Background: BceS-like histidine kinases strictly require BceAB-like ABC transporters for sensing of peptide antibiotics.
Results: BceAB of Bacillus subtilis interacted with BceS in vivo and in vitro and specifically bound the substrate peptide bacitracin.
Conclusion: Complex formation with the ABC transporter affects the activity of the histidine kinase.
Significance: Histidine kinase and ABC transporter form a sensory complex for the detection of peptide antibiotics.
Keywords: ABC Transporter, Antimicrobial Peptide (AMP), Histidine Kinase, Membrane Protein, Protein-Protein Interaction, Bacitracin
Abstract
Resistance against antimicrobial peptides in many Firmicutes bacteria is mediated by detoxification systems that are composed of a two-component regulatory system (TCS) and an ATP-binding cassette (ABC) transporter. The histidine kinases of these systems depend entirely on the transporter for sensing of antimicrobial peptides, suggesting a novel mode of signal transduction where the transporter constitutes the actual sensor. The aim of this study was to investigate the molecular mechanisms of this unusual signaling pathway in more detail, using the bacitracin resistance system BceRS-BceAB of Bacillus subtilis as an example. To analyze the proposed communication between TCS and the ABC transporter, we characterized their interactions by bacterial two-hybrid analyses and could show that the permease BceB and the histidine kinase BceS interact directly. In vitro pulldown assays confirmed this interaction, which was found to be independent of bacitracin. Because it was unknown whether BceAB-type transporters could detect their substrate peptides directly or instead recognized the peptide-target complex in the cell envelope, we next analyzed substrate binding by the transport permease, BceB. Direct and specific binding of bacitracin by BceB was demonstrated by surface plasmon resonance spectroscopy. Finally, in vitro signal transduction assays indicated that complex formation with the transporter influenced the autophosphorylation activity of the histidine kinase. Taken together, our findings clearly show the existence of a sensory complex composed of TCS and ABC transporters and provide the first functional insights into the mechanisms of stimulus perception, signal transduction, and antimicrobial resistance employed by Bce-like detoxification systems.
Introduction
In recent years, a number of cases were described where transport proteins act as co-sensors for bacterial signal transduction systems. Such transporters are able to interfere with signal transduction processes, for example by transporting effector molecules into the cytoplasm or by interacting directly with sensory components (1). The latter process is based on regulatory protein-protein interactions between the sensing unit, which harbors specificity for certain substrates, and the signaling unit, which transfers the information into the cytoplasmic compartment of the cell.
One well known example is the widespread Pst/Pho system, which senses environmental phosphate. Transcription of the genes for bacterial high affinity phosphate transport systems is usually regulated by a two-component regulatory system (TCS),2 PhoBR in Gram-negative bacteria (2), PhoPR in Gram-positive bacteria (3, 4), and SenX3-RegX3 in mycobacteria (5), where PhoR or SenX3 act as the histidine kinase (HK) and PhoB, PhoP, or RegX3 act as the cognate response regulator. The TCS further requires the phosphate-specific ATP-binding cassette (ABC) transporter PstSCAB and the peripheral membrane protein PhoU for signal-dependent activation (6). Together, transporter and HK are thought to form a membrane-bound repressor complex under phosphate-replete conditions (2), and mutations in the pstSCAB operon have been shown to lead to constitutive activation of the Pho regulon genes in a number of bacteria such as Escherichia coli (7), Sinorhizobium meliloti (8), and Mycobacterium smegmatis (9). Furthermore, early indications that membrane transport and sensory transduction processes could be coupled in bacteria came from studies of the phosphoenolpyruvate-dependent sugar transport and chemotactic sensory system of E. coli (10). Regulation of C4-dicarboxylate uptake in E. coli is mediated by the TCS DcuS/DcuR and the secondary transporters DctA or DcuB. The HK DcuS is able to bind C4-dicarboxylates directly but additionally requires the transporters DctA or DcuB as co-sensors under aerobic or anaerobic conditions, respectively (11). In dctA- or dcuB-deficient strains, DcuS is deregulated and permanently active even in the absence of C4-dicarboxylates (12). DcuS and the transporter DctA were shown to interact physically, suggesting the formation of a DctA-DcuS sensory complex to inhibit DcuS activity in the absence of its substrate (13). A similar setup was recently shown to exist in Bacillus subtilis (14). The CadC/LysP system of E. coli presents a further sensory complex consisting of a one-component signaling system and a secondary transporter. The central component of this system is the membrane-integrated pH sensor and transcriptional activator CadC, which regulates induction of the cadBA operon under low pH. CadC activity is also dependent on the presence of lysine (15), and this lysine-dependent activation of CadC requires the co-sensor LysP, a lysine-specific permease (16).
In all these signal transduction systems, the accessory transporters act as inhibitors of their respective signal transduction system in the absence of the stimulus. In contrast, in the antimicrobial peptide detoxification systems studied here, activation of signaling depends entirely on a sensory transporter, and the system remains in an inactive state in the absence of the transporter (17–22). These systems are found widely spread among low GC Gram-positive bacteria and consist of a TCS, where the HK lacks an obvious input domain, and an unusual ABC transporter of 10 TM helices with a large extracellular domain (23, 24). All examples characterized to date are involved in resistance against peptide antibiotics, and the requirement for the transporter in signaling appears to be a conserved characteristic (25).
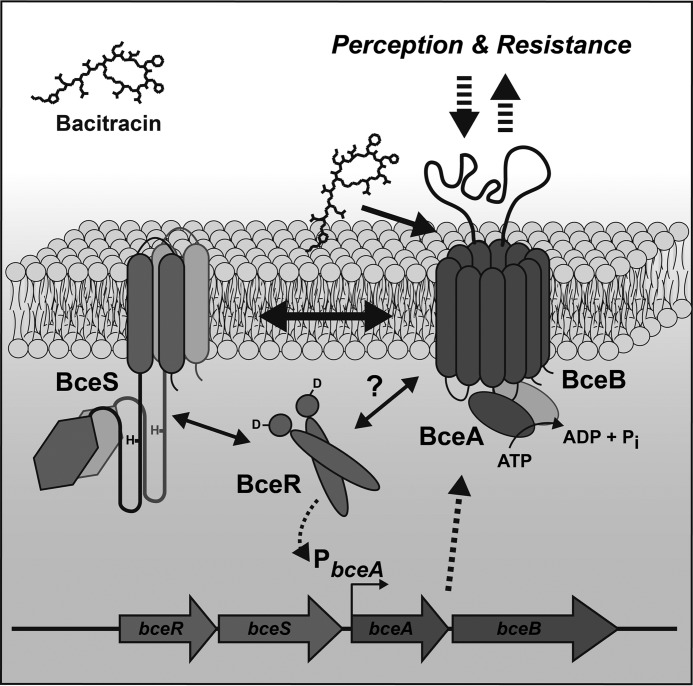
The paradigm example for this is the BceRS-BceAB system of B. subtilis, which mediates resistance against bacitracin (Fig. 1) (17, 26). The HK BceS was found to be unable to detect bacitracin in the absence of the transporter BceAB, which led to the assumption that the transporter constitutes the sensory component of the system (17, 27). Additionally, ATP hydrolysis by the ATPase BceA, i.e. active transport, was shown to be essential for signaling (17). However, the mechanism by which the transporter and TCS communicate is not known. A comparative phylogenetic analysis of Bce-like modules showed a co-evolution between the transport permeases and HKs, suggesting direct interactions between the proteins (24), but clear experimental evidence for this is missing to date. A second open question concerns the mechanism of substrate binding and resistance by the transporter itself. Bacitracin inhibits cell wall synthesis by binding to undecaprenyl-pyrophosphate (UPP), the phosphorylated form of the carrier molecule for peptidoglycan precursors and thus prevents its recycling (28). Other peptide antibiotics, such as the lantibiotic nisin, often bind to the lipid II intermediate of peptidoglycan synthesis (29). Both UPP and lipid II are found on the surface of the cell, and it is therefore not immediately obvious how a transporter can provide protection against antibiotics targeting these structures. Possible scenarios that have been discussed include removal of cell-associated peptides to the culture supernatant or import of the antibiotics for subsequent degradation (17, 18, 25). More recently, it was proposed that BceAB might in fact not transport bacitracin at all but rather flip UPP to the cytoplasmic face of the membrane to prevent bacitracin binding (30). Addressing this important question is hampered by the lack of knowledge on the transporter's true substrate; it is unclear whether BceAB is able to bind free bacitracin as its substrate or whether it instead recognizes a membrane-associated UPP-bacitracin complex.
FIGURE 1.
Working model for the BceRS-BceAB bacitracin resistance system of B. subtilis. Bacitracin is bound directly by the transporter BceAB. BceAB and BceS interact to form a sensory complex in the membrane. ATP hydrolysis by the transporter triggers the activation of BceS, which in turn leads to phosphorylation of BceR. Activation of the target promoter (PbceA) by BceR then induces increased production of BceAB to ensure resistance. Interactions between proteins are marked by double-headed arrows; events relating to transcription are labeled with dotted arrows; the potential interaction of BceR with the sensory complex of BceS and BceAB is indicated with a question mark.
To gain a better understanding of the molecular mechanisms of these unusual resistance determinants, we here set out to functionally characterize the Bce system of B. subtilis. Using both in vivo and in vitro approaches, we could demonstrate that the transporter BceAB is indeed able to interact directly with the TCS BceRS, with the permease and HK components providing the interaction scaffold. Additionally, we showed that BceB bound its substrate bacitracin directly, providing first insights into the mechanism of resistance. Furthermore, we show for the first time that complex formation with the transporter can influence the autophosphorylation activity of the HK in vitro.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
E. coli and B. subtilis were routinely grown in lysogeny broth (LB) medium (31) at 37 °C with agitation (200 rpm). During cloning of bacterial two-hybrid constructs, all media for E. coli were supplemented with 0.4% (w/v) glucose. Selective media contained kanamycin (50 μg/ml), chloramphenicol (5 μg/ml), ampicillin (100 μg/ml), or erythromycin (1 μg/ml) and lincomycin (25 μg/ml) (macrolide-lincosamide-streptogramin B (mls)). Solid media contained 1.5% (w/v) agar. All strains used in this study are listed in Table 1; all primer sequences are listed in Table 2.
TABLE 1.
Plasmids and strains used in this study
| Plasmid or strain | Descriptiona | Ref. or source |
|---|---|---|
| Plasmids | ||
| pASK-IBA5 plus | Vector for tetracycline-inducible gene expression; carries an N-terminal Strep-tag II® sequence; Ampr | IBA |
| pTrc99a | Vector for IPTG-inducible gene expression; Ampr | 42 |
| pET16b | Vector for IPTG-inducible gene expression; carries an N-terminal His10 tag sequence; Ampr | Novagen |
| pBAD24 | Vector for arabinose-inducible gene expression; Ampr | 43 |
| pBS2E | Empty vector for integration at amyE; Ampr; mlsr | 44 |
| pKT25 | Vector for translational fusions of cyaA T25 fragment to N terminus of insert polypeptide; lac promoter; Kanr | 32 |
| pKTN25 | Vector for translational fusions of cyaA T25 fragment to C terminus of insert polypeptide; lac promoter; Kanr | 32 |
| pUT18 | Vector for translational fusions of cyaA T18 fragment to C terminus of insert polypeptide; lac promoter; Ampr | 32 |
| pUT18C | Vector for translational fusions of cyaA T18 fragment to N terminus of insert polypeptide; lac promoter; Ampr | 32 |
| pSDIBA501 | pASK-IBA5C-bceAB | This study |
| pSD2402 | pBAD24-bceAB-Strep | This study |
| pRU2401 | pTrc99a-bceS-His8 | This study |
| pCF120 | pET16b-bceR-His10 | This study |
| pNT2E07 | pBS2E-Pxyl-bceS | This study |
| pNT2E01 | pBS2E-Pxyl-bceAB | This study |
| pSD2E01 | pBS2E-Pxyl-bceS-His8 | This study |
| pSD2E02 | pBS2E-Pxyl-Strep-bceAB | This study |
| pAS1803 | pUT18-bceS | 33 |
| pAS2503 | pKT25-bceS | This study |
| pCF18C01 | pUT18C-bceR | This study |
| pCF2501 | pKT25-bceR | This study |
| pCF18C02 | pUT18C-bceRS | This study |
| AS1804 | pUT18-bceA | 33 |
| pAS2504 | pKT25-bceA | This study |
| pAS1805 | pUT18-bceB | This study |
| pAS2505 | pKT25-bceB | 33 |
| pHF1804 | pUT18-bceAB | This study |
| pHF2509 | pKT25-bceAB | This study |
| E. coli strains | ||
| XL1-blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 Tetr] | Stratagene |
| DH5α | fhuA2 lac (Δ)U169 phoA glnV44 Φ80′ lacZ (Δ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 | 45 |
| BTH101 | F−, cya-99, araD139, galE15, galK16, rpsL1, hadR2, mcrA1, mcrB1 | Euromedex |
| C43(DE3) | F− ompT gal dcm hsdSB (rB− mB−) (DE3) | 46 |
| SGE217 | C43 (DE3); arnC::kan | This study |
| B. subtilis strains | ||
| TMB1557 | ΔbceS; sacA::PbceA-luxABCDE | This study |
| SGB79 | bceAB::kan; sacA::PbceA-luxABCDE | This study |
| SGB276 | ΔbceS; sacA::PbceA-luxABCDE; lacA::Pxyl-bceS-His8 | This study |
| SGB277 | bceAB::kan; sacA::PbceA-luxABCDE; lacA::Pxyl-Strep-bceAB | This study |
| TMB1650 | ΔbceS; sacA::PbceA-luxABCDE; lacA::Pxyl-BceS | This study |
| TMB1636 | bceAB::kan; sacA::PbceA-luxABCDE; lacA::Pxyl-bceAB | This study |
| SGB276 | ΔbceS; sacA::PbceA-luxABCDE; lacA::Pxyl-BceS-His8 | This study |
| SGB277 | bceAB::kan; sacA::PbceA-luxABCDE; lacA::Pxyl-Strep-bceAB | This study |
a The abbreviations used are as follows: Ampr, ampicillin resistance; kanr, kanamycin resistance; mlsr, macrolide-lincosamide-streptogramin B resistance.
TABLE 2.
Primers used in this study
| Name | Sequence (5̍′ to 3̍′)a | Used for amplification of |
|---|---|---|
| 2256 | AATTGGTCTCAGCGCCATGGTGATTTTAGAAGCG | Strep-bceAB (fwd) |
| 2257 | AATTGGTCTCGTATCACAACGACGATTTAATG | Strep-bceAB (rev) |
| 2007 | ATCGCTCGAGTTGTTTAAACTTTTGCTGATTG | bceR-His10 (fwd) |
| 2008 | ATCGGGATCCTTAATCATAGAACTTGTCCTC | bceR-His10 (rev) |
| 1895 | AATTCCATGGTTAAAGCATTCCTTATCGAAAGG | bceS-His8 (fwd) |
| 1905 | AATTCTGCAGTCAGTGATGGTGATGGTGATGGTGATGCACGCTTATGACATGTTCAAATTG | bceS-His8 (rev) |
| 1525 | AATTCCATGGTGATTTTAGAAGCGAA | bceAB-Strep (fwd) |
| 2201 | AATTGTCGACTTATTTTTCGAACTGCGGGTGGCTCCAGCCACCGCCACCGCCACCCAACGACGATTTAATGACC | bceAB-Strep (rev) |
| 1355 | GTCATCTAGAGATGATTAAAGCATTCCTTATCG | bceS (fwd) BACTH |
| 1356 | GTCAGGTACCTGCACGCTTATGACATGTTC | bceS (rev) BACTH |
| 1359 | GTCATCTAGAGATGAACATTAATCAGCTCATCC | bceB (fwd) BACTH |
| 1360 | GTCAGGTACCTGCAACGACGATTTAATGACC | bceB (rev) BACTH |
| 1357 | GTCATCTAGAGATGGTGATTTTAGAAGCG | bceA (fwd) BACTH |
| 1358 | GTCAGGTACCTGATGTTCATGCTGCACC | bceA (rev) BACTH |
| 3218 | AATTTCTAGAGTTGTTTAAACTTTTGCTGATTGAAG | bceR (fwd) BACTH |
| 3219 | AATTGGTACCTGTTAATCATAGAACTTGTCCTCTTC | bceR (rev) BACTH |
a Restriction sites are shown in bold; nucleotides encoding the His8 tag are shown underlined; nucleotides encoding the Strep-tag are shown in italics; fwd, forward; rev, reverse.
Bacterial Two-hybrid Assays
To test protein-protein interactions in the BceRS-BceAB module, translational fusions of the T18 and T25 domains of the adenylate cyclase CyaA of Bordetella pertussis were constructed for each Bce module protein individually. Additionally, BceAB was fused to the N-terminal end of T18 or the C-terminal ends of T18 and T25. Furthermore, we generated a BceRS fusion to the C-terminal end of T18 (Table 1). Fusions were tested in pairwise combinations in E. coli BTH101 (32). Data are shown for the optimal pair for each protein combination. Of each transformation mixture, 10 μl were spotted onto LB agar plates containing 0.5 mm isopropyl 1-thio-β-d-galactopyranoside and 40 μg/ml 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (X-Gal) with selection for ampicillin and kanamycin resistance. Plates were incubated at 30 °C for 48 h. Formation of blue colonies was scored as a positive interaction result. To test for effects of bacitracin on the interaction between BceB and BceS, the corresponding pairs were also spotted on LB plates containing a linear gradient from 0 to 800 μg/ml bacitracin.
In Vivo Assays of Bce Module Functionality
To test for signaling activity, luciferase activities of strains harboring the PbceA-luxABCDE (pSDlux101) reporter, deletions of bceS or bceAB, and respective complementation constructs were assayed as described previously (33).
The sensitivity of B. subtilis strains to bacitracin was determined as the minimal inhibitory concentration (MIC). For this, serial 2-fold dilutions of Zn2+-bacitracin from 32 to 2 μg/ml were prepared in Mueller-Hinton medium containing 0.2% (w/v) xylose. For each concentration, 2 ml of media were inoculated at 1:500 from overnight cultures grown in Mueller-Hinton medium with xylose and selective antibiotics. Each culture was scored for growth after 20–24 h of incubation at 37 °C with agitation. The MIC was determined as the lowest bacitracin concentration where no growth was detected.
Protein Production
All proteins were produced in E. coli C43(DE3). Because BceAB consistently co-purified with a 38-kDa protein identified as ArnC by mass spectrometry analysis, the expression host strain for BceAB and BceS was deleted for the corresponding gene.
The bceS coding region was amplified using primers 1895/1905, resulting in addition of a C-terminal His8 tag, and was cloned into the NcoI and PstI sites of pTrc99a, yielding pRU2401. To produce BceS-His8, cells were grown at 37 °C with agitation until the culture reached A600 = 0.5, induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside, and incubated for a further 2.5 h. Cells were then harvested by centrifugation at 5,400 × g for 15 min. The cell pellet was washed in buffer A (50 mm KPi (pH 7.5), 150 mm NaCl, 5 mm β-mercaptoethanol (β-ME), and 10% (w/v) glycerol) and stored at −20 °C until use. To purify BceS, cells were resuspended in buffer A supplemented with 0.1 mm phenylmethylsulfonyl fluoride (PMSF) and 2 mg of DNase I and disrupted by three passages through a French pressure cell (Thermo Fisher) at 20,000 p.s.i. Unbroken cells were removed by centrifugation at 12,000 × g for 30 min, and the membranes were collected from the cell-free supernatant by ultracentrifugation at 180,000 × g for 1 h. Membranes were resuspended in buffer A and stored at −80 °C. Solubilization of membrane proteins was performed at a protein concentration of ∼5 mg/ml in buffer A containing 0.5% (w/v) n-dodecyl β-d-maltoside (DDM) with gentle stirring at 4 °C for 1.5 h. The mixture was ultracentrifuged at 180,000 × g for 1 h. The supernatant (solubilized BceS) was then loaded onto a 1-ml Ni2+-NTA column (Qiagen) pre-equilibrated with 4 column volumes (CVs) buffer A containing 0.05% (w/v) DDM. Loading was followed by washing with 5 CVs of buffer A and 5 CVs of buffer A containing 50 mm imidazole. BceS was eluted with buffer A containing 200 mm imidazole. All washing and elution steps were performed in the presence of 0.05% (w/v) DDM. Fractions containing BceS were pooled; protein concentrations were determined with Roti®-Nanoquant (Carl Roth), and the proteins were stored at 4 °C on ice until use.
Construction of the bceAB expression plasmid pSDIBA501 was achieved by amplifying the bceAB coding region using primers 2256/2257 and cloning the product into pASK-IBA5 via BsaI sites, resulting in an N-terminal fusion of a Strep-tag® II to BceA. To produce Strep-BceAB, cells were grown at 37 °C with agitation for 1 h, followed by a shift in temperature to 30 °C and continued incubation until the culture reached A600 = 0.5. The cells were induced with 50 ng/ml tetracycline, incubated for a further 2.5 h, and harvested by centrifugation at 5,400 × g for 15 min. The cell pellet was washed in buffer B (100 mm Tris/HCl (pH 8.0), 150 mm NaCl, 5 mm β-ME, and 10% (w/v) glycerol) and stored at −20 °C until use. To purify BceAB, cells were disrupted and membranes isolated as described above. Membranes were resuspended in buffer B and stored at −80 °C. Solubilization of membrane proteins was performed in buffer B containing 0.5% (w/v) DDM as mentioned before. The supernatant (solubilized BceAB) was loaded onto a 1-ml Strep-Tactin® column (IBA) pre-equilibrated with 4 CVs of buffer B containing 0.05% (w/v) DDM. Loading was followed by washing with 4 CVs of buffer B. BceAB was eluted with buffer B supplemented with 2.5 mm d-desthiobiotin. All washing and elution steps were performed in the presence of 0.05% (w/v) DDM. Fractions containing BceAB were pooled and concentrated ∼2-fold using a Vivaspin® 500 centrifugal concentrator (Sartorius); protein concentrations were determined with Roti®-Nanoquant (Carl Roth); and the proteins were stored at 4 °C on ice until use. To purify BceB alone, we generated pSD2401 by amplification of the bceAB coding region with primers 1525/2201. The product was digested with NcoI and SalI and cloned into pBAD24, resulting in the fusion of a Strep-tag® II to the C terminus of BceB. To produce BceAB-Strep, cells were grown as described above, but here the expression was induced with 0.02% arabinose. To purify BceB, the cells were treated as described for BceAB; BceA was completely removed during the washing step, and only BceB found in the eluted fractions.
Construction of the bceR expression plasmid pCF120 was achieved by amplifying the bceR coding region using primers 2007/2008. The product was digested with XhoI and BamHI and cloned into pET16b, resulting in an N-terminal fusion to a His10 tag. To produce BceR, cells were treated as described for BceS production. The cell pellet was washed in buffer C (20 mm KPi (pH 7.5), 100 mm NaCl, 5 mm β-ME, and 10% (w/v) glycerol) and stored at −20 °C until use. To purify BceR, cells were resuspended in buffer D (50 mm KPi (pH 7.5), 500 mm NaCl, 5 mm β-ME, 10 mm imidazole, and 10% (w/v) glycerol) supplemented with 0.1 mm PMSF and 2 mg of DNase I, and disrupted by three passages through a French pressure cell (Thermo Fisher) at 20,000 p.s.i. Unbroken cells were removed by centrifugation at 12,000 × g for 20 min, and the cell-free supernatant was filtered through a 0.45-μm syringe filter before loading onto a 1-ml Ni2+-NTA resin column (Qiagen) pre-equilibrated with 5 CVs of buffer A. Loading was followed by washing with 5 CVs of buffer A, 5 CVs of buffer D, and then 5 CVs of buffer D containing 100 mm imidazole. BceR was eluted with buffer D supplemented with 250 mm imidazole. Fractions containing BceR were pooled, and protein concentrations were determined with Roti®-Nanoquant (Carl Roth), and the proteins were stored at 4 °C on ice until use.
Size Exclusion Chromatography of BceAB
Chromatography was performed using an ÄKTA FPLC system (GE Healthcare). Preparations of purified Strep-BceAB were incubated either alone or with 10 μg/ml bacitracin at 4 °C on ice for 20 min followed by centrifugation at 13,000 rpm and 4 °C for 10 min prior to resolution on a Superdex 200 10/30 gel filtration column (GE Healthcare). The proteins were resolved at a flow rate of 0.4 ml/min using buffer B containing 0.05% (w/v) DDM. Fractions (500 μl) were collected and analyzed by SDS-PAGE (12.5% gel). The molecular weights were calculated from the elution volumes in comparison with a gel filtration LMW/HMW calibration kit (GE Healthcare).
ATP Hydrolysis Assays
ATPase activity was determined by a phosphate-release assay essentially as described by Monk et al. (34). In brief, 320 μl of assay mixture contained 100 mm Tris/HCl (pH 7.5), 150 mm NaCl, 5 mm β-ME, 10% (w/v) glycerol, 0.05% (w/v) DDM, 10 mm MgCl2, 1 mm ATP, and 1.2–12 μg of purified BceAB. Reactions were started by the addition of ATP. After incubation at room temperature for 0–20 min, 40-μl aliquots of the reaction were transferred to 96-well microplates, and the reaction was stopped by adding 5 μl of SDS (4% (w/v)). To each well, 65 μl of developing reagent (34) was added, and the absorbance at 660 nm was measured in a microtiter plate reader (Tecan Instruments) after 10 min of incubation at room temperature. The inorganic phosphate released from ATP was quantified in comparison with a standard curve of 0–100 nmol of KPi in the reaction mixture, and suitable blanks were used to correct for nonspecific hydrolysis.
Bacitracin Binding by BceB Using Surface Plasmon Resonance (SPR) Spectroscopy
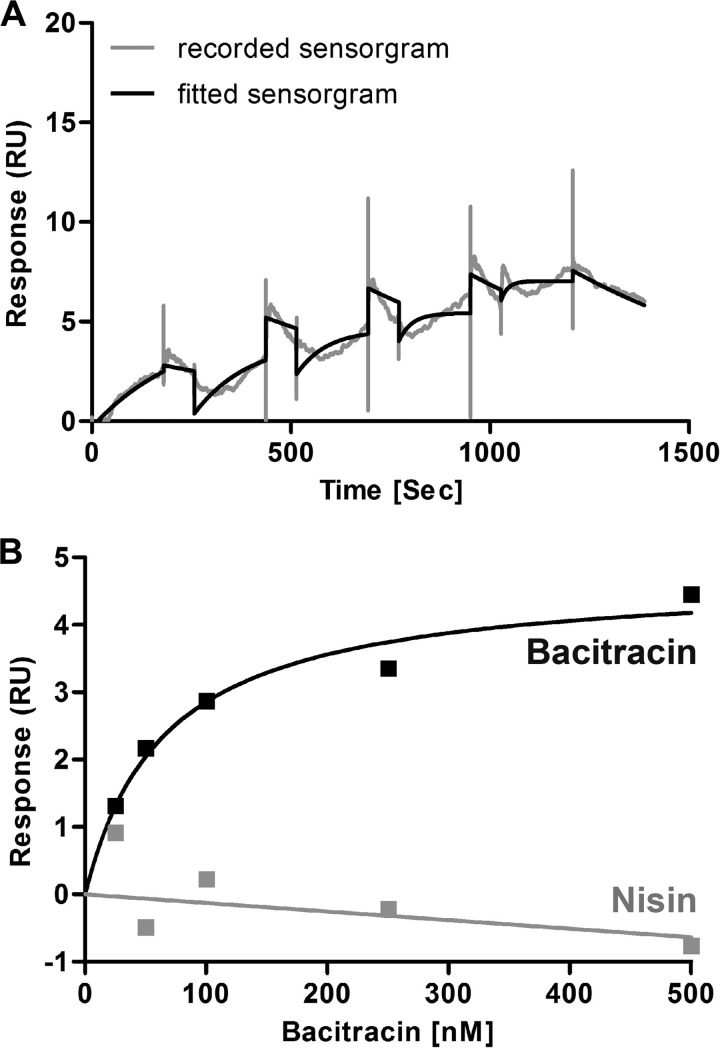
SPR assays were performed in a BIAcore T200 using carboxymethyl dextran sensor chips (Xantec CMD200-L) that were coated with Strep-Tactin® XT resin (IBA, Göttingen, Germany). This is the first material that allows the complete regeneration of Strep-tagged molecules from a sensor chip in a capturing SPR approach. First, the chips were equilibrated with HBS-EP buffer (10 mm HEPES (pH 7.4), 150 mm NaCl, 3 mm EDTA, 0.005% (v/v) detergent P20) until the dextran matrix was swollen. Then, two of the four flow cells of the sensor chips were activated by injecting a 1:1 mixture of N-ethyl-N-(3-dimethylaminopropyl) carbodiimide hydrochloride and N-hydroxysuccinimide using the standard amine-coupling protocol. Both flow cells were loaded with a final concentration of 10 μg/ml Strep-Tactin® XT in 10 mm acetate (pH 5.5) using a contact time of 420 s, so that the surfaces contained densities of 5,000–6,000 resonance units. Free binding sites of the flow cells were saturated by injection of 1 m ethanolamine/HCl (pH 8.0). Preparation of chip surfaces was carried out at a flow rate of 10 μl/min. Binding between BceB and bacitracin was analyzed using a single cycle kinetic approach (35) with HBS-DDM buffer (10 mm HEPES (pH 7.4), 150 mm NaCl buffer, 0.05% (w/v) DDM). BceB carrying a C-terminal Strep-tag® II (20 μg/ml) was captured onto the second flow cell using a contact time of 300 s at a constant flow rate of 10 μl/min, followed by a stabilization time of 120 s so that ∼600–1,000 resonance units of BceB were captured. Single cycle kinetics using Zn2+-bacitracin or nisin (control) were performed at a flow rate of 30 μl/min. Increasing concentrations (25, 50, 100, 250, and 500 nm) of Zn2+-bacitracin or nisin, respectively, were then sequentially injected onto both flow cells without interim regeneration using a contact time of 180 s each and a final dissociation of 180 s. Then, the chip was regenerated by injection of 10 mm glycine (pH 1.5) for 60 s at a flow rate of 30 μl/min over both flow cells, which completely removed BceB from the Strep-Tactin® XT surface. Furthermore, blank single cycle kinetics were recorded by sequentially injecting buffer instead of increasing concentrations of bacitracin or nisin after capturing BceB-Strep. Each single cycle kinetic was performed four times in a row. All experiments were performed at 25 °C. Sensorgrams were recorded using the BIAcore T200 Control software 1.0 and analyzed with the BIAcore T200 Evaluation software 1.0. The surface of flow cell 1 was used to obtain blank sensorgrams for subtraction of bulk refractive index background. Buffer controls on the second surface were subtracted from the sensorgrams obtained with bacitracin or nisin to normalize drifts on the surface. The referenced sensorgrams were then normalized to a baseline of 0. Peaks in the sensorgrams at the beginning and the end of the injections emerged from the runtime difference between the flow cells of each chip.
In Vitro Pulldown Assay
For in vitro interaction assays, 300 μg of purified BceS or a control protein were conjugated to N-hydroxysuccinimide (NHS)-activated Sepharose beads (GE Healthcare). After blocking of unconjugated NHS groups through extensive washing in buffer B containing 0.05% (w/v) DDM, conjugated beads were stored at 4 °C in the same buffer as a 50% (v/v) bead slurry. A typical experiment involved resuspending 50 μl of the bead slurry in buffer B containing 0.05% (w/v) DDM and supplementing the suspension with 30 μg of test protein. After extensive washing, the beads were resuspended in 30 μl of 1× SDS-PAGE sample buffer, and beads were sedimented, and 20 μl of sample buffer containing dissolved proteins were analyzed by SDS-PAGE using a 12.5% polyacrylamide gel. For each experiment, beads conjugated with bovine serum albumin (BSA) or empty beads blocked by Tris through incubation in buffer B were used to control for incidental protein carry-over and nonspecific binding of test proteins to conjugated protein or the Sepharose bead matrix.
Phosphorylation Assays
To test phosphorylation of Bce module proteins, equimolar mixtures (∼300 pmol per protein in a 190-μl reaction) of purified, detergent-solubilized protein were incubated with a mixture of [γ-32P]ATP (0.01 mCi; 3000 Ci/mmol) and unlabeled ATP yielding a final concentration of 40 μm in phosphorylation buffer (100 mm Tris/HCl (pH 8), 150 mm NaCl, 15 mm MgCl2, 5 mm β-ME, 0.05% (w/v) DDM, and 10% (w/v) glycerol) at room temperature. At different time points, 20-μl aliquots (∼30 pmol per protein) were removed and mixed with SDS sample buffer. The sample for the t0 time point was taken immediately after addition of ATP. All samples were subjected to SDS-PAGE. Gels were dried, and phosphorylated proteins were detected by PhosphorImager (Typhoon TrioTM, GE Healthcare). The band intensities of the phosphorylated proteins were quantified using ImageJ by measuring the peak area of the corresponding peaks after subtraction of the local background intensity for each band.
RESULTS
In Vivo Protein Interactions within the Bce System
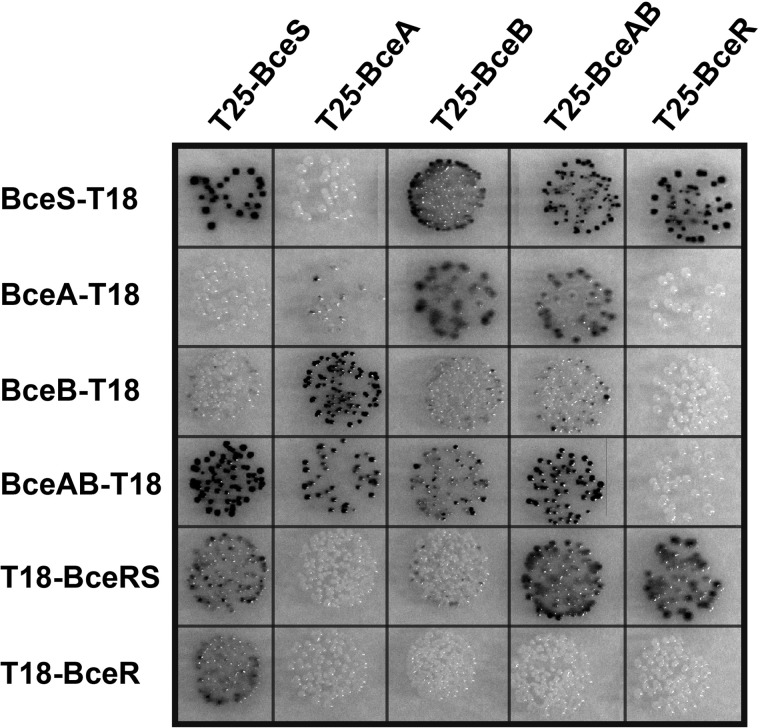
Because the signaling pathway within Bce-like systems involves both the transporter and the HK (17, 27), and because the protein families of BceB-like permeases and BceS-like HKs have co-evolved (24), we proposed direct interactions between the two proteins. Initial experimental indication for this was obtained previously using a bacterial two-hybrid (BACTH) assay of BceS and BceB (33). Here, we first wanted to test whether additional protein-protein interactions between the transporter and TCS occurred. For this, we applied a comprehensive BACTH analysis of the entire system, based on the in vivo reconstitution of adenylate cyclase activity from separate T18 and T25 domains of B. pertussis CyaA in E. coli (BTH101) (32). We generated fusions to the CyaA T18 and T25 domains of each protein individually (BceR, BceS, BceA, and BceB). Additionally, plasmids were generated containing the complete transporter (bceAB) or TCS (bceRS) operon, to yield CyaA fusions of one component with concomitant production of the second untagged protein. Pairwise combinations were then introduced into E. coli BTH101 (Fig. 2). The T18-BceS fusion showed positive results (i.e. blue colonies on indicator agar) with T25-BceS, as expected for homodimer formation. Moreover, an interaction of BceR with BceS was observed as expected, whereas homodimerization of BceR was not found unless BceS was also supplied in the cell (T18-BceRS construct), suggesting that dimerization of the HK brought the BceR monomers into close proximity or that phosphorylation of BceR by BceS induced dimerization of the regulator. We further found the expected association of the permease BceB with its cognate ATPase BceA in all combinations tested. The lack of a signal for BceB homodimerization is discussed in more detail below.
FIGURE 2.
Bacterial two-hybrid analysis of the BceRS-BceAB module. Hybrids consisting of B. pertussis CyaA T18 or T25 domains and each individual Bce protein or the complete transporter (BceAB) or TCS (BceRS) were introduced into E. coli BTH101. Cells were spotted onto LB plates containing X-Gal (40 μg/ml), isopropyl 1-thio-β-d-galactopyranoside (0.5 mm), and antibiotics for selection. Pictures were taken after 48 h of incubation at 30 °C. The blue colonies indicating positive results for interaction are depicted as dark gray in the grayscale image.
As predicted from previous findings, we clearly observed interactions between BceS and BceB or BceAB. Addition of 0–800 μg/ml bacitracin to the indicator plates did not result in any changes of color intensity, suggesting that complex formation was not affected by the substrate antibiotic. More quantitative assays in liquid cultures were unsuccessful, most likely due to toxicity of BceAB overproduction in E. coli. Next, we wanted to test interactions between the cytoplasmic and membrane-bound proteins of the system. For the ATPase BceA, no homodimerization was found when produced alone, but in a combination where the permease was also present (BceA-T18 with T25-BceAB), interactions between the ATPase domains were clearly observed. No interactions of BceA with the components of the TCS (BceS, BceR, or BceRS) were observed. When BceR was produced in the absence of the HK, no interaction with the transporter (BceA, BceB, and BceAB) was found. Interestingly, however, when the T18-BceR fusion was co-expressed with the HK, positive results for interaction with BceAB and weak results with BceB were obtained. These data indicate that the regulator may be part of a membrane-associated protein complex of BceRS and BceAB, but this interaction requires the presence of both BceS and BceAB. Taken together, our data consistently show interactions between BceS and BceAB and further suggest that together the HK and permease may form a scaffold for BceR to also interact with the complex.
Purification of All Protein Components of the Bce Module
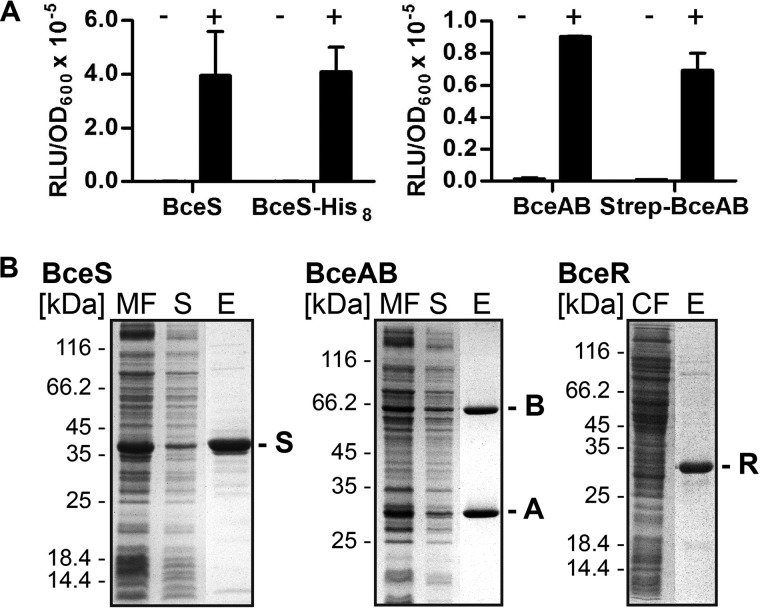
To further investigate the Bce system, we next wanted to analyze its activities in vitro. For this, we generated overproduction plasmids for the full-length proteins carrying affinity tags. For BceS production, we chose a construct resulting in a C-terminal His8 tag fusion of BceS (Table 1). To produce BceAB, we cloned the corresponding operon into a vector that facilitated addition of an N-terminal Strep-tag to the ATPase (Table 1). To test whether the affinity tags affected protein function, similar constructs harboring the tagged and untagged versions of bceS and bceAB were also introduced into B. subtilis and tested for functionality in bacitracin-dependent signal transduction by complementation of the respective deletion strains. After introduction of bceS-His8 into TMB1557 (ΔbceS; PbceA-luxABCDE), BceS-His had comparable activities to the untagged protein (Fig. 3A, left panel), thus showing full functionality despite the tag. Introducing Strep-bceAB into SGB79 (bceAB::kan; PbceA-luxABCDE) resulted in a slightly reduced promoter activity compared with the untagged version (Fig. 3A, right panel). Furthermore, the Strep-BceAB derivative was able to increase the resistance of a bceAB deleted strain (MIC = 2 μg/ml) 4-fold to 8 μg/ml, compared with a 16-fold (32 μg/ml) increase by the untagged protein, showing that the transporter retained at least partial activity despite the tag.
FIGURE 3.
A, signal transduction activities of affinity-tagged BceAB and BceS. Exponentially growing cells of strains carrying the PbceA-luxABCDE reporter were challenged with 30 μg/ml Zn2+-bacitracin (+) or left untreated (−). Luminescence (relative luminescence units, RLU) was measured at 48 min post-induction. Luminescence was normalized to cell density and is expressed as relative luminescence units/A600. The left graph shows complementation of bceS deletion by untagged (left) and His-tagged (right) BceS. The right graph shows complementation of bceAB deletion by untagged (left) and Strep-tagged (right) BceAB. Data are shown as the mean ± S.D. of three to four independently performed experiments. B, affinity purification of BceAB, BceS, and BceR from E. coli C43(DE3) cells. Left panel, BceS with a C-terminal His8 tag was purified with a Ni2+-NTA column. Center panel, BceAB carrying an N-terminal Strep II®-tag on the ATPase domain was purified via a Strep-Tactin® column. Right panel, BceR carrying a C-terminal His10 tag was purified with a Ni2+-NTA column. Proteins were analyzed using SDS-PAGE, and gels were stained with Coomassie Brilliant Blue. Purified proteins are indicated on the right by the last letter of their name. A molecular size marker is indicated on the left in kDa. MF, membrane fraction; S, solubilized fraction; E, elution; CF, cytoplasmic fraction.
After confirmation of functionality, both BceS-His8 and Strep-BceAB were overproduced in E. coli, where they were both found in the respective membrane fractions. Induction of BceS production resulted in the accumulation of large amounts of BceS, which migrated as a prominent 36-kDa band during SDS-PAGE analysis (Fig. 3B). This is consistent with the predicted molecular mass for BceS of 38 kDa. Induction of bceAB expression resulted in a moderate overproduction of BceA and BceB, which could be detected after SDS-PAGE as bands at 60 kDa for BceB, which is slightly below its expected molecular mass (72 kDa) but consistent with previous observations (33), and 27 kDa for BceA (expected mass: 27 kDa). Both BceS and BceAB could be solubilized using DDM and purified to high yields and apparent homogeneity using the respective affinity matrix. Additionally, BceR carrying a C-terminal His10 tag was produced in and purified from cytoplasmic factions of E. coli. The expected mass of BceR is 27 kDa, which is consistent with its observed migration on SDS-polyacrylamide gels.
In Vitro Characterization of BceAB Activity
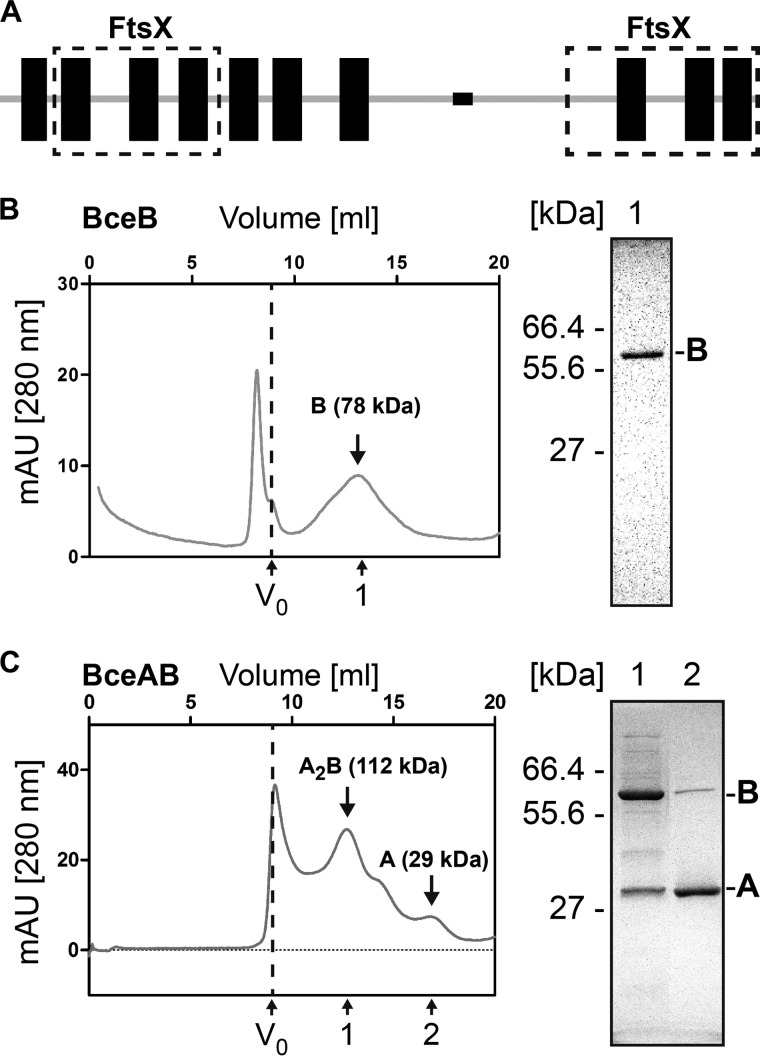
As mentioned above, we observed no dimerization of BceB in the BACTH analysis (Fig. 2). Dimerization would have been expected for an ABC transporter, which generally consists of two membrane-bound permeases that provide a passageway for the cargo and two cytoplasmic nucleotide-binding domains that bind and hydrolyze ATP (36). However, the permease of the Bce system, BceB, possesses 10 TM helices, and a conserved domain analysis showed the presence of two FtsX domains, one encompassing TM helices 2–4 and the second encompassing TM helices 8–10 (Fig. 4A). FtsX itself is the permease of an ABC transporter involved in regulation of cell wall hydrolases (37) and is predicted to consist of 4 TM helices. It is therefore conceivable that BceB originated from a fusion of two smaller FtsX domain permeases and thus does not require further dimerization. To test this in more detail, we determined the molecular mass of the native transporter by size exclusion chromatography of detergent-solubilized BceB and BceAB. BceB alone eluted in a single peak corresponding to a molecular mass of 78 kDa, which is consistent with the theoretical mass of a BceB monomer of 72 kDa (Fig. 4B). Analysis of the BceAB complex showed two elution peaks. The second, smaller peak contained the ATPase BceA alone, which eluted at a volume corresponding to a mass of 29 kDa (theoretical mass, 27 kDa). The first peak contained the BceAB complex and eluted at a mass of 112 kDa (Fig. 4C). In both analyses, the peak eluting with the void volume most likely contained aggregated protein and was not analyzed further. Importantly, no peak corresponding to the theoretical mass of 200 kDa for a BceA2B2 complex was observed. Our results are therefore much more consistent with a transporter composed of a single permease domain, thus confirming the observed lack of dimerization of the permease domains in the BACTH analysis. To further analyze the composition of the transporter complex, we quantified the band intensities for BceB and BceA from SDS-PAGE analysis of the peak fractions of two separate size exclusion analyses and obtained a BceB/BceA intensity ratio of 1.09 ± 0.08. Taking into account the 2.6-fold difference in mass between the permease (72 kDa) and the ATPase (27 kDa), the ratio in band intensities supports a composition of the BceAB transporter of one permease and two ATPase domains (theoretical intensity ratio of 1.3).
FIGURE 4.
Size exclusion analysis of the BceAB complex. A, conserved domain analysis of BceB. TM helices are shown as black rectangles. FtsX domains are indicated by dashed boxes. Predictions were done using the SMART database (41). B and C, size exclusion analysis of BceB (B) and BceAB (C) on a Superdex 200 10/30 column. Left panels show the chromatograms. The vertical dashed line indicates the void volume (V0) of the column. The calculated molecular mass corresponding to the major eluted peaks is shown in parentheses, and eluted proteins are given by the last letter of their name. The right panels show the corresponding SDS-PAGE analysis of the fractions indicated by numbers below the chromatograms. A molecular size marker is indicated on the left in kDa; proteins are indicated on the right by last letter of their name. mAU, milli absorbance units.
Next, we tested the ATP hydrolysis activity of the purified transporter; however, no hydrolysis was observed either in the presence or absence of the substrate bacitracin (data not shown), indicating that the transporter was not able to cleave ATP in the detergent-solubilized form.
We then wanted to test whether BceAB directly interacted with its substrate peptide bacitracin. To test for direct binding of bacitracin by BceAB, we applied SPR spectroscopy. Because the BceAB complex was unstable when coupled to the sensor chip and both subunits dissociated over time, binding assays were performed with the isolated permease BceB. We applied a single cycle kinetic approach, where increasing concentrations of Zn2+-bacitracin were sequentially injected on a sensor chip with BceB captured to the surface. Nisin as a nonsubstrate peptide was used as control. Bacitracin showed a rapid association to BceB already at low nanomolar concentrations. The response units sequentially increased when the higher Zn2+-bacitracin concentrations were injected. The kinetic approach showed an association rate (ka) of 1.1 × 105 m·s, and a dissociation (kd) rate of 0.0015/s of Zn2+-bacitracin and BceB (Fig. 5A). The affinity of Zn2+-bacitracin to BceB at steady state was determined to be KD = 60 nm (Fig. 5B). The nonsubstrate peptide nisin showed no binding to the BceB (Fig. 5B). Furthermore, the absence of Zn2+ prohibited binding of bacitracin to BceB (data not shown), providing further evidence that the interaction of BceB is specific for Zn2+-bacitracin, the active form of the peptide (38). These data clearly show that the BceB subunit of the BceAB transporter directly and specifically interacts with free bacitracin with high affinity.
FIGURE 5.
Binding of bacitracin to BceB. SPR spectroscopy was used to quantify interactions between BceB and Zn2+-bacitracin or nisin. BceB-Strep was captured onto a Strep-Tactin® XT-coated chip before increasing concentrations of Zn2+-bacitracin or nisin (control) were injected. A, single cycle binding kinetic of bacitracin binding to BceB. The gray line shows the recorded sensorgram, and the black line shows the fitted sensorgram. B, steady-state affinity of bacitracin (black) and nisin (gray) to BceB. RU, response units.
BceAB and BceS Interact in Vitro
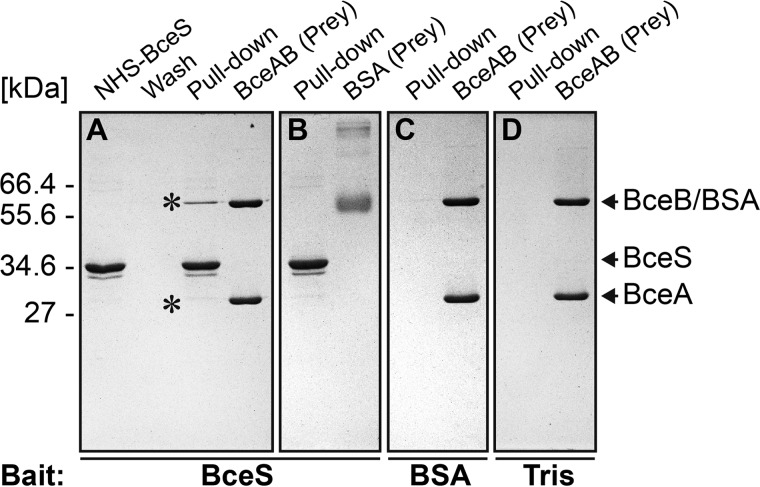
Although the BACTH assays indicated complex formation between BceS and BceB (Fig. 2), these assays were performed under conditions where membrane proteins of a Gram-positive bacterium were overproduced in a Gram-negative host. To confirm these results, we therefore wanted to further characterize the interaction in vitro. For this, we chose an approach where full-length BceS was linked to a Sepharose bead matrix via a chemically stable amide bond with an NHS group. When BceS-coupled Sepharose beads were mixed with a denaturing buffer and applied to SDS-PAGE, a distinct band corresponding to BceS was observed (Fig. 6A, NHS-BceS). This showed that not all BceS molecules were covalently bound to the beads but that a fraction of the protein was attached via protein-protein interactions, most likely due to the typical homodimer formation of HKs. We then incubated the BceS-coupled beads with purified BceAB transporter (prey). After extensive washing, BceAB remained bound to the beads and could be dislodged under denaturing conditions and detected via SDS-PAGE (Fig. 6A, Pull-down). This result shows that BceAB is able to bind to BceS in vitro. To exclude nonspecific interactions, we repeated the same experiment using BSA as prey protein, which showed no interaction with BceS beads (Fig. 6B). When BSA was coupled to the beads or empty beads were blocked with Tris buffer, no binding of BceAB was observed (Fig. 6, C and D). We also tested whether the complex formation is affected by the addition of bacitracin, but no differences were observed (data not shown). Together, these results clearly show that the BceAB transporter and BceS HK specifically interact in vitro and that formation of this complex appears to be independent of the signal.
FIGURE 6.
In vitro pull-down assay using NHS-activated Sepharose. Each panel shows the fraction containing all proteins eluted from the beads (Pull-down), as well as the purified protein tested for interaction with the beads (Prey). A, NHS-conjugated BceS (NHS-BceS) and the last washing step before elution (Wash) are also shown. The protein conjugated to the beads (Bait) is given below each panel. A and B, NHS-conjugated BceS incubated with purified BceAB (A) or BSA (B). C, NHS-conjugated BSA incubated with purified BceAB. D, NHS-Sepharose blocked with Tris and incubated with purified BceAB. BceAB eluting specifically from NHS-BceS beads is indicated by asterisk. Proteins were analyzed using SDS-PAGE, and gels were stained with Coomassie Brilliant Blue. A molecular size marker is indicated on the left in kDa, and the protein bands are labeled on the right.
Complex Formation Affects the Autophosphorylation Activity of BceS
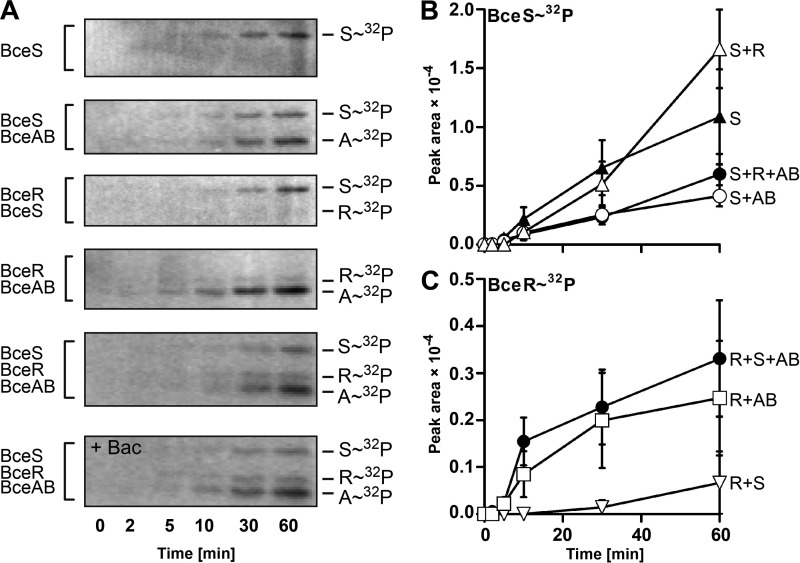
As mentioned in the Introduction, genetic data previously showed that the transporter triggers signal transduction in response to its substrate antibiotic (17). We therefore wanted to test whether the transporter had an effect on the signaling activity of the TCS in vitro. For this, we first tested the autophosphorylation activity of detergent-solubilized BceS. During incubation of BceS with [γ-32P]ATP, a radiolabeled band of the expected size appeared after 5–10 min and further increased in intensity over time (Fig. 7, A, top panel, and B), showing that BceS had a low basal autophosphorylation activity in vitro. Addition of bacitracin had no effect (data not shown), consistent with in vivo data where BceS alone is unable to respond to the antibiotic. In the presence of the transporter, a prominent radiolabeled band appeared at a size of 27 kDa (Fig. 7A, 2nd panel). This corresponds to the molecular mass of the ATPase, and we could indeed also observe this band when BceAB alone was incubated with [γ-32P]ATP (data not shown). It therefore appears that the ATPase is able to bind ATP despite its inactivity to hydrolyze it in vitro. The band intensity of the HK was reduced by about 50% in the presence of the transporter (Fig. 7, A and C), indicating a lower autophosphorylation rate in the complex compared with BceS alone (p = 0.009, two-way analysis of variance). The difference in phosphorylation activity of BceS is unlikely to be simply due to reduced access to ATP caused by the observed ATP binding by BceA, because the protein concentration (1.4 μm) is too low to significantly impact the total ATP concentration (40 μm) in the assay, even if every BceA molecule were to bind an ATP molecule. We therefore interpret the data as a true reduction of HK activity in its uninduced state by the transporter as discussed in more detail below.
FIGURE 7.
In vitro phosphorylation of Bce module proteins. Purified, detergent-solubilized BceS-His8 was mixed in equimolar ratios with BceAB, BceR, or both as indicated on the left. A, phosphorylation was started at t = 0 min by adding [γ-32P]ATP. At the indicated times, reactions were stopped; the samples were subjected to SDS-PAGE, and phosphorylated proteins were detected by phosphorimaging. Representative autoradiographs of three to four independently performed experiments are shown. B and C, band intensities of BceS-32P (B) and BceR-32P (C) were quantified and plotted over time. The protein combinations in each assay are given on the right by the last letter of their names. Data are shown as the mean ± S.D. of three to four independently performed experiments.
Phosphotransfer from BceS to BceR only occurred at a very low rate as seen by the weak signal for BceR after 30–60 min when BceS and BceR were both incubated in an equimolar ratio (Fig. 7, A and C). Surprisingly, when the regulator was incubated with the transporter BceAB in the absence of the HK, a weak but distinct band of BceR-32P was observed (Fig. 7, A and C). However, this observed HK-independent in vitro phosphorylation of BceR in the presence of BceAB is not likely to be of biological significance, because in a strain of B. subtilis carrying BceR and BceAB but not BceS, no target promoter activities above background were detected.3
When all module components were incubated together, BceS had the same phosphorylation rate as in a combination of only BceS and BceAB (Fig. 7, A and B), showing no further effect on autophosphorylation. The same was true for the BceR band, which showed the same intensity as when incubated with BceAB alone (Fig. 7, A and C). This indicates that BceAB was not able to influence phosphotransfer under the chosen assay conditions. Furthermore, the addition of bacitracin showed no effect on band intensities (Fig. 7A, bottom panel). This inability of the system to respond to its stimulus in vitro is consistent with the lack of ATP hydrolysis by the transporter under the same conditions, because it was shown previously that signaling is abolished with an ATPase-defective transporter (17). Therefore, our assay conditions monitor only the ground state of the signaling pathway. Nevertheless, the significant reduction of the BceS autophosphorylation rate in the presence of BceAB indicated that the transporter was indeed able to directly influence HK's basal activity.
DISCUSSION
In former studies, it was shown that BceS-like HKs have an absolute requirement for BceAB-type transporters in perception of antimicrobial peptides, not only in the model organism B. subtilis but also in medically and biotechnologically relevant Gram-positive species, such as Staphylococcus aureus, Enterococcus faecalis, or Lactobacillus casei (18, 21, 22). Besides the unusual mode of signaling, the mechanism of resistance provided by a transporter against antibiotics that are active on the cell surface remains enigmatic. The aim of this study was to investigate the molecular mechanisms underpinning these resistance systems in more detail, using the bacitracin resistance system BceRS-BceAB of B. subtilis as an example.
BceAB Forms a Sensory Complex with BceS
The observation that signaling in Bce-like systems requires an active transporter could be explained by different conceivable scenarios. One explanation might be that the transporter is required to actively move the substrate peptide to a location where it can then be sensed directly by the HK. An alternative hypothesis is that the transporter and TCS communicate directly via protein-protein interactions. A direct interaction of BceS-like HKs and BceB-like transporters was proposed previously based on protein co-evolution (24). A BACTH approach in S. aureus provided the first experimental evidence for this, showing interactions between the HK GraS, the transport permease VraG, and the accessory membrane protein GraX (20). Here, we showed that in the Bce system of B. subtilis, which lacks a homologue of the GraX protein, BceB and BceS could also interact in a BACTH assay (Fig. 2). We further confirmed these results by in vitro pulldown of BceAB by BceS (Fig. 6), showing a direct and specific interaction between the purified HK and ABC transporter. We could not find any effect of bacitracin on this interaction, suggesting that the complex is formed constitutively, independent of the signal. Together, our data clearly demonstrate that BceRS and BceAB form a sensory complex through the direct interaction between BceS and BceB. The BACTH analysis appeared to indicate that BceR could also interact with this complex as long as all membrane-bound components were present, suggesting that the signaling complex may be comprised of all four proteins of the system. As mentioned under “Experimental Procedures,” BceAB consistently co-purified with the E. coli protein ArnC, which catalyzes an undecaprenyl-phosphate-dependent transfer reaction during lipid A modification (39). Because this modification pathway is involved in resistance of E. coli against polymyxin, which, like bacitracin, is a nonribosomally synthesized cyclic peptide antibiotic, this may point to a more general significance of the protein-protein interactions studied here beyond Firmicutes bacteria.
Direct Binding of Bacitracin by BceB
BceAB-like transporters confer resistance against antimicrobial peptides that bind to targets in the cytoplasmic membrane (28, 40), leading to the speculation that such transporters may not directly recognize the peptides as substrates, but rather the target-peptide complex in the membrane (17, 27). Taking this hypothesis one step further, it was recently proposed that BceAB of B. subtilis does not transport bacitracin at all, but instead it acts by flipping the target molecule UPP to the cytoplasmic side of the membrane (30). To address this important question, here we used SPR spectroscopy to test whether the permease BceB could bind bacitracin in the absence of the native membrane environment. Our results clearly showed high affinity binding of bacitracin with a steady-state KD of 60 nm. This interaction was specific for the active form, i.e. the Zn2+-complex, of bacitracin, as no binding was observed in the absence of Zn2+ or with the nonsubstrate peptide antibiotic nisin (Fig. 5). Moreover, the KD obtained in vitro for the purified permease very closely matches the in vivo threshold concentration of bacitracin required for activation of the target promoter, PbceA, of 70 nm,4 supporting the physiological relevance of the in vitro data. Although the results obtained here do not exclude the possibility that a bacitracin-UPP complex can also be recognized by the transporter, they certainly lend weight to a model for a resistance mechanism involving translocation of the antimicrobial peptide directly, rather than flipping of the cellular target molecule.
BceAB Affects the Autophosphorylation of BceS
To study the influence of BceAB on signaling, we reconstructed the signal transduction pathway in vitro using the purified Bce components and monitored protein phosphorylation from radiolabeled [γ-32P]ATP over time (Fig. 7). The results showed that the activity of BceS was reduced in the presence of the transporter, suggesting that in the complex the transporter inhibited signal-independent autophosphorylation of BceS, presumably via the direct protein-protein interactions shown above (Figs. 2 and 6). Activation of signaling by bacitracin could not be shown in vitro, probably due to the observed inability of the transporter to hydrolyze ATP under the test conditions, which may be a consequence of the absence of a phospholipid environment for the transporter and lacking lateral pressure in detergent micelles. This finding is consistent with earlier observations that ATP hydrolysis is absolutely required for signaling in vivo (17, 18). We therefore concluded that here we assayed the ground state of the system, reflecting phosphorylation activities in the absence of active signaling. Explanation of the results from the phosphorylation assays is further complicated by the observed HK-independent phosphorylation of the response regulator in the presence of the ABC transporter, which cannot currently be explained and demands some caution in interpretation of the data. Nevertheless, BceAB was able to reduce the signal-independent autophosphorylation activity of BceS in vitro, which suggested that the transporter was able to directly influence the activity of its cognate HK. Future efforts will be directed at elucidating the molecular details of this effect and how it is influenced by the presence of bacitracin.
Based on the data obtained in this study and in previous genetic and comparative genomic analyses (17, 24, 33), we here propose a working model for the mechanism of signal transduction within Bce-like models (Fig. 1). The signaling complex is comprised of the HK and the transporter, which most likely consists of a single permease subunit and two ATPase domains (Fig. 4). A potential involvement of the response regulator has been discussed above. In the presence of bacitracin, the peptide is bound directly by the transporter. The resulting transport activity, as implied from the previously reported requirement for ATP hydrolysis by the transporter (17), then triggers activation of the HK. The exact mechanism for this remains unknown to date, but based on the constitutive complex formation observed here, it appears likely to involve direct communication via protein conformational changes. Autophosphorylation of the HK leads to phosphorylation of the response regulator and in turn to an increased transcription from the target promoter, PbceA. The resulting elevated cellular levels of the ABC transporter ultimately lead to resistance, most likely by active removal of the antimicrobial peptide from its site of action.
Taken together, this study provides first functional insights into the mechanisms of stimulus perception, signal transduction, and resistance by Bce-like systems, which represent widely spread resistance determinants against peptide antibiotics in Firmicutes bacteria.
Acknowledgments
We thank Hannah Ulm and Tanja Schneider for valuable suggestions on heterologous production of the ABC transporter and Shawn MacLellan for expert advice regarding the in vitro pull-down assays. We are also grateful to IBA for generously supplying the Strep-Tactin® XT resin. SPR spectroscopy was performed in the Bioanalytics Core Facility of the Ludwig-Maximilians-Universität München Biocenter.
This work was supported by Deutsche Forschungsgemeinschaft Grant GE2164/3-1 and the Fonds der Chemischen Industrie (to the S. G. laboratory) and Deutsche Forschungsgemeinschaft Grant Exc114-2 (to the K. J. laboratory).
T. Mascher and C. Fang, personal communication.
S. Dintner, T. Mascher, and S. Gebhard, unpublished observations.
- TCS
- two-component system
- ABC
- ATP-binding cassette
- HK
- histidine kinase
- UPP
- undecaprenyl-pyrophosphate
- MIC
- minimal inhibitory concentration
- DDM
- n-dodecyl β-d-maltoside
- SPR
- surface plasmon resonance
- NHS
- N-hydroxysuccinimide
- BACTH
- bacterial two-hybrid
- β-ME
- β-mercaptoethanol
- Ni2+-NTA
- Ni2+-nitrilotriacetic acid
- TM
- transmembrane.
REFERENCES
- 1. Tetsch L., Jung K. (2009) The regulatory interplay between membrane-integrated sensors and transport proteins in bacteria. Mol. Microbiol. 73, 982–991 [DOI] [PubMed] [Google Scholar]
- 2. van Veen H. W. (1997) Phosphate transport in prokaryotes: molecules, mediators and mechanisms. Antonie van Leeuwenhoek. 72, 299–315 [DOI] [PubMed] [Google Scholar]
- 3. Qi Y., Kobayashi Y., Hulett F. M. (1997) The pst operon of Bacillus subtilis has a phosphate-regulated promoter and is involved in phosphate transport but not in regulation of the Pho regulon. J. Bacteriol. 179, 2534–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sola-Landa A., Rodríguez-García A., Franco-Domínguez E., Martín J. F. (2005) Binding of PhoP to promoters of phosphate-regulated genes in Streptomyces coelicolor: identification of PHO boxes. Mol. Microbiol. 56, 1373–1385 [DOI] [PubMed] [Google Scholar]
- 5. Glover R. T., Kriakov J., Garforth S. J., Baughn A. D., Jacobs W. R., Jr. (2007) The two-component regulatory system senX3-regX3 regulates phosphate-dependent gene expression in Mycobacterium smegmatis. J. Bacteriol. 189, 5495–5503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Steed P. M., Wanner B. L. (1993) Use of the rep technique for allele replacement to construct mutants with deletions of the pstSCAB-phoU operon: evidence of a new role for the PhoU protein in the phosphate regulon. J. Bacteriol. 175, 6797–6809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wanner B. L. (1996) Signal transduction in the control of phosphate-regulated genes of Escherichia coli. Kidney Int. 49, 964–967 [DOI] [PubMed] [Google Scholar]
- 8. Yuan Z.-C., Zaheer R., Finan T. M. (2006) Regulation and properties of PstSCAB, a high-affinity, high-velocity phosphate transport system of Sinorhizobium meliloti. J. Bacteriol. 188, 1089–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gebhard S., Cook G. M. (2008) Differential regulation of high-affinity phosphate transport systems of Mycobacterium smegmatis: identification of PhnF, a repressor of the phnDCE operon. J. Bacteriol. 190, 1335–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lux R., Jahreis K., Bettenbrock K., Parkinson J. S., Lengeler J. W. (1995) Coupling the phosphotransferase system and the methyl-accepting chemotaxis protein-dependent chemotaxis signaling pathways of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 92, 11583–11587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Scheu P. D., Liao Y. F., Bauer J., Kneuper H., Basché T., Unden G., Erker W. (2010) Oligomeric sensor kinase DcuS in the membrane of Escherichia coli and in proteoliposomes: chemical cross-linking and FRET spectroscopy. J. Bacteriol. 192, 3474–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kleefeld A., Ackermann B., Bauer J., Krämer J., Unden G. (2009) The fumarate/succinate antiporter DcuB of Escherichia coli is a bifunctional protein with sites for regulation of DcuS-dependent gene expression. J. Biol. Chem. 284, 265–275 [DOI] [PubMed] [Google Scholar]
- 13. Witan J., Bauer J., Wittig I., Steinmetz P. A., Erker W., Unden G. (2012) Interaction of the Escherichia coli transporter DctA with the sensor kinase DcuS: presence of functional DctA/DcuS sensor units. Mol. Microbiol. 85, 846–861 [DOI] [PubMed] [Google Scholar]
- 14. Graf S., Schmieden D., Tschauner K., Hunke S., Unden G. (2014) The sensor kinase DctS forms a tripartite sensor unit with DctB and DctA for sensing C4-dicarboxylates in Bacillus subtilis. J. Bacteriol. 196, 1084–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haneburger I., Fritz G., Jurkschat N., Tetsch L., Eichinger A., Skerra A., Gerland U., Jung K. (2012) Deactivation of the E. coli pH stress sensor CadC by cadaverine. J. Mol. Biol. 424, 15–27 [DOI] [PubMed] [Google Scholar]
- 16. Tetsch L., Koller C., Haneburger I., Jung K. (2008) The membrane-integrated transcriptional activator CadC of Escherichia coli senses lysine indirectly via the interaction with the lysine permease LysP. Mol. Microbiol. 67, 570–583 [DOI] [PubMed] [Google Scholar]
- 17. Rietkötter E., Hoyer D., Mascher T. (2008) Bacitracin sensing in Bacillus subtilis. Mol. Microbiol. 68, 768–785 [DOI] [PubMed] [Google Scholar]
- 18. Hiron A., Falord M., Valle J., Débarbouillé M., Msadek T. (2011) Bacitracin and nisin resistance in Staphylococcus aureus: a novel pathway involving the BraS/BraR two-component system (SA2417/SA2418) and both the BraD/BraE and VraD/VraE ABC transporters. Mol. Microbiol. 81, 602–622 [DOI] [PubMed] [Google Scholar]
- 19. Gebhard S., Mascher T. (2011) Antimicrobial peptide sensing and detoxification modules: unravelling the regulatory circuitry of Staphylococcus aureus. Mol. Microbiol. 81, 581–587 [DOI] [PubMed] [Google Scholar]
- 20. Falord M., Karimova G., Hiron A., Msadek T. (2012) GraXSR proteins interact with the VraFG ABC transporter to form a five-component system required for cationic antimicrobial peptide sensing and resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 56, 1047–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gebhard S., Fang C., Shaaly A., Leslie D. J., Weimar M. R., Kalamorz F., Carne A., Cook G. M. (2014) Identification and characterization of a bacitracin resistance network in Enterococcus faecalis. Antimicrob. Agents Chemother. 58, 1425–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Revilla-Guarinos A., Gebhard S., Alcántara C., Staron A., Mascher T., Zúñiga M. (2013) Characterization of a regulatory network of peptide antibiotic detoxification modules in Lactobacillus casei BL23. Appl. Environ. Microbiol. 79, 3160–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mascher T., Helmann J. D., Unden G. (2006) Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 70, 910–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dintner S., Staron A., Berchtold E., Petri T., Mascher T., Gebhard S. (2011) Coevolution of ABC transporters and two-component regulatory systems as resistance modules against antimicrobial peptides in Firmicutes bacteria. J. Bacteriol. 193, 3851–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gebhard S. (2012) ABC transporters of antimicrobial peptides in Firmicutes bacteria–phylogeny, function and regulation. Mol. Microbiol. 86, 1295–1317 [DOI] [PubMed] [Google Scholar]
- 26. Ohki R., Giyanto, Tateno K., Masuyama W., Moriya S., Kobayashi K., Ogasawara N. (2003) The BceRS two-component regulatory system induces expression of the bacitracin transporter, BceAB, in Bacillus subtilis. Mol. Microbiol. 49, 1135–1144 [DOI] [PubMed] [Google Scholar]
- 27. Bernard R., Guiseppi A., Chippaux M., Foglino M., Denizot F. (2007) Resistance to bacitracin in Bacillus subtilis: unexpected requirement of the BceAB ABC transporter in the control of expression of its own structural genes. J. Bacteriol. 189, 8636–8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stone K. J., Strominger J. L. (1971) Mechanism of action of bacitracin: complexation with metal ion and C55-isoprenyl pyrophosphate. Proc. Natl. Acad. Sci. U.S.A. 68, 3223–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hasper H. E., de Kruijff B., Breukink E. (2004) Assembly and stability of nisin-Lipid II pores. Biochemistry 43, 11567–11575 [DOI] [PubMed] [Google Scholar]
- 30. Kingston A. W., Zhao H., Cook G. M., Helmann J. D. (2014) Accumulation of heptaprenyl diphosphate sensitizes Bacillus subtilis to bacitracin: implications for the mechanism of resistance mediated by the BceAB transporter. Mol. Microbiol. 93, 37–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bertani G. (1951) Studies on lysogenesis I: the mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62, 293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Karimova G., Pidoux J., Ullmann A., Ladant D. (1998) A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kallenberg F., Dintner S., Schmitz R., Gebhard S. (2013) Identification of regions important for resistance and signalling within the antimicrobial peptide transporter BceAB of Bacillus subtilis. J. Bacteriol. 195, 3287–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Monk B. C., Kurtz M. B., Marrinan J. A., Perlin D. S. (1991) Cloning and characterization of the plasma membrane H+-ATPase from Candida albicans. J. Bacteriol 173, 6826–6836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Karlsson R., Katsamba P. S., Nordin H., Pol E., Myszka D. G. (2006) Analyzing a kinetic titration series using affinity biosensors. Anal. Biochem. 349, 136–147 [DOI] [PubMed] [Google Scholar]
- 36. Biemans-Oldehinkel E., Doeven M. K., Poolman B. (2006) ABC transporter architecture and regulatory roles of accessory domains. FEBS Lett. 580, 1023–1035 [DOI] [PubMed] [Google Scholar]
- 37. Yang D. C., Peters N. T., Parzych K. R., Uehara T., Markovski M., Bernhardt T. G. (2011) An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. Proc. Natl. Acad. Sci. U.S.A. 108, E1052–E1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Storm D. R., Strominger J. L. (1973) Complex formation between bacitracin peptides and isoprenyl pyrophosphates. The specificity of lipid-peptide interactions. J. Biol. Chem. 248, 3940–3945 [PubMed] [Google Scholar]
- 39. Breazeale S. D., Ribeiro A. A., McClerren A. L., Raetz C. R. (2005) A formyltransferase required for polymyxin resistance in Escherichia coli and the modification of lipid A with 4-amino-4-deoxy-l-arabinose: identification and function of UDP-4-deoxy-4-formamido-l-arabinose. J. Biol. Chem. 280, 14154–14167 [DOI] [PubMed] [Google Scholar]
- 40. Breukink E., de Kruijff B. (2006) Lipid II as a target for antibiotics. Nat. Rev. Drug Discov. 5, 321–332 [DOI] [PubMed] [Google Scholar]
- 41. Schultz J., Milpetz F., Bork P., Ponting C. P. (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U.S.A. 95, 5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Amann E., Ochs B., Abel K. J. (1988) Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69, 301–315 [DOI] [PubMed] [Google Scholar]
- 43. Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promotor. J. Bacteriol. 177, 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Radeck J., Kraft K., Bartels J., Cikovic T., Dürr F., Emenegger J., Kelterborn S., Sauer C., Fritz G., Gebhard S., Mascher T. (2013) The Bacillus BioBrick Box: generation and evaluation of essential genetic building blocks for standardized work with Bacillus subtilis. J. Biol. Eng. 7, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Taylor R. G., Walker D. C., McInnes R. R. (1993) E. coli host strains significantly affect the quality of small scale plasmid DNA preparations used for sequencing. Nucleic Acids Res. 21, 1677–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Miroux B., Walker J. E. (1996) Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260, 289–298 [DOI] [PubMed] [Google Scholar]