Abstract
Aims: The neural cell adhesion molecule (NCAM) has been reported to be involved in the development of the central nervous system and its mRNA level might decrease in the serum of autistic patients. However, there was no evidence of the association of the NCAM1 gene polymorphisms with autism. In the present study, we enrolled 237 children with autism and 451 healthy control subjects. Then, we used the direct DNA sequencing for genotyping five tag single-nucleotide polymorphisms (SNPs) in the NCAM1 gene. Results: By using case–control association analyses, we found that three SNPs at the NCAM1 gene were associated with autism (rs4937786, p=0.015; rs12418058, p=0.0076; rs1436109, p=0.0023). Two of them remained significant after the Bonferroni multiple testing correction (rs12418058, pcorrcted=0.038; rs1436109, pcorrcted=0.012). Moreover, two of the SNPs were associated with the parental age at conception in autism (rs12418058, p=0.037; rs1436109, p=0.01). Conclusion: These results showed that NCAM1 might play an important role in the pathogenesis of autism.
Introduction
Autism is a kind of common pervasive neurodevelopmental disorder. The typical clinical characteristics of autism include deficits in social interaction, communication, and the presence of repetitive or stereotypic behaviors (Cooper, 1995). These clinical symptoms become obvious in the first 3 years of life in childhood (Lord et al., 2000). Previous research suggested that strong genetic components might be involved in the susceptibility to autism (Rutter, 2000; Folstein and Rosen-Sheidley, 2001; Veenstra-Vanderweele et al., 2003). Although the etiology of autism is unknown, family and twin studies have indicated autism as a highly heritable neuropsychiatry disorder with the heritability of about ∼90% (Steffenburg et al., 1989; Folstein and Rosen-Sheidley, 2001).
Up to now, hundreds of susceptibility genes have been implicated in the etiology of autism and this disease has also been identified as genetically heterogeneous (Miyauchi and Voineagu, 2013). Therefore, few of the susceptibility genes could be validated among various populations. Therefore, many studies focused on some special neurodevelopmental processes, such as synaptic transmission and neuronal cell adhesion. Accumulating evidence has suggested that synaptic genes might be involved in susceptibility to autism (Kim et al., 2008; Peca et al., 2011). Association studies and mutation analysis of candidate genes have implicated the substantial roles of the synaptic genes Neurexin 1 (NRXN1), Neuroligin 3 (NLGN3), Neuroligin 4 (NLGN4), SH3 and multiple ankyrin repeat domains 3 (SHANK3), and contactin-associated protein-like 2 (CNTNAP2) in autism (Kumar et al., 2009; Kenny et al., 2013).
The neural cell adhesion molecule (NCAM) has been reported to be involved in the development of the central nervous system and its mRNA level might decrease in the serum of autistic individuals (Plioplys et al., 1990). However, another study showed that NCAM mRNA levels were not altered either in serum or postmortem brain samples (Purcell et al., 2001) of autistic children. It has been reported that autism might be associated with the neuronal cell adhesion molecule (NRCAM, 7q31.1) gene polymorphisms (Marui et al., 2009). The CAMs are immunoglobin (Ig) superfamily members, which exist in the nervous systems of both vertebrates and invertebrates. CAMs usually act as surface membrane proteins and they include multiple Ig domains at the N termini and a transmembrane intracellular domain or a glycophosphatidylinositol-linked membrane anchor at the C terminus (Lane et al., 1996).
However, to our knowledge, no genetic study previously focused on the gene coding for the neuronal cell adhesion molecule 1 (NCAM1), as a susceptibility gene of autism. The NCAM1 gene has been implicated in many psychiatric disorders such as bipolar disorder and schizophrenia (Vawter, 2000). The NCAM1 (11q23.1) gene contains 18 exons and it spans approximately 314 kb. It exerts a number of important functions in the development of the central nervous system (Fujita et al., 2000) and it is involved in the plasticity of the adult brain (Doherty et al., 1995; Gascon et al., 2007). Interestingly, studies on animal models found that mice deficient for the NCAM showed behavioral abnormalities in the adulthood, including increased intermale aggression and neuroendocrine response (Stork et al., 1997). NCAM deletion in rats was found to be related to a cognitive and behavioral phenotype reflective of impulsivity, which may be one of the typical clinical characteristics of autism (Matzel et al., 2008). Thus, in the present study, we investigated a panel of markers in NCAM1 (rs4937786, rs12418058, rs1436109, rs584427, and rs605843) in association with autism as well as the parental age at conception in a Chinese Han sample of 237 patients with autism and 451 healthy control subjects.
Moreover, a body of evidence has suggested that advanced paternal age (APA) might increase the risk of autism, schizophrenia, and other neuropsychiatric disorders (Crow, 2003; Lopez-Castroman et al., 2010; Buizer-Voskamp et al., 2011; Hehir-Kwa et al., 2011; Kong et al., 2012; Buizer-Voskamp et al., 2013). Therefore, we also intended to explore the potential relationship between NCAM1 polymorphisms and the paternal age at conception in autistic children.
Materials and Methods
Subjects
The sample for this study consisted of 237 children affected with autism and 451 adult healthy control subjects. These probands and controls were recruited at the Beijing Children's Hospital, China. Among the 237 patients with autism, 207 were male and 30 were female. The age of the children at the time of testing ranged from 2 to 17 years. The assessments of autism were established by two senior psychiatrists using the Diagnostic and Statistical Manual of Mental Disorders, 4th Edition (DSM-IV) criteria, Autism Behavior Checklist (ABC) (Krug et al., 1980), and Childhood Autism Rating Scale (CARS) (Schopler et al., 1980). Excluded criteria included children with fragile X syndrome, tuberous sclerosis, a previously identified chromosomal abnormality by karyotyping analysis, and non-Han Chinese ancestry. The paternal ages at conception (20–42 years) were reported by parents and recorded in the medical documents of autistic children.
Healthy control subjects were eligible for inclusion if they and their parents denied any history of psychiatric disorders. The healthy control subjects consisted of 451 (408 males and 43 females, aged 18–48 years old) Chinese Han subjects. All the healthy control subjects and cases came from the Northern regions of China.
Ethics statements
This study was approved by the Ethics Committee of the Beijing Children's Hospital. All subjects provided written informed consent for participation in this study. Written informed consents for children were obtained from their legal guardians.
Genotyping
Venous blood samples were obtained from children with autism and healthy control subjects. Genomic DNA was extracted using the QIAamp DNA Blood Mini Kit (Qiagen). Information on single-nucleotide polymorphisms (SNPs) was obtained from the dbSNP (www.ncbi.nlm.nih.gov/SNP/) and the international HapMap project (www.hapmap.org/).
PCR amplification was performed in a 25 μL volume containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 mM of each dNTP, 0.3 mM of each primer, 0.6 U of Hotstart Taq DNA polymerase, and 30 ng of the genomic DNA. The conditions used for PCR amplification were an initial denaturation phase at 94°C for 5 min, followed by 36 cycles at 94°C for 30 s, annealing at 55°C–61°C for 30 s, and extension at 72°C for 40 s, followed by a final extension phase at 72°C for 10 min.
PCR products were sequenced, respectively, by DNA sequencing after cleaning the PCR products using a BigDye Terminator Cycle Sequencing Ready Reaction Kit with AmpliTaq DNA polymerase (PE Biosystem). The fragments were separated by electrophoresis on an ABI PRISM genetic analyzer (Applied Biosystem).
Statistical analyses
Deviation from the Hardy–Weinberg equilibrium (HWE) for genotype frequency distributions was tested using the chi-square goodness-of-fit test. All those with frequencies of minor alleles greater than 5% were used as genetic markers in this study. The Haploview (version 4.0) program was used to calculate pairwise linkage disequilibrium (LD) and haplotype-based association analysis using the option of determining blocks based on the criteria defined by Gabriel et al. (Gabriel et al., 2002; Barrett et al., 2005). Allele and genotype frequencies for each polymorphism were compared between patients and controls using chi-square tests. The one-way analysis of variance (ANOVA) tests were used to compare the paternal age at conception among various genotype carriers by using the Statistical Product and Service Solutions (SPSS) software version 13.0. The power of sample size for association tests was evaluated using the Genetic Power Calculator program (http://pngu.mgh.harvard.edu/∼purcell/gpc/) (Purcell et al., 2003). For the disease locus, the minor allele frequency of 0.15, the prevalence of 0.006, the genotype relative risk of Aa=1.5 and AA=1.5, and a D-prime=1 were used to perform the analyses. The significant level was set at p<0.05 (two sided).
Results
The genotype distributions of all five SNPs at the NCAM1 gene detected met the criteria of the Hardy–Weinberg equilibrium (p>0.05). The statistical power of the common SNPs in our study was about ∼0.73.
By using the case–control association analyses, we found that three SNPs at the NCAM1 gene were associated with autism (rs4937786, p=0.015; rs12418058, p=0.0076; rs1436109, p=0.0023; Table 1). Two of them remained significant after the Bonferroni multiple testing correction (rs12418058, pcorrcted=0.038; rs1436109, pcorrcted=0.012).
Table 1.
Association of NCAM1 Polymorphisms with Autism
| Minor allele frequency | ||||||||
|---|---|---|---|---|---|---|---|---|
| SNP | Location in gene | Chromosome position | Minor/major alleles | Case | Control | Chi-square | p-Value | OR (95% CI) |
| rs4937786 | 5′UTR | 112258317 | C/A | 0.414 | 0.377 | 5.863 | 0.015 | 1.17 (1.01–1.35) |
| rs12418058 | Intron | 112413264 | G/A | 0.131 | 0.161 | 7.103 | 0.0076 | 0.79 (0.65–0.94) |
| rs1436109 | Intron | 112496828 | A/C | 0.150 | 0.118 | 9.274 | 0.0023 | 1.32 (1.08–1.57) |
| rs584427 | Coding | 112609206 | A/C | 0.284 | 0.292 | 0.361 | 0.547 | 0.96 (0.80–1.09) |
| rs605843 | Intron | 112630444 | G/A | 0.325 | 0.340 | 0.963 | 0.326 | 0.94 (0.79–1.08) |
SNP, single-nucleotide polymorphism; 5′UTR, 5′ untranslated region; OR, odds ratio; 95% CI, 95% confidence interval.
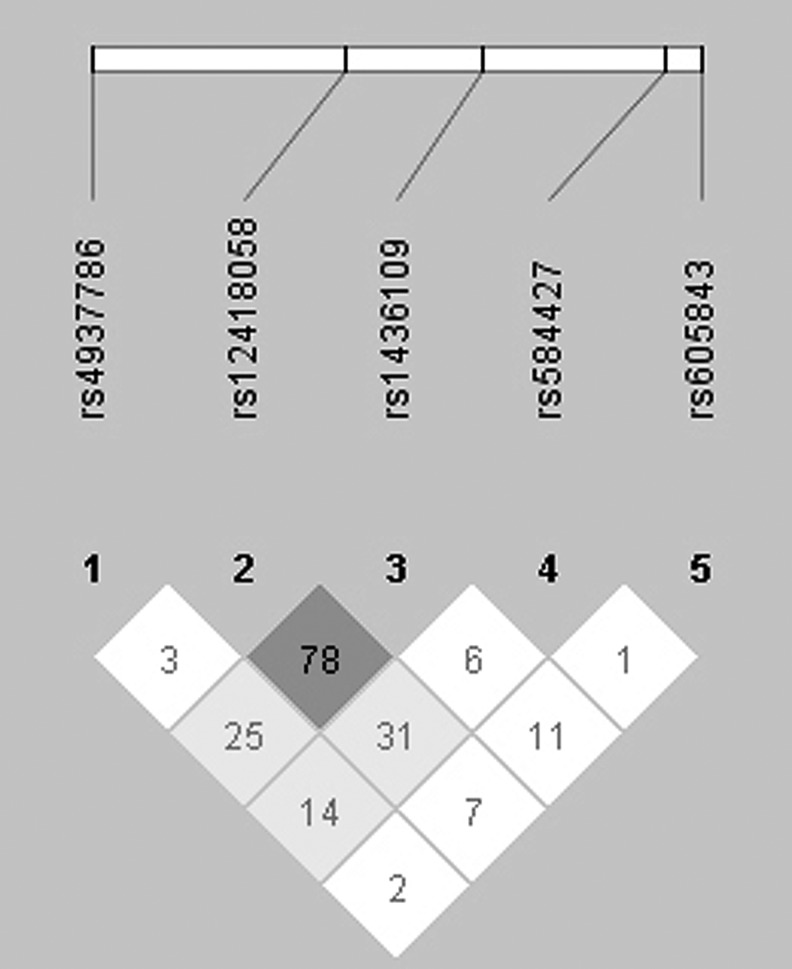
The pairwise LD showed that D′-value between rs12418058 and rs1436109 was 0.78 (Fig. 1). Then, we made a haplotype-based association test and found that two haplotypes constructed by rs12418058-rs1436109 indicated an association with autism (CC, p=0.0291, Ppermutation=0.0719; TA, p=0.0035, Ppermutation=0.0116; Table 2). The results suggested that the rs12418058-rs1436109 haplotype TA remained associated with autism, even after 10,000 times of permutation tests.
FIG. 1.
The linkage disequilibrium (LD) structure of the regions of five tag SNPs of NCAM1 gene, according to the Haploview (solid spine of LD, D′>0.7; www.broad.mit.edu/mpg/haploview/). Markers with LD (D′<1 and LOD>2) are shown in black through grey (color intensity decreases with decreasing D′ value). Regions of low LD and low LOD scores (D′<1 and LOD>2) are shown in white.
Table 2.
Haplotype-Based Association of RS12418058-RS1436109 in NCAM1 with Autism
| Minor allele frequency | ||||
|---|---|---|---|---|
| Haplotype | Case | Control | Chi-square | p-Value |
| TC | 0.722 | 0.729 | 0.213 | 0.6444 |
| CC | 0.128 | 0.153 | 4.761 | 0.0291 |
| TA | 0.146 | 0.115 | 8.513 | 0.0035 |
Moreover, by using the ANOVA tests, we found that two of the SNPs were associated with the parental age at conception (rs12418058, p=0.037; rs1436109, p=0.01; Table 3). Both the risk allele carriers (A of rs12418058 and A of rs1436109) showed association with the APA at conception. These findings suggested obvious dose-dependent effects of genotypes of rs12418058 and rs1436109 in the NCAM1 gene on the parental age at conception in autistic children. That means the advanced parental age at conception may also be one of the risk factors for autism.
Table 3.
Association of NCAM1 Polymorphisms with Parental Age at Conception in Autistic Children
| SNP | Genotype | Parental age at conception (year) | F-value | p-Value |
|---|---|---|---|---|
| rs12418058 | GG | 26.44±4.91 | ||
| GA | 32.68±6.12 | 3.32 | 0.037 | |
| AA | 33.25±6.59 | |||
| rs1436109 | CC | 25.44±4.91 | ||
| CA | 31.68±6.12 | 6.66 | 0.01 | |
| AA | 34.09±6.23 |
Discussion
The present study aimed to investigate a possible relationship between autism and the NCAM1 gene polymorphisms (rs4937786, rs12418058, rs1436109, rs584427, and rs605843). To our knowledge, this is the first study investigating these associations. According to our results, NCAM1 polymorphisms might be associated with autism, especially in the Chinese Han population. In the present study, we used the direct DNA sequencing for genotyping five tag SNPs (rs4937786, rs12418058, rs1436109, rs584427, and rs605843) at the NCAM1 gene. As a result, two of them remained significant after the Bonferroni multiple testing correction (rs12418058, pcorrcted=0.038; rs1436109, pcorrcted=0.012). The LD and haplotype analyses also suggested the association of autism and NCAM1 (rs12418058-rs1436109, haplotype CC, Ppermutation=0.0116). Considering our relatively small sample size, these findings need to be replicated in much larger samples or other populations.
Autism has been described as a kind of neural development disorder that is associated with synaptic dysfunction. Genetic studies have reported several susceptibility genes of autism such as neurexins (NRXN1 and NRXN3, Yan et al., 2008), neuroligins (NLGN3 and NLGN4, Feng et al., 2006), cadherins (CDH8, CDH9, CDH10, CDH13, and CDH15, Wang et al., 2009; Sanders et al., 2011), contactins (CNTN4, CNTN5, and CNTN6, Cottrell et al., 2011; van Daalen et al., 2011), also known as the CAMs. Accumulating evidence has suggested some association between mutations in different CAM genes and autism. However, up to now, it remains unknown as to what is the potential functional implication of the CAMs on behavioral abnormalities in autistic children, such as impairments in social interaction and communication, and stereotypic or repetitive behaviors.
The NCAM, as one of the immunoglobulin superfamily members, have been reported to be engaged in multiple neurodevelopmental processes, including the neuron migration, axonal and/or dendritic projection, and synaptic targeting (Kenny et al., 2013; Minhas et al., 2013). The NCAM has been reported to accumulate in both the pre- and postsynaptic membranes and regulates synapse formation, maturation, and function through intra- and extracellular scaffold proteins (Bukalo and Dityatev, 2012). The presynaptic NCAM has been reported to be involved in the vesicle recycling in both neuronal and endocrine cells (Chan et al., 2005). The postsynaptic NCAM recruits the N-methyl-D-aspartate receptors (NMDARs) and Ca2+/calmodulin-dependent protein kinase II alpha (CaM-Kllalpha), and other postsynaptic density components for both synaptic formation and plasticity (Muller et al., 1996; Bukalo et al., 2004; Sytnyk et al., 2006). On the other hand, NCAM can also control axonal branching in GABAergic synapses of the interneurons (Chattopadhyaya et al., 2013).
Marui et al. (2009) have reported that the NRCAM, another gene coding the NCAM, might be associated with autism Overall, these findings suggested that the early neurodevelopmental process, including NCAM, might play an important role in the pathogenesis of autism.
Moreover, our study also found that two of the NCAM1 SNPs were associated with the parental age at conception in autism (rs12418058, p=0.037; rs1436109, p=0.01). On the other hand, the findings of Buizer-Voskamp et al. and others suggested that the level of global variation burden there is no influence of increased paternal age (Crow, 2003; Malaspina et al., 2005; Lopez-Castroman et al., 2010; Buizer-Voskamp et al., 2011; Hehir-Kwa et al., 2011; Krishnaswamy et al., 2011; Buizer-Voskamp et al., 2013). Recent studies have also reported the association between APA at conception and de novo in schizophrenia and autism (Buizer-Voskamp et al., 2011; Buizer-Voskamp et al., 2013; Gulsuner et al., 2013). Along with our findings, these studies suggested that some early neurodevelopmental processes might involve abnormalities, which may be related to the APA at conception. However, the potential mechanism should be explored in the future.
Nevertheless, two limitations might influence the results we obtained such as a small sample size and the heterogenous genetic backgrounds among different populations. Therefore, to control false-positive findings, many more samples should be genotyped and the replication in other populations should also be implemented in the future.
In summary, our findings have provided preliminary evidence for a significant association of NCAM1 polymorphisms with autism as well as paternal age at conception. Considering the important role of the NCAM1 gene in brain development, our results therefore indicated that the NCAM1 gene is a strong candidate gene for autism.
Acknowledgments
The authors thank all subjects who participated in this study and their colleagues for their assistance in recruiting patients in the study. This research was supported by research grants from the National Natural Science Foundation (grant numbers 81222017, 30870897).
Author Disclosure Statement
No competing financial interests exist.
References
- Barrett JC, Fry B, Maller J, Daly MJ. (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265 [DOI] [PubMed] [Google Scholar]
- Buizer-Voskamp JE, Blauw HM, Boks MP, et al. (2013) Increased paternal age and the influence on burden of genomic copy number variation in the general population. Hum Genet 132:443–450 [DOI] [PubMed] [Google Scholar]
- Buizer-Voskamp JE, Laan W, Staal WG, et al. (2011) Paternal age and psychiatric disorders: findings from a Dutch population registry. Schizophr Res 129:128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukalo O, Dityatev A. (2012) Synaptic cell adhesion molecules. Adv Exp Med Biol 970:97–128 [DOI] [PubMed] [Google Scholar]
- Bukalo O, Fentrop N, Lee AY, et al. (2004) Conditional ablation of the neural cell adhesion molecule reduces precision of spatial learning, long-term potentiation, and depression in the CA1 subfield of mouse hippocampus. J Neurosci 24:1565–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SA, Polo-Parada L, Landmesser LT, Smith C. (2005) Adrenal chromaffin cells exhibit impaired granule trafficking in NCAM knockout mice. J Neurophysiol 94:1037–1047 [DOI] [PubMed] [Google Scholar]
- Chattopadhyaya B, Baho E, Huang ZJ, et al. (2013) Neural cell adhesion molecule-mediated Fyn activation promotes GABAergic synapse maturation in postnatal mouse cortex. J Neurosci 33:5957–5968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JE. (1995) On the publication of the Diagnostic and Statistical Manual of Mental Disorders: 427 Fourth Edition (DSM-IV). Br J Psychiatry166:4–8 [DOI] [PubMed] [Google Scholar]
- Cottrell CE, Bir N, Varga E, et al. (2011) Contactin 4 as an autism susceptibility locus. Autism Res 4:189–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow JF. (2003) Development. There's something curious about paternal-age effects. Science 301:606–607 [DOI] [PubMed] [Google Scholar]
- Doherty P, Fazeli MS, Walsh FS. (1995) The neural cell adhesion molecule and synaptic plasticity. J Neurobiol 26:437–446 [DOI] [PubMed] [Google Scholar]
- Feng J, Schroer R, Yan J, et al. (2006) High frequency of neurexin 1beta signal peptide structural variants in patients with autism. Neurosci Lett 409:10–13 [DOI] [PubMed] [Google Scholar]
- Folstein SE, Rosen-Sheidley B. (2001) Genetics of autism: complex aetiology for a heterogeneous disorder. Nat Rev Genet 2:943–955 [DOI] [PubMed] [Google Scholar]
- Fujita N, Saito R, Watanabe K, Nagata S. (2000) An essential role of the neuronal cell adhesion molecule contactin in development of the Xenopus primary sensory system. Dev Biol 221:308–320 [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, et al. (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229 [DOI] [PubMed] [Google Scholar]
- Gascon E, Vutskits L, Kiss JZ. (2007) Polysialic acid-neural cell adhesion molecule in brain plasticity: from synapses to integration of new neurons. Brain Res Rev 56:101–118 [DOI] [PubMed] [Google Scholar]
- Gulsuner S, Walsh T, Watts AC, et al. (2013) Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell 154:518–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehir-Kwa JY, Rodríguez-Santiago B, Vissers LE, et al. (2011) De novo copy number variants associated with intellectual disability have a paternal origin and age bias. J Med Genet 48:776–778 [DOI] [PubMed] [Google Scholar]
- Kenny EM, Cormican P, Furlong S, et al. (2014) Excess of rare novel loss-of-function variants in synaptic genes in schizophrenia and autism spectrum disorders. Mol Psychiatry 19:872–879 [DOI] [PubMed] [Google Scholar]
- Kim HG, Kishikawa S, Higgins AW, et al. (2008) Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet 82:199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Frigge ML, Masson G, et al. (2012) Rate of de novo mutations and the importance of father's age to disease risk. Nature 488:471–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug DA, Arick J, Almond P. (1980) Behavior checklist for identifying severely handicapped individuals with high levels of autistic behavior. J Child Psychol Psychiatry 21:221–229 [DOI] [PubMed] [Google Scholar]
- Kumar RA, Christian SL. (2009) Genetics of autism spectrum disorders. Curr Neurol Neurosci Rep 9:188–197 [DOI] [PubMed] [Google Scholar]
- Lane RP, Chen XN, Yamakawa K, et al. (1996) Characterization of a highly conserved human homolog to the chicken neural cell surface protein Bravo/Nr-CAM that maps to chromosome band 7q31. Genomics 35:456–465 [DOI] [PubMed] [Google Scholar]
- Lopez-Castroman J, Gómez DD, Belloso JJ, et al. (2010) Differences in maternal and paternal age between schizophrenia and other psychiatric disorders. Schizophr Res 116:184–190 [DOI] [PubMed] [Google Scholar]
- Lord C, Cook EH, Leventhal BL, Amaral DG. (2000) Autism spectrum disorders. Neuron 28:355–363 [DOI] [PubMed] [Google Scholar]
- Marui T, Funatogawa I, Koishi S, et al. (2009) Association of the neuronal cell adhesion molecule (NRCAM) gene variants with autism. Int J Neuropsychopharmacol 12:1–10 [DOI] [PubMed] [Google Scholar]
- Matzel LD, Babiarz J, Townsend DA, et al. (2008) Neuronal cell adhesion molecule deletion induces a cognitive and behavioral phenotype reflective of impulsivity. Genes Brain Behav 7:470–480 [DOI] [PubMed] [Google Scholar]
- Minhas HM, Pescosolido MF, Schwede M, et al. (2013) An unbalanced translocation involving loss of 10q26.2 and gain of 11q25 in a pedigree with autism spectrum disorder and cerebellar juvenile pilocytic astrocytoma. Am J Med Genet A 161A:787–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyauchi S, Voineagu I. (2013) Autism susceptibility genes and the transcriptional landscape of the human brain. Int Rev Neurobiol 113:303–318 [DOI] [PubMed] [Google Scholar]
- Muller D, Wang C, Skibo G, et al. (1996) PSA-NCAM is required for activity-induced synaptic plasticity. Neuron 17:413–422 [DOI] [PubMed] [Google Scholar]
- Peca J, Ting J, Feng G. (2011) SnapShot: autism and the synapse. Cell 147:706:e1. [DOI] [PubMed] [Google Scholar]
- Plioplys AV, Hemmens SE, Regan CM. (1990) Expression of a neural cell adhesion molecule serum fragment is depressed in autism. J Neuropsychiatry Clin Neurosci 2:413–417 [DOI] [PubMed] [Google Scholar]
- Purcell AE, Rocco MM, Lenhart JA, et al. (2001) Assessment of neural cell adhesion molecule (NCAM) in autistic serum and postmortem brain. J Autism Dev Disord 31:183–194 [DOI] [PubMed] [Google Scholar]
- Purcell S, Cherny SS, Sham PC. (2003) Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 19:149–150 [DOI] [PubMed] [Google Scholar]
- Rutter M. (2000) Genetic studies of autism: from the 1970s into the millennium. J Abnorm Child Psychol 28:3–14 [DOI] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, et al. (2011) Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron 70:863–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopler E, Reichler RJ, DeVellis RF, Daly K. (1980) Toward objective classification of childhood autism: Childhood Autism Rating Scale (CARS). J Autism Dev Disord 10:91–103 [DOI] [PubMed] [Google Scholar]
- Steffenburg S, Gillberg C, Hellgren L, et al. (1989) A twin study of autism in Denmark, Finland, Iceland, Norway and Sweden. J Child Psychol Psychiatry 30:405–416 [DOI] [PubMed] [Google Scholar]
- Stork O, Welzl H, Cremer H, Schachner M. (1997) Increased intermale aggression and neuroendocrine response in mice deficient for the neural cell adhesion molecule (NCAM). Eur J Neurosci 9:1117–1125 [DOI] [PubMed] [Google Scholar]
- Sytnyk V, Leshchyns'ka I, Nikonenko AG, Schachner M. (2006) NCAM promotes assembly and activity-dependent remodeling of the post-synaptic signaling complex. J Cell Biol 174:1071–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Daalen E, Kemner C, Verbeek NE, et al. (2011) Social Responsiveness Scale-aided analysis of the clinical impact of copy number variations in autism. Neurogenetics 12:315–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vawter MP. (2000) Dysregulation of the neural cell adhesion molecule and neuropsychiatric disorders. Eur J Pharmacol 405:385–395 [DOI] [PubMed] [Google Scholar]
- Veenstra-Vanderweele J, Cook E, Jr, Lombroso PJ. (2003) Genetics of childhood disorders: XLVI. Autism, part 5: genetics of autism. J Am Acad Child Adolesc Psychiatry 42:116–118 [DOI] [PubMed] [Google Scholar]
- Wang K, Zhang H, Ma D, et al. (2009) Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature 459:528–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Noltner K, Feng J, et al. (2008) Neurexin 1alpha structural variants associated with autism. Neurosci Lett 438:368–370 [DOI] [PubMed] [Google Scholar]