Abstract
Aims: Genetic variations in DNA repair genes may impact repair functions, DNA damage, and breast cancer risk. This study is aimed to assess the associations of genetic polymorphisms in excision repair cross-complementing group 2 (ERCC2) with the risk of developing breast cancer. Materials and Methods: In total, 101 histopathologically confirmed breast cancer cases and 101 age/region-matched healthy controls were genotyped for rs3916840, rs1799793, and rs238416 in ERCC2 by polymerase chain reaction–restriction fragment length polymorphism. Results: The rs238416 heterozygous GA genotype combined with the rs238416 genotypes (GA+AA) showed a significant association with breast cancer susceptibility (corrected p<0.01, odds ratio [OR]=0.29, 95% confidence interval [CI]=0.15–0.54; corrected p<0.01, OR=0.31, 95% CI=0.17–0.56, respectively). The rs238416 GA genotype carriers had a decreased risk of breast cancer. However, we observed no significant association between the rs3916840 and rs1799793 polymorphisms in ERCC2 and breast cancer risk. Moreover, haplotype analysis showed that the ACG haplotype was associated with a significantly decreased risk of breast cancer, whereas the GCG haplotype was associated with a significantly increased risk of breast cancer (corrected p=0.004 and p=0.002, respectively). Multifactor dimensionality reduction analysis demonstrated that the interactions between rs3916840 and rs238416 were significantly synergistic. Conclusion: To the best of our knowledge, this study is the first to demonstrate that the rs238416 heterozygous genotype likely has a higher DNA repair capacity and, thus, can be protective against breast cancer in Chinese Han women.
Introduction
A 2012 epidemiological report of cancer distribution in the Chinese population found that female breast cancer is the most commonly diagnosed cancer (He and Chen, 2013). Breast cancer is the fifth leading cause of cancer-related deaths among Chinese women, and the incidence rate (42.55/100,000) continues to increase (He and Chen, 2013). Molecular epidemiological studies of cancer have identified a number of breast cancer susceptibility genes (e.g., BRCA1, BRCA2, ATM, PTEN, and TP53). Mutations in BRCA1 and BRCA2 account for up to 50% of all hereditary and familial breast cancers (Bennett et al., 2000). Moreover, genetic models show that the susceptibility to breast cancer is likely to be conferred by a large number of loci (Pharoah et al., 2002). To test the predictive value for defining cancer risk groups, association studies have been used to look for genetic variation across many loci in the population. Recently, genome-wide association studies have suggested that polymorphic variants may influence the susceptibility to breast cancer (Easton et al., 2007).
DNA repair systems are essential for responding to damage caused by endogenous and exogenous carcinogens as well as mutagens. Functioning DNA repair systems play a central role in reducing the risk of all cancers (Berwick and Vineis, 2000), including breast cancer. Women with breast cancer have been reported to have significantly reduced DNA repair proficiencies (Helzlsouer et al., 1995). The accumulation of DNA damage may contribute to the initiation of aberrant cell growth and carcinogenesis development (Helzlsouer et al., 1996). In addition, several enzymes involved in the nucleotide excision repair (NER) pathway are known to be associated with breast cancer. These enzymes may also partake in other regulatory processes in the cell, including NER, transcription initiation (Spitz et al., 2001), cell cycle progression (Robles et al., 1999), and apoptosis (Barnes and Camplejohn, 1996). The DNA helicase encoded by the excision repair cross-complementing group 2 (ERCC2) gene (also known as XPD) is a key NER enzyme that intervenes in the transcription-coupled NER sub-pathway and can cause Xeroderma pigmentosum when mutated in the germline (Coin et al., 1998). Recent studies of DNA repair and breast cancer have focused on the association between single nucleotide polymorphisms (SNPs) in genes involved in DNA repair and the likelihood of developing breast cancer. Justenhoven et al. (2004) found a highly significant association between ERCC2 Asp312Asp and breast cancer risk in a German population; with GG homozygote individuals having a two-fold increase in risk. A huge review also found some statistically significant associations between XPD/ERCC2 SNPs and skin, breast, and lung cancers (Manuguerra et al., 2006). More recently, Samson et al. (2011) reported that the XPD Gln/Gln genotype is significantly associated with an increased risk of breast cancer in south Indian women. Similarly, Roberts et al. (2011) concluded that variants in base excision repair and NER genes might influence a person's risk of developing breast cancer. Smith et al. (2008) also suggested that individual DNA repair genotypes may have a small effect on breast cancer risk; however, there is a combined effect of multiple DNA repair genotypes from different pathways on the breast cancer risk. Nevertheless, contradictory results have been observed due to differences in the genetic background, the environment, where the study population resides in, and the sample size (Pabalan et al., 2010; Yao et al., 2010).
The aim of this study is to determine whether common polymorphisms (frequency ≥5%) in the ERCC2/XPD gene are involved in breast cancer susceptibility. We used HapMap data to identify SNPs for genotyping in a breast cancer case–control study of Chinese women. The HapMap data represent a key resource for researchers to identify genes that affect health, disease, and responses to drugs and environmental factors. The goal of the International HapMap Project is to develop a haplotype map of the human genome, which could help refine association studies of common disease variants (The International HapMap Consortium, 2005; Frazer et al., 2007). We selected three tag SNPs (rs3916840, rs1799793, and rs238416) in ERCC2 using a functional analysis tool and then investigated the associations between the genotype and both the risk and clinicopathological features of breast cancer among Han women in the Gansu Province, a less developed area in Northwest China.
Materials and Methods
Study subjects and clinical data collection
In total, 106 histopathologically confirmed breast cancer cases (5 cases voluntarily withdrew within 1 month of entry) were recruited from the Department of Breast Surgery between August 2011 and August 2013 at the Gansu Provincial Cancer Hospital, Gansu Province, China. All cases were previously untreated (before chemotherapy or radiotherapy) for cancer. In total, 101 cancer-free controls were selected at random from the Health Examination Surveys of the hospital and were age- and region-matched to the cases. The eligibility criteria for the controls included normal mammography results and no prior cancer history. All covariate data were obtained from medical charts and questionnaires. All study participants provided a 5-mL peripheral blood sample drawn into EDTA tubes. The ethics committee of the Gansu Provincial Medical Science Institute approved the study, and a written informed consent was obtained from all enrolled patients.
SNP identification and selection
Tagging SNPs (tag SNPs) can capture the common genetic variations within a gene. Tag SNPs serve as markers to detect associations between a particular region and a disease, regardless of whether the tag SNPs themselves have a functional effect (Gabriel et al., 2002). Using tagger pairwise selection approaches, we selected tag SNPs from the HapMap database (www.hapmap.org, HapMap Data Rel 24/phaseII Nov08, on NCBI B36 assembly, dbSNP b126) using the tag SNPs software available online using the following criteria: r2 cutoff of 0.8 and minor allele frequency cutoff of 0.05 in samples of Han Chinese in Beijing. The HapMap data showed that there were seven tag SNPs that spanned the ERCC2 region. Then, we used the FastSNP software online (http://fastsnp.ibms.sinica.edu.tw/pages/input_CandidateGeneSearch.jsp), a functional analysis and selection tool for SNPs, to analyze the putative functional effects for each tag SNP through their particular risk ranking. The details of the FastSNP method have been published previously (Yuan et al., 2006). Briefly, FastSNP is a web server that allows users to efficiently identify and prioritize high-risk SNPs according to their phenotypic risks and putative functional effects, and the functional effect information used for SNP prioritization is always up-to-date. FastSNP provides a decision tree to assess the risk of a SNP. The decision tree classifies a SNP into 1 of 13 types of functional effects, each of which is assigned a risk ranking number between 0 and 5. A high risk rank implies a high risk level. Finally, three tag SNPs with high risk ranks (rs3916840, rs1799793, and rs238416) were selected.
Genotyping
Genomic DNA was extracted from 5-mL peripheral blood samples using the universal genomic DNA Extraction Kit VER.3.0 (TaKaRa Biotechnology Co., Ltd., Dalian, China). The tag SNPs (rs3916840, rs1799793, and rs238416) were detected using the polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) method. The primer sequences and restriction endonucleases used are shown in Table 1. The primers used for PCR-RFLP were synthesized by Sangon Biotechnology Co., Ltd. (Shanghai, China). Restriction endonucleases were obtained from TaKaRa Biotechnology Co., Ltd. PCR amplification was performed with a S1000TM Thermal Cycler (Bio-Rad Laboratories, Hercules, CA) in a 30-μL reaction solution containing 0.1 μg of genomic DNA, 15 μL of 2× GoTaq® Green Master Mix (Promega Corporation, Madison, WI), and a pair of primers at a final concentration of 100 nM. The PCR cycle conditions were a hot start at 95°C for 5 min, followed by 32 cycles of denaturation at 95°C for 30 s, annealing (58°C for rs3916840, 58°C for rs1799793, and 50°C for rs238416) for 30 s and extension at 72°C for 20 s. Finally, the products were incubated at 72°C for 10 min. Then, 10 μL of PCR products was digested with restriction endonucleases in a 20-μL reaction mixture according to the manufacturer's instructions, and the digested products were analyzed by electrophoresis on a 3.0% agarose gel. Negative controls (no template controls) and controls with a known genotype were included in the assays. Genotyping of 20% of the samples selected at random was repeated and yielded 100% accurate genotyping results.
Table 1.
Primer Sequences and Restriction Endonucleases for Three SNPs
| SNPs | Base change | Allele frequencies in HapMap CHB | Primersa | Enzymes | Digested fragments (bp) |
|---|---|---|---|---|---|
| rs3916840 | C/T | C 0.942/T 0.058 | F: GTCTGTCTCCTACTGGACTGCGTA | Csp6I | C (21,198) |
| R: ATAAGTTCTGGGGGGTTAGGGATG | T (219) | ||||
| rs1799793 | G/A | G 0.933/A 0.067 | F: ACCTGGCCAACCCCGTGCTGCTC | TaqI | G (22,199) |
| R: TCTCCTGCACCACATGCTGCACAC | A (221) | ||||
| rs238416 | G/A | G 0.512/A 0.488 | F: GGGGGTCAAGGTTCATTTTTTGGTTCCATCCG | MspI | G (30,182) |
| R: TGACTGTGAGGCAGGCAGAGCCAATCAGAG | A (212) |
Sequences for primers and probes in 5′ to 3′ order.
CHB, Han Chinese in Beijing, China; F, forward; R, reverse; SNP, single nucleotide polymorphism.
Statistical analysis
To ensure that the controls were representative of the general population, the deviation of the genotype frequencies of all three ERCC2 tag SNPs in the control subjects from those expected under Hardy–Weinberg equilibrium was assessed using the chi-square goodness-of-fit test. Pearson's chi-square test or Fisher's exact test (when the expected number in any cell was <5) was used to compare the distribution of the ERCC2 genotypes between the cases and controls. The association of ERCC2 gene polymorphisms with the clinicopathological features of breast cancer was estimated by calculating the odds ratios (ORs) and 95% confidence intervals (CIs) using unconditional logistic regression adjusted for age at the onset of breast cancer. The associations were analyzed using different genetic models (codominant, dominant, and recessive models) as previously described (Xu et al., 2012). The SHEsis online software (http://analysis.bio-x.cn/myanalysis.php) (Shi and He, 2005; Li et al., 2009) was used to calculate the frequency distributions of the ERCC2 haplotypes in the cases and controls.
In addition, the effects of SNP–SNP interactions were explored using the multifactor dimensionality reduction (MDR) software (available free at www.epistasis.org) to analyze the cross action of multilocus genotype combinations related to breast cancer susceptibility. MDR is a novel and powerful statistical method that has reasonable power for the detection and characterization of nonlinear interactions among discrete genetic and environmental attributes (Namkung et al., 2009). The MDR method combines attribute selection, attribute construction, classification, cross-validation, and visualization to provide a comprehensive and powerful data mining approach to the detection, characterization, and interpretation of gene–gene and gene–environment interactions (Hahn et al., 2003). With MDR, multilocus genotypes are pooled into high-risk and low-risk groups, effectively reducing the dimensionality of the genotype predictors from N dimensions to one dimension. The new one-dimensional multilocus genotype variable is evaluated for its ability to classify and predict disease status using cross-validation and permutation testing. Furthermore, the best MDR model is selected, which is the model with the maximum testing accuracy and highest cross-validation consistency.
Haplotype of frequencies >3% in the combined cases and controls were examined. Bonferroni correction was used to adjust for multiple testing. Statistical significance was uniformly set at p<0.05, and the statistical analyses were performed using SPSS 16.0 software (SPSS, Inc., Chicago, IL).
Results
Demographic and pathological characteristics
All demographic and pathological characteristics of the breast cancer cases and controls are shown in Table 2. The mean ages of the breast cancer cases and controls were 45.2 years (SD=7.4) and 45.5 years (SD=7.6), respectively. The age distribution and mean ages were similar between the cases and controls. No deviation from Hardy–Weinberg equilibrium was found in the genotype frequencies for all three SNPs in the control subjects (p>0.05).
Table 2.
Characteristics Between Breast Cancer Cases and Controls
| Characteristics | Cases (%) | Controls (%) | p-Value |
|---|---|---|---|
| Age (years) | |||
| Mean (±SD) | 45.2 (±7.4) | 45.5 (±7.6) | 0.83a |
| ≤40 | 28 (27.72) | 22 (21.78) | |
| 41–50 | 50 (49.50) | 58 (57.43) | |
| 51–60 | 19 (18.81) | 18 (17.82) | |
| >60 | 4 (3.96) | 3 (2.97) | 0.69b |
| Histological type | |||
| Infiltrating duct carcinoma | 88 (87.13) | ||
| Others | 13 (12.87) | ||
| Tumor size (cm) | |||
| ≤3 | 55 (54.46) | ||
| >3 | 36 (35.64) | ||
| Unknown | 10 (9.90) | ||
| LN involvement | |||
| Positive | 22 (21.78) | ||
| Negative | 48 (47.53) | ||
| Unknown | 31 (30.69) | ||
| ER | |||
| Positive | 56 (55.45) | ||
| Negative | 42 (41.58) | ||
| Unknown | 3 (2.97) | ||
| PR | |||
| Positive | 52 (51.49) | ||
| Negative | 46 (45.54) | ||
| Unknown | 3 (2.97) | ||
| P53 | |||
| Positive | 50 (49.51) | ||
| Negative | 43 (42.57) | ||
| Unknown | 8 (7.92) | ||
| Her-2 | |||
| Positive | 30 (29.70) | ||
| Negative | 68 (67.33) | ||
| Unknown | 3 (2.97) | ||
| Ki-67 | |||
| Positive | 92 (91.09) | ||
| Negative | 5 (4.95) | ||
| Unknown | 4 (3.96) | ||
For paired-sample t-test.
For chi-square test (two-sided).
ER, estrogen receptor; Her-2, human epidermal growth factor receptor 2; Ki-67, monoclonal antibody Ki-67; LN, lymph node; PR, progesterone receptor.
The association of ERCC2 gene polymorphisms with breast cancer risk
The genetic models of the three SNPs are shown in Table 3. When the homozygous genotypes of the higher frequency allele of the three tag SNPs were used as the reference group, the rs238416 heterozygous GA genotype and A allele showed an association with breast cancer susceptibility (p<0.001, OR=0.29, 95% CI=0.15–0.54; p=0.003, OR=0.54, 95% CI=0.36–0.81, respectively). After performing multiple tests by a stricter traditional Bonferroni adjustment, these associations were still significant (corrected p<0.01 and p=0.027, respectively). Furthermore, in the dominant model, the combined rs238416 genotypes (GA+AA) conferred a decreased risk of breast cancer (p<0.001, OR=0.31, 95% CI=0.17–0.56). This association was statistically significant after correction by the Bonferroni test (corrected p<0.01). However, for the overall genotype frequencies of rs3916840 and rs1799793, no significant difference was observed between the breast cancer patients and the control population.
Table 3.
Genotype Frequencies of ERCC2 Gene Polymorphisms in Controls and Cases and Their Associations with Breast Cancer
| SNPs | Genotype | Case n (%) | Control n (%) | OR (95% CI)a | p-Value | pc |
|---|---|---|---|---|---|---|
| rs3916840 | CC | 91 (90.1) | 90 (89.1) | Reference | ||
| CT | 10 (9.9) | 11 (10.9) | 0.889 (0.364–2.222) | 0.818 | NS | |
| TT | 0 | 0 | ||||
| C | 192 (95.1) | 191 (94.6) | Reference | |||
| T | 10 (4.9) | 11 (5.4) | 0.904 (0.375–2.179) | 0.823 | NS | |
| rs1799793 | GG | 84 (83.2) | 89 (88.1) | Reference | ||
| GA | 17 (16.8) | 12 (11.9) | 1.501 (0.667–3.330) | 0.316 | NS | |
| AA | 0 | 0 | ||||
| G | 185 (91.6) | 190 (94.1) | Reference | |||
| A | 17 (8.4) | 12 (5.9) | 1.455 (0.676–3.130) | 0.335 | NS | |
| rs238416 | GG | 52 (51.5) | 25 (24.7) | Reference | ||
| GA | 37 (36.6) | 62 (61.4) | 0.287 (0.153–0.537) | <0.001 | <0.01 | |
| AA | 12 (11.9) | 14 (13.9) | 0.412 (0.166–1.020) | 0.052 | NS | |
| G | 141 (69.8) | 112 (55.4) | Reference | |||
| A | 61 (30.2) | 90 (44.6) | 0.538 (0.358–0.810) | 0.003 | 0.027 | |
| Dominantb | 0.310 (0.171–0.563) | <0.001 | <0.01 | |||
| Recessivec | 0.838 (0.367–1.913) | 0.674 | NS |
ORs were adjusted for age of onset.
The dominant model: comparing the combination of heterozygotes and minor allele homozygotes with the major allele homozygotes.
The recessive model: comparing minor allele homozygotes with the combination of heterozygotes and major allele homozygotes.
CI, confidence interval; ERCC2, excision repair cross-complementing group 2; NS, not significant; OR, odds ratio; pc, corrected p-value (after Bonferroni multiple adjustment).
ERCC2 gene polymorphisms and clinicopathological features
ERCC2 gene polymorphisms were also analyzed to investigate their associations with clinicopathological features, including lymph node metastasis, staging, histologic classification, the statuses of the estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (Her-2), P53 protein, Ki-67 antibody, and the triple-negative phenotype. The significant results are shown in Table 4. However, associations were only found for rs1799793 and rs238416. For rs1799793, the frequency of the GA genotype was decreased in PR-positive cases compared with the GG genotype (p=0.038, OR=0.30, 95% CI=0.10–0.94). Meanwhile, women carrying the GA genotype were extremely likely to have triple-negative breast cancer compared with women with the GG genotype (p=0.037, OR=0.28, 95% CI=0.09–0.93). For rs238416, compared with the GG genotype, the GA genotype had a lower frequency in P53-positive cases (p=0.026, OR=0.35, 95% CI=0.14–0.88), and a significant association was also found in the dominant model of P53-positive cases (p=0.018, OR=0.36, 95% CI=0.16–0.84). Furthermore, the rs238416 SNP in the dominant models was associated with ER-positivity (p=0.043, OR=2.33, 95% CI=1.03–5.29). However, these associations were not statistically significant after correction by the Bonferroni test.
Table 4.
Clinicopathological Features and ERCC2 Gene Polymorphisms
| n (%) | |||||||
|---|---|---|---|---|---|---|---|
| Clinical features | SNP | Genotype | Positive | Negative | OR (95% CI)a | p-Value | pc |
| ER | rs238416 | GG | 23 (41.1) | 26 (61.9) | Reference | ||
| GA | 25 (44.6) | 12 (28.6) | 2.40 (0.98–5.85) | 0.055 | NS | ||
| AA | 8 (14.3) | 4 (9.5) | 2.29 (0.61–8.65) | 0.220 | NS | ||
| Dominantb | 2.33 (1.03–5.29) | 0.043 | NS | ||||
| Recessivec | 1.58 (0.44–5.66) | 0.479 | NS | ||||
| PR | rs1799793 | GG | 47 (90.4) | 34 (73.9) | Reference | ||
| GA | 5 (9.6) | 12 (26.1) | 0.30 (0.10–0.94) | 0.038 | NS | ||
| AA | 0 | 0 | — | ||||
| P53 | rs238416 | GG | 31 (62.0) | 16 (37.2) | Reference | ||
| GA | 14 (28.0) | 20 (46.5) | 0.35 (0.14–0.88) | 0.026 | NS | ||
| AA | 5 (10.0) | 7 (16.3) | 0.36 (0.10–1.32) | 0.124 | NS | ||
| Dominant | 0.36 (0.16–0.84) | 0.018 | NS | ||||
| Recessive | 0.57 (0.17–1.95) | 0.372 | NS | ||||
| TNBC | rs1799793 | GG | 11 (64.7) | 70 (86.4) | Reference | ||
| GA | 6 (35.3) | 11 (13.6) | 0.28 (0.09–0.93) | 0.037 | NS | ||
| AA | 0 | 0 | — | ||||
ORs were adjusted for age of onset.
The dominant model: comparing the combination of heterozygotes and minor allele homozygotes with the major allele homozygotes.
The recessive model: comparing minor allele homozygotes with the combination of heterozygotes and major allele homozygotes.
TNBC, triple-negative breast cancer.
Haplotype analyses
We further analyzed the distribution of haplotypes in the cases and controls. Seven haplotypes were constructed for the ERCC2 gene based on the three tag SNPs (rs3916840, rs1799793, and rs238416), and all haplotypes with a frequency >3% were selected for analysis. The main haplotype frequencies and distributions are summarized in Table 5. Two haplotypes were found to be significantly different between the cases and controls. The ACG haplotype had a lower frequency in the cases than in the controls (p=8.71×10−4). Moreover, the GCG haplotype had a higher frequency in the cases (p=3.51×10−4). After correction by the Bonferroni test, the two haplotypes, ACG and GCG, were still significantly associated with breast cancer susceptibility (corrected p=0.004 and p=0.002, respectively). However, no significant differences were found for the other ERCC2 haplotypes.
Table 5.
Frequency Distributions of Haplotypes of ERCC2 in Cases and Controls
| Haplotype | Case freq (%) | Control freq (%) | p-Valuea | OR (95% CI) | pc |
|---|---|---|---|---|---|
| ACA | 3.40 (0.017) | 3.32 (0.016) | — | — | — |
| ACG | 52.80 (0.261) | 86.67 (0.429) | 8.71e-04 | 0.490 (0.321–0.747) | 0.004 |
| GCA | 11.94 (0.059) | 8.68 (0.043) | 0.414 | 1.451 (0.591–3.562) | NS |
| GCG | 123.86 (0.613) | 92.33 (0.457) | 3.51e-04 | 2.089 (1.392–3.137) | 0.002 |
| GTG | 3.54 (0.018) | 11.00 (0.054) | 0.054 | 0.320 (0.095–1.079) | NS |
Frequencies <0.03 in both case and control have been excluded. The order of SNPs in ERCC2 is rs1799793, rs3916840, and rs238416.
p-Value calculated by Fisher's exact test.
SNP–SNP interactions reveal moderate synergistic effects
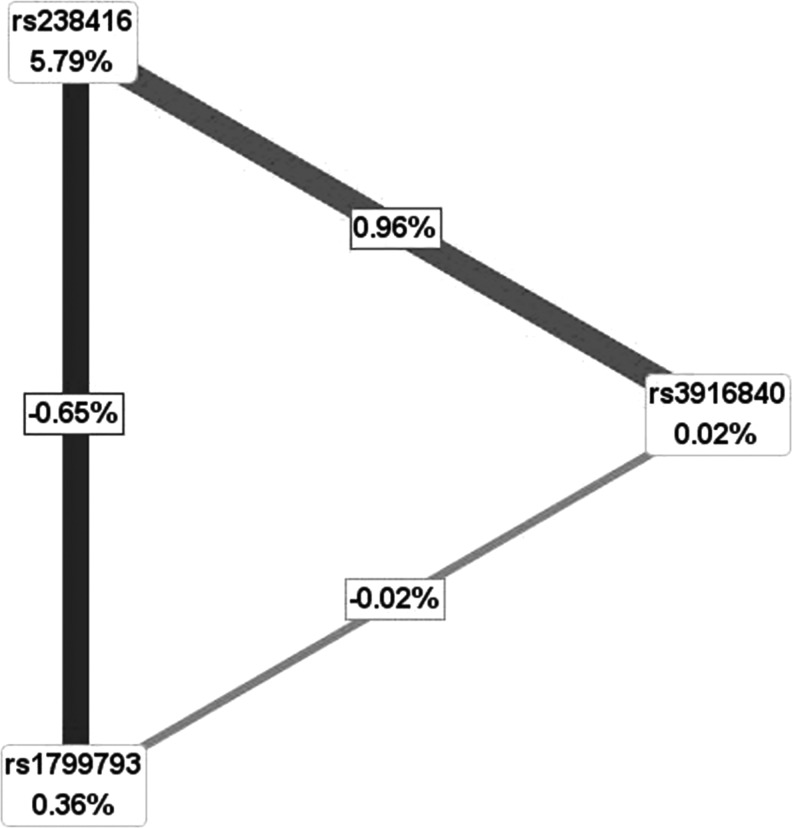
We performed MDR analysis to reveal the SNP–SNP interactions in this general population and compared our results with the available data on breast cancer. All possible interactions were exhaustively examined. The numerical results and gene information for the identified best model of interactions by MDR are summarized in Table 6. We found the most potent interaction in breast cancer compared with the controls to be rs3916840, rs1799793, and rs238416 with a testing balance accuracy of 0.6287 and cross validation consistency of 10 (p=0.003). Furthermore, we applied interaction entropy algorithms to support this interpretation of the relationship among the variables. However, in this case, the three-way interaction model did not seem to bring any meaningful improvement upon the other best model. As described in Figure 1, rs238416 is the strongest factor (entropy explained: 5.79%) for carcinomatosis development. The entropy of the rs3916840–rs238416 is 0.96%, in contrast, the entropies of the rs1799793–rs238416 and rs1799793–rs3916840 interactions are both negative.
Table 6.
MDR Interaction Analysis Between SNP–SNP
| MDR models | Training balance accuracy | Testing balance accuracy | CVC | p-Valuea |
|---|---|---|---|---|
| rs238416 | 0.6337 | 0.5941 | 10/10 | 0.039 |
| rs3916840 rs238416 | 0.6436 | 0.6139 | 10/10 | 0.008 |
| rs3916840 rs1799793 rs238416 | 0.6535 | 0.6287 | 10/10 | 0.003 |
p-Values as calculated after 1000 permutations.
CVC, cross validation consistency; MDR, multifactor dimensionality reduction.
FIG. 1.
Single nucleotide polymorphism (SNP)–SNP interaction model. The model describes the percent of entropy explanation by interactions to support interpretation of the relationship among SNPs. The boxes describe the SNPs with the percentage of entropy explained. Interaction is represented by lines. Interaction model is constructed on cases versus controls.
Discussion
Polymorphic variants in ERCC2 are good candidates for evaluating breast cancer susceptibility because of its key role in the NER pathway. The DNA repair capacity is well known to be an important determinant of the susceptibility to carcinogenesis (Parshad et al., 1996). The NER pathway is a crucial mechanism for repairing DNA damage and protecting against gene mutations. Moreover, ERCC2 is a key component of this pathway. The protein encoded by this gene is involved in transcription-coupled NER and an integral member of the BTF2/TFIIH basal transcription factor complex (Schaeffer et al., 1994). In this study, we estimated the breast cancer risks associated with three polymorphisms in ERCC2 using a HapMap-based case–control study among Han women in the Gansu Province, a less developed area in Northwest China. An association was observed for the rs238416 heterozygous GA genotype and combined genotype (GA+AA) compared with the homozygous GG genotype in breast cancer patients. We found that the rs238416 GA genotype carriers had a decreased risk of breast cancer, indicating that being heterozygous at rs238416 likely provides higher DNA repair capacity and thus be protective against breast cancer. However, this protective effect was not found for the rs3916840 and rs1799793 polymorphisms. Our current results for the rs1799793 polymorphism are consistent with prior studies showing that this SNP is not associated with breast cancer (Kuschel et al., 2005; Frolova et al., 2009; Pabalan et al., 2010). To the best of our knowledge, the loci (rs3916840 and rs238416) of these polymorphisms have not been evaluated previously with respect to breast cancer. Nevertheless, we cannot exclude the possibility that the associations found in the current study occurred by chance, and further verification in larger samples and different regional population groups, as well as functional studies are required to confirm these results.
In addition, the analysis between ERCC2 gene polymorphisms and clinicopathological features confirmed the association between these three SNPs and prognostic factors. As a result, the rs238416 polymorphism was associated with the clinicopathological features of breast cancer. Women carrying the GA genotype had a lower frequency of P53-positive breast cancer. P53 is a vital regulator of genomic stability that controls the cell cycle and induces apoptosis when cell damage is beyond repair (Lara et al., 2011). P53 protein expression has been related to poor outcomes in breast cancer (Kroger et al., 2006; Yamashita et al., 2006). A mutant P53 protein not only loses its tumor suppressive function, but also gains new abilities that promote carcinogenesis (Brosh and Rotter, 2009). Therefore, our current results indicate that rs238416 GA genotype deficiency may possibly, more easily, lead to oncogenesis.
We further analyzed the associations between haplotypes and breast cancer risk. Based on this analysis, the ACG haplotype was determined to have a higher frequency in the controls, whereas the GCG haplotype was found to have a lower frequency. Hence, we infer that the ACG haplotype may play an important role in decreasing the breast cancer risk, but that the GCG haplotype is associated with a significantly increased risk of breast cancer. The MDR analysis identified a three-locus interaction that is significantly associated with breast cancer risk. However, in this case, the three-way interaction model did not seem to bring any meaningful improvement upon the other model. The best main effect model of rs238416 plays a more active role. The rs238416 polymorphism is the strongest risk factor for breast cancer, and it may interact synergistically with the other SNPs for carcinomatosis development. This particular phenomenon was also observed with the rs238416 polymorphism in DNA repair genes in bladder cancer (Andrew et al., 2006). The hypothesis of SNP–SNP interactions lacked evidence to support these models. Moreover, the entropies of the rs1799793–rs238416 and rs1799793–rs3916840 interactions are both negative, indicating that on its own, rs1799793 plays a minimally antagonistic role or has no effect. This result is similar to our previous result that rs1799793 is not associated with the risk of breast cancer. Although the interaction of rs238416 and rs3916840 was shown to be significantly synergistic, rs3916840 did not display any correlation with the risk of breast cancer. Hence, whether such a synergistic effect does exist needs to be further investigated. Nevertheless, it is apparent that rs238416 plays a dominant role in the mutual interactions of these three genes. Thus, the DNA repair capacity contributed by different repair machinery and the synergistic interaction between SNPs associated with the most potent risk of breast cancer can be suggested to be the ultimate determinants of breast cancer in an individual.
In conclusion, our study indicated that the three analyzed ERCC2 polymorphisms and some of the corresponding haplotypes are likely associated with the breast cancer risk in Han women of Northwest China. It is worth noting that the rs238416 heterozygous genotype likely has a higher DNA repair capacity and, thus, may be protective against breast cancer. To the best of our knowledge, this study is the first to demonstrate that the ERCC2 genotype encoding rs238416 is correlated with an increased risk of breast cancer. This finding calls attention to genetic differences in DNA repair genes that can lead to differences in cancer susceptibility. Our current understanding of this mechanism will contribute to improving the prevention and individualized treatment of breast cancer. However, there are several limitations in the current study. These limitations include the small sample size and the insufficient availability of information on the person's family history of cancer and lifestyle. In the future, available cryopreserved lymphocytes for further functional assays are needed to confirm our results.
Acknowledgments
The authors wish to thank all patients and healthy volunteers for providing blood samples. The authors also thank the nurses, clinical researchers, and regulatory coordinators at the Department of Breast Surgery and Research Center of Translational Medicine at Gansu Provincial Cancer Hospital for their collaboration. This study was supported by the Key Project of the Eleventh-Five Year Research Program of Gansu Province of China (Grant No. 1011FKCA089).
Author Disclosure Statement
No competing financial interests exist.
References
- Andrew AS, Nelson HH, Kelsey KT, et al. (2006) Concordance of multiple analytical approaches demonstrates a complex relationship between DNA repair gene SNPs, smoking and bladder cancer susceptibility. Carcinogenesis 27:1030–1037 [DOI] [PubMed] [Google Scholar]
- Barnes DM, Camplejohn RS. (1996) P53, apoptosis, and breast cancer. J Mammary Gland Biol Neoplasia 1:163–175 [DOI] [PubMed] [Google Scholar]
- Bennett IC, Gattas M, Teh BT. (2000) The management of familial breast cancer. Breast 9:247–263 [DOI] [PubMed] [Google Scholar]
- Berwick M, Vineis P. (2000) Markers of DNA repair and susceptibility to cancer in humans: an epidemiologic review. J Natl Cancer Inst 92:874–897 [DOI] [PubMed] [Google Scholar]
- Brosh R, Rotter V. (2009) When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer 9:701–713 [DOI] [PubMed] [Google Scholar]
- Coin F, Marinoni JC, Rodolfo C, et al. (1998) Mutations in the XPD helicase gene result in XP and TTD phenotypes, preventing interaction between XPD and the p44 subunit of TFIIH. Nat Genet 20:184–188 [DOI] [PubMed] [Google Scholar]
- Easton DF, Pooley KA, Dunning AM, et al. (2007) Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 447:1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Ballinger DG, Cox DR, et al. (2007) A second generation human haplotype map of over 3.1 million SNPs. Nature 449:851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova N, Edmonds MD, Bodenstine TM, et al. (2009) A shift from nuclear to cytoplasmic breast cancer metastasis suppressor 1 expression is associated with highly proliferative estrogen receptor-negative breast cancers. Tumour Biol 30:148–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, et al. (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229 [DOI] [PubMed] [Google Scholar]
- Hahn LW, Ritchie MD, Moore JH. (2003) Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics 19:376–382 [DOI] [PubMed] [Google Scholar]
- He J, Chen W. (2013) 2012 Chinese Cancer Registry Annual Report, 1st ed. Military Medical Science Press, Beijing, pp. 27–36 [Google Scholar]
- Helzlsouer KJ, Harris EL, Parshad R, et al. (1995) Familial clustering of breast cancer: possible interaction between DNA repair proficiency and radiation exposure in the development of breast cancer. Int J Cancer 64:14–17 [DOI] [PubMed] [Google Scholar]
- Helzlsouer KJ, Harris EL, Parshad R, et al. (1996) DNA repair proficiency: potential susceptiblity factor for breast cancer. J Natl Cancer Inst 88:754–755 [DOI] [PubMed] [Google Scholar]
- Justenhoven C, Hamann U, Pesch B, et al. (2004) ERCC2 genotypes and a corresponding haplotype are linked with breast cancer risk in a German population. Cancer Epidemiol Biomarkers Prev 13:2059–2064 [PubMed] [Google Scholar]
- Kroger N, Milde-Langosch K, Riethdorf S, et al. (2006) Prognostic and predictive effects of immunohistochemical factors in high-risk primary breast cancer patients. Clin Cancer Res 12:159–168 [DOI] [PubMed] [Google Scholar]
- Kuschel B, Chenevix-Trench G, Spurdle AB, et al. (2005) Common polymorphisms in ERCC2 (Xeroderma pigmentosum D) are not associated with breast cancer risk. Cancer Epidemiol Biomarkers Prev 14:1828–1831 [DOI] [PubMed] [Google Scholar]
- Lara JF, Thor AD, Dressler LG, et al. (2011) p53 Expression in node-positive breast cancer patients: results from the Cancer and Leukemia Group B 9344 Trial (159905). Clin Cancer Res 17:5170–5178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang Z, He Z, et al. (2009). A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn). Cell Res 19:519–523 [DOI] [PubMed]
- Manuguerra M, Saletta F, Karagas MR, et al. (2006) XRCC3 and XPD/ERCC2 single nucleotide polymorphisms and the risk of cancer: a HuGE review. Am J Epidemiol 164:297–302 [DOI] [PubMed] [Google Scholar]
- Namkung J, Kim K, Yi S, et al. (2009) New evaluation measures for multifactor dimensionality reduction classifiers in gene-gene interaction analysis. Bioinformatics 25:338–345 [DOI] [PubMed] [Google Scholar]
- Pabalan N, Francisco-Pabalan O, Sung L, et al. (2010) Meta-analysis of two ERCC2 (XPD) polymorphisms, Asp312Asn and Lys751Gln, in breast cancer. Breast Cancer Res Treat 124:531–541 [DOI] [PubMed] [Google Scholar]
- Parshad R, Price FM, Bohr VA, et al. (1996) Deficient DNA repair capacity, a predisposing factor in breast cancer. Br J Cancer 74:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pharoah PD, Antoniou A, Bobrow M, et al. (2002) Polygenic susceptibility to breast cancer and implications for prevention. Nat Genet 31:33–36 [DOI] [PubMed] [Google Scholar]
- Roberts MR, Shields PG, Ambrosone CB, et al. (2011) Single-nucleotide polymorphisms in DNA repair genes and association with breast cancer risk in the web study. Carcinogenesis 32:1223–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles AI, Wang XW, Harris CC. (1999) Drug-induced apoptosis is delayed and reduced in XPD lymphoblastoid cell lines: possible role of TFIIH in p53-mediated apoptotic cell death. Oncogene 18:4681–4688 [DOI] [PubMed] [Google Scholar]
- Samson M, Singh SS, Rama R, et al. (2011) XPD Lys751Gln increases the risk of breast cancer. Oncol Lett 2:155–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer L, Moncollin V, Roy R, et al. (1994) The ERCC2/DNA repair protein is associated with the class II BTF2/TFIIH transcription factor. EMBO J 13:2388–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YY, He L. (2005) SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res 15:97–98 [DOI] [PubMed] [Google Scholar]
- Smith TR, Levine EA, Freimanis RI, et al. (2008) Polygenic model of DNA repair genetic polymorphisms in human breast cancer risk. Carcinogenesis 29:2132–2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz MR, Wu X, Wang Y, et al. (2001) Modulation of nucleotide excision repair capacity by XPD polymorphisms in lung cancer patients. Cancer Res 61:1354–1357 [PubMed] [Google Scholar]
- The International HapMap Consortium (2005) A haplotype map of the human genome. Nature 437:1299–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Li D, Zhang Q, et al. (2012) Association of CD27 and CD70 gene polymorphisms with risk of sporadic breast cancer in Chinese women in Heilongjiang Province. Breast Cancer Res Treat 133:1105–1113 [DOI] [PubMed] [Google Scholar]
- Yamashita H, Toyama T, Nishio M, et al. (2006) p53 protein accumulation predicts resistance to endocrine therapy and decreased post-relapse survival in metastatic breast cancer. Breast Cancer Res 8:R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao L, Qiu LX, Yu L, et al. (2010) The association between ERCC2 Asp312Asn polymorphism and breast cancer risk: a meta-analysis involving 22,766 subjects. Breast Cancer Res Treat 123:227–231 [DOI] [PubMed] [Google Scholar]
- Yuan HY, Chiou JJ, Tseng WH, et al. (2006) FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res 34:W635–W641 [DOI] [PMC free article] [PubMed] [Google Scholar]