Abstract
Background:
The absence of BRCA1-associated protein 1 (BAP1) expression in uveal melanoma (UM) is associated with metastatic progression and reduced survival. In this study, we examine nuclear BAP1 (nBAP1) protein expression in primary UMs (PUMs) that show both ‘typical' and ‘atypical' clinical courses according to their chromosome 3 status, and secondary hepatic metastatic UM (MUM), correlating the results with histological, clinical and survival data.
Methods:
Nuclear BAP1 expression was immunohistochemically assessed in tissue microarrays (TMAs) of: (a) 68 PUM patients, who had been treated surgically; and (b) 13 MUM patients, with 5 cases being paired with primary tumour tissue. All cases were fully annotated. The percentage of tumour cell nuclei staining positively for BAP1 was scored by independent observers.
Results:
Nuclear BAP1 protein expression was absent in 35 out of 68 (51%) PUM patients, correlating strongly with poor prognostic clinicopathological and genetic parameters and reduced survival (Log rank, P<0.001). Lack of nBAP1 expression importantly identified a subset of ‘atypical' PUM patients with disomy of chromosome 3 but with unexpected metastatic relapse. Nuclear BAP1 expression was absent in 10 out of 13 (77%) MUM and expression was concordant in all paired PUM and MUM patients.
Conclusions:
Absent nBAP1 protein expression is an independent survival predictor for UM patients, easily examined using immunohistochemistry.
Keywords: BAP1, uveal melanoma, metastasis, tissue microarray, immunohistochemistry, survival, prognosis
Approximately 50% of patients with uveal melanoma (UM) develop metastatic disease, which usually involves the liver. Metastatic disease is rarely detectable at the time of treatment of the primary UM (PUM), developing varyingly months to years after diagnosis.
The development of metastatic disease correlates strongly with chromosomal abnormalities in UM. We and others have shown that disease-specific mortality is intimately linked with loss of one copy of chromosome 3 and gains of chromosome 8q (Prescher et al, 1996; White et al, 1998; Mensink et al, 2008; Damato et al, 2010; Abdel-Rahman et al, 2011a; Thomas et al, 2012). Attempts to identify a minimal region of loss on chromosome 3, contributing to metastasis, have yielded differing results (Blasi et al, 1999; Tschentscher et al, 2001; Parrella et al, 2003; Zeschnigk et al, 2003; Trolet et al, 2009). Mutually exclusive activating mutations in GNAQ and GNA11, located on chromosomes 9q and 19p, respectively, occur in most UM but are not associated with metastatic disease (Van Raamsdonk et al, 2009, 2010). In 2010, Harbour and co-workers identified inactivating mutations in the BAP1 gene located on chromosome 3p21.1, which occurred almost exclusively in metastasising ‘class 2' UM with monosomy 3 (Harbour et al, 2010). This finding, together with data showing frequent loss of heterozygosity for the BAP1 gene in human tumours, demonstrates the importance of BAP1 as a tumour-suppressor gene (Jensen et al, 1998; Bott et al, 2011; Testa et al, 2011; Wiesner et al, 2011; Abdel-Rahman et al, 2011b).
The BAP1 gene encodes a nuclear protein belonging to a subfamily of deubiquitinating enzymes called ubiquitin C-terminal hydrolases, which was initially discovered through its interaction with the tumour-suppressor protein BRCA1 (Jensen et al, 1998). BRCA1-associated protein 1 (BAP1) protein is reported to be involved in a variety of cellular processes, including cell cycle regulation, DNA repair and protein trafficking. A loss of or mis-localised protein expression may result in severe deregulation of these processes contributing to tumour development and/or progression (Machida et al, 2009; Misaghi et al, 2009; Yu et al, 2010). In PUM, an absence of BAP1 protein was recently shown to be strongly correlated with BAP1 gene mutations, and was significantly associated with metastatic progression and reduced survival in these patients (Koopmans et al, 2014). Furthermore, the two recent studies examining BAP1 protein expression in PUM have advocated the use of immunohistochemistry (IHC) for its testing in the routine work-up of UM cases where possible (Shah et al, 2013; Koopmans et al, 2014).
The purpose of this study was to examine BAP1 protein expression in ‘typical' and ‘atypical' cases of PUM, previously classified as such by chromosome 3 status and survival time, and in metastatic liver lesions from UM patients, and also to correlate these data with histological, clinical and survival information. To achieve this, two tissue microarrays (TMAs) of clinically, histomorphologically and genetically well-defined primary and metastatic UM (PUM-TMA and MUM-TMA, respectively) were used.
Materials and Methods
UM specimens and TMA construction
Seventy histologically confirmed PUM treated by primary enucleation or local resection between 1996 and 2007 at the Liverpool Ocular Oncology Centre were used to construct PUM-TMA using the Manual Tissue Arrayer 1 (Beecher Instruments, Sun Prairie, WI, USA). The UM samples on PUM-TMA represented ‘typical' (1 and 2) and ‘atypical' (3 and 4) subgroups of patients originally selected on the basis of their chromosome 3 status and survival time as follows:
Monosomy 3 UM causing metastatic death within 6 years (yr) of diagnosis (M3⩽6yr; n=21);
Disomy 3 UM with survival exceeding 6 years (D3>6yr; n=13);
Monosomy 3 UM with survival exceeding 6 years (M3>6yr; n=17); and
Disomy 3 UM with metastatic death within 6 years of diagnosis (D3⩽6yr; n=15; Lake et al, 2013).
The typical UM subgroups account for >90% of all UMs (Damato et al, 2007). Two UM with unknown chromosome 3 status were also present on PUM-TMA and were included in the analyses. Triplicate 0.6 mm cores from each tumour were randomly distributed across the TMA alongside non-tumour reference tissues, which were obtained from tonsil, duodenum and pancreas. Whenever possible, the tumour cores were taken from different regions of the UM. Histological, clinical and survival data were available for all patients. Chromosome 3 copy number had previously been determined by fluorescence in-situ hybridisation as described by Damato et al (2007). In the case of subgroup 4 (D3⩽6yr), the disomy 3 status was also confirmed by Multiplex Ligation-Dependent Probe Amplification, as previously described (Lake et al, 2013).
MUM-TMA contained duplicate or triplicate 0.6 mm cores from the liver metastases of 17 UM patients, who had not received any other systemic treatment, along with tonsil and liver as non-tumour reference tissues. Seven of these cases were matched with primary tumour material on PUM-TMA.
All patients were treated in accordance with the Declaration of Helsinki. Ethical approval from the National Research Ethics Service had been provided for all work included in this study (NRES study numbers: 10/H1015/56 and 11/NW/0759).
Immunohistochemistry
Immunohistochemistry was performed on 4 μm sections cut from the formalin-fixed, paraffin wax-embedded TMA tissue blocks. Antigen retrieval and IHC were performed using the Dako PT Link and Autostainer Plus systems according to the standard manufacturers' procedures (Dako UK Ltd, Cambridgeshire, UK). A mouse anti-human BAP1 antibody was used at a previously optimised concentration of 1 μg ml−1 (C-4, Santa Cruz, Insight Biotechnology Ltd, Middlesex, UK). The sections were then counterstained with haematoxylin, dehydrated, cleared and mounted. Positive nuclear BAP1 (nBAP1) staining in pancreas and/or tonsil cores in PUM-TMA and MUM-TMA demonstrated a ‘valid' IHC run. Additional whole tonsil and pancreas sections were treated with a mouse IgG1 isotype control (Dako) at the same concentration as the BAP1 primary antibody and served as negative controls. Neither nuclear nor cytoplasmic staining was observed in mouse IgG1-treated sections.
Scoring
Stained TMA slides were evaluated for nBAP1 protein expression by three independent observers (SEC, HK and SF) with no prior knowledge of the chromosome 3 status of the individual cases. Specimens were given a final score for the percentage of tumour nuclei positive for BAP1 protein expression based on data across the TMA cores. The presence or absence of cytoplasmic BAP1 (cBAP1) staining was also recorded.
Statistical analyses
All data were processed in SPSS (ver.20.0; SPSS Science, Chicago, IL, USA) for statistical analyses. Correlation of BAP1 expression with known risk factors of UM metastatic progression was made using Chi-Square for categorical variables and either Student's t-test or Mann–Whitney for linear variables. Multivariate analyses were performed using Cox Regression and backward likelihood ratio analysis. Survival analyses were performed with Kaplan–Meier.
Results
TMA assessment
Of the 70 PUMs included in PUM-TMA, 63 had three tumour cores that were assessable and 5 had only two assessable tumour cores, which was considered the minimum requirement for scoring. Two tumours in PUM-TMA were lost due to missing or inadequate cores following sectioning.
For the 17 liver metastases arising from a PUM included on MUM-TMA, three tumour cores were assessable in 11 cases and two tumour cores were assessable in 2 cases. Four tumours on MUM-TMA were lost due to missing or inadequate cores following sectioning. Of the 13 assessable cases, 5 were matched with a PUM on PUM-TMA.
Clinicopathological data
PUM-TMA comprised 24 males and 44 females, with a mean age at diagnosis of 60 years (median, 63 years; range, 21–89 years; Supplementary Table 1). The tumours had a mean largest basal diameter of 16.5 mm (median, 17.0 mm; range, 10.8–21.5 mm). Largest basal diameter measurements were not significantly different between the four tumour subgroups included on PUM-TMA (P=0.09; one-way ANOVA), suggesting that lead time bias had not influenced the overall survival time. Histological examination previously performed on full tumour sections as part of the routine diagnostic work-up, classified 21 UMs as predominantly spindle and 47 UMs of mixed cell type, with variable proportions of epithelioid cells. Periodic acid Schiff+ closed connective tissue loops were found in 39 UMs, and the mitotic count exceeded 5 per 40 high-power fields (HPF) in 40 tumours. Genetic analysis of all UMs examined classified 28 of these tumours as D3 and 38 as M3. In two patients, the chromosome 3 status was not known. Follow-up information was available for all patients at the close of study, 31 December 2013: 22 patients were still alive; 43 had died of metastatic melanoma; and 3 had died of other causes (i.e., ischemic heart disease, myocardial infarction and chronic obstructive pulmonary disease). After excluding these three patients, the mean survival time was 6.2 years (median 5.1 years; range 0.5–14.4 years).
Of the 13 hepatic metastases assessed on MUM-TMA, 12 were from UM patients who had undergone surgical resection, and one from a patient who had undergone staging laparoscopy and then found to have non-resectable disease. Information was not available regarding the number and size of the hepatic metastases; however, resection was incomplete following pathological examination in two cases (MUM1 and MUM4). Pathological examination of the tumour cores in MUM-TMA reported epithelioid cells to be present in nine cases. Clinical information regarding primary management of the intraocular UM was available in all cases. Details of primary tumour size, ciliary body involvement and chromosome 3 status were available in the majority of cases (Supplementary Table 2).
Expression of BAP1
PUM
BAP1 protein expression was localised predominantly to the nuclei of tumour cells in 33 out of 68 (49%) cases of PUM (Figures 1A–C). Weak to moderate cytoplasmic expression was also observed in 30 out of 68 (44%) of the UM analysed, and occurred predominantly in cases for which nBAP1 was also present. For all cases in PUM-TMA, the percentage of nBAP1-positive tumour cells in the individual cores of each UM did not differ by more than 10%, indicating no significant heterogeneity in the presence or absence of nBAP1 protein expression across any individual tumour. The percentage of nBAP1+ tumour cell nuclei was: ⩾80% across the cores in 32 cases, and was completely absent in all cores for 35 cases. A single case showed staining in 70% of tumour nuclei. For all further statistical analyses, nBAP1 protein expression was considered as either present or completely absent.
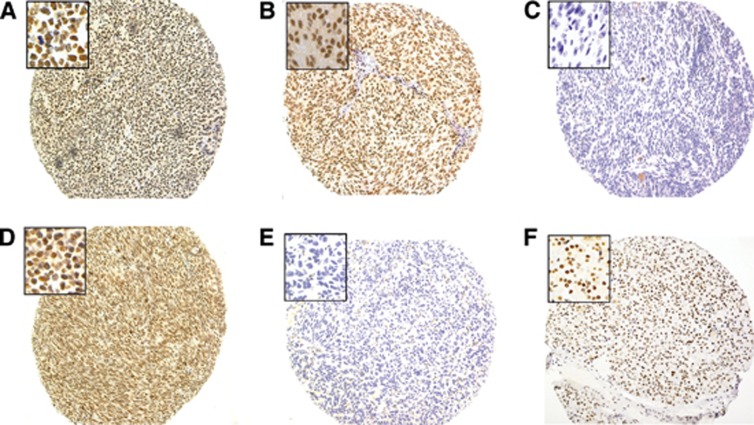
Figure 1.
Representative images of BAP1 protein expression in primary tumours (A–C) and metastatic liver lesions (D and E) from patients with UM ( × 10 magnification). Insets show nBAP1 at × 40 magnification. Images A, B and D show strong nBAP1 protein expression in 100% of the tumour cells together with weak (A) or moderate cytoplasmic staining (B and D). Images C and E show no detectable nBAP1 protein. Image F shows nBAP1 protein expression in internal positive control pancreatic tissue.
MUM
nBAP1 protein was absent in all cores of 10 out of 13 (77%) cases (Figures 1D and E). In the remaining three cases, nBAP1 staining was positive in MUM cell nuclei in every tissue core. The five pairs of matched primary and secondary MUMs showed complete concordance between the nBAP1 score in the primary tumour and the hepatic metastasis; nBAP1 was absent in four of these cases and present in one case.
Correlation of nBAP1 protein expression with clinicopathological and genetic features of metastatic risk
PUM
Univariate analysis of all cases demonstrated that lack of nBAP1 protein expression was strongly associated with clinicopathological and genetic features of increased metastatic risk, namely: increased age at primary management (P=0.002); ciliary body involvement (P=0.001); presence of periodic acid Schiff+ closed connective tissue loops (P=0.001); presence of epithelioid cells (P=0.011) and M3 (P=0.001; Table 1).
Table 1. Association of nBAP1 status with clinical, pathological and genetic risk factors in UM.
| Variable | nBAP1 absent | nBAP1 present | P-value |
|---|---|---|---|
|
Age at PM (years) | |||
| Mean | 65.4 | 55.2 | 0.002 |
| Median | 65.0 | 57.0 | |
| Range |
38.0–89.0 |
21.0–79.0 |
|
|
Gender | |||
| Female | 22 (50%) | 22 (50%) | 0.747 |
| Male |
13 (54%) |
11 (46%) |
|
|
LBD (mm) | |||
| Mean | 16.9 | 16.0 | 0.151 |
| Median | 17.5 | 15.9 | |
| Range |
10.8–21.1 |
12.2–21.5 |
|
|
UH (mm) | |||
| Mean | 9.6 | 8.6 | 0.157 |
| Median | 9.7 | 9.0 | |
| Range |
3.4–15.7 |
2.4–14.0 |
|
|
Ciliary body involvement | |||
| Yes | 17 (81%) | 4 (19%) | 0.001 |
| No |
18 (38%) |
29 (62%) |
|
|
Epithelioid cells present | |||
| Yes | 29 (62%) | 18 (32%) | 0.011 |
| No |
6 (29%) |
15 (71%) |
|
|
Closed loops present | |||
| Yes | 27 (69%) | 12 (31%) | 0.001 |
| No |
8 (28%) |
21 (72%) |
|
|
Mitotic count per 40 HPF | |||
| Mean | 8.3 | 6.7 | 0.235 |
| Median | 7.0 | 5.0 | |
| Range |
1.0–23.0 |
1.0–16.0 |
|
|
Monosomy 3 | |||
| Yes | 25 (66%) | 13 (34%) | 0.001 |
| No | 8 (29%) | 20 (71%) | |
| Not available | 2 (100%) | 0 | |
Abbreviations: HPF=high-power fields; LBD=largest basal diameter; PM=primary management; nBAP1=nuclear BRCA1-associated protein 1; UH=ultrasound height.
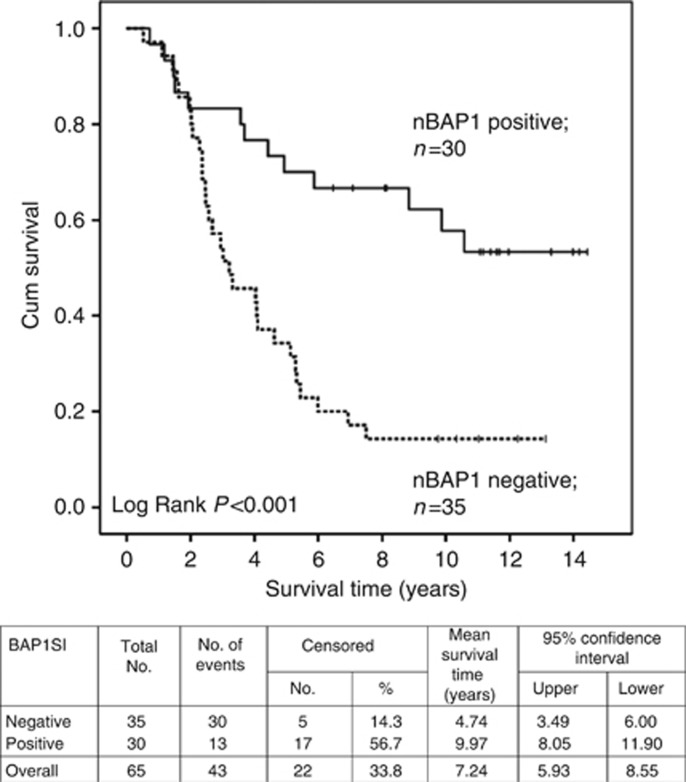
Multivariate analysis using the Cox regression model was performed on all UM cases (‘typical' and ‘atypical' subgroups, excluding only those who had died of causes other than metastatic melanoma) to determine independent risk factors associated with survival. Lack of nBAP1 protein (P=0.002) and an increased mitotic rate (P=0.046) remained in the Cox regression model as significant independent predictors of reduced survival time. Kaplan–Meier analysis further confirmed the significant association between a lack of nBAP1 protein and reduced survival time (Figure 2; Log Rank, P<0.001).
Figure 2.
Kaplan–Meier survival curve and table for all primary UM stratified according to nBAP1 protein expression. UM patients who had died of causes other than metastatic melanoma were excluded in the analyses. BAPSI indicates whether the nBAP1 protein expression was scored as positive or negative. No. of events indicates the number of deaths.
When only the D3 UM were considered, the atypical D3⩽6yr cases (subgroup 4) were more likely than D3>6yr cases (subgroup 2) to have: ciliary body involvement (P=0.009); epithelioid cells present (P=0.007); lack of nBAP1 protein (P=0.001) and mitotic count ⩾5/40 HPF (P=0.002; Table 2). Nuclear BAP1 protein expression was absent in 8 out of 15 (53%) D3⩽6yr PUM; however, it was detected in all 13 D3>6yr PUMs (Table 2). Multivariate Cox regression analysis of all D3 cases demonstrated the presence of epithelioid cells (P=0.019) and a high mitotic count (P=0.001) as significant independent predictors of reduced survival time (Table 3).
Table 2. Associations of the chromosome 3 subgroups with clinical and pathological variables.
| Variable | Subgroup 1 (M3⩽6yr) | Subgroup 2 (D3>6yr) | Subgroup 3 (M3>6yr) | Subgroup 4 (D3⩽6yr) | P-value* | P-value |
|---|---|---|---|---|---|---|
|
Age at PM (yr) | ||||||
| Mean |
66.3 |
51.3 |
62.4 |
58.3 |
0.450 |
0.277 |
|
Gender | ||||||
| Female | 15 | 11 | 8 | 10 | 0.291 | 0.133 |
| Male |
6 |
2 |
9 |
5 |
|
|
|
LBD | ||||||
| <16 mm | 4 | 8 | 9 | 4 | 0.067 | 0.029 |
| ⩾16 mm |
17 |
5 |
8 |
11 |
|
|
|
CBI | ||||||
| Yes | 10 | 0 | 4 | 6 | 0.009 | 0.133 |
| No |
11 |
13 |
13 |
9 |
|
|
|
Epithelioid cells | ||||||
| Yes | 17 | 5 | 11 | 13 | 0.007 | 0.270 |
| No |
4 |
8 |
6 |
2 |
|
|
|
Closed loops | ||||||
| Yes | 18 | 5 | 7 | 8 | 0.450 | 0.003 |
| No |
3 |
8 |
10 |
7 |
|
|
|
Mitotic count | ||||||
| <5/40_HPF | 6 | 10 | 6 | 3 | 0.002 | 0.668 |
| ⩾5/40_HPF |
15 |
3 |
11 |
12 |
|
|
|
nBAP1 | ||||||
| Absent | 18 | 0 | 7 | 8 | 0.001 | 0.003 |
| Present | 3 | 13 | 10 | 7 | ||
Abbreviations: CBI=ciliary body involvement; HPF=high-power fields; LBD=largest basal diameter; PM=primary management; P-value*=subgroup 2 vs subgroup 4; P-value=subgroup 1 vs subgroup 3; yr, year.
Table 3. Multivariate Cox regression of survival in all patients classified as D3.
| Variable | Hazard ratio | 95% CI | P-value |
|---|---|---|---|
| Epithelioid cells |
6.605 |
1.371–31.819 |
0.019 |
| Mitotic count ⩾5/40 HPF |
10.246 |
2.526–41.560 |
0.001 |
| ‘Closed' loops |
— |
— |
— |
| LBD ⩾16 mm |
— |
— |
— |
| Ciliary body involvement |
— |
— |
— |
| Absent nBAP1 | — | — | — |
Abbreviations: CI=confidence interval; HPF=high-power fields; LBD=largest basal diameter; nBAP1=nuclear BRCA1-associated protein 1. ‘–' Indicates that variable did not remain in the Cox regression model.
When only the M3 UM were considered, M3⩽6yr PUMs (subgroup 1) were less likely to have nBAP1 protein positivity (P=0.003) and more likely to have closed connective tissue loops (P=0.003) and a largest basal diameter ⩾16 mm (P=0.029) when compared with M3>6yr (subgroup 3) PUM (Table 2). Nuclear BAP1 protein was absent in 18 out of 21 (86%) M3⩽6yr and 7 out of 17 (41%) M3>6yr PUMs (Table 2). Multivariate Cox regression analysis of all M3 cases highlighted an absence of nBAP1 as the only significant independent predictor of reduced survival time in this group (P=0.05).
MUM
Of the 13 patients with assessable cores in MUM-TMA, all 10 with undetectable nBAP1 protein expression had died at the close of study. The mean survival time of this cohort of patients was 6.5 years (median, 4.2 years; range, 1.9–19.9 years). Of particular note was that three patients with nBAP1 protein staining in 100% of UM cells in the hepatic metastasis were alive at the close of study (MUM3, MUM12, MUM13; Supplementary Table 2). The time period from primary management to the development of metastatic disease tended to be longer in these individuals (7.9, 11.4 and 6.2 years, respectively), compared with patients who had hepatic metastases in which nBAP1 protein was absent.
Discussion
In this study, we found a significant independent association between lack of nBAP1 protein expression in PUM and reduced survival time, irrespective of chromosome 3 status. Moreover, we report the results of nBAP1 protein expression in MUM to the liver, demonstrating not only complete concordance between matched PUM and metastatic liver lesions, but also that the presence of nBAP1 positivity tended to be associated with increased metastatic latency in M3 tumours.
Low or absent nBAP1 protein expression is associated with a poor prognosis in non-small-cell lung carcinoma (NSCLC) and clear cell renal carcinomas with data demonstrating a shorter median survival time and increased metastatic spread to lymph nodes in NSCLC (Fan et al, 2012; Pena-Llopis et al, 2012). In UM, two recent studies similarly demonstrated that loss of BAP1 protein, examined in 30% (Koopmans et al, 2014) and 58% (Shah et al, 2013) of UM cases, is strongly associated with an aggressive metastatic phenotype and poor prognosis. Consistent with these reports, we observed absent nBAP1 protein in 51% of the PUM cases, and also demonstrated its significance as an independent predictor of reduced overall survival.
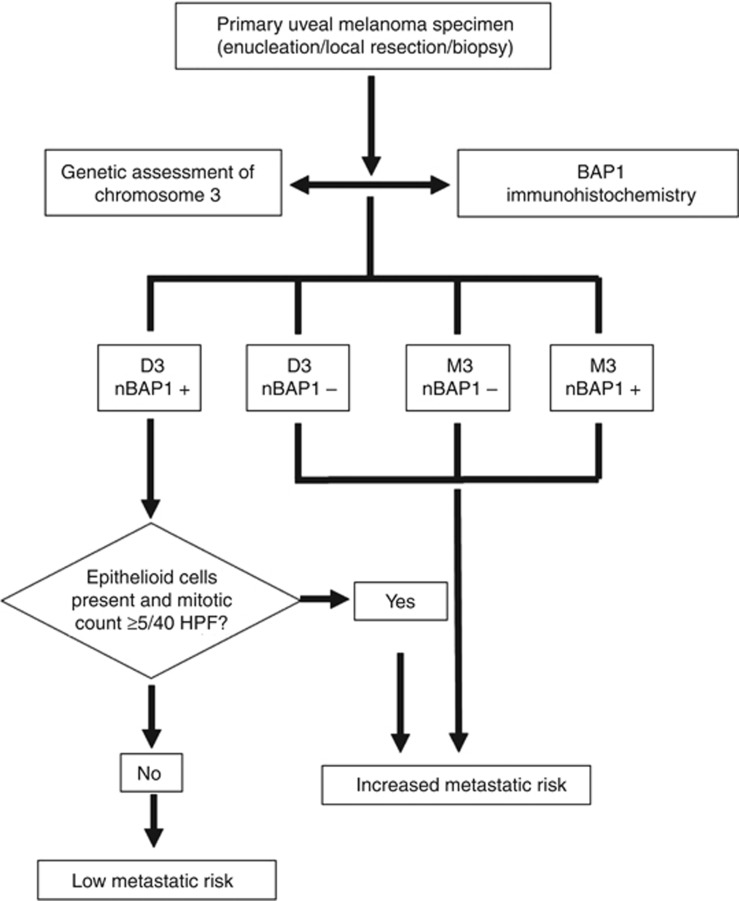
Unlike the two studies recently reported for UM, our cohort contained UM samples with both ‘typical' (i.e., M3 UM with short survival (subgroup 1) and D3 UM with prolonged survival (subgroup 2)) and ‘atypical' (i.e., M3 UM with prolonged survival (subgroup 3) and D3 UM with short survival (subgroup 4)) clinical courses. The ‘atypical' subgroups, based on chromosome 3 status and survival time following diagnosis of a PUM, represent approximately 10% of all UM (Damato et al, 2007, 2010): although rare, it is essential to examine these cases to fully understand the molecular pathogenesis of UM. Of particular, clinical relevance are those UM cases with D3 and ‘unexpected' development of metastatic disease (subgroup 4). Interestingly, approximately 50% of subgroup 4 patients completely lacked nBAP1 protein staining. It is possible that loss of heterozygosity of a small region containing the BAP1 gene coupled with a mutation on the remaining BAP1 allele in these individuals is responsible for this phenomenon, as was reported recently for three UMs analysed by SNP array and BAP1 sequencing (Koopmans et al, 2014). We have previously published, however, the SNP results for these cases, which did not identify the BAP1 locus as a common region of deletion (Lake et al, 2010). Similar UM cases that suggest alternative mechanisms influencing the metastatic process have been reported. Koopmans et al (2014) show 17% (3 out of 18) of monosomy 3 cases with short survival, for which BAP1 mutations are absent and BAP1 protein is detected. In another study, three cases without metastases (disease-free survival of 32, 40 and 81 months) despite monosomy 3 and BAP1 mutation are described (Dono et al, 2014). Newly identified driver mutations in splicing factor 3b subunit 1 (SF3B1) and eukaryotic translation initiation factor 1A, X-linked (EIF1AX) have been described in about 20–30% and 48% of UM patients with disomy 3, respectively, and are associated with a favourable prognosis (Furney et al, 2013; Martin et al, 2013). Although it is not yet clear how these mutations promote UM, it is of interest that BAP1, SF3B1 and EIF1AX mutations occur in a largely mutually exclusive manner. Moreover, although the majority of SF3B1 mutations detected result in a missense change at Arg625, distinct SF3B1 mutations in 30% (3 out of 10) of disomy 3 patients who developed metastatic disease were also reported (Martin et al, 2013). Further subgrouping of the UM according to SF3B1 and EIF1AX mutations may enhance prognostication for these patients and also be incorporated into the prognostic workflow proposed in Figure 3.
Figure 3.
Proposed prognostic test workflow and risk decision outcome for primary UM specimens, based on genetic analysis of chromosome 3 and BAP1 immunohistochemistry.
Nuclear BAP1 protein was absent in the majority of ‘typical' M3 UMs with short survival (subgroup 1) and tended to be associated with a reduced overall survival time across all M3 cases examined (40 months) compared with tumours with positive nBAP1 staining (143 months). These data are consistent with those of Koopmans et al (2014) for both patient survival based on the BAP1 mutational status (32 vs 133 months for BAP1 mutation-positive vs BAP1 mutation-negative tumours) or BAP1 protein expression (31 vs 133 months for tumours showing negative BAP1 expression by IHC vs those with positive BAP1 expression) and thus the proposed role of BAP1 in the pathogenesis of UM with an aggressive phenotype. Nonetheless, three patients within subgroup 1 displayed nBAP1 staining and yet died within 5 years of primary management of their UM. Clinically, at the Liverpool Ocular Oncology Centre, all patients with M3 are classified as ‘high risk' with respect to the development of metastases, and as such would be followed-up with regular screening for metastatic disease irrespective of BAP1 protein expression (Marshall et al, 2013).
Previous studies have reported the presence of BAP1 mutations in 84% of poor prognosis class 2 UMs (Harbour et al, 2010). More recently, BAP1 mutations were identified in 58% (11 out of 19) of UM patients who had developed metastasis, and loss of BAP1 immunohistochemical staining was observed in the tumour cells of all but 2 of those 11 cases (Koopmans et al, 2014). We were unable to conduct mutational analysis of the BAP1 gene in this cohort of patient samples because of insufficient material remaining for this study; however, there is mounting evidence in the literature that lack of BAP1 protein expression is strongly associated with the presence of inactivating BAP1 mutations in UM (Koopmans et al, 2014), mesothelioma (Yoshikawa et al, 2012) and Spitz melanomas (Wiesner et al, 2011, 2012). Moreover, work conducted in a human NSCLC cell line in nude mice, implies that both de-ubiquinating activity and nuclear localisation are necessary for the tumour-suppressive activity of BAP1 (Ventii et al, 2008). Our finding that BAP1 protein (both nuclear and cytoplasmic) was absent in 70% of patients who developed metastatic disease supports this concept. Our data demonstrate that such UM cases can be easily identified at primary management using BAP1 IHC, in particular to enhance the decision regarding the frequency of liver screening in a subset of ‘atypical' disomy 3 cases, as highlighted in the proposed prognostic workflow shown in Figure 3. It is important to acknowledge, however, that in a human NSCLC cell line, missense mutations introduced into the catalytic domain of BAP1 reduced its tumour-suppressor function, but BAP1 protein expression was unaltered (Ventii et al, 2008). Furthermore, in two UM samples with loss of one copy of chromosome 3 and a BAP1 mutation, BAP1 protein could be detected by IHC (Koopmans et al, 2014). Ongoing, prospective analysis of BAP1 protein expression, assessed in all UMs during routine pathological work-up, together with other clinicopathological and genetic features is therefore necessary.
The inclusion of MUM tissue in this study allowed us to demonstrate that differences in BAP1 protein expression occurring in the PUM are retained in the hepatic metastasis. Interestingly, we observed two cases of MUM that displayed positive nBAP1 staining, which is consistent with a recent report detecting BAP1 protein in 3 out of 16 metastatic lesions (Griewank et al, 2014). This is of key importance not only to enhance our understanding of the biology of these tumours but also when considering therapeutic intervention, which at present is likely to be determined based on the results of tumour sampling from the PUM. It should be recognised, however, that the MUM is likely to have undergone additional genetic alterations (Trolet et al, 2009) that could further influence the therapeutic response, as seen in other tumours (Swanton, 2012).
Although the loss of BAP1 function may be linked to metastasis, the precise mechanism of tumour dissemination in UM is still unclear. In functional studies, depletion of BAP1 protein in the 92.1 UM cell line using siRNAs led to a change in cell morphology from spindle to a more epithelioid phenotype, loss of melanocytic differentiation and transition from a ‘class 1' to a ‘class 2' gene expression profile (Harbour et al, 2010). BAP1 is also reported to interact with host cell factor-1 resulting in the regulation of cell cycle progression at the G1/S checkpoint by controlling the transcription of genes regulating cell growth and proliferation (Machida et al, 2009; Misaghi et al, 2009; Yu et al, 2010). Although the in vitro morphological changes are consistent with the significant association between absence of nBAP1 protein and the presence of epithelioid cells reported for PUM in this study, we observed no correlation between nBAP1 protein expression and changes in the mitotic count. Evidence linking BAP1 with differentiation, however, has importantly identified histone deacetylase inhibitors as therapeutic compounds that could revert UM cells with BAP1 loss to a more differentiated phenotype (Landreville et al, 2012). Further studies are essential to understand how these therapeutic strategies may target specific pathways in MUM.
In conclusion, our data demonstrate that the absence of nBAP1 protein expression is strongly associated with a poor outcome in patients with UM and that this can be easily (and cost efficiently) detected using IHC. Moreover, our finding that lack of BAP1 protein can identify a subgroup of ‘atypical' poor prognosis D3 patients warrants the introduction of this test into the routine pathological work-up of all UM specimens. Ongoing collection and further detailed molecular analyses of all ‘atypical' UM are required to fully understand the aetiology of this aggressive disease.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Abdel-Rahman MH, Christopher BN, Faramawi MF, Said-Ahmed K, Cole C, McFaddin A, Ray-Chaudhury A, Heerema N, Davidorf FH. Frequency, molecular pathology and potential clinical significance of partial chromosome 3 aberrations in uveal melanoma. Mod Pathol. 2011;24 (7:954–962. doi: 10.1038/modpathol.2011.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Rahman MH, Pilarski R, Cebulla CM, Massengill JB, Christopher BN, Boru G, Hovland P, Davidorf FH. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J Med Genet. 2011;48 (12:856–859. doi: 10.1136/jmedgenet-2011-100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasi MA, Roccella F, Balestrazzi E, Del Porto G, De Felice N, Roccella M, Rota R, Grammatico P. 3p13 region: a possible location of a tumor suppressor gene involved in uveal melanoma. Cancer Genet Cytogenet. 1999;108 (1:81–83. doi: 10.1016/s0165-4608(98)00114-9. [DOI] [PubMed] [Google Scholar]
- Bott M, Brevet M, Taylor BS, Shimizu S, Ito T, Wang L, Creaney J, Lake RA, Zakowski MF, Reva B, Sander C, Delsite R, Powell S, Zhou Q, Shen R, Olshen A, Rusch V, Ladanyi M. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet. 2011;43 (7:668–672. doi: 10.1038/ng.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damato B, Dopierala JA, Coupland SE. Genotypic profiling of 452 choroidal melanomas with multiplex ligation-dependent probe amplification. Clin Cancer Res. 2010;16 (24:6083–6092. doi: 10.1158/1078-0432.CCR-10-2076. [DOI] [PubMed] [Google Scholar]
- Damato B, Duke C, Coupland SE, Hiscott P, Smith PA, Campbell I, Douglas A, Howard P. Cytogenetics of uveal melanoma: a 7-year clinical experience. Ophthalmology. 2007;114 (10:1925–1931. doi: 10.1016/j.ophtha.2007.06.012. [DOI] [PubMed] [Google Scholar]
- Dono M, Angelini G, Cecconi M, Amaro A, Esposito AI, Mirisola V, Maric I, Lanza F, Nasciuti F, Viaggi S, Gualco M, Bandelloni R, Truini M, Coviello DA, Zupo S, Mosci C, Pfeffer U. Mutation frequencies of GNAQ, GNA11, BAP1, SF3B1, EIF1AX and TERT in uveal melanoma: detection of an activating mutation in the TERT gene promoter in a single case of uveal melanoma. Br J Cancer. 2014;110 (4:1058–1065. doi: 10.1038/bjc.2013.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan LH, Tang LN, Yue L, Yang Y, Gao ZL, Shen Z. BAP1 is a good prognostic factor in advanced non-small cell lung cancer. Clin Invest Med. 2012;35 (4:E182–E189. doi: 10.25011/cim.v35i4.17146. [DOI] [PubMed] [Google Scholar]
- Furney SJ, Pedersen M, Gentien D, Dumont AG, Rapinat A, Desjardins L, Turajlic S, Piperno-Neumann S, de la Grange P, Roman-Roman S, Stern MH, Marais R. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov. 2013;3 (10:1122–1129. doi: 10.1158/2159-8290.CD-13-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griewank KG, van de Nes J, Schilling B, Moll I, Sucker A, Kakavand H, Haydu LE, Asher M, Zimmer L, Hillen U, Thompson JF, Scolyer RA, Schadendorf D, Murali R. Genetic and clinico-pathologic analysis of metastatic uveal melanoma. Mod Pathol. 2014;27 (2:175–183. doi: 10.1038/modpathol.2013.138. [DOI] [PubMed] [Google Scholar]
- Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, Council ML, Matatall KA, Helms C, Bowcock AM. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330 (6009:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen DE, Proctor M, Marquis ST, Gardner HP, Ha SI, Chodosh LA, Ishov AM, Tommerup N, Vissing H, Sekido Y, Minna J, Borodovsky A, Schultz DC, Wilkinson KD, Maul GG, Barlev N, Berger SL, Prendergast GC, Rauscher FJ., 3rd BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16 (9:1097–1112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- Koopmans AE, Verdijk RM, Brouwer RW, van den Bosch TP, van den Berg MM, Vaarwater J, Kockx CE, Paridaens D, Naus NC, Nellist M, van Ijcken WF, Kiliç E, de Klein A.2014Clinical significance of immunohistochemistry for detection of BAP1 mutations in uveal melanoma Mod Pathole-pub ahead of print 14 March 2014;doi: 10.1038/modpathol.2014.43 [DOI] [PubMed]
- Lake SL, Coupland SE, Taktak AF, Damato BE. Whole-genome microarray detects deletions and loss of heterozygosity of chromosome 3 occurring exclusively in metastasizing uveal melanoma. Invest Ophthalmol Vis Sci. 2010;51 (10:4884–4891. doi: 10.1167/iovs.09-5083. [DOI] [PubMed] [Google Scholar]
- Lake SL, Damato BE, Kalirai H, Dodson AR, Taktak AF, Lloyd BH, Coupland SE. Single nucleotide polymorphism array analysis of uveal melanomas reveals that amplification of CNKSR3 is correlated with improved patient survival. Am J Pathol. 2013;182 (3:678–687. doi: 10.1016/j.ajpath.2012.11.036. [DOI] [PubMed] [Google Scholar]
- Landreville S, Agapova OA, Matatall KA, Kneass ZT, Onken MD, Lee RS, Bowcock AM, Harbour JW. Histone deacetylase inhibitors induce growth arrest and differentiation in uveal melanoma. Clin Cancer Res. 2012;18 (2:408–416. doi: 10.1158/1078-0432.CCR-11-0946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida YJ, Machida Y, Vashisht AA, Wohlschlegel JA, Dutta A. The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. J Biol Chem. 2009;284 (49:34179–34188. doi: 10.1074/jbc.M109.046755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall E, Romaniuk C, Ghaneh P, Wong H, McKay M, Chopra M, Coupland SE, Damato BE. MRI in the detection of hepatic metastases from high-risk uveal melanoma: a prospective study in 188 patients. Br J Ophthalmol. 2013;97 (2:159–163. doi: 10.1136/bjophthalmol-2012-302323. [DOI] [PubMed] [Google Scholar]
- Martin M, Masshofer L, Temming P, Rahmann S, Metz C, Bornfeld N, van de Nes J, Klein-Hitpass L, Hinnebusch AG, Horsthemke B, Lohmann DR, Zeschnigk M. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet. 2013;45 (8:933–936. doi: 10.1038/ng.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mensink HW, Kilic E, Vaarwater J, Douben H, Paridaens D, de Klein A. Molecular cytogenetic analysis of archival uveal melanoma with known clinical outcome. Cancer Genet Cytogenet. 2008;181 (2:108–111. doi: 10.1016/j.cancergencyto.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Misaghi S, Ottosen S, Izrael-Tomasevic A, Arnott D, Lamkanfi M, Lee J, Liu J, O'Rourke K, Dixit VM, Wilson AC. Association of C-terminal ubiquitin hydrolase BRCA1-associated protein 1 with cell cycle regulator host cell factor 1. Mol Cell Biol. 2009;29 (8:2181–2192. doi: 10.1128/MCB.01517-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrella P, Fazio VM, Gallo AP, Sidransky D, Merbs SL. Fine mapping of chromosome 3 in uveal melanoma: identification of a minimal region of deletion on chromosomal arm 3p25.1-p25.2. Cancer Res. 2003;63 (23:8507–8510. [PubMed] [Google Scholar]
- Pena-Llopis S, Vega-Rubin-de-Celis S, Liao A, Leng N, Pavia-Jimenez A, Wang S, Yamasaki T, Zhrebker L, Sivanand S, Spence P, Kinch L, Hambuch T, Jain S, Lotan Y, Margulis V, Sagalowsky AI, Summerour PB, Kabbani W, Wong SW, Grishin N, Laurent M, Xie XJ, Haudenschild CD, Ross MT, Bentley DR, Kapur P, Brugarolas J. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet. 2012;44 (7:751–759. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescher G, Bornfeld N, Hirche H, Horsthemke B, Jockel KH, Becher R. Prognostic implications of monosomy 3 in uveal melanoma. Lancet. 1996;347 (9010:1222–1225. doi: 10.1016/s0140-6736(96)90736-9. [DOI] [PubMed] [Google Scholar]
- Shah AA, Bourne TD, Murali R. BAP1 protein loss by immunohistochemistry: a potentially useful tool for prognostic prediction in patients with uveal melanoma. Pathology. 2013;45 (7:651–656. doi: 10.1097/PAT.0000000000000002. [DOI] [PubMed] [Google Scholar]
- Swanton C. Intratumor heterogeneity: evolution through space and time. Cancer Res. 2012;72 (19:4875–4882. doi: 10.1158/0008-5472.CAN-12-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa JR, Cheung M, Pei J, Below JE, Tan Y, Sementino E, Cox NJ, Dogan AU, Pass HI, Trusa S, Hesdorffer M, Nasu M, Powers A, Rivera Z, Comertpay S, Tanji M, Gaudino G, Yang H, Carbone M. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet. 2011;43 (10:1022–1025. doi: 10.1038/ng.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S, Putter C, Weber S, Bornfeld N, Lohmann DR, Zeschnigk M. Prognostic significance of chromosome 3 alterations determined by microsatellite analysis in uveal melanoma: a long-term follow-up study. Br J Cancer. 2012;106 (6:1171–1176. doi: 10.1038/bjc.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trolet J, Hupe P, Huon I, Lebigot I, Decraene C, Delattre O, Sastre-Garau X, Saule S, Thiery JP, Plancher C, Asselain B, Desjardins L, Mariani P, Piperno-Neumann S, Barillot E, Couturier J. Genomic profiling and identification of high risk uveal melanoma by array-CGH analysis of primary tumors and liver metastases. Invest Ophthalmol Vis Sci. 2009;50 (6:2572–2580. doi: 10.1167/iovs.08-2296. [DOI] [PubMed] [Google Scholar]
- Tschentscher F, Prescher G, Horsman DE, White VA, Rieder H, Anastassiou G, Schilling H, Bornfeld N, Bartz-Schmidt KU, Horsthemke B, Lohmann DR, Zeschnigk M. Partial deletions of the long and short arm of chromosome 3 point to two tumor suppressor genes in uveal melanoma. Cancer Res. 2001;61 (8:3439–3442. [PubMed] [Google Scholar]
- Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O'Brien JM, Simpson EM, Barsh GS, Bastian BC. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457 (7229:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, Obenauf AC, Wackernagel W, Green G, Bouvier N, Sozen MM, Baimukanova G, Roy R, Heguy A, Dolgalev I, Khanin R, Busam K, Speicher MR, O'Brien J, Bastian BC. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363 (23:2191–2199. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventii KH, Devi NS, Friedrich KL, Chernova TA, Tighiouart M, Van Meir EG, Wilkinson KD. BRCA1-associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Cancer Res. 2008;68 (17:6953–6962. doi: 10.1158/0008-5472.CAN-08-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White VA, Chambers JD, Courtright PD, Chang WY, Horsman DE. Correlation of cytogenetic abnormalities with the outcome of patients with uveal melanoma. Cancer. 1998;83 (2:354–359. [PubMed] [Google Scholar]
- Wiesner T, Murali R, Fried I, Cerroni L, Busam K, Kutzner H, Bastian BC. A distinct subset of atypical spitz tumors is characterized by BRAF mutation and loss of BAP1 expression. Am J Surg Pathol. 2012;36 (6:818–830. doi: 10.1097/PAS.0b013e3182498be5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner T, Obenauf AC, Murali R, Fried I, Griewank KG, Ulz P, Windpassinger C, Wackernagel W, Loy S, Wolf I, Viale A, Lash AE, Pirun M, Socci ND, Rutten A, Palmedo G, Abramson D, Offit K, Ott A, Becker JC, Cerroni L, Kutzner H, Bastian BC, Speicher MR. Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet. 2011;43 (10:1018–1021. doi: 10.1038/ng.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa Y, Sato A, Tsujimura T, Emi M, Morinaga T, Fukuoka K, Yamada S, Murakami A, Kondo N, Matsumoto S, Okumura Y, Tanaka F, Hasegawa S, Nakano T, Hashimoto-Tamaoki T. Frequent inactivation of the BAP1 gene in epithelioid-type malignant mesothelioma. Cancer Sci. 2012;103 (5:868–874. doi: 10.1111/j.1349-7006.2012.02223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Mashtalir N, Daou S, Hammond-Martel I, Ross J, Sui G, Hart GW, Rauscher FJ, 3rd, Drobetsky E, Milot E, Shi Y, Affar el B. The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Mol Cell Biol. 2010;30 (21:5071–5085. doi: 10.1128/MCB.00396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeschnigk M, Tschentscher F, Lich C, Brandt B, Horsthemke B, Lohmann DR. Methylation analysis of several tumour suppressor genes shows a low frequency of methylation of CDKN2A and RARB in uveal melanomas. Comp Funct Genomics. 2003;4 (3:329–336. doi: 10.1002/cfg.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.