant HCLase
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1. Poole A. R. (1986) Proteoglycans in health and disease: structures and functions. Biochem. J. 236, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Faissner A., Clement A., Lochter A., Streit A., Mandl C., Schachner M. (1994) Isolation of a neural chondroitin sulfate proteoglycan with neurite outgrowth promoting properties. J. Cell Biol. 126, 783–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clement A. M., Nadanaka S., Masayama K., Mandl C., Sugahara K., Faissner A. (1998) The DSD-1 carbohydrate epitope depends on sulfation, correlates with chondroitin sulfate D motifs, and is sufficient to promote neurite outgrowth. J. Biol. Chem. 273, 28444–28453 [DOI] [PubMed] [Google Scholar]
- 4. Penc S. F., Pomahac B., Winkler T., Dorschner R. A., Eriksson E., Herndon M., Gallo R. L. (1998) Dermatan sulfate released after injury is a potent promoter of fibroblast growth factor-2 function. J. Biol. Chem. 273, 28116–28121 [DOI] [PubMed] [Google Scholar]
- 5. Trowbridge J. M., Rudisill J. A., Ron D., Gallo R. L. (2002) Dermatan sulfate binds and potentiates activity of keratinocyte growth factor (FGF-7). J. Biol. Chem. 277, 42815–42820 [DOI] [PubMed] [Google Scholar]
- 6. Hsiao J. C., Chung C. S., Chang W. (1999) Vaccinia virus envelope D8L protein binds to cell surface chondroitin sulfate and mediates the adsorption of intracellular mature virions to cells. J. Virol. 73, 8750–8761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Williams R. K., Straus S. E. (1997) Specificity and affinity of binding of herpes simplex virus type 2 glycoprotein B to glycosaminoglycans. J. Virol. 71, 1375–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bergefall K., Trybala E., Johansson M., Uyama T., Naito S., Yamada S., Kitagawa H., Sugahara K., Bergström T. (2005) Chondroitin sulfate characterized by the E-disaccharide unit is a potent inhibitor of herpes simplex virus infectivity and provides the virus binding sites on gro2C cells. J. Biol. Chem. 280, 32193–32199 [DOI] [PubMed] [Google Scholar]
- 9. Nandi S., Akhter M. P., Seifert M. F., Dai X. M., Stanley E. R. (2006) Developmental and functional significance of the CSF-1 proteoglycan chondroitin sulfate chain. Blood 107, 786–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Taylor K. R., Gallo R. L. (2006) Glycosaminoglycans and their proteoglycans: host-associated molecular patterns for initiation and modulation of inflammation. FASEB J. 20, 9–22 [DOI] [PubMed] [Google Scholar]
- 11. Klüppel M., Wight T. N., Chan C., Hinek A., Wrana J. L. (2005) Maintenance of chondroitin sulfation balance by chondroitin-4-sulfotransferase 1 is required for chondrocyte development and growth factor signaling during cartilage morphogenesis. Development 132, 3989–4003 [DOI] [PubMed] [Google Scholar]
- 12. Hwang H. Y., Olson S. K., Esko J. D., Horvitz H. R. (2003) Caenorhabditis elegans early embryogenesis and vulval morphogenesis require chondroitin biosynthesis. Nature 423, 439–443 [DOI] [PubMed] [Google Scholar]
- 13. Mizuguchi S., Uyama T., Kitagawa H., Nomura K. H., Dejima K., Gengyo-Ando K., Mitani S., Sugahara K., Nomura K. (2003) Chondroitin proteoglycans are involved in cell division of Caenorhabditis elegans. Nature 423, 443–448 [DOI] [PubMed] [Google Scholar]
- 14. Izumikawa T., Kitagawa H., Mizuguchi S., Nomura K. H., Nomura K., Tamura J., Gengyo-Ando K., Mitani S., Sugahara K. (2004) Nematode chondroitin polymerizing factor showing cell-/organ-specific expression is indispensable for chondroitin synthesis and embryonic cell division. J. Biol. Chem. 279, 53755–53761 [DOI] [PubMed] [Google Scholar]
- 15. Kitagawa H., Izumikawa T., Uyama T., Sugahara K. (2003) Molecular cloning of a chondroitin polymerizing factor that cooperates with chondroitin synthase for chondroitin polymerization. J. Biol. Chem. 278, 23666–23671 [DOI] [PubMed] [Google Scholar]
- 16. Izumikawa T., Uyama T., Okuura Y., Sugahara K., Kitagawa H. (2007) Involvement of chondroitin sulfate synthase-3 in chondroitin polymerization through its interaction with chondroitin synthase-1 or chondroitin polymerizing factor. Biochem. J. 403, 545–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mizumoto S., Ikegawa S., Sugahara K. (2013) Human genetic disorders caused by mutations in the genes encoding biosynthetic enzymes for sulfated glycosaminoglycans. J. Biol. Chem. 288, 10953–10961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kusche-Gullberg M., Kjellén L. (2003) Sulfotransferases in glycosaminoglycan biosynthesis. Curr. Opin. Struct. Biol. 13, 605–611 [DOI] [PubMed] [Google Scholar]
- 19. Silbert J. E., Sugumaran G. (2002) Biosynthesis of chondroitin/dermatan sulfate. IUBMB Life. 54, 177–186 [DOI] [PubMed] [Google Scholar]
- 20. Maccarana M., Olander B., Malmström J., Tiedemann K., Aebersold R., Lindahl U., Li J. P., Malmström A. (2006) Biosynthesis of dermatan sulfate: chondroitin-glucuronate C5-epimerase is identical to SART2. J. Biol. Chem. 281, 11560–11568 [DOI] [PubMed] [Google Scholar]
- 21. Cheng F., Heinegård D., Malmström A., Schmidtchen A., Yoshida K., Fransson L. A. (1994) Patterns of uronosyl epimerization and 4-/6-O-sulphation in chondroitin/dermatan sulphate from decorin and biglycan of various bovine tissues. Glycobiology 4, 685–696 [DOI] [PubMed] [Google Scholar]
- 22. Sugahara K., Mikami T., Uyama T., Mizuguchi S., Nomura K., Kitagawa H. (2003) Recent advances in the structural biology of chondroitin sulfate and dermatan sulfate. Curr. Opin. Struct. Biol. 13, 612–620 [DOI] [PubMed] [Google Scholar]
- 23. Linhardt R. J., Avci F. Y., Toida T., Kim Y. S., Cygler M. (2006) CS Lyases: structure, activity, and applications in analysis and the treatment of diseases. Adv. Pharmacol. 53, 187–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li F., Nandini C. D., Hattori T., Bao X., Murayama D., Nakamura T., Fukushima N., Sugahara K. (2010) Structure of pleiotrophin- and hepatocyte growth factor-binding sulfated hexasaccharide determined by biochemical and computational approaches. J. Biol. Chem. 285, 27673–27685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nadanaka S., Clement A., Masayama K., Faissner A., Sugahara K. (1998) Characteristic hexasaccharide sequences in octasaccharides derived from shark cartilage chondroitin sulfate D with a neurite outgrowth promoting activity. J. Biol. Chem. 273, 3296–3307 [DOI] [PubMed] [Google Scholar]
- 26. Garron M.-L., Cygler M. (2010) Structural and mechanistic classification of uronic acid-containing polysaccharide lyases. Glycobiology 20, 1547–1573 [DOI] [PubMed] [Google Scholar]
- 27. Yamagata T., Saito H., Habuchi O., Suzuki S. (1968) Purification and properties of bacterial chondroitinases and chondrosulfatases. J. Biol. Chem. 243, 1523–1535 [PubMed] [Google Scholar]
- 28. Hiyama K., Okada S. (1975) Crystallization and some properties of chondroitinase from Arthrobacter aurescens. J. Biol. Chem. 250, 1824–1828 [PubMed] [Google Scholar]
- 29. Bradbury E. J., Moon L. D., Popat R. J., King V. R., Bennett G. S., Patel P. N., Fawcett J. W., McMahon S. B. (2002) Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature 416, 636–640 [DOI] [PubMed] [Google Scholar]
- 30. Ramachandra R., Namburi R. B., Ortega-Martinez O., Shi X., Zaia J., Dupont S. T., Thorndyke M. C., Lindahl U., Spillmann D. (2014) Brittlestars contain highly sulfated chondroitin sulfates/dermatan sulfates that promote fibroblast growth factor 2-induced cell signaling. Glycobiology 24, 195–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vieira R. P., Mulloy B., Mourão P. A. (1991) Structure of a fucose-branched chondroitin sulfate from sea cucumber. Evidence for the presence of 3-O-sulfo-β-d-glucuronosyl residues. J. Biol. Chem. 266, 13530–13536 [PubMed] [Google Scholar]
- 32. Li F., Shetty A. K., Sugahara K. (2007) Neuritogenic activity of chondroitin/dermatan sulfate hybrid chains of embryonic pig brain and their mimicry from shark liver: involvement of the pleiotrophin and hepatocyte growth factor signaling pathways. J. Biol. Chem. 282, 2956–2966 [DOI] [PubMed] [Google Scholar]
- 33. Nandini C. D., Itoh N., Sugahara K. (2005) Novel 70-kDa chondroitin sulfate/dermatan sulfate hybrid chains with a unique heterogenous sulfation pattern from shark skin, which exhibit neuritogenic activity and binding activities for growth factors and neurotrophic factors. J. Biol. Chem. 280, 4058–4069 [DOI] [PubMed] [Google Scholar]
- 34. Nandini C. D., Mikami T., Ohta M., Itoh N., Akiyama-Nambu F., Sugahara K. (2004) Structural and functional characterization of oversulfated chondroitin sulfate/dermatan sulfate hybrid chains from the notochord of hagfish: neuritogenic and binding activities for growth factors and neurotrophic factors. J. Biol. Chem. 279, 50799–50809 [DOI] [PubMed] [Google Scholar]
- 35. Marchesi J. R., Sato T., Weightman A. J., Martin T. A., Fry J. C., Hiom S. J., Dymock D., Wade W. G. (1998) Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 64, 795–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hyatt D., Chen G.-L., Locascio P. F., Land M. L., Larimer F. W., Hauser L. J. (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pati A., Ivanova N. N., Mikhailova N., Ovchinnikova G., Hooper S. D., Lykidis A., Kyrpides N. C. (2010) GenePRIMP: a gene prediction improvement pipeline for prokaryotic genomes. Nat. Methods 7, 455–457 [DOI] [PubMed] [Google Scholar]
- 39. Bairoch A., Apweiler R., Wu C. H., Barker W. C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., Martin M. J., Natale D. A., O'Donovan C., Redaschi N., Yeh L. S. (2005) The universal protein resource (UniProt). Nucleic Acids Res. 33, D154–D159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Haft D. H., Selengut J. D., White O. (2003) The TIGRFAMs database of protein families. Nucleic Acids Res. 31, 371–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kanehisa M., Goto S. (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Apweiler R., Attwood T. K., Bairoch A., Bateman A., Birney E., Biswas M., Bucher P., Cerutti L., Corpet F., Croning M. D., Durbin R., Falquet L., Fleischmann W., Gouzy J., Hermjakob H., Hulo N., Jonassen I., Kahn D., Kanapin A., Karavidopoulou Y., Lopez R., Marx B., Mulder N. J., Oinn T. M., Pagni M., Servant F., Sigrist C. J., Zdobnov E. M. (2001) The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29, 37–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tatusov R. L., Fedorova N. D., Jackson J. D., Jacobs A. R., Kiryutin B., Koonin E. V., Krylov D. M., Mazumder R., Mekhedov S. L., Nikolskaya A. N., Rao B. S., Smirnov S., Sverdlov A. V., Vasudevan S., Wolf Y. I., Yin J. J., Natale D. A. (2003) The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Claudel-Renard C., Chevalet C., Faraut T., Kahn D. (2003) Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res. 31, 6633–6639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 46. Lowe T. M., Eddy S. R. (1997) TRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lagesen K., Hallin P., Rødland E. A., Staerfeldt H. H., Rognes T., Ussery D. W. (2007) RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sambrook J., Russel D. W. (2001) in Molecular Cloning: A Laboratory Manual, 3rd Ed., pp. A8.40–A8.47, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 49. Bramhall S., Noack N., Wu M., Loewenberg J. R. (1969) A simple colorimetric method for determination of protein. Anal. Biochem. 31, 146–148 [DOI] [PubMed] [Google Scholar]
- 50. Bigge J. C., Patel T. P., Bruce J. A., Goulding P. N., Charles S. M., Parekh R. B. (1995) Nonselective and efficient fluorescent labeling of glycans using 2-amino benzamide and anthranilic acid. Anal. Biochem. 230, 229–238 [DOI] [PubMed] [Google Scholar]
- 51. Kinoshita A., Sugahara K. (1999) Microanalysis of glycosaminoglycan-derived oligosaccharides labeled with a fluorophore 2-aminobenzamide by high-performance liquid chromatography: application to disaccharide composition analysis and exosequencing of oligosaccharides. Anal. Biochem. 269, 367–378 [DOI] [PubMed] [Google Scholar]
- 52. Nielsen M., Lundegaard C., Lund O., Petersen T. N. (2010) CPHmodels-3.0-remote homology modeling using structure-guided sequence profiles. Nucleic Acids Res. 38, W576–W581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Guex N., Peitsch M. C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 54. Guo X., Shi Y., Sheng J., Wang F. (2014) A novel hyaluronidase produced by Bacillus sp. A50. PLoS One 9, e94156–e94156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hashimoto W., Miki H., Tsuchiya N., Nankai H., Murata K. (2001) Polysaccharide lyase: molecular cloning, sequencing, and overexpression of the xanthan lyase gene of Bacillus sp. strain GL1. Appl. Environ. Microbiol. 67, 713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ernst S., Langer R., Cooney C. L., Sasisekharan R. (1995) Enzymatic degradation of glycosaminoglycans. Crit. Rev. Biochem. Mol. Biol. 30, 387–444 [DOI] [PubMed] [Google Scholar]
- 57. Mizumoto S., Murakoshi S., Kalayanamitra K., Deepa S. S., Fukui S., Kongtawelert P., Yamada S., Sugahara K. (2013) Highly sulfated hexasaccharide sequences isolated from chondroitin sulfate of shark fin cartilage: insights into the sugar sequences with bioactivities. Glycobiology 23, 155–168 [DOI] [PubMed] [Google Scholar]
- 58. Ohya T., Kaneko Y. (1970) Novel hyaluronidase from streptomyces. Biochim. Biophys. Acta 198, 607–609 [DOI] [PubMed] [Google Scholar]
- 59. Stern R., Jedrzejas M. J. (2006) Hyaluronidases: their genomics, structures, and mechanisms of action. Chem. Rev. 106, 818–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kaneiwa T., Mizumoto S., Sugahara K., Yamada S. (2010) Identification of human hyaluronidase-4 as a novel chondroitin sulfate hydrolase that preferentially cleaves the galactosaminidic linkage in the trisulfated tetrasaccharide sequence. Glycobiology 20, 300–309 [DOI] [PubMed] [Google Scholar]
- 61. Kushner D. J., Kamekura M. (1988) in Halophilic Bacteria (Rodríguez-Valera F., ed) pp. 109–138, CRC Press, Boca Raton, FL [Google Scholar]
- 62. Tam Y. C., Chan E. C. (1985) Purification and characterization of hyaluronidase from oral Peptostreptococcus species. Infect. Immun. 47, 508–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yoshida K., Arai M., Kohno Y., Maeyama K., Miyazono H., Kikuchi H., Morikawa K., Tawada A., Suzuki S. (1993) in Dermatan Sulphate Proteoglycans (Scott J., ed.) pp. 55–80, Portland Press, London [Google Scholar]
- 64. Lee A., Grummer S. E., Kriegel D., Marmur E. (2010) Hyaluronidase. Dermatol. Surg. 36, 1071–1077 [DOI] [PubMed] [Google Scholar]
- 65. Sharma K., Selzer M. E., Li S. (2012) Scar-mediated inhibition and CSPG receptors in the CNS. Exp. Neurol. 237, 370–378 [DOI] [PMC free article] [PubMed] [Google Scholar]