Background: Modification of cysteine residues, including glutathionylation, commonly occurs in peptides bound to and presented by MHC molecules.
Results: Glutathionylation of a coronavirus-specific T cell epitope results in diminished CD8 T cell recognition.
Conclusion: Cysteine modification of a T cell epitope negatively impacts the host immune response.
Significance: Cross-talk between virus-induced oxidative stress and the T cell response probably occurs, diminishing host cell recognition of infected cells.
Keywords: Antigen Presentation, Antigen Processing, Glutathionylation, Mass Spectrometry (MS), Oxidation-Reduction (Redox), Redox Regulation, T-cell, Viral Immunology
Abstract
Cysteine-containing peptides represent an important class of T cell epitopes, yet their prevalence remains underestimated. We have established and interrogated a database of around 70,000 naturally processed MHC-bound peptides and demonstrate that cysteine-containing peptides are presented on the surface of cells in an MHC allomorph-dependent manner and comprise on average 5–10% of the immunopeptidome. A significant proportion of these peptides are oxidatively modified, most commonly through covalent linkage with the antioxidant glutathione. Unlike some of the previously reported cysteine-based modifications, this represents a true physiological alteration of cysteine residues. Furthermore, our results suggest that alterations in the cellular redox state induced by viral infection are communicated to the immune system through the presentation of S-glutathionylated viral peptides, resulting in altered T cell recognition. Our data provide a structural basis for how the glutathione modification alters recognition by virus-specific T cells. Collectively, these results suggest that oxidative stress represents a mechanism for modulating the virus-specific T cell response.
Introduction
Small fragments of proteins (peptides) derived from both intracellular and extracellular sources are displayed on the surface of cells by molecules encoded within the major histocompatibility complex (MHC). These peptides are recognized by T lymphocytes and provide the immune system with a surveillance mechanism for the detection of pathogens and cancer cells. The fidelity with which antigen presentation communicates changes in the intracellular proteome is critical for immune surveillance. Not only do antigens expressed at vastly different abundances need to be represented within the array of peptides selected and presented at the cell surface (collectively termed the immunopeptidome (1, 2)), but changes in their post-translational state also need to be conveyed within this complex mixture of peptides. For example, changes in antigen phosphorylation have been linked to cancer, and the presentation of phosphorylated peptides has been shown to communicate the cancerous state of cells to the immune system (3–6). Other types of post-translational modification play a central role in the pathogenesis of autoimmune diseases (7), such as arginine citrullination in arthritis (8–10), deamidation of glutamine residues in wheat proteins in celiac disease (11–15), and cysteine oxidation in type 1 diabetes (16, 17). Cysteine is predicted to be present in up to 14% of potential T cell epitopes based on its prevalence in various pathogen and host proteomes (18). However, reports of cysteine-containing epitopes are much less frequent due to technical difficulties associated with synthesis and handling of cysteine-containing peptides and their subsequent avoidance in many epitope mapping studies (19). Cysteine can be modified in numerous ways, including cysteinylation (the disulfide linkage of free cysteine to peptide or protein cysteine residues), oxidation to cysteine sulfenic (oxidation), sulfinic (dioxidation) and sulfonic acids (trioxidation), S-nitrosylation, and S-glutathionylation. Such modifications may occur prior to or during antigen processing; however, the role of cysteine modification in T-cell-mediated immunity has not been systematically addressed.
In addition to constitutive presentation of a subset of oxidatively modified peptides, it is anticipated that changes in the proportion of these ligands will occur upon infection because oxidative stress, triggering of the unfolded protein response, and modulation of host cell synthesis by the virus are hallmarks of this process (20–27). For example, host cell stress responses modulate expression, localization, and function of Toll-like receptors, a key event in the initiation of the immune response (28). Oxidative stress would also be predicted to affect protein function through post-translational modification of amino acids, such as cysteine. Indeed, because of the reactive nature of cysteine and the requirements for cells to regulate the redox state of proteins to maintain function, a number of scavenging systems for redox-reactive intermediates exist. The tripeptide glutathione (GSH) is one of the key intracellular antioxidants, acting as a scavenger for reactive oxygen species. Reduced GSH is equilibrated with its oxidized form, GSSG, with normal cytosolic conditions being that of the reduced state in a ratio of ∼50:1 (GSH/GSSG) (29). Modification of proteins and peptides with GSH (termed S-glutathionylation) occurs following reaction of GSSG with the thiol group of cysteine in a reaction catalyzed by the detoxifying enzyme, glutathione S-transferase (GST). A variety of cellular processes and signaling pathways, such as the induction of innate immunity, apoptosis, redox homeostasis, and cytokine production, are modulated by this GST-catalyzed post-translational modification (30–32). S-Glutathionylation can eventuate via oxidative stress, whereby the intracellular levels of GSSG increase.
Given that viruses are known to induce oxidative stress (33–35), the intracellular environment of viral infection may lead to an increase in S-glutathionylated cellular proteins and viral antigens. For instance, HSV infection induces an early burst of reactive oxygen species, resulting in S-glutathionylation of TRAF family members, which in turn is linked to downstream signaling and interferon production (36). The potential for modification of viral antigens subsequent to reactive oxygen species production is highlighted by S-glutathionylation of several retroviral proteases, leading to host modulation of protease function (37). Indeed large scale changes in protein S-glutathionylation are observed in HIV-infected T cell blasts (38), suggesting that functional modulation of both host and viral proteins occurs via this mechanism. Whether these S-glutathionylated proteins inhibit or enhance immune responses to the unmodified epitope or generate novel T-cell epitopes that are subsequently recognized by the adaptive immune system is unclear.
Here, we investigate the frequency of modification of cysteine-containing MHC-bound peptides by interrogating a large database of naturally processed self-peptides derived from B-lymphoblastoid cells, murine tissues, and cytokine-treated cells. In addition, the functional consequences of Cys modification of T cell epitopes was investigated using an established model of infection that involves an immunodominant cysteine-containing epitope derived from a neurotropic strain of mouse hepatitis virus, strain JHM (JHMV)8 (39–41). We describe S-glutathionylation of this viral T cell epitope and the functional and structural implications of redox-modulated antigen presentation. Collectively our studies suggest that S-glutathionylation plays a key, previously unappreciated role in adaptive immune recognition.
EXPERIMENTAL PROCEDURES
Mice
6–8-week-old pathogen-free C57BL/6 (B6) mice were purchased (NCI-Frederick, National Institutes of Health). NOD mice were purchased and housed in the Bio21 Institute animal facility (Melbourne, Australia). Studies were carried out in accordance with accepted standards of humane animal care and were approved by the animal ethics committee (University of Melbourne). Tissues were snap-frozen in liquid nitrogen and stored at −80 °C.
Viruses
Wild-type JHMV was propagated and titered as described previously (42). VACV-S510 was engineered as described previously (40). Mice were infected intraperitoneally with 2 × 107 pfu of VACV-S510, and CD8+ T cell responses were measured in the spleen.
Glutathione Levels in Naive and JHMV-infected Brain Tissue
Brains were harvested from either naive mice or mice infected intranasally with JHMV at day 7 p.i. Brains were directly homogenized into 5% (w/v) 5-sulfosalicylic acid. The soluble fraction was used for thiol analysis. Total GSH (GSH + GSSG) and GSSG were determined using a spectrophotometric recycling assay as described previously (43, 44). Glutathione measurements were normalized to the total amount of precipitated tissue protein in the 5-sulfosalicylic acid-treated samples as determined using a bicinchoninic acid protein assay with a Micro BCA protein assay reagent kit (Pierce).
Staining with H-2Db·S510 and H-2Db·GSH-S510 Tetramer
Tetramers were obtained either from the University of Melbourne Tetramer Facility (Parkville, Australia) or the NIAID, National Institutes of Health, MHC Tetramer Core Facility (Atlanta, GA). Cells were stained with CD8, CD16/CD32, and tetramer and analyzed as described previously (45).
H-2Db Binding Assay
Binding assays were performed as described previously (46, 47). Briefly, RMA-S cells grown to a density of 106 cells/ml were incubated at 26 °C for 20 h in order to allow stabilization of “empty” MHC complexes at the cell surface. Cells were pulsed with graded concentrations of peptide, incubated for 1 h at 26 °C, and incubated for an additional 2 h at 37 °C. After washing, cell surface expression of H-2Db was detected using the mAb 28-14-8S, which binds properly conformed Db-peptide complexes on the cell surface, followed by secondary detection with an FITC-conjugated sheep anti-mouse Ig antibody (Chemicon/Millipore, catalog no. A326; 1:500 dilution). Cells were analyzed using a CyAn ADP flow cytometer (Beckman Coulter) in conjunction with FloJo software version 7.2.5.
Purification of MHC·Peptide Complexes
EBV-transformed B lymphoblastoid cells, cytokine-treated NIT-1 cells (100 IU/ml IFN-γ for 48 h), and murine tissues (pooled thymus and spleens from 12-week-old female NOD mice, n = 10) were harvested and snap-frozen in liquid nitrogen. DC2.4 cells were infected with VACV-S510 at 5 pfu/cell and incubated for 4 h before snap freezing. Frozen cell pellets or tissues were ground in a Retsch Mixer Mill MM 400 (2 min at 30 Hz) under cryogenic conditions, resuspended in a lysis buffer containing 0.5% IGEPAL (Sigma), 50 mm Tris, pH 8, 150 mm NaCl, and protease inhibitors (Complete Protease Inhibitor Mixture t; Roche Applied Science). For DC2.4, 1 × 108 DC2.4 cells were disrupted by gentle resuspension in a total of 5 ml of lysis buffer without cryogenic milling. Lysates were incubated with rotation for 1 h at 4 °C and cleared by centrifugation. MHC·peptide complexes were immunoaffinity-purified using specific monoclonal antibodies 28-14-8S (anti-H-2Db), SF1.1.10 (anti-H-2Kd), w632 (pan-HLA class I), LB3.1 (anti-HLA-DR), SPV-L3 (anti-HLA-DQ), or B721 (anti-HLA-DP) bound to protein A-Sepharose, as described previously (47, 48). Bound complexes were eluted by acidification with 10% acetic acid. The mixture of peptides and MHC protein chains was fractionated on a 4.6-mm internal diameter × 50-mm-long reversed-phase C18 HPLC column (Chromolith Speed Rod, Merck), using an ÄKTAmicroTM HPLC system (GE Healthcare) running on a mobile phase buffer A of 0.1% trifluoroacetic acid (TFA) and buffer B of 80% acetonitrile, 0.1% TFA and at a flow rate of 1 ml/min.
Liquid Chromatography-Multiple Reaction Monitoring-Mass Spectrometry
Following peptide elution, samples were concentrated and immediately analyzed by mass spectrometry. An AB SCIEX QTRAP® 5500 mass spectrometer was used for multiple reaction monitoring (MRM) detection, coupled on-line to a Tempo nano-LC and nanoflex cHiPLC manifold (Eksigent). 20-μl samples were injected and loaded onto a trap column (200 μm × 0.5-mm ChromXP C18-CL 3 μm 120 Å) at a flow rate of 10 μl/min in 98% buffer A for 10 min. For on-line fractionation of samples onto the mass spectrometer, samples were eluted from the trap column and over a cHiPLC column (75 μm × 15-cm ChromXP C18-CL 3 μm 120 Å) at 300 nl/min under the following buffer B (95% acetonitrile, 0.1% formic acid in water) gradient conditions: 0–3 min, 2–10% B; 3–33 min, 10–40% B; 33–36 min, 40–80% B; 36–38 min, hold at 80% B; 38–39 min, 80–2% B, followed by equilibration at 2% B until the end of the run at 48 min. The QTRAP® 5500 was operated in MRM mode in unit resolution for Q1 and Q3, coupled to an information-dependent acquisition criterion set to trigger an EPI scan (10,000 Da/s; rolling CE; unit resolution) following any MRM transition exceeding 500 counts. Optimal MRM Q1 → Q3 transition conditions were designed through analysis of synthetic peptides and are listed in Table 1. Data analysis was performed using Analyst version 1.5.2 (AB SCIEX).
TABLE 1.
MRM transitions for the detection of S510 and GSH-S510
| Epitope | Sequence | Q1 (charge) | Q3 (ion) | Optimal CE |
|---|---|---|---|---|
| S510 | CSLWNGPHL | 513.7 (+2) | 723.4 (y6) | 20 |
| 537.3 (y5) | 20 | |||
| 423.2 (y4) | 20 | |||
| GSH-S510 | GSH-CSLWNGPHL | 444.5 (+3) | 723.4 (y6) | 17 |
| 609.2 (b3) | 17 | |||
| 537.3 (y5) | 17 | |||
| 423.2 (y4) | 17 |
Identification of MHC-bound Peptides Using LC-MS/MS
Peptide-containing fractions were concentrated and loaded onto a microfluidic trap column packed with ChromXP C18-CL 3-μm particles (300 Å nominal pore size; equilibrated in 0.1% formic acid, 5% acetonitrile) at 5 μl/min using an Eksigent NanoUltra cHiPLC system. An analytical (15 cm × 75-μm ChromXP C18-CL 3) microfluidic column was then switched in line, and peptides were separated using linear gradient elution of 0–80% acetonitrile over 90 min (300 nl/min). Separated peptides were analyzed using an AB SCIEX 5600 TripleTOF mass spectrometer equipped with a Nanospray III ion source and accumulating up to 30 MS/MS spectra per second. Data were analyzed with ProteinPilotTM software, and peptide identities were determined subject to strict bioinformatic criteria that included the use of a decoy database to calculate the false discovery rate. A false discovery rate cut-off of 5% was applied, and the filtered data set was further analyzed manually to exclude redundant peptides and known contaminants.
Intracellular Cytokine Staining and Functional Avidity
6–8-week-old C57BL/6 mice were inoculated intranasally with 4 × 104 pfu of JHMV. Mononuclear cells were harvested from the brains of acutely ill mice 7 days p.i. and analyzed for expression of IFN-γ after peptide stimulation by an intracellular cytokine assay, as described previously (49). Unless otherwise noted, peptides corresponding to S510 or GSH-S510 epitope were used at a final concentration of 1 μm. GSH-510 peptide was generated by incubation of the native peptide in 5 mm reduced glutathione (GSSG) in PBS for 4 h, followed by acidification and purification by reversed-phase HPLC. For measurements of functional avidity, brain-derived cells were stimulated in the presence of antigen-presenting cells (CHB3 cells, B cell line, H-2Db, H-2Kb) and pulsed with 10-fold dilutions of the relevant peptide for 6 h at 37 °C in the presence of brefeldin A. Cells were analyzed for IFN-γ production. Cells were analyzed by flow cytometry using a FACSCalibur (BD Biosciences). Data sets were analyzed using FlowJo software (Tree Star, Inc., Ashland, OR). For each epitope-specific population, data were normalized to the maximum frequency of epitope-specific CD8+ T cells detected. Peptide concentration curves were fit to sigmoid concentration-response curves and used to calculate EC50 values. All antibodies and reagents were purchased from BD Pharmingen (San Diego, CA).
X-ray Crystallographic Studies
The expression, purification, and refolding of H-2Db with murine β-2 microglobulin and peptide were performed as described (39). Crystals of H-2Db·GSH-S510 were grown at 21 °C in 0.1 m citrate, pH 7.5, 28% PEG 3,350, 0.2 m LiSO4, using a protein concentration of 3 mg/ml. Crystals were cryoprotected by equilibration against mother liquor containing 5% glycerol and flash-frozen by placing in a nitrogen stream at 100 K. The 2.8 Å data set was collected on an in-house x-ray generator and processed using HKL2000 (50). The structure was solved by molecular replacement in PHASER (51), using the protein coordinates of a previously solved H-2Db complex (Protein Data Bank entry 1YN6) as a search model. The structure was initially subjected to simulated annealing and coordinate/atomic B-factor refinement in PHENIX (52), followed by iterative cycles of model building in COOT (53) and refinement in REFMAC (54) with TLS parameterization. Structure validation was carried out using MOLPROBITY (55) and tools in the PHENIX package. Structure analysis was carried out using programs in CCP4i (56). A summary of data collection and refinement statistics is provided in Table 2.
TABLE 2.
H2Db·GSH-S510 crystallographic statistics
a Values in parentheses refer to the highest resolution bin.
bRmerge = Σ(hkl) Σi|Ii(hkl) − 〈I(hkl)〉|/Σ(hkl)Σi〈I(hkl)〉, where I is the observed intensity, and 〈I〉 is the average intensity of multiple observations from symmetry-related reflections.
cRp.i.m. = Σ(hkl) (1/(N − 1))1/2 Σi|Ii(hkl) − 〈I(hkl)〉|/Σ(hkl)Σi〈I(hkl)〉, where N is the redundancy of the hkl reflection.
dRfactor = (Σ‖Fo| − |Fc‖)/(Σ|Fo|).
e 10.0% of data were used for the Rfree calculation.
Despite being crystallized in the same condition, the S-glutathionylated H-2Db·S510 structure is characterized by a crystal symmetry different from yet related to that observed for the non-S-glutathionylated complex (Protein Data Bank entry 2ZOK), which translates into small variations in the packing environment of individual pMHC molecules. These differences were taken into account when comparing the two structures. Two loop regions of the α1/α2 domain, in particular, which are involved in crystal contacts (Gly16–Glu19 and Gly104–Asp106) and display large conformational differences between the two structures were excluded from root mean square deviation calculations. The α1/α2 domain of the glutathionylated structure superimposes with individual pMHC copies of 2ZOK with an average root mean square deviation of 0.28 Å for CA atoms and 0.92 Å for all atoms. The coordinates of the H-2Db·GSH-S510 complex have been deposited in the Protein Data Bank database (entry 4PG2).
RESULTS
Modification of Cysteine Residues Is Common in T Cell Epitopes
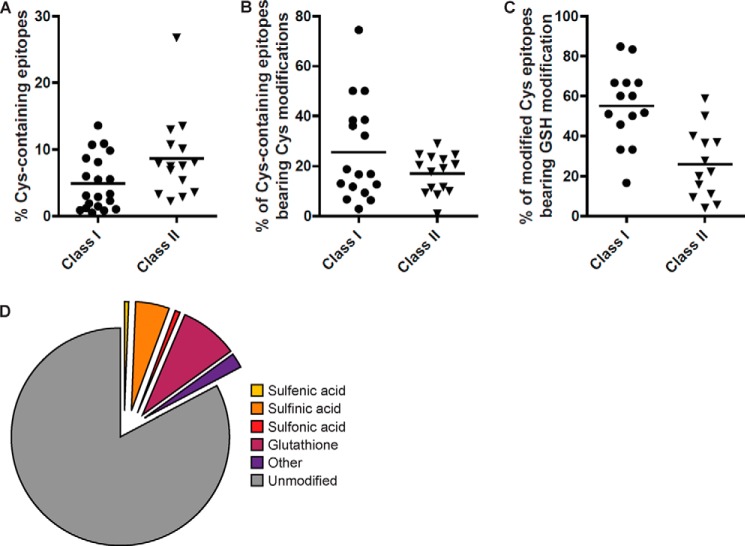
Due to technical challenges of working with cysteine-containing peptides, their importance in immunity remains underappreciated. To address this, we generated a large data set of peptide sequences identified by mass spectrometry from immunoaffinity-purified MHC class I and class II molecules that consists of over 70,000 high confidence peptide identities. A subset of these peptides arises from cells expected to be undergoing some degree of oxidative stress due to transformation (57) or cytokine exposure (22, 28, 58, 59). These peptides were isolated from a panel of EBV-transformed human B lymphoblastoid cells expressing a variety of MHC alleles (12 different class I and 10 different class II alleles), murine NIT-1 cells (a pancreatic β cell line that is highly sensitive to IFN-γ treatment that expresses H-2Kd and H-2Db), and various murine tissues (see Table 3 for a list of cell lines and alleles). Global peptide repertoires were assessed by eluting peptides from these cells and analysis by tandem mass spectrometry, with epitope sequences determined from spectra by the Protein Pilot algorithm (60). Within this data set, numerous cysteine-containing peptides were identified (Fig. 1 and supplemental Table 1). The frequency of cysteine-containing epitopes was influenced by the immunogenetics of the cell line with between 1 and 15% of class I restricted ligands and between 3 and 28% of class II ligands found to contain at least one cysteine residue (Fig. 1A). The higher percentage of cysteine-containing class II peptides was anticipated, given their increased average length. The proportion of these ligands that contained a modified cysteine residue also varied, with 25% of class I and 18% of class II ligands, on average, having a modified cysteine residue (Fig. 1B). A high proportion contained modifications that were enzyme-mediated, which are anticipated to occur in vivo. For instance, S-glutathionylation was found in a proportion of epitopes (Fig. 1C and supplemental Table 1) and was present at between 30 and 50% of the cysteine-containing epitopes that were presented in modified form (Fig. 1D). Many of these peptides were derived from proteins that are known physiological targets of S-glutathionylation, such as ubiquitin ligases (61) and actin (62).
TABLE 3.
DNA typing and serological profiles (boldface type) of cell types used for peptide elution
Note that C1R cells express low levels of Cw4.
| Cell line | Class I | Class II |
|---|---|---|
| Jesthom (BLCL)a | A*02:01, B*27:05, C*01:02 | DRA*01:01, DRB1*01:01, DQA1*01:01, DQB1*05:01, DPA1*01, DPB1*04:01 |
| COX (BLCL) | (A2, B27, Cw1) | |
| BOLETH (BLCL) | A*01:01, B*08:01, C*07:01 | (DR1, DQ5, DP4) |
| SCHU (BLCL) | (A1, B8, Cw7) | DRA*01:02, DRB1*03:01, DRB3*01:01, DQA1*05:01, DQB1*02:01, DPA1*01, DPB1*03:01 |
| STEINLIN (BLCL) | A*02:01,B*1501, C*03:04, | (DR3, DR52, DQ2, DP3) |
| (A2, B62, Cw10) | DRA1*01:01, DRB1*04:01, DRB4*01:03, DQA1*03:01, DQB1*03:02, DPA1*01:03, DPB1*04:01 | |
| A*03:01, B*07:02, C*07:02 | (DR4, DR53, DQ8, DP4) | |
| (A3, B7, Cw7) | DRA*01:02, DRB1*15:01, DRB5*01:01, DQA1*01:02, DQB1*06:02, DPA1*01, DPB1*04:02 | |
| A*01:01, B*08:01, C*07:01 | (DR15, DR51, DQ6, DP4) | |
| (A1, B8, Cw7) | DRA*01:02, DRB1*03:01, DRB3*01:01, DQA1*05:01, DQB1*02:01, DPA1*01, DPB1*03:01/04:01 | |
| 721.221 transfectant | A*01:01 (A1) | (DR3, DR52, DQ2, DP3/4) |
| CIR transfectant | B*1502 (B75, Cw4*) | NDb |
| C1R transfectant | B*2705 (B27, Cw4*) | ND |
| C1R transfectant | B*5701 (B57, Cw4*) | ND |
| C1R transfectant | B*5703 (B57, Cw4*) | ND |
| C1R transfectant | B*5801 (B58, Cw4*) | ND |
| C1R transfectant | B*4403 (B44, Cw4*) | ND |
| NIT-1 | Kd Db | |
| NOD mouse | Kd Db | IAg7 |
a BLCL, B lymphoblastoid cell line.
b ND, not determined.
FIGURE 1.
Identification of cysteine-containing peptides in the immunopeptidome. A, proportion of cysteine-containing epitopes isolated and identified from a panel of B-lymphoblastoid cell lines, NIT-1 cells, and murine tissues. A large data set of peptide sequences (over 70,000 high confidence peptide identities) was identified from immunoaffinity-purified MHC class I and class II molecules. A subset of these peptides originated from cells undergoing some degree of oxidative stress induced by transformation or cytokine exposure. These peptides were isolated from a panel of EBV-transformed B lymphoblastoid cells spanning a variety of MHC alleles (including International Histocompatibility Workshop consanguineous cell lines Jesthom, 9022, 9013, 9031, 9087, and transfectants expressing defined HLA class I allotypes CIR B*27:05 (47), CIR B*44:03 (78), CIR B*57:01 (79), CIR B*58:01 (79), C1R B*57:03 (79), CIR B*15:02 (79); 721.221 A*01:01), murine NIT-1 cells, a pancreatic β cell line that is highly sensitive to IFN-γ treatment, and various murine tissues from NOD mice (1, 2). Global peptide repertoires were assessed by peptide elution and analysis by tandem mass spectrometry, with epitope sequences determined from spectra by the Protein Pilot algorithm. Each point represents a different cell line (see supplemental Table 1). B, proportion of cysteine-containing epitopes bearing oxidative modifications. C, proportion of modified epitopes with S-glutathionylation. D, a pie chart highlighting the proportion of cysteine-containing epitopes bearing specific oxidative modifications.
Viral Infection Creates an Environment for Viral Antigen Modification
The functional consequences of Cys modification in MHC epitopes are ideally studied from the starting point of Cys-containing peptides that are known to induce strong T cell responses. A well characterized example is that of the immunodominant murine H-2Db-restricted epitope CSLWNGPHL, spanning residues 510–518 of the JHMV spike glycoprotein (epitope S510) (39–41, 63). Infection with JHMV results in an acute, fatal encephalitis or a chronic demyelinating disease, depending on the experimental protocol (63, 64). This epitope is prone to oxidation in vitro, necessitating the use of a variant of S510 (bearing a Cys to aminobutyric acid substitution at position 1) in functional and structural studies (39). Given that viral infection can induce cellular oxidative stress (20–27), we initially examined the redox state of JHMV-infected brains to determine whether the infected brain milieu would favor cysteine modification with oxidized glutathione. GSH and GSSG levels were assayed in brains harvested from either naive mice or mice at day 7 post intranasal infection with JHMV (Fig. 2). Infected mice were found to have significantly reduced ratios of reduced (GSH) to oxidized (GSSG) glutathione compared with uninfected controls, suggesting that this is an environment that is conducive for cysteine modification with oxidized glutathione. Therefore, we next investigated whether the S510 epitope was S-glutathionylated during infection using an in vitro system, whereby DC2.4 cells (H-2b-restricted) were infected with a vaccinia virus (VACV) expressing the S510 epitope (VACV-S510) using a sensitive targeted mass spectrometry approach (MRM) (48). This approach was chosen to generate enough material for the analysis because only a small number of antigen presenting cells are infected in the brain, and VACV-S510 reproduces aspects of the infection in mice (40), with ∼4% (Fig. 3A) of splenic CD8+ T cells labeled with a H-2Db·S510-specific tetramer, a percentage similar to that detected after intraperitoneal infection with JHMV (65). Initially, the MRM methodology was validated for the detection of both native S510 and modified GSH-S510 by exogenously loading H-2b+ve DC2.4 cells with synthetic S510 or GSH-S510 (typically with a limit of detection of 500 amol) and incubating cells for 1 h, followed by extensive washing to remove unbound peptide. H-2Db·peptide complexes were immunoprecipitated, and bound peptides were eluted using mild acid. Subsequent LC-MRM-MS analysis confirmed that both forms of S510 could be detected from their respective samples (Fig. 3B), thus establishing the methodology for examining virus-infected samples. Under these conditions, a small proportion of the native peptide was modified by GSH, reflecting endogenous S-glutathionylation. These analyses also suggest that, once modified, GSH-S510 was stable.
FIGURE 2.
Elevated levels of oxidized glutathione in infected mouse brains. Shown are reduced and oxidized glutathione levels and the GSG/GSSG ratio in naive and JHMV-infected brain tissue at day 7 p.i. Measurements were obtained as described under “Experimental Procedures.” Data are from one experiment representative of three independent experiments (mean and S.E. (error bars) with 3 mice/group).
FIGURE 3.
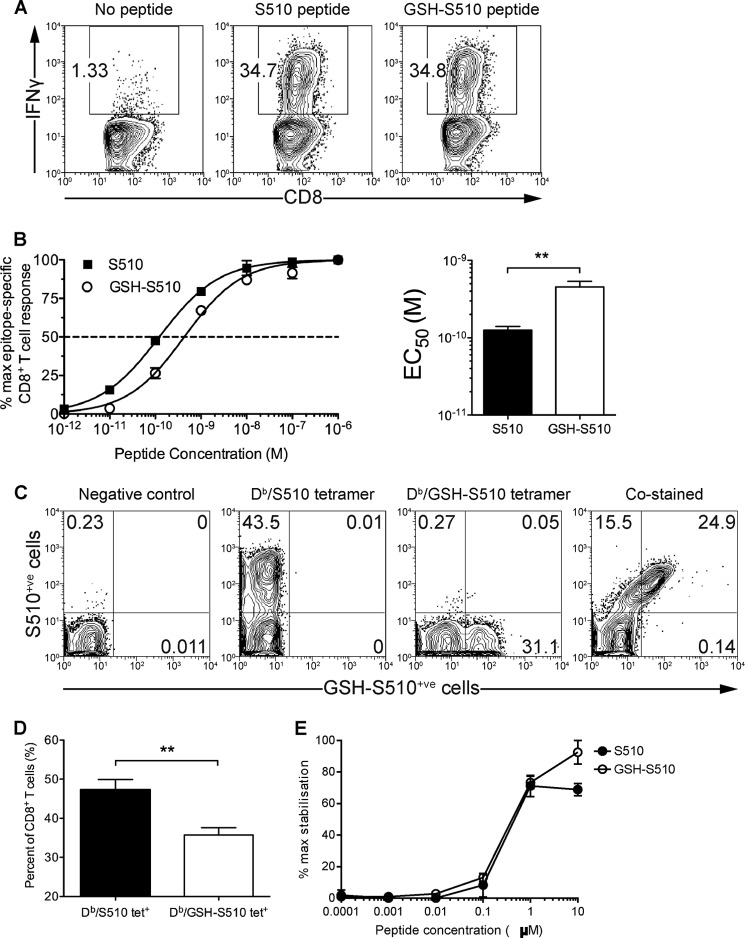
Use of VACV-S510 as a surrogate for the study of S510 presentation and MRM-based detection of native and GSH-modified S510 presentation. A, representative FACS plots of the frequencies of S510-specific T cells elicited in mice infected with recombinant VACV expressing epitope S510 (VACV-S510). Splenocytes were harvested from B6 mice intraperitoneally infected with 2 × 107 pfu of VACV-S510 at day 7 p.i. and stained with a H-2Db·S510 tetramer. Data from one of three mice analyzed are shown. B, detection of S510 and GSH-S510 from DC2.4 cells exogenously loaded with the indicated peptides (1 μm) or unpulsed as a negative control. Data show MRM traces (Q1 → Q3 transitions as indicated) for both forms of the peptide. A small amount of native S510 was also detected in the GSH-S510-pulsed sample because the in vitro conversion of synthetic S510 to GSH-S510 is an incomplete reaction. C, detection of S510 and GSH-S510 eluted from H-2Db·peptide complexes at 4 h p.i. with VACV-S510. MRM trace transitions are labeled as in B. Data are from one of three independent experiments. D, further confirmation of sequence and modification of S510 was obtained through MRM-triggered MS/MS sequencing scans (bottom panels) compared with those from the corresponding synthetic peptide (top panels). Representative sequencing ions are labeled.
Analysis of peptides eluted from DC2.4 cells at 4 h p.i. by targeted LC-MRM-MS revealed the presence of significant quantities of both native S510 and modified GSH-S510 in a roughly 50:1 proportion (Fig. 3C). Further confirmation of the presence of these two peptide species was achieved by an MRM-triggered MS/MS sequencing scan (Fig. 3D), where the MS/MS spectra obtained for the endogenously processed peptides was compared with that obtained for synthetic S510 and GSH-S510.
GSH-S510 Is Only Recognized by a Proportion of Brain-derived CD8 T Cells
Following confirmation that VACV-S510 infection leads to native and modified S510 epitope presentation in vitro, and given that T cell responses to S510 can be generated by the same virus in vivo, we reasoned that JHMV infection would indeed lead to presentation of GSH-S510. Therefore, we determined whether CD8+ T cells in the brains of JHMV-infected mice recognized the H-2Db·GSH-S510 epitope and, in doing so, assessed what impact S-glutathionylation of S510 had upon T cell recognition. We observed that a similar percentage of CD8+ T cells produced IFN-γ upon direct ex vivo stimulation with saturating concentrations of either GSH-S510 or S510 peptide (Fig. 4A). The response of CD8+ T cells to stimulation with saturating concentrations of peptide does not reflect the responsiveness (functional avidity) of the cells to physiological levels of presented peptide. Therefore, we determined the responsiveness of the brain-derived CD8+ T cells to titrated amounts of GSH-S510 or S510 peptide by measuring IFN-γ production upon stimulation and calculating the concentration of each peptide needed to reach a half-maximum response (EC50) (Fig. 4B). GSH-S510 peptide was less potent than S510 peptide in inducing IFN-γ responses (EC50 = 1.3 × 10−10 m versus 4.6 × 10−10 m, respectively) (Fig. 4B), suggesting that S-glutathionylation of S510 moderately reduced CD8+ T cell recognition.
FIGURE 4.
Overlapping subsets of CD8+ T cells in the JHMV-infected brain recognize epitopes S510 and GSH-S510. A, representative FACS plots of the frequencies of brain-derived CD8+ T cells producing IFN-γ upon stimulation with either S510 or GSH-S510 peptide. Brain-derived mononuclear cells were isolated from the brains of mice infected with JHMV (4 × 104 pfu intranasally) at day 7 p.i. Cells were stimulated ex vivo with either 1 μm S510 or GSH-S510 peptide and analyzed for IFN-γ production. B, functional avidities of brain-derived CD8+ T cells specific for H-2Db·S510 or H-2Db·GSH-S510. Brain-derived cells were stimulated ex vivo with 10-fold dilutions of either S510 or GSH-S510 peptide and analyzed for intracellular IFN-γ production. Data were normalized to the maximum frequency of epitope-specific CD8+ T cells detected. Peptide concentration curves were analyzed to fit to sigmoidal concentration-response curves and used to calculate EC50 values. Data are from one experiment representative of three independent experiments (mean and S.E. (error bars) with 4 mice/group; **, p < 0.01). C, representative FACS plots of the frequencies of brain-derived CD8+ T cells specific for H-2Db·S510 and H-2Db·GSH-S510. CNS leukocytes isolated from JHMV-infected brains at day 7 p.i. were incubated without tetramer (negative control), stained with H-2Db·S510 tetramer or H-2Db·GSH-S510 tetramer, or co-stained with both tetramers. D, frequencies of H-2Db·S510 tetramer-positive or H-2Db·GSH-S510 tetramer-positive CD8+ T cells isolated form JHMV-infected brains at day 7 p.i. (mean and S.E. with 8 mice/group; **, p < 0.01). E, RMA-S epitope stabilization assay examining S510 and GSH-S510 shows equivalent binding to H-2Db.
Because it is possible that the JHMV-specific CD8+ T cell response consisted of cells specific to S510 or GSH-S510 or dually specific to both forms of S510, we examined whether CD8+ T cells were composed of discrete populations by co-staining cells with H-2Db·GSH-S510 and H-2Db·S510 tetramers. Initially, we stained CNS leukocytes isolated from JHMV-infected brains with allophycocyanin- or phycoerythrin-conjugated H-2Db tetramers loaded with either GSH-S510 or S510 peptide, respectively. On average, 48% of the CD8+ T cells present in the brain labeled with H-2Db·S510 tetramer, whereas only 36% stained with H-2Db·GSH-S510 tetramer, again suggesting that S-glutathionylation of S510 diminished CD8+ T cell recognition (Fig. 4, C and D). Moreover, when CD8+ T cells were co-stained with the tetramers, ∼40% of H-2Db·S510 tetramer+ cells were not labeled with H-2Db·GSH-S510 tetramer, indicating that a considerable fraction of the cells recognize only the native form of S510. Virtually no cells were labeled solely with the H-Db·GSH-S510 tetramer (Fig. 4C). Notably, there was also no distinct population of cells labeled with H-2Db·S510 tetramer alone, suggesting that S-glutathionylation did not completely abrogate recognition by S510-specific CD8 T cells. Rather, the modification variably affected H-2Db·S510 interaction with a heterogeneous population of S510-specific CD8 T cells. This incomplete loss of binding after S-glutathionylation may also explain the apparently discordant results observed in Fig. 4, A and C, because the saturating amounts of peptide used in Fig. 4A could compensate for diminished H-2Db·S510/TCR avidity, despite both peptides exhibiting equivalent binding to H-2Kb (Fig. 4E). Differences between numbers of cells detected by MHCI/peptide binding and IFN-γ expression after peptide stimulation have been observed in other settings. For example, the subdominant CD8 T cell epitope recognized in B6 mice elicits a low functional avidity response (39, 66, 67) that is detected by IFN-γ expression after peptide stimulation. However, the corresponding H-2Kb·S598 is able to detect only a minor fraction of these cells (data not shown).
The Structure of GSH-S510 Bound to H-2Db
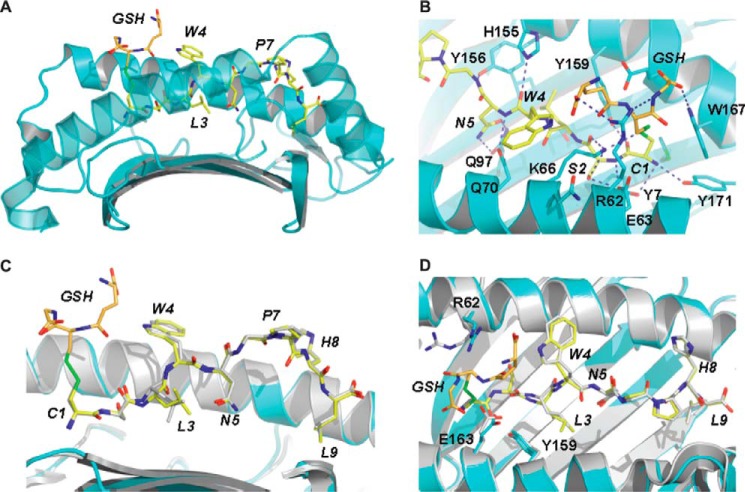
To evaluate the structural consequences of the presentation of glutathionylated S510, the structure of the H-2Db·GSH-S510 binary complex was determined to 2.8 Å resolution (see Table 2 for data collection and refinement statistics). Initial refinement revealed clear unbiased density for the peptide with a covalent modification at P1. In complex with H-2Db, the GSH-S510 peptide adopts an extended conformation equivalent to that observed for the native peptide complex (Protein Data Bank entry 2ZOK) (39), with a kink about the proline at position 7. Briefly, the side chains of Asn(P)5 and Leu(P)9 point down toward the floor of the cleft, whereas those of Cys(P)1, Trp(P)4, Pro(P)7, and His(P)8 point out of the cleft, and those of Ser(P)2 and Leu(P)3 are directed toward the α1 and α2 helices, respectively. The side chains of Trp(P)4, Pro(P)7, and His(P)8 are highly solvent-exposed and therefore in a position to play important roles in TCR recognition. Intramolecular contacts between the peptide side chains of Leu(P)3 and Asn(P)5 are consistent with a Type III constrained conformation (68) about the prominent TCR contact residue Trp(P)4 (39). The GSH molecule is covalently linked to Cys(P)1 via a disulfide bond and extends prominently out of the cleft, with its backbone lying roughly parallel to that of the peptide but with its N and C termini adopting the opposite orientation to the peptide termini (Fig. 5A). In this position, GSH masks Cys(P)1–Leu(P)3 of the peptide and dominates the landscape of the N-terminal region of the complex. Anchoring of the GSH-S510 peptide to the cleft of H-2Db is mediated primarily by Cys(P)1, Ser(P)2, Asn(P)5, and Leu(P)9 (see Table 4), which, in addition to multiple van der Waals contacts, also form a number of hydrogen bond interactions with the heavy chain (Table 4 and Fig. 5B). The glutathione tripeptide forms multiple van der Waals contacts with the heavy chain of H-2Db as well as with the peptide residues Cys(P)1 and Trp(P)4. In addition, its C-terminal glycine residue forms hydrogen bond contacts with Arg62 and Trp167, whereas its N-terminal carboxylate side chain is able to form a salt bridge with the guanidinium group of Arg62.
FIGURE 5.
Overview of the structure of H-2Db·GSH-S510 and comparison against the native H-2Db·S510 complex. A, the antigen binding cleft of H-2Db in complex with the GSH-CSLWNGPHL peptide. The heavy chain is drawn in schematic format and colored cyan. Glutathione and peptide residues are represented as orange and yellow sticks, respectively. B, detail of the antigen-binding cleft, highlighting the contacts formed between H-2Db and the N-terminal half of the glutathionylated peptide. Heavy chain residues interacting with the ligand are shown in stick format. Hydrogen bonds are indicated by blue dashes. C and D, superposition of the structures of H-2Db in complex with the glutathionylated and non-glutathionylated forms of the CSLWNGPHL peptide. The superposition is based on main chain atoms of the peptide and residues 3–175 of the heavy chain of H-2Db. The large conformational changes displayed by the peptide residue Leu-3 and H-2Db residues Arg62 and Glu163 can be accounted for in terms of the glutathione modification. For clarity, the α2 helix has been rendered in semitransparent mode in A and removed in C. In all panels, the ligand residues are labeled in italic type.
TABLE 4.
Ligand contacts in the H2Db·GSH-S510 binary structure
| Ligand residue | Interaction | Contacta,b |
|---|---|---|
| GSH | VDWc | Cys-1, Trp-4, Arg-62, Glu-63, Lys-66, Glu-163, Trp-167 |
| GSHN3, O32 | HBd | Arg-62Nη2, Trp-167Nϵ1 |
| GSHO11, O12 | SBe | Arg-62Nη1 |
| GSHSγ2 | SSf | Cys-1Sγ |
| Cys-1 | VDW | GSH, Met-5, Tyr-7, Glu-63, Lys-66, Tyr-159, Glu-163, Trp-167, Tyr-171 |
| Cys-1N,O | HB | Tyr-7Oη, Tyr-159Oη, Tyr-171Oη |
| Cys-1Sγ | HB | Lys-66Nζ |
| Cys-1Sγ | SS | GSHSγ2 |
| Ser-2 | VDW | Tyr-7, Tyr-45, Glu-63, Lys-66, Tyr-159 |
| Ser-2N,O | HB | Glu-63Oϵ1, Lys-66Nζ |
| Ser-2Oγ | HB | Glu-63Oϵ1, Lys-66Nζ |
| Leu-3 | VDW | Asn-5, Gln-70, Gln-97, Ser-99, Leu-114, Tyr-156, Tyr-159 |
| Trp-4 | VDW | GSH, Gln-65, Lys-66, Gly-69, Gln-70, His-155 |
| Trp-4O | HB | His-155Nϵ2 |
| Asn-5 | VDW | Leu-3, Gln-70, Trp-73, Gln-97, Phe-116, Tyr-156 |
| Asn-5N | HB | Gln-70Oϵ1 |
| Asn-5Oδ1, Nδ2 | HB | Gln-97Oϵ1, Nϵ2 |
| Gly-6 | VDW | Trp-73, Tyr-156 |
| Gly-6N,O | HB | Trp-73Nϵ1, Tyr-156Oη |
| Pro-7 | VDW | Trp-73, Trp-147, Ser-150, Ala-152 |
| Pro-7O | HB | Trp-73Nϵ1 |
| Pro-7O | Smg | Tyr-156Oη |
| His-8 | VDW | Trp-73, Val-76, Ser-77, Lys-146, Trp-147 |
| His-8O | HB | Lys-146Nζ, Trp-147Nϵ1 |
| His-8Nδ1 | Sm | Asn-80Nδ2, Lys-146Nζ |
| Leu-9 | VDW | Trp-73, Ser-77, Asn-80, Leu-81, Tyr-84, Leu-95, Tyr-123, Thr-143, Lys-146, Trp-147 |
| Leu-9N,O,OXT | HB, SB | Ser-77Oγ, Asn-80Nδ2, Tyr-84Oη, Thr-143Oγ1, Lys-146Nζ |
a Atomic contacts determined using the CCP4i implementation of CONTACT and a cut-off of 4.0 Å.
b Selected contacts between ligand residues are marked in italic type.
c VDW, Van der Waals' interactions, defined as non-hydrogen bond contact distances of 4.0 Å or less.
d HB, hydrogen bonds, defined as contact distances of 3.5 Å or less between suitable atoms at appropriate angles.
e SB, ionic interactions, defined as contact distances of 4.0 Å or less between suitable atoms.
f SS, disulfide bond.
g Sm, hydrogen bond mediated by one solvent molecule.
The antigen binding cleft of H-2Db in the glutathionylated structure adopts a conformation that is highly conserved with respect to that observed in the non-glutathionylated S510 complex (Protein Data Bank entry 2ZOK) (39), and the α1/α2 domain of the glutathionylated structure superimposes with individual pMHC copies of 2ZOK with an average root mean square deviation of 0.28 Å for CA atoms. Moreover, in the two structures, the CSLWNGPHL peptide adopts an equivalent conformation, with an average root mean square deviation between copies of 0.51 Å for all peptide atoms (Fig. 5, C and D). The only significant conformational difference in the peptide is observed at position Leu3, the side chain of which displays maximum deviations of up to 2.4 Å. This large side chain movement appears to be the result of the glutathione modification, which through interactions with the side chain of Trp(P)4 forces the peptide backbone down into the cleft. The conformational change in the side chain of Leu(P)3 prevents steric clashes with the heavy chain's Ser99. The GSH modification also directly induces side chain movements in the H-2Db residues Arg62 and Glu163 that are up to several Å in magnitude (Fig. 5D).
In addition to these local conformational changes in the pMHC, the glutathione modification caps the N-terminal region of the cleft, creating a TCR epitope that is unique to that observed in the native H-2Db·S510 structure, both in terms of shape and electrostatic properties. Despite this prominent structural change, not all T cell recognition of the H-2Db·GSH-S510 was perturbed, with 60% of native epitope-specific T cells capable of cross-reacting, as assessed by tetramer binding (Fig. 4D), reflecting the predominantly central focus of anti-S510 TCRs (39, 41). The remaining 40% of cytotoxic T lymphocytes presumably have a more N-terminal focus, and their recognition was ablated, demonstrating that a previously unrecognized potential consequence of virus-induced stress is evasion of the T cell response.
DISCUSSION
Cysteine residues are highly susceptible to oxidative modifications that are manifested through a variety of redox reactions and that are largely dependent on the nature of the antioxidant the protein encounters. For example, in the presence of reactive oxygen species, mono-, di-, and trioxidation to sulfenic, sulfinic, and sulfonic acid, respectively, are prevalent. In the presence of reactive nitrogen species, S-nitrosylation or S-glutathionylation is predominant, with the latter arising from the detoxifying action of the abundant natural antioxidant glutathione. In recent years, it has become apparent that these modifications are not merely the footprint of oxidative stress but are also involved in the redox modulation of protein structure and function. Here we show that these dynamic modifications of cysteine residues are represented within the immunopeptidome, presumably following processing of modified antigens. Thus, the presentation of cysteine-containing peptides by MHC class I and class II molecules provides an opportunity to communicate the redox state of the cell to the immune system. By interrogating a large database of naturally processed and presented peptides bound to various MHC allotypes, we demonstrated that the prevalence of cysteine-containing peptides varies between alleles but, on average, largely reflects the cysteine usage within the proteome of host cells and pathogens. Importantly, 20–30% of these peptides contained modified cysteine residues, a high proportion of which bore a specific enzyme-catalyzed modification: S-glutathionylation. This represents the first large scale and systematic investigation of the cysteine-containing immunopeptidome and reflects the chemical diversity achieved through oxidative modification of cysteine residues.
Upon infection, a complex interplay of host-mediated defense and virus-mediated countermechanisms are elicited that dictate the outcome of the infection. Viral infection induces the unfolded protein response and oxidative stress. Although dictating the productivity of viral replication, little is known about how such generic responses to virus-induced stress impact the ability of adaptive immunity to respond to virally infected cells. Depletion of antioxidants is a common theme during viral infection (30, 33, 34, 36, 37, 57), leading to an imbalance of cellular GSH/GSSG and, under some circumstances, an increase in protein S-glutathionylation (30). Here we demonstrate that during infection with JHMV, a marked depletion in GSH is observed, leading to conditions in the brain of infected mice conducive to S-glutathionylation of antigens. Modifications in GSH and GSSG resulting from increased oxidative stress have been reported previously in cells infected with hepatitis C virus (30, 31), but to our knowledge, our study is the first to describe a difference in infected animals. Our results show that there is a statistically significant increase in the GSSG/GSH ratio in the brains of JHMV-infected mice, but additional work will be required to determine quantitatively the difference in this ratio that is predictive of a biologically significant effect. However, our results suggest such an effect because the Cys-containing immunodominant T cell epitope, S510, derived from the spike glycoprotein (S) of JHMV is presented in an unmodified (S510) or glutathionylated (S510-GSH) form in a redox-sensitive manner. This indicates that presentation of this epitope to T cells informs the host of not only viral infection but also of the redox state of the infected antigen presenting cells. Antigen pulsing experiments showed a requirement for intracellular processing of the antigen for this modification to occur because the addition of native S510 peptide to cells in culture did not lead to significant production of the GSH-S510 epitope.
T cells can be detected in infected mice that recognize the native form of this epitope or that cross-react toward both, suggesting a complex relationship between antigen presentation and oxidative stress in infected cells. We could not detect CD8 T cells that recognized only glutathionylated S510. One explanation for this is that glutathionylation occurs during peptide processing and loading onto the MHCI molecule. If epitope presentation occurred in JHMV-infected mice primarily via cross-presentation, cell stress and subsequent glutathionylation might occur to a lesser degree than in infected cells. The net result would be priming only to the native epitope, resulting in the lack of outgrowth of T cells responding only to GSH-S510.
The cross-recognition of both S510 and GSH-S510 by T cells can be appreciated from our structure and the previously reported bias in T cell recognition of the central regions of the S510 epitope (39, 41). Approximately 15–35% of the T cell response, as assessed by MHC class I/peptide tetramer binding, is blunted by this modification; one would expect that modifications of cysteines that directly interacted with TCR would even more significantly impact recognition by native epitope-specific cells, possibly contributing to evasion of the T cell response. CD8 T cell escape is common in JHMV and other viral infections, including those caused by HIV and hepatitis C virus (69, 70), and generally involves mutations that inhibit epitope recognition or processing. Our data indicate that induction of the stress response, with subsequent post-translational modification of Cys residues, may have consequences for T cell escape. We speculate that this may be a common theme in viral infections and other pathologies, such as tumors. If the redox state of a cell is altered such that Cys-containing epitopes are modified, then cognate T cell responses may be diminished if there is insufficient cross-reactivity. Indeed, previous reports indicated that the oxidation state of Cys residues in candidate tumor peptide vaccines has influenced the anti-peptide CD4 and CD8 responses. However, these effects vary from demonstrating reduced responses due to oxidation (71) to those showing recognition of both the monomeric or Cys-Cys dimerized epitope (72). In one study, Campos-Perez et al. (73) argued that the use of DNA vaccines circumvented the concerns over spontaneous modification of synthetic epitopes, with any post-translational modifications mirroring those that would be found on the endogenous tumor epitope.
T cell epitopes bearing other cysteine modifications have been reported (74, 75), yet the mechanism leading to their generation has remained elusive. Cysteinylation of T cell epitopes has been documented (18, 75–77); however, its precise biological role is uncertain, with the possibility that oxidized cysteine, present in cell culture media, results in this modification. Unlike cysteinylation, S-glutathionylation is a common modification of cysteine thiols in response to oxidative stress.
In summary, we have mapped the subset of the immunopeptidome that contains redox-sensitive cysteine residues and show that a high proportion of cysteine-containing peptides are oxidatively modified. The extent to which cysteine-containing peptides are selected for presentation and the extent to which they bear modifications are allele-dependent. The prevalence and chemical diversity of this subset of MHC ligands reiterates the importance of this often neglected class of peptides in immunity. In addition, we demonstrate that viral infection modulates the redox balance and skews the GSH/GSSG ratio, promoting S-glutathionylation, an observation consistent with other infectious systems. This represents an effective mechanism for the cell to communicate oxidative stress to the immune system and to impact the host-virus axis. Consequent modulation of antigen presentation may have important implications for not only antiviral immunity and vaccine development but also immunity to other pathogens that provoke stress responses and to cancers where oxidative stress is induced during tumorigenesis.
Supplementary Material
Acknowledgments
Measurements of glutathione levels were performed by the Radiation and Free Radical Research Core Facility at the University of Iowa.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 NS036592. This work was also supported by an infrastructure grant (Grant LE100100036) from the Australian Research Council (ARC) and a project grant from the Juvenile Diabetes Research Foundation (17-2012-134).

This article contains supplemental Table 1.
- JHMV
- JHM strain of mouse hepatitis virus
- MRM
- multiple reaction monitoring
- p.i.
- postinfection
- VACV
- vaccinia virus
- TCR
- T cell receptor.
REFERENCES
- 1. Dudek N. L., Tan C. T., Gorasia D. G., Croft N. P., Illing P. T., Purcell A. W. (2012) Constitutive and inflammatory immunopeptidome of pancreatic beta-cells. Diabetes 61, 3018–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Scull K. E., Dudek N. L., Corbett A. J., Ramarathinam S. H., Gorasia D. G., Williamson N. A., Purcell A. W. (2012) Secreted HLA recapitulates the immunopeptidome and allows in-depth coverage of HLA A*02:01 ligands. Mol. Immunol. 51, 136–142 [DOI] [PubMed] [Google Scholar]
- 3. Mohammed F., Cobbold M., Zarling A. L., Salim M., Barrett-Wilt G. A., Shabanowitz J., Hunt D. F., Engelhard V. H., Willcox B. E. (2008) Phosphorylation-dependent interaction between antigenic peptides and MHC class I: a molecular basis for the presentation of transformed self. Nat. Immunol. 9, 1236–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Petersen J., Wurzbacher S. J., Williamson N. A., Ramarathinam S. H., Reid H. H., Nair A. K., Zhao A. Y., Nastovska R., Rudge G., Rossjohn J., Purcell A. W. (2009) Phosphorylated self-peptides alter human leukocyte antigen class I-restricted antigen presentation and generate tumor-specific epitopes. Proc. Natl. Acad. Sci. U.S.A. 106, 2776–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zarling A. L., Polefrone J. M., Evans A. M., Mikesh L. M., Shabanowitz J., Lewis S. T., Engelhard V. H., Hunt D. F. (2006) Identification of class I MHC-associated phosphopeptides as targets for cancer immunotherapy. Proc. Natl. Acad. Sci. U.S.A. 103, 14889–14894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Depontieu F. R., Qian J., Zarling A. L., McMiller T. L., Salay T. M., Norris A., English A. M., Shabanowitz J., Engelhard V. H., Hunt D. F., Topalian S. L. (2009) Identification of tumor-associated, MHC class II-restricted phosphopeptides as targets for immunotherapy. Proc. Natl. Acad. Sci. U.S.A. 106, 12073–12078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Petersen J., Purcell A. W., Rossjohn J. (2009) Post-translationally modified T cell epitopes: immune recognition and immunotherapy. J. Mol. Med. 87, 1045–1051 [DOI] [PubMed] [Google Scholar]
- 8. Suzuki A., Yamada R., Chang X., Tokuhiro S., Sawada T., Suzuki M., Nagasaki M., Nakayama-Hamada M., Kawaida R., Ono M., Ohtsuki M., Furukawa H., Yoshino S., Yukioka M., Tohma S., Matsubara T., Wakitani S., Teshima R., Nishioka Y., Sekine A., Iida A., Takahashi A., Tsunoda T., Nakamura Y., Yamamoto K. (2003) Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat. Genet. 34, 395–402 [DOI] [PubMed] [Google Scholar]
- 9. van Venrooij W. J., van Beers J. J., Pruijn G. J. (2011) Anti-CCP antibodies: the past, the present and the future. Nat. Rev. Rheumatol. 7, 391–398 [DOI] [PubMed] [Google Scholar]
- 10. Scally S. W., Petersen J., Law S. C., Dudek N. L., Nel H. J., Loh K. L., Wijeyewickrema L. C., Eckle S. B., van Heemst J., Pike R. N., McCluskey J., Toes R. E., La Gruta N. L., Purcell A. W., Reid H. H., Thomas R., Rossjohn J. (2013) A molecular basis for the association of the HLA-DRB1 locus, citrullination, and rheumatoid arthritis. J. Exp. Med. 210, 2569–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Anderson R. P., Degano P., Godkin A. J., Jewell D. P., Hill A. V. (2000) In vivo antigen challenge in celiac disease identifies a single transglutaminase-modified peptide as the dominant A-gliadin T-cell epitope. Nat. Med. 6, 337–342 [DOI] [PubMed] [Google Scholar]
- 12. Henderson K. N., Tye-Din J. A., Reid H. H., Chen Z., Borg N. A., Beissbarth T., Tatham A., Mannering S. I., Purcell A. W., Dudek N. L., van Heel D. A., McCluskey J., Rossjohn J., Anderson R. P. (2007) A structural and immunological basis for the role of human leukocyte antigen DQ8 in celiac disease. Immunity 27, 23–34 [DOI] [PubMed] [Google Scholar]
- 13. Qiao S. W., Sollid L. M., Blumberg R. S. (2009) Antigen presentation in celiac disease. Curr. Opin. Immunol. 21, 111–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Broughton S. E., Petersen J., Theodossis A., Scally S. W., Loh K. L., Thompson A., van Bergen J., Kooy-Winkelaar Y., Henderson K. N., Beddoe T., Tye-Din J. A., Mannering S. I., Purcell A. W., McCluskey J., Anderson R. P., Koning F., Reid H. H., Rossjohn J. (2012) Biased T cell receptor usage directed against human leukocyte antigen DQ8-restricted gliadin peptides is associated with celiac disease. Immunity 37, 611–621 [DOI] [PubMed] [Google Scholar]
- 15. Petersen J., Montserrat V., Mujico J. R., Loh K. L., Beringer D. X., van Lummel M., Thompson A., Mearin M. L., Schweizer J., Kooy-Winkelaar Y., van Bergen J., Drijfhout J. W., Kan W. T., La Gruta N. L., Anderson R. P., Reid H. H., Koning F., Rossjohn J. (2014) T-cell receptor recognition of HLA-DQ2-gliadin complexes associated with celiac disease. Nat. Struct. Mol. Biol. 21, 480–488 [DOI] [PubMed] [Google Scholar]
- 16. Mannering S. I., Harrison L. C., Williamson N. A., Morris J. S., Thearle D. J., Jensen K. P., Kay T. W., Rossjohn J., Falk B. A., Nepom G. T., Purcell A. W. (2005) The insulin A-chain epitope recognized by human T cells is posttranslationally modified. J. Exp. Med. 202, 1191–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mannering S. I., Pang S. H., Williamson N. A., Naselli G., Reynolds E. C., O'Brien-Simpson N. M., Purcell A. W., Harrison L. C. (2009) The A-chain of insulin is a hot-spot for CD4+ T cell epitopes in human type 1 diabetes. Clin. Exp. Immunol. 156, 226–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Webb A. I., Dunstone M. A., Chen W., Aguilar M. I., Chen Q., Jackson H., Chang L., Kjer-Nielsen L., Beddoe T., McCluskey J., Rossjohn J., Purcell A. W. (2004) Functional and structural characteristics of NY-ESO-1-related HLA A2-restricted epitopes and the design of a novel immunogenic analogue. J. Biol. Chem. 279, 23438–23446 [DOI] [PubMed] [Google Scholar]
- 19. Purcell A. W., McCluskey J., Rossjohn J. (2007) More than one reason to rethink the use of peptides in vaccine design. Nat. Rev. Drug Discov. 6, 404–414 [DOI] [PubMed] [Google Scholar]
- 20. Ambrose R. L., Mackenzie J. M. (2011) West Nile virus differentially modulates the unfolded protein response to facilitate replication and immune evasion. J. Virol. 85, 2723–2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Burnett H. F., Audas T. E., Liang G., Lu R. R. (2012) Herpes simplex virus-1 disarms the unfolded protein response in the early stages of infection. Cell Stress Chaperones 17, 473–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. DeDiego M. L., Nieto-Torres J. L., Jiménez-Guardeño J. M., Regla-Nava J. A., Alvarez E., Oliveros J. C., Zhao J., Fett C., Perlman S., Enjuanes L. (2011) Severe acute respiratory syndrome coronavirus envelope protein regulates cell stress response and apoptosis. PLoS Pathog. 7, e1002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hassan I. H., Zhang M. S., Powers L. S., Shao J. Q., Baltrusaitis J., Rutkowski D. T., Legge K., Monick M. M. (2012) Influenza A viral replication is blocked by inhibition of the inositol-requiring enzyme 1 (IRE1) stress pathway. J. Biol. Chem. 287, 4679–4689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Merquiol E., Uzi D., Mueller T., Goldenberg D., Nahmias Y., Xavier R. J., Tirosh B., Shibolet O. (2011) HCV causes chronic endoplasmic reticulum stress leading to adaptation and interference with the unfolded protein response. PLoS One 6, e24660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Qian Z., Xuan B., Chapa T. J., Gualberto N., Yu D. (2012) Murine cytomegalovirus targets transcription factor ATF4 to exploit the unfolded-protein response. J. Virol. 86, 6712–6723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saeed M., Suzuki R., Watanabe N., Masaki T., Tomonaga M., Muhammad A., Kato T., Matsuura Y., Watanabe H., Wakita T., Suzuki T. (2011) Role of the endoplasmic reticulum-associated degradation (ERAD) pathway in degradation of hepatitis C virus envelope proteins and production of virus particles. J. Biol. Chem. 286, 37264–37273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Trujillo-Alonso V., Maruri-Avidal L., Arias C. F., López S. (2011) Rotavirus infection induces the unfolded protein response of the cell and controls it through the nonstructural protein NSP3. J. Virol. 85, 12594–12604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lawless M. W., Greene C. M. (2012) Toll-like receptor signalling in liver disease: ER stress the missing link? Cytokine 59, 195–202 [DOI] [PubMed] [Google Scholar]
- 29. Gelderman K. A., Hultqvist M., Olsson L. M., Bauer K., Pizzolla A., Olofsson P., Holmdahl R. (2007) Rheumatoid arthritis: the role of reactive oxygen species in disease development and therapeutic strategies. Antioxid. Redox Signal. 9, 1541–1567 [DOI] [PubMed] [Google Scholar]
- 30. Mieyal J. J., Chock P. B. (2012) Posttranslational modification of cysteine in redox signaling and oxidative stress: focus on S-glutathionylation. Antioxid. Redox Signal. 16, 471–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mieyal J. J., Gallogly M. M., Qanungo S., Sabens E. A., Shelton M. D. (2008) Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxid. Redox Signal. 10, 1941–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shelton M. D., Mieyal J. J. (2008) Regulation by reversible S-glutathionylation: molecular targets implicated in inflammatory diseases. Mol. Cells 25, 332–346 [PMC free article] [PubMed] [Google Scholar]
- 33. Buhl R., Jaffe H. A., Holroyd K. J., Wells F. B., Mastrangeli A., Saltini C., Cantin A. M., Crystal R. G. (1989) Systemic glutathione deficiency in symptom-free HIV-seropositive individuals. Lancet 2, 1294–1298 [DOI] [PubMed] [Google Scholar]
- 34. Gong G., Waris G., Tanveer R., Siddiqui A. (2001) Human hepatitis C virus NS5A protein alters intracellular calcium levels, induces oxidative stress, and activates STAT-3 and NF-κB. Proc. Natl. Acad. Sci. U.S.A. 98, 9599–9604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Abdalla M. Y., Ahmad I. M., Spitz D. R., Schmidt W. N., Britigan B. E. (2005) Hepatitis C virus-core and non structural proteins lead to different effects on cellular antioxidant defenses. J. Med. Virol. 76, 489–497 [DOI] [PubMed] [Google Scholar]
- 36. Gonzalez-Dosal R., Horan K. A., Rahbek S. H., Ichijo H., Chen Z. J., Mieyal J. J., Hartmann R., Paludan S. R. (2011) HSV infection induces production of ROS, which potentiate signaling from pattern recognition receptors: role for S-glutathionylation of TRAF3 and 6. PLoS Pathog. 7, e1002250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Davis D. A., Brown C. A., Newcomb F. M., Boja E. S., Fales H. M., Kaufman J., Stahl S. J., Wingfield P., Yarchoan R. (2003) Reversible oxidative modification as a mechanism for regulating retroviral protease dimerization and activation. J. Virol. 77, 3319–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ghezzi P., Romines B., Fratelli M., Eberini I., Gianazza E., Casagrande S., Laragione T., Mengozzi M., Herzenberg L. A., Herzenberg L. A. (2002) Protein glutathionylation: coupling and uncoupling of glutathione to protein thiol groups in lymphocytes under oxidative stress and HIV infection. Mol. Immunol. 38, 773–780 [DOI] [PubMed] [Google Scholar]
- 39. Butler N. S., Theodossis A., Webb A. I., Dunstone M. A., Nastovska R., Ramarathinam S. H., Rossjohn J., Purcell A. W., Perlman S. (2008) Structural and biological basis of CTL escape in coronavirus-infected mice. J. Immunol. 180, 3926–3937 [DOI] [PubMed] [Google Scholar]
- 40. Castro R. F., Perlman S. (1995) CD8+ T-cell epitopes within the surface glycoprotein of a neurotropic coronavirus and correlation with pathogenicity. J. Virol. 69, 8127–8131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pewe L., Perlman S. (1999) Immune response to the immunodominant epitope of mouse hepatitis virus is polyclonal, but functionally monospecific in C57Bl/6 mice. Virology 255, 106–116 [DOI] [PubMed] [Google Scholar]
- 42. Anghelina D., Pewe L., Perlman S. (2006) Pathogenic role for virus-specific CD4 T cells in mice with coronavirus-induced acute encephalitis. Am. J. Pathol. 169, 209–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Anderson M. E. (1985) Determination of glutathione and glutathione disulfide in biological samples. Methods Enzymol. 113, 548–555 [DOI] [PubMed] [Google Scholar]
- 44. Griffith O. W. (1980) Determination of glutathione and glutathione disulfide using glutathione reductase and 2-vinylpyridine. Anal. Biochem. 106, 207–212 [DOI] [PubMed] [Google Scholar]
- 45. Zhao J., Zhao J., Perlman S. (2009) De novo recruitment of antigen-experienced and naive T cells contributes to the long-term maintenance of antiviral T cell populations in the persistently infected central nervous system. J. Immunol. 183, 5163–5170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Webb A. I., Dunstone M. A., Williamson N. A., Price J. D., de Kauwe A., Chen W., Oakley A., Perlmutter P., McCluskey J., Aguilar M. I., Rossjohn J., Purcell A. W. (2005) T cell determinants incorporating β-amino acid residues are protease resistant and remain immunogenic in vivo. J. Immunol. 175, 3810–3818 [DOI] [PubMed] [Google Scholar]
- 47. Purcell A. W., Gorman J. J., Garcia-Peydró M., Paradela A., Burrows S. R., Talbo G. H., Laham N., Peh C. A., Reynolds E. C., López De Castro J. A., McCluskey J. (2001) Quantitative and qualitative influences of tapasin on the class I peptide repertoire. J. Immunol. 166, 1016–1027 [DOI] [PubMed] [Google Scholar]
- 48. Tan C. T., Croft N. P., Dudek N. L., Williamson N. A., Purcell A. W. (2011) Direct quantitation of MHC-bound peptide epitopes by selected reaction monitoring. Proteomics 11, 2336–2340 [DOI] [PubMed] [Google Scholar]
- 49. Wu G. F., Dandekar A. A., Pewe L., Perlman S. (2000) CD4 and CD8 T cells have redundant but not identical roles in virus-induced demyelination. J. Immunol. 165, 2278–2286 [DOI] [PubMed] [Google Scholar]
- 50. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 51. Storoni L. C., McCoy A. J., Read R. J. (2004) Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 60, 432–438 [DOI] [PubMed] [Google Scholar]
- 52. Afonine P. V., Grosse-Kunstleve R. W., Adams P. D. (2005) A robust bulk-solvent correction and anisotropic scaling procedure. Acta Crystallogr. D Biol. Crystallogr. 61, 850–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 54. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 55. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Collaborative Computational Project, Number 4 (1994) The CCP4 Suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 57. Lassoued S., Ben Ameur R., Ayadi W., Gargouri B., Ben Mansour R., Attia H. (2008) Epstein-Barr virus induces an oxidative stress during the early stages of infection in B lymphocytes, epithelial, and lymphoblastoid cell lines. Mol. Cell. Biochem. 313, 179–186 [DOI] [PubMed] [Google Scholar]
- 58. Viswanathan K., Dhabhar F. S. (2005) Stress-induced enhancement of leukocyte trafficking into sites of surgery or immune activation. Proc. Natl. Acad. Sci. U.S.A. 102, 5808–5813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. La Gruta N. L., Kedzierska K., Stambas J., Doherty P. C. (2007) A question of self-preservation: immunopathology in influenza virus infection. Immunol. Cell Biol. 85, 85–92 [DOI] [PubMed] [Google Scholar]
- 60. Shilov I. V., Seymour S. L., Patel A. A., Loboda A., Tang W. H., Keating S. P., Hunter C. L., Nuwaysir L. M., Schaeffer D. A. (2007) The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell Proteomics 6, 1638–1655 [DOI] [PubMed] [Google Scholar]
- 61. Kim K., Kim S. H., Kim J., Kim H., Yim J. (2012) Glutathione S-transferase ω1 activity is sufficient to suppress neurodegeneration in a Drosophila model of Parkinson disease. J. Biol. Chem. 287, 6628–6641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sakai J., Li J., Subramanian K. K., Mondal S., Bajrami B., Hattori H., Jia Y., Dickinson B. C., Zhong J., Ye K., Chang C. J., Ho Y. S., Zhou J., Luo H. R. (2012) Reactive oxygen species-induced actin glutathionylation controls actin dynamics in neutrophils. Immunity 37, 1037–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bergmann C. C., Lane T. E., Stohlman S. A. (2006) Coronavirus infection of the central nervous system: host-virus stand-off. Nat. Rev. Microbiol. 4, 121–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Perlman S., Dandekar A. A. (2005) Immunopathogenesis of coronavirus infections: implications for SARS. Nat. Rev. Immunol. 5, 917–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pewe L. L., Netland J. M., Heard S. B., Perlman S. (2004) Very diverse CD8 T cell clonotypic responses after virus infections. J. Immunol. 172, 3151–3156 [DOI] [PubMed] [Google Scholar]
- 66. Butler N. S., Theodossis A., Webb A. I., Nastovska R., Ramarathinam S. H., Dunstone M. A., Rossjohn J., Purcell A. W., Perlman S. (2008) Prevention of cytotoxic T cell escape using a heteroclitic subdominant viral T cell determinant. PLoS Pathog. 4, e1000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Trujillo J. A., Gras S., Twist K. A., Croft N. P., Channappanavar R., Rossjohn J., Purcell A. W., Perlman S. (2014) Structural and functional correlates of enhanced antiviral immunity generated by heteroclitic CD8 T cell epitopes. J. Immunol. 192, 5245–5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Theodossis A., Guillonneau C., Welland A., Ely L. K., Clements C. S., Williamson N. A., Webb A. I., Wilce J. A., Mulder R. J., Dunstone M. A., Doherty P. C., McCluskey J., Purcell A. W., Turner S. J., Rossjohn J. (2010) Constraints within major histocompatibility complex class I restricted peptides: presentation and consequences for T-cell recognition. Proc. Natl. Acad. Sci. U.S.A. 107, 5534–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bowen D. G., Walker C. M. (2005) Mutational escape from CD8+ T cell immunity: HCV evolution, from chimpanzees to man. J. Exp. Med. 201, 1709–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Goulder P. J., Watkins D. I. (2004) HIV and SIV CTL escape: implications for vaccine design. Nat. Rev. Immunol. 4, 630–640 [DOI] [PubMed] [Google Scholar]
- 71. Kittlesen D. J., Thompson L. W., Gulden P. H., Skipper J. C., Colella T. A., Shabanowitz J., Hunt D. F., Engelhard V. H., Slingluff C. L., Jr., Shabanowitz J. A. (1998) Human melanoma patients recognize an HLA-A1-restricted CTL epitope from tyrosinase containing two cysteine residues: implications for tumor vaccine development [published erratum appears in J. Immunol. (1999) 162, 3106]. J. Immunol. 160, 2099–2106 [PubMed] [Google Scholar]
- 72. Harada M., Gohara R., Matsueda S., Muto A., Oda T., Iwamoto Y., Itoh K. (2004) In vivo evidence that peptide vaccination can induce HLA-DR-restricted CD4+ T cells reactive to a class I tumor peptide. J. Immunol. 172, 2659–2667 [DOI] [PubMed] [Google Scholar]
- 73. Campos-Perez J., Rice J., Escors D., Collins M., Paterson A., Savelyeva N., Stevenson F. K. (2013) DNA fusion vaccine designs to induce tumor-lytic CD8+ T-cell attack via the immunodominant cysteine-containing epitope of NY-ESO 1. Int. J. Cancer 133, 1400–1407 [DOI] [PubMed] [Google Scholar]
- 74. Meadows L., Wang W., den Haan J. M., Blokland E., Reinhardus C., Drijfhout J. W., Shabanowitz J., Pierce R., Agulnik A. I., Bishop C. E., Hunt D. F., Goulmy E., Engelhard V. H. (1997) The HLA-A*0201-restricted H-Y antigen contains a posttranslationally modified cysteine that significantly affects T cell recognition. Immunity 6, 273–281 [DOI] [PubMed] [Google Scholar]
- 75. Chen W., Yewdell J. W., Levine R. L., Bennink J. R. (1999) Modification of cysteine residues in vitro and in vivo affects the immunogenicity and antigenicity of major histocompatibility complex class I-restricted viral determinants. J. Exp. Med. 189, 1757–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Pierce R. A., Field E. D., den Haan J. M. M., Caldwell J. A., White F. M., Marto J. A., Wang W., Frost L. M., Blokland E., Reinhardus C., Shabanowitz J., Hunt D. F., Goulmy E., Engelhard V. H. (1999) Cutting edge: the HLA-A*0101-restricted HY minor histocompatibility antigen originates from DFFRY and contains a cysteinylated cysteine residue as identified by a novel mass spectrometric technique. J. Immunol. 163, 6360–6364 [PubMed] [Google Scholar]
- 77. Haque M. A., Hawes J. W., Blum J. S. (2001) Cysteinylation of MHC class II ligands: peptide endocytosis and reduction within APC influences T cell recognition. J. Immunol. 166, 4543–4551 [DOI] [PubMed] [Google Scholar]
- 78. Macdonald W. A., Purcell A. W., Mifsud N. A., Ely L. K., Williams D. S., Chang L., Gorman J. J., Clements C. S., Kjer-Nielsen L., Koelle D. M., Burrows S. R., Tait B. D., Holdsworth R., Brooks A. G., Lovrecz G. O., Lu L., Rossjohn J., McCluskey J. (2003) A naturally selected dimorphism within the HLA-B44 supertype alters class I structure, peptide repertoire, and T cell recognition. J. Exp. Med. 198, 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Illing P. T., Vivian J. P., Dudek N. L., Kostenko L., Chen Z., Bharadwaj M., Miles J. J., Kjer-Nielsen L., Gras S., Williamson N. A., Burrows S. R., Purcell A. W., Rossjohn J., McCluskey J. (2012) Immune self-reactivity triggered by drug-modified HLA-peptide repertoire. Nature 486, 554–558 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.