Abstract
Abstract: Recent studies have increasingly linked microRNAs to colorectal cancer (CRC). MiR-194 has been reported deregulated in different tumor types, whereas the function of miR-194 in CRC largely remains unexplored. Here we investigated the biological effects, mechanisms and clinical significance of miR-194. Functional assay revealed that overexpression of miR-194 inhibited CRC cell viability and invasion in vitro and suppressed CRC xenograft tumor growth in vivo. Conversely, block of miR-194 in APCMin/+ mice promoted tumor growth. Furthermore, miR-194 reduced the expression of AKT2 both in vitro and in vivo. Clinically, the expression of miR-194 gradually decreased from 20 normal colorectal mucosa (N-N) cases through 40 colorectal adenomas (CRA) cases and then to 40 CRC cases, and was negatively correlated with AKT2 and pAKT2 expression. Furthermore, expression of miR-194 in stool samples was gradually decreased from 20 healthy cases, 20 CRA cases, then to 28 CRC cases. Low expression of miR-194 in CRC tissues was associated with large tumor size (P=0.006), lymph node metastasis (P=0.012) and shorter survival (HR =2.349, 95% CI = 1.242 to 4.442; P=0.009). In conclusion, our data indicated that miR-194 acted as a tumor suppressor in the colorectal carcinogenesis via targeting PDK1/AKT2/XIAP pathway, and could be a significant diagnostic and prognostic biomarker for CRC.
Keywords: colorectal cancer, miR-194, AKT2 pathway, biomarker, carcinogenesis.
Introduction
Colorectal cancer (CRC) is the most prevalent cancer and the fourth leading cause of cancer death worldwide1. Although fecal occult blood test and endoscopy have contributed to a great reduction in CRC mortality2, 70 % of newly discovered CRC patients are still detected at an advanced stage, resulting in poor prognosis3. The ability to distinguish between indolent and aggressive tumors during surgery or biopsy may facilitate more accurate and disease-specific operations. Identification of biomarkers which are sensitive and specific for CRC opens up many avenues for diagnosis and therapeutic exploitation. Therefore, new biomarkers which can discover the tendency of poor prognosis earlier and simpler are urgently needed4.
Mature microRNAs (miRNA) are a group of small non-coding RNAs with 20-23 nucleotides. They regulate target genes at post-transcription level via binding with microRNA response element (MRE) in 3' untranslated regions (3'-UTRs) of target mRNAs 5-6. Increasing studies demonstrated that miRNAs play critical roles in cancer pathogenesis7, through regulation of different cellular processes such as proliferation, differentiation, and apoptosis and so on. The pattern of miRNA expression is also a promising biomarker associated with tumor subtypes and clinical outcomes 8-12. Therefore, identification of CRC-associated miRNA as biomarkers for early tumor detection, prognostication and treatment is of great importance.
The expression pattern and function of miR-194 in various cancers has been widely studied recently. Decreased miR-194 has been reported in liver13, lung14, gastric cancer15 and Multiple Myeloma16, suggesting its tumor-suppressive function. Chiang et al also confirmed miR-194 as a tumor-suppressor gene in CRC patients17, while another study revealed a pro-angiogenesis function of miR-194 in HCT116 cells 18. To date, the function of miR-194 in CRC still remains largely unknown and controversial.
Our study was designed to identify the potential biological role of miR-194 in CRC and its potential target genes. We found that miR194 contributes to CRC cell proliferation via suppressing the expression of PDK1/AKT2/XIAP pathway, and low expression of miR-194 in both tissues and stool samples not only strongly associated with occurrence of CRA or CRC but also related to the poor survival of CRC patients. MiR-194 could be used as a promising biomarker for CRC diagnosis and treatment.
Methods and Materials
Clinical specimen collection
Fresh samples of 20 normal colorectal mucosa (N-N), 40 CRC tissues, 40 colorectal adenomas were obtained from colorectal patients who underwent surgery at Shanghai Ren Ji Hospital. The study protocol was approved by the ethics committee of Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University. Written informed consents were obtained from all participants in this study. All the research was carried out in accordance with the provisions of the Helsinki Declaration of 1975. None of these patients had received radiotherapy or chemotherapy. Stool samples were also collected from the former patients (20 normal individuals, 20 patients with CRA, 28 patients with CRC). Moreover, 90 CRC samples with clinicopathological information and their corresponding adjacent normal mucosa (N-C) were obtained from Ren Ji Hospital from Feb 1999 to May 2005.
Cell culture
Two human colon cancer cell lines, SW480, SW1116 (ATCC, Manassas, VA) were maintained in RPMI 1640 medium (Gibco, Carlsbad, CA) and HT29, HCT116 were maintained in McCoy's 5A medium(Gibco, Carlsbad, CA), respectively, supplemented with 10% fetal bovine serum. Caco2 was maintained in DMEM medium (Gibco, Carlsbad, CA) and supplemented with 20% fetal bovine serum. Cells were then plated in fibronectin- and collagen type I- coated T-25 flasks. All the cells were incubated at 37°C in humidified 5% CO2 atmosphere. The miR-194 mimics (5'-UGUAACAGCAACUCCAUGUGGA-3', 5'-CACAUGGAGUUGCUGUUACAUU-3') and the control miRNA (5'-UUCUCCGAACGUGUCACGUTT-3', 5'-ACGUGACACGUUCGGAGAATT-3') were purchased from Dharmacon RNA Technology (Lafayette, CO, USA). Twenty-four hours before transfection at 30-40% confluence, cells were transferred to 6-well plates. Transfection of miRNA was carried out with LipofectamineTM 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Cells were collected for analysis 48 h or 72h after transfection.
Cell viability assays
Cell viability was assessed using atetrazolium salt (WST-8)-based colorlmetric assay provided by the Cell Counting Kit 8 (cck-8, Dojindo, Japan). Briefly, HCT116 and SW1116 cells transfected with control miRNA or miR-194 mimics were seeded into 96-well plates (3.5×103 cells/well) with 100μl culture medium. At specific time points, 10μl of CCK-8 solution was added to wells and the plated were incubated for 2hr in 37°C. Cell viability was determined from absorbance reading at 450nm. Data was expressed as relative viability (%) calculated as follows: [A450(treated)-A450(blank)].
Flow cytometry assays
After 48hr transfected with miRNA, HCT116 and SW1116 cells were collected and stained using the FITC annexinV Apoptosis Detection Kit I (BD Pharmigen, San Diegs, CA, USA) and performed in accordance with the manufacture's protocol. Cell cycle analysis was performed after staining with propidium iodide. Both apoptosis and cell cycle distribution were quantified using a flow cytometry.
In vitro invasion and migration assays
Cell invasion assays were performed as follow. 8μm-pored polycarbonate membranes were coated with Matrigel on the upper side (Becton-Dickinson, San Diego, CA, USA). HCT116 (1×105) and SW1116 cells (2×105) in serum-free medium were seeded into the upper chamber and medium supplemented with 30% fetal bovine serum was applied to the lower chamber. After 48h, cells remaining on the upper chamber of the filter were removed with a cotton swab. Cells that had migrated to the bottom surface of the filter were fixed, stained and counted. For in vitro migration assays, the chamber membranes were not coated with Matrigel on the upper side and counted after 24h.
RNA extraction and quantitative real-time PCR
Total RNA was extracted by Trizol reagent (Invitrogen), and 2 μg of total RNA from cultured cells was reverse-transcribed using the All-in-One MiRNA Q-PCR Detection Kit (GeneCopoeia, Rockville, USA) according to manufacturer's instructions in a total reaction volume of 25 uL. Quantitative real-time PCR was performed on an Applied Biosystem 7900 quantitative PCR system (Applied Biosystems, Foster City, CA). Quantitative real-time PCR for mature miR-194 and U6 was performed according to the manufacturer's instructions and specific primers designed by GeneCopoeia, China. The amplified transcript level of each specific gene was normalized to that of 18S. The primers used were: IGF1-F (5'-TACAACTACGCCCTGGTCATC-3'), IGF1-R (5'-CTTCTCACACATCGGCTTCTC-3'); FZD4-F (5'-GGTGGCTCCCCTCTTTACTTAT-3'), FZD4-R (5'-TCTGTCTTTGTCCCATCCTTTT-3'); DNMT3A-F (5'-GTGATGATTGATGCCAAAGAAG-3'), DNMT3A-R (5'-ATGAAGACAGGAAAATGCTGGT-3'); AKT2-F (5'-ACCCAACACCTTTGTCATACG-3'), AKT2-R (5'-ACTTCCATCTCCTCAGTCGTG-3'); PDK1-F(5'-CACCAGGACAGCCAATACAA-3'), PDK1-R(5'-CTCATTACCCAGCGTGACAT-3'); XIAP-F(5'-CGGTGCTTTAGTTGTCATGC-3'), XIAP-R(5'-AGCCGTTGATAAATCTGCAA-3'); 18S-F(5'-CGGACAGGATTGACAGATTGATAGC-3') and 18S-R (5'-TGCCAGAGTCTCGTTCGTTATCG-3').
Luciferase assay
Designated combinations of pmirGLO-AKT2-3'UTR or pmirGLO-AKT2-3'UTR mutant with other siRNA and 100 ng phRL (Renilla luciferase) TK plasmid (Promega) for monitoring transfection efficiency were transiently transfected in triplicate with Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer's directions. Twenty-four hours later, luciferase activity was detected using the Dual-Luciferase Reporter Assay System (Promega). Luciferase activity was measured using a BD Monolight 3010 luminometer (BD Biosciences, San Jose, CA).
Western Blot
Western blot analysis was performed using standard techniques as described previously 19. Briefly, 100 μg of protein was electropharesed through 10% SDS polyacrylamide gels and were then transferred to a PVDF membrane (Millipone). The membrane was incubated into AKT2 antibody (1:1000, #2964, CST), pAKT2 antibody (1:1000, #8599, CST), cleaved caspase3 (1:1000, #9664, CST), β-actin (1:1000, #8456, CST), PDK1 (1:1000, #3062,CST), XIAP(1:1000, #2042, CST) at 4℃ overnight. Secondary antibodies were labeled with HRP (1:5000, KangChen, China) and the signals were detected using ECL Kit (Pierce Biotech, Rockford, IL, USA). Subsequently, the images were analyzed by ImageJ 1.43 software. The protein expression was normalized to an endogenous reference (β-actin) and relative to the control.
TUNEL assay
The in situ cell death kit (Roche Molecular Biochemicals, Mannheim, Germany) based on TUNEL technology was performed to detect individual apoptotic cells in formalin fixed tissue sections. Dewax and rehybrate tissue section according to standard protocols. Endogenous peroxidase activity was blocked by incubated in 0.3% hydrogen peroxide for 10 min. Slides were made permeable with pereabilisation on solution. After washing, slides were incubated with TUNEL reaction mixture for 60 min at 37℃ in a humified atmosphere in the dark. The negative control was incubated with label solution (without forminal transferase) instead of TUNEL reaction mixture. After washing, samples can be analyzed in a drop of PBS under a fluorescence microscope. Use an excitation wave length in the range of 450-500nm and detection in the range of 515-565nm.
Adenovirus generation
The Ad-control miRNA adenovirus (Ad-control miRNA), miR-194 overexpression adenovirus (Ad-miR194), miR-194 inhibitor overexpression adenovirus (Ad-miR194 inhibitor), lenti-control virus and lenti-miR-194 virus were constructed by Shanghai SBO medical biotechnology company, Shanghai, China.
Animal experiments
24 male nude mice (5-weeks-old) were purchased from the Chinese Academy of Sciences (Shanghai, China). Fifteen male APCMin/+ mice were purchased from Model Animal Research Center of Nanjing University and fed with a high-fat and high-glucose diet. To establish CRC xenografts, HCT116 (1×107) were subcutaneously injected into the axillary region of nude mice. One week after injection, the mice were randomly separated into three groups (eight mice in each group): The Ad-miR-194 group, Ad-control miRNA group and PBS group. Ad-miR-194 was an adenovirus overexpressing mature miR-194, Ad-control miRNA was an adenovirus expressing control miRNA, and PBS as the negative control. Ad-miR-194, Ad-control miRNA and PBS were directly injected into the subcutaneous tumors of each group once every other day. Tumor volumes were measured before each injection in two dimensions, and calculated according to the formula: v=0.5× ( length×width2). After 14 days (seven times of injection), all nude mice were sacrificed and the subcutaneous tumors were removed and measured the weight. Some of the samples were frozen immediately in liquid nitrogen and the remaining samples were fixed in formalin solution and embedded in paraffin blocks.
APCMin/+ mice were randomly and equally separated into 3 groups: Ad-miR-194 inhibitor, Ad-control miRNA and PBS. Ad-miR-194 inhibitor was an adenovirus dysfunction of mature miR-194, and Ad-control miRNA was an adenovirus having no effect on miRNAs function, and PBS as the negative control. Mice were fed with adenovirus and PBS separately every three day. After 12 weeks, mice were sacrificed, and colons removed, and flushed with 1×phosphate-buffered saline (PBS). The tumors which could be observed by human eyes were counted and removed. Parts were frozen immediately in liquid nitrogen and remaining parts were fixed in formalin solution and embedded in paraffin blocks. The left colons were fixed in formalin solution for 24h and were stained with 0.1% methylene blue (Sigma-Aldrich, St. Louis, Mo) dissolved in PBS and tumor number was counted at 4×magnification using a dissecting microscope.
To further investigate the effect of miR-194 on tumor invasion in vivo, we developed a CRC metastasis model in nude mice. The HCT116 cells (2.5 × 106) per mouse in 200μl PBS were subcutaneous injected through the right flank of 4-week-old BALB/C nude mice. Mice were inspected daily and tumors measured with a caliper every 4d. Tumor volume (V) was calculated as V=π/6×a×b2, where a is the longer and b is the shorter of two orthogonal diameters. About ten days later, all the tumors were grown up to a size of ~25 mm3. All the animals with tumors were randomly divided into three groups. One group was intratumoral injected with PBS, another group was intratumoral injected with lenti-control virus and the last group was intratumoral injected with lenti-miR-194 virus (1×108 Tu/mouse, twice a week) for 10 weeks. For lung metastasis examination, mice were sacrificed in 10 weeks after injection of PBS, lenti-control virus and lenti-miR-194 virus. Lungs were removed and fixed in 4% PFA, embedded in paraffin, and sections of lungs were produced and H&E stained. The nodules numbers of each sections (=metastasis per animal) was determined under a binocular microscope (Leica, DM 300). All the H&E staining lung tissue sections were examined and the numbers of mice with or without lung metastasis were calculated by Chi-square test, Fisher's exact test. All experimental procedures were approved by the Institutional Animal Care and Use Committee.
Immunofluorescence
Pretreated CRC cells were plated in four-well chamber slides (Nunc, Denmark) 24h before experiment. The cells were probed with corresponding antibodies for 1h at RT and incubated with Alexa 546-conjugated donkey anti-mouse IgG, followed by Alexa-488-conjugated donkey anti-rabbit IgG (Invitrogen). Afterwards the slides were mounted in DAPI Fluoromount-G (SouthernBiotech, Birmingham, USA) and images were captured using a laser-scanning confocal microscope (LSM-710, Zeiss, Germany).
In situ hybridization(ISH) and Immunohistochemistry (IHC)
The in situ detection of miR-194 was performed on 6μm formalin-fixed, paraffin-embeded (FFPE) section. Briefly, the sides were hybrided with 10 pmol probe (LNA-modified and DIG-labeled oligonucleotide; Exiqon) complementary to miR-194 and after incubation with anti-DIG-AP Fab fragments conjugated to alkaline phosphatase, and the hybridized probes were detected by applying nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate color substrate (Roche) to the slide. Positive control (U6, Exiqon) and no-probe controls (scramble) were included for each hybridization procedure.
Immunohistochemistry (IHC) was performed using a standard streptavidin-biotin-peroxidase complex method. Briefly paraffin-embeded section were deparaffinized and rehydrated. Endogenous peroxidase activity was blocked by incubated in 0.3% hydrogen peroxide for 15 min, then the slides were microwaved and boiled in 10mm citrate buffer(pH 6.0) for antigen retrieval. Nonspecific antigens were blocked by incubation in sheep serum for 30min. Slides were incubated overnight at 4°C with AKT2 antibody (1:100, #ab66129, abcam); pAKT2 antibody (1:100, #ab 38513, abcam); Ki67 antibody (1:100, #ab 15580, abcam). Sample incubated with PBS instead of primary antibody were used as negative control.
The slides were examined independently by two investigators, who were blinded to the clinical and pathological data. The staining intensity was quantified using a visual grading system based on the extent of staining (graded from 0 to 4: 0, none; 1, 1-25%; 2,26-50%; 3,51-75%; 4,76-100%) and the intensity of staining (grade from 0 to 3: 0, no staining; 1, weak staining; 2, moderate staining; 3, strong staining). An index value was calculated as a product of the extent of staining and intensity of staining scores to evaluate the expression of miR-194, U6, AKT2, pAKT2, Ki67. Then the expression level of miR-194 was furthermore valued by miR-194/U6 and a relative score of ≤0.75 or >0.75 were considered as low or high expression of miR-194, respectively, dividing the patients into two groups of approximately equal size.
Statistical analysis
All quantified data represent an average of at least triplicate samples or as indicated. Data were expressed as mean ± standard deviation (SD). The difference in tumor growth rate between the two groups of mice was determined by repeated-measures analysis of variance. The χ2-test was used for comparison of patient characteristics. HRs of death associated with low expression of miR-194 and other predictor variables were estimated from univariate Cox proportional hazards model first. Multivariate Cox models also were constructed to estimate the HR of low expression of miR-194. Overall survival in relation to the expression of miR-194 was evaluated by the Kaplan-Meier survival curve. The sensitivity and specificity of miR-194 as a diagnosis biomarker was calculated by receiver operating characteristic (ROC) analysis. All analyses were performed using SPSS for Windows 17.0.1 software (SPSS Inc., Chicago, IL, USA). P<0.05 was considered statistically significant.
Result
MiR-194 acts as a tumor suppressor gene in CRC cells
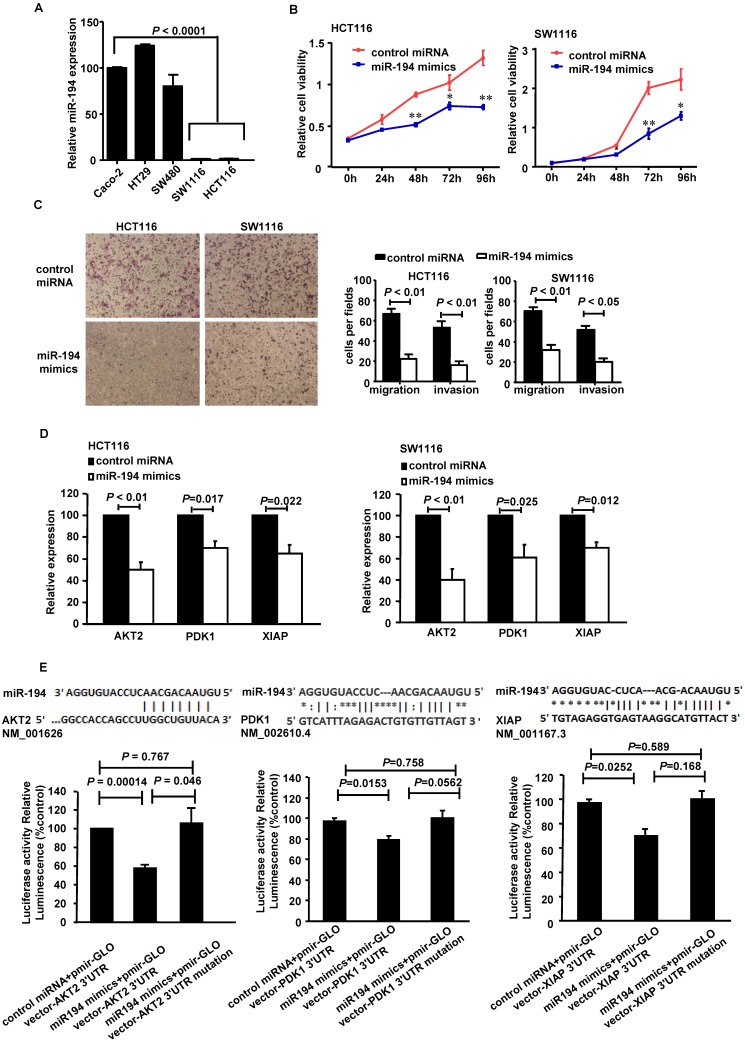
It has been reported that miR-194 is deregulated in CRC patients17. In this study, we further investigated its expression in different CRC cell lines. As Fig.1A showed, the expression level of miR-194 was 75 to 125 times lower in the low differentiated HCT116 and SW1116 cells, compared with the high differentiated cells from Caco-2, HT29 and SW480 cell lines. Therefore, we chose HCT116 and SW1116 cells for further functional assays.
Figure 1.
Expression, functions and the targets of miR-194 in CRC cells. (A) Real-time PCR quantification of miR-194 in seven human CRC cell lines. (B) CCK-8 assay was performed in HCT116 and SW1116 cells transfected with miR-194 mimics or control miRNA. Cell viability was measured from absorbance reading at 450nm. Data was expressed as relative viability (%) calculated: [A450(treated)-A450(blank)]. (C) In vitro invasion and migration assays were performed using HCT116 and SW1116 cells. Cells were photographed under a light microscope and counted from five random microscopic fields (200×) per insert in triplicate. (D) mRNA levels of PDK1, AKT2 and XIAP after miR-194 mimics transfection in HCT116 and SW1116 cells. (E) Schematic illustration of the predicted miR-194-binding sites in PDK1, AKT2 and XIAP 3'-UTR. MiR-194 mimics and report constructs, containing a wild-type or a mutated PDK1, AKT2 or XIAP 3'-UTR, were co-transfected into HCT116 cells respectively. Relative repression of firefly luciferase expression was standardized to a transfection control. n=3, ANOVA. All experiments were repeated at least three separate times, Data are shown as mean ± SD, *P<0.05, **P<0.01(Student's t-test), compared with the control miRNA.
Firstly, we introduced miR-194 mimics into HCT116 cells (Additional File 1: Supplementary Fig.1A), Transfection of the mimics successfully up-regulated miR-194 in the cells, compared with control miRNA trasfected cells and control CRC cells.
CCK8 cell variability assay was completed in HCT116 and SW1116 cells with miR-194 or control miRNA transfection. Upregulation of miR-194 significantly reduced cellular viability in both cells (Fig. 1B), suggesting that miR-194 may play an important role in CRC cell growth. To explore the reason for the decrease in cell viability, we examined the effects of miR-194 on cell cycle progression, apoptosis, migration and invasion. As illustrated in Supplementary Fig.1B, over-expression of miR-194 induced a slight G0/G1 phase arrest in HCT116 cell and an S phase arrest in SW1116 cell. Apoptosis assay with Annexin V-staining was completed in HCT116 and SW1116 cells. The miR-194 transfection induced 1.5-folds increase of apoptosis in HCT116 cells but did not affect cell apoptosis in SW1116 cells, indicating that there may be cell specific mechanisms responsible for miR-194 induced functional changes. The effect of miR-194 on cell apoptosis was further confirmed by western blot, miR-194 overexpression significantly increased expression of cleaved caspase-3 in HCT116 cells (Supplementary Fig. 1C). Moreover, miR-194 overexpression dramatically decreased the migration and invasion ability of HCT116 and SW1116 cells (Fig. 1C), indicating that miR-194 expression also plays a critical role in CRC cell motility and metastasis. These data suggested that miR-194 has an in vitro tumor suppressive feature in CRC.
MiR-194 blocks AKT2 pathway activation through directly targeting PDK1/AKT2/XIAP 3'UTR
We have found that miR-194 could inhibit CRC cell proliferation, apoptosis, migration and invasion, as a tumor suppressor gene. So, the putative target genes of miR-194 should be related to these functions of cells. Focusing on these genes, we predicted the putative targets using the target prediction programs, miRanda and TargetScan. Our analyses revealed that IGF1R, Fizzled-4, DNMT3A, PDK1, AKT2 and XIAP were the potential targets of miR-194. The 3'-UTR of IGF1R, Fizzled-4, DNMT3A, PDK1, AKT2 and XIAP mRNA contains a complementary site for the seed region of miR-194 (Fig. 1E, Additional File 1: Supplementary Fig.2A). We first performed real-time PCR assay with miR-194 transfected HCT116 and SW1116 cells to verify whether these six genes were regulated by miR-194. The results showed that transfection of miR-194 mimics significantly decreased the mRNA level of PDK1/AKT2/XIAP in HCT116 and SW1116 cells (Fig. 1D), while there is no significant change in the expressions of any other molecules (Supplementary Fig.2A).
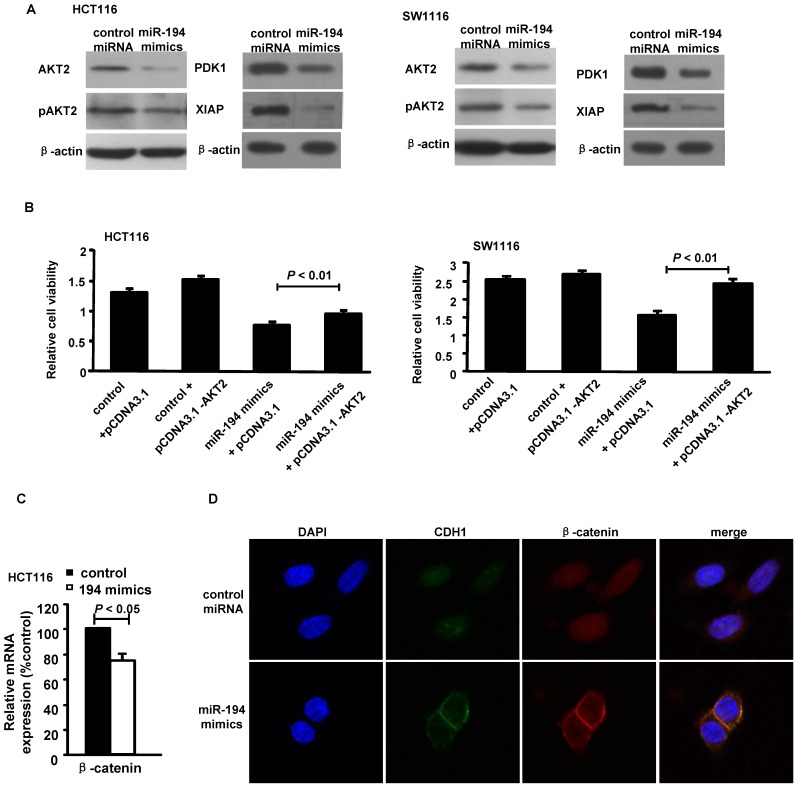
We then carried out luciferase expression analyses to detect whether there is a direct interaction between PDK1/AKT2/XIAP and miR-194. The fragment of PDK1/AKT2/XIAP 3'UTR with wild type or mutant MREs of miR-194 were cloned into downstream of the luciferase open reading frame to generate the luciferase report vectors. CRC cells were cotransfected with the reporters and miR-194 mimics. The results of luciferase activity assay demonstrated that miR-194 significantly decreased the luciferase expression. Conversely, the luciferase activity of the mutant reporter was not significantly affected in response to miR-194 mimics treatment (Fig. 1E). The data suggests that miR-194 may negatively regulate AKT2 pathway in CRC cells through a direct interaction with the 3'UTR regions of PDK1, AKT2 and XIAP. We further confirmed these data in protein level. Western blot and summarized data revealed that overexpression of miR-194 dramatically decreased the expression of PDK1, AKT2, pAKT2 and XIAP in CRC cells (Fig. 2A).
Figure 2.
MiR-194 blocked AKT2 pathway in CRC cells. (A) Western blot analysis and summarized data showed that expression of PDK1, AKT2, pAKT2 and XIAP were deregulated in cells transfected with miR-194 mimics compared with transfected with control miRNA. (B) CCK-8 assay performed in HCT116 and SW1116 cells which were co-transfected miR-194 mimcs or control miRNA with pcDNA-AKT2 or pcDNA-control. Cell viability was measured from absorbance reading at 450nm. Data was expressed as relative viability (%) calculated: [A450 (treated)-A450 (blank)]. (C) The mRNA levels of β-catenin after miR-194 mimics transfection in HCT116 cells. (D) Immunofluorescence data showed that CDH1 and β-catenin interacts with each other and colocalized together on the cell membrane after transfection of miR-194 mimics in CRC cells, compared with control, n = 3. All experiments were repeated at least three separate times, Data are shown as mean ± SD.
Moreover, upregulation of AKT2 restored the miR-194-induced decrease in cell viability in HCT116 and SW1116 cells (Fig. 2B), suggesting that miR-194 may exert its function via suppressing AKT2 in CRC cells. As AKT2 may activate β-catenin by inhibition of GSK-3β 20 and Wnt/β-catenin signal pathway is very important in cancer cell progression, including CRC21, we next to detect whether miR-194 inhibits CRC cell invasion via blocking Wnt/β-catenin pathway. The real time PCR data showed that introduction of miR-194 mimics significantly decreased the expression of β-catenin (Fig.2C). However, we can not find any seed region of miR-194 in the 3'UTR region of β-catenin. The data indicate that miR-194 may inhibit the expression of β-catenin via inhibiting of the AKT2 pathway. The cell-cell adhesion molecule E-cadherin sequesters β-catenin to the cell membrane and the Wnt/β-catenin pathway can be activated through accumulation of β-catenin in the cytoplasm and its translocation to the nucleus21. We next found that introduction of miR-194 mimics significantly increased the connection of β-catenin/E-cadherin on the cell membrane of CRC cells (Fig.2D). The data indicates that miR-194 may inhibit the invasion of CRC cells via blocking the Wnt/β-catenin pathway.
Overexpression of miR-194 suppresses CRC xenograft tumor growth and metastasis
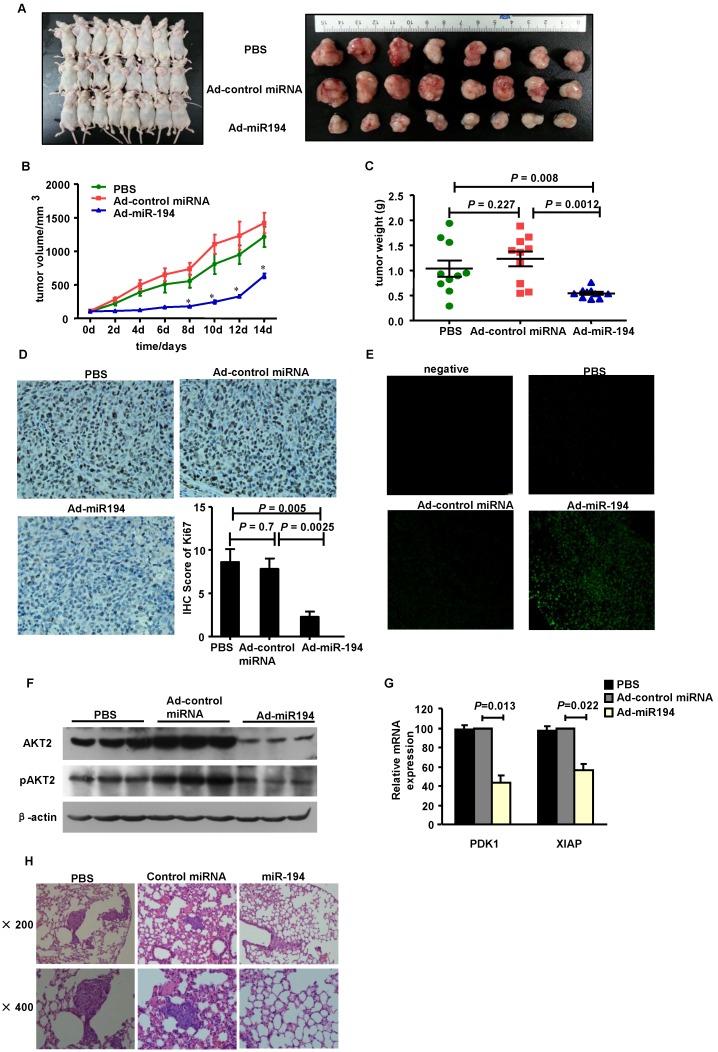
To determine the functional impact of miR-194 on CRC cell proliferation in vivo, we established a tumor model in nude mice bearing HCT116 xenografts. As shown in Fig. 3A-C, overexpression of miR-194 significantly suppressed the growth of xenograft tumors as compared with Ad-control miRNA and PBS groups.
Figure 3.
Overexpression of miR-194 inhibited CRC xenograft growth and metastasis. (A) Mice carrying xenograft tumors treated with Ad-miR-194, Ad control miRNA and PBS at the last time point (14 days after first injection) (B) Tumors growth curves of xenograft tumors in different groups. Data represent mean with 95% confidence interval (C) Comparison of excised tumor weight in three groups. (D) Ki67 staining showed lower percentage of growing cells in tumors treated with Ad-miR-194 compared with the other two groups, original magnification: 200×. (E) TUNEL assay revealed higher percentage of apoptotic cells in tumors treated with Ad-miR-194 compared with the other two groups, original magnification: 100×. (F) Representative Western blot detected the expression of AKT2 and pAKT2 in xenograft tumors of three groups. β-actin was used as the loading control. (G) Real time PCR examined the expression of DPK1 and XIAP in xenograft tumors of three groups. All values were normalized to 18s expression. Data are shown as mean ± SD, n = 24, ANOVA. (H) Representative hematoxylin-eosin staining showed that less metastatic CRC cells were observed in the lungs of nude mice at 10 weeks after injection of lenti-miR-194 virus, while most of the mice injected with lenti-control virus or PBS displayed significant lung metastases.
Ki67 staining data showed that the number of Ki67+ proliferating cells was much less in tumors treated with Ad-miR-194 than that of Ad-control miRNA and PBS groups (Fig. 3D). The apoptotic cells in tumors detected by TUNEL staining were significantly increased in Ad-miR-194-treated mice compared with other groups (Fig. 3E), suggesting that miR-194 increased cell apoptosis in vivo.
We then validated the expression level of miR-194, PDK1, AKT2, pAKT2 and XIAP. Real-time PCR data showed that the expression of miR-194 was dramatically increased in Ad-miR-194 group compared with Ad-control miRNA and PBS groups (Supplementary Fig. 2B). Western bolt analysis revealed that the expression levels of AKT2, pAKT2 were decreased in Ad-miR-194 group compared with Ad-miR-control and PBS groups (Fig. 3F). Furthermore, real time PCR data showed that the expression levels of PDK1 and XIAP were decreased in Ad-miR-194 group compared with Ad-miR-control and PBS groups (Fig. 3G). These finding suggested that miR-194 inhibited cell growth via inhibition of AKT2 pathway in vivo.
We next developed a CRC metastasis model in BALB/c nude mice. Hematoxylin-eosin staining showed that less metastatic CRC cells were observed in the lungs of mice at 10 weeks after injection of lenti-miR-194 virus, while most of the mice injected with lenti-control virus or PBS displayed significant lung metastases (Fig. 3H, Additional File 1: Supplementary Table 1). These data suggest that miR-194 may play a critical role in CRC cell metastasis in vivo.
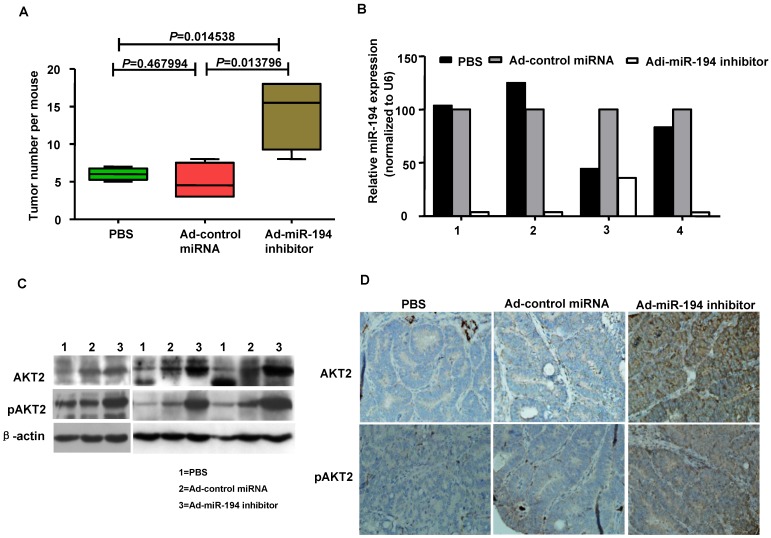
Downregulation of miR-194 promoted tumor formation in APCMin/+ mice
The ApcMin/+ (multiple intestinal neoplasia) mouse model harbors a germline mutation in the Apc tumor suppressor gene and exhibits multiple tumors in the small intestine and colon 22. We explored whether colonic carcinogenesis could be enforced by the loss of miR-194 in tumor formation process of APCMin/+ mice. As showing in Fig. 4A, Ad-miR-194 inhibitor treatment induces 3-fold increase in the number of tumors compared with Ad-control miRNA and PBS treatments in APCMin/+ mice. The data of real-time PCR analysis showed that the expression of miR-194 was decreased in Ad-miR-194 inhibitor group compared with Ad-control miRNA and PBS groups (Fig. 4B). Moreover, the expression levels of AKT2, pAKT2 were dramatically increased in Ad-miR-194 inhibitor group (Fig. 4C-D). These result indicated that downregulation of miR-194 partially promoted tumor formation in APCMin/+ mice.
Figure 4.
Attenuation of miR-194 partially promoted the process of tumor formation in APCMin/+ mice. (A) Ad -miR-194 inhibitor-treated group burdened significantly more tumors in colon than Ad-control miRNA-treated group and PBS-treated group, Data are shown as mean ± SD, n = 12, ANOVA. (B) Real time PCR examined the expression of miR-194 in tumors of three groups. All values were normalized to U6 expression. (C) Western blot detected the expression of AKT2, pAKT2 in tumors of three groups. β-actin was used as the loading control. (D) Immunohistochemical analysis of consecutive tissue sections revealed expression of AKT2, pAKT2 in tumors of three groups.
Expression of miR-194 was decreased in CRA and CRC patients, and might be a prognostic marker in CRC
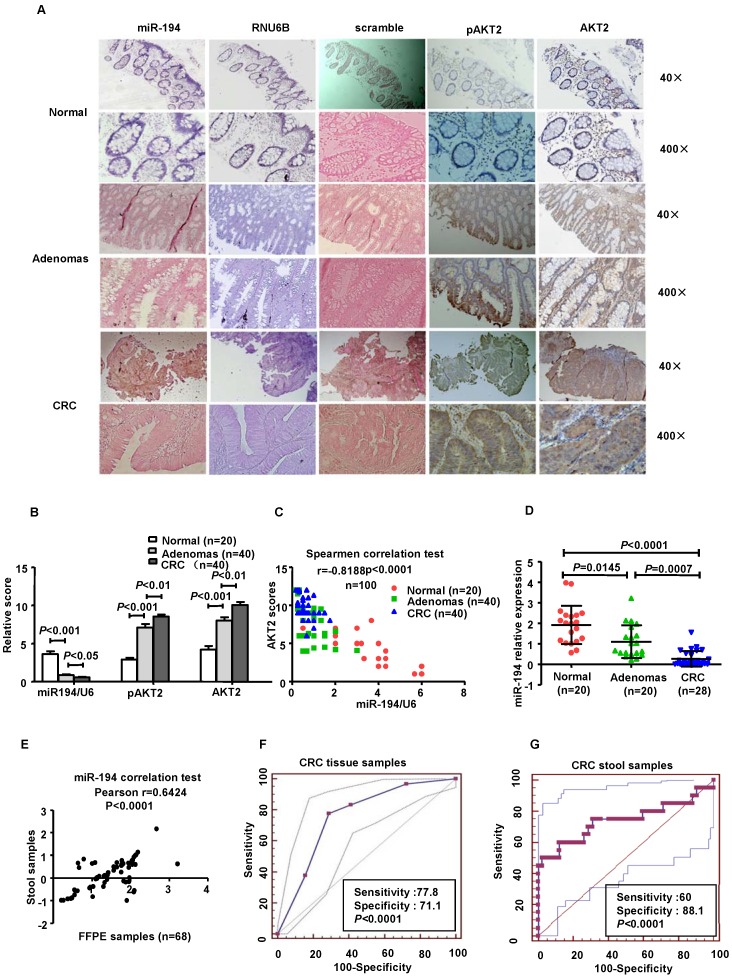
Clinical reports have identified downregulation of miR-194 in various tumors. We explored the expression of miR-194 in human colorectal tissues from 20 normal colorectal mucosa (N-N) tissues, 40 CRC tissues and 40 colorectal adenomas (CRA) by tissues ISH. The results showed that miR-194 was expressed only in the colonic epithelial cells in N-N, which was consistent with the previous study 21. The consecutive section data and summarized analyses data suggested that expression of miR-194 was gradually decreased during progression from N-N to CRC via CRA (Fig. 5A), indicating that downregulation of miR-194 may be an early event in colorectal carcinogenesis. Conversely, the expression of AKT2 and pAKT2 gradually increased during the CRC progression (Fig. 5B-C).
Figure 5.
Expression of miR-194 was decreased in CRC patients' tissues and stools, and inversely correlated with AKT2, pAKT2 in human colorectal tissues. Expression of miR-194 in colorectal epithelial cells using in situ hybridization (ISH) analysis. Consecutive tissue sections were incubated with a full length DIG-labeled LNA probe to miR-194. The positive staining of epithelium cells was expressed as blue-violet. The scramble control probe shows no significant staining at low or high magnification from normal colorectal tissues to CRC tissues. Immunohistochemical staining showed that the expression of AKT2/pAKT2 is significantly increased from normal colorectal tissues to CRC tissues. Original magnification: 40×, 400×. (B) Statistical analysis of miR-194, AKT2, pAKT2 in N-N, CRA, and CRC. Data are shown as mean ± SD, n=100, ANOVA. (C) Correlation between the expression of miR-194 and AKT2 (r = -0.8188; P<0.0001). (D) Real time PCR examined the expression of miR-194 in stool samples from patients burdening N-N, CRA and CRC. All values were normalized to U6 expression, n=68, ANOVA. (E) Scatter plot of miR-194 expression correlation between stool samples and FFPE samples in 68 pairs samples. n=68. (F) ROC curve analysis showing the ability of miR-194 expression to discriminate between normal tissues and CRC tissues. (G) ROC curve analysis showing the ability of miR-194 expression to discriminate stools samples from normal individuals and CRC patients.
These data suggest that miR-194 levels are inversely correlated with the expressions of AKT2 and pAKT2 from N-N to CRC via CRA tissues (Spearmen coefficient, r= -0.8188, P<0.0001, n=100).
We further confirmed these data in corresponding stool samples from 20 normal individuals, 20 patients with CRA, 28 patients with CRC and found that the expression of miR-194 in stool was decreased from health individuals to CRC via CRA (Fig. 5D). Pearson correlation test demonstrated that miR-194 expression between stool samples and paired FFPE specimens were strongly positive correlated (Pearson r= 0.6424, 95% CI: 0.45676-0.7688, P<0.0001; Fig. 5E). To evaluate the potential capability of miR-194 as a diagnostic biomarker for colorectal cancer, receiver operating characteristic (ROC) analyses was conducted in the tissues samples (Fig.5F) and stool samples respectively (Fig.5G). ROC analyses of tissues showed that miR-194 expression can be a single significant parameter to discriminate between normal and tumor tissues with an area under the ROC curve (AUC) of 0.889 (sensitivity=77.78%, specificity=71.11%; P < 0.0001). ROC analyses of 48 stool samples showed an AUC of 0.739 (sensitivity=60%, specificity=88.1%; P < 0.0001).
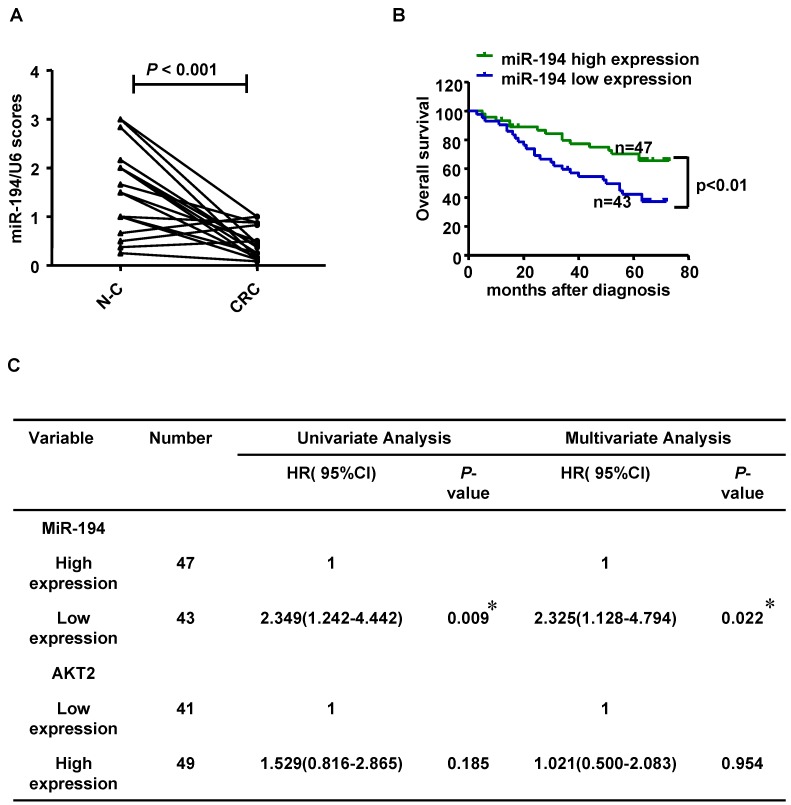
We further validated the expression level of miR-194 in 90 CRC cases with corresponding adjacent normal mucosa (N-C). The expression of miR-194 was dramatically decreased in CRC than N-C in ISH analysis (P<0.001, Fig.6A). Moreover, we found an association between miR-194 expression and several clinicopathological features in CRC. Expression of miR-194 was inversely correlated with tumor size (P<0.001) and lymph node metastasis (P<0.05, Table 1). In univariate Cox regression analysis, low expression of miR-194 in CRC was associated with an increased risk of cancer-related death (HR=0.438; 95% CI, 0.231-0.828; P=0.009, Table 2). In the multivariate model (Table 3), lymph node metastasis but not low expression of miR-194 was an independent predictor for overall survival in CRC. Furthermore, univariate and multivariate analyses were used to analyze whether miR-194 or AKT2 was associated with patients' survival in human CRC (Fig. 6C). In univariate analysis, miR-194 was significantly associated with poor survival, while AKT2 showed no significance with patients' prognosis. In the multivariate model, after the adjustment for AKT2, miR-194 expression was still associated with patients' survival, indicating that miR-194 is a promising prognostic biomarker. We then compared the survival time in patients with miR-194 low expressing tumors (n=43) with that of patients with miR-194 high expressing tumors (n=47). The cumulative survival rate was significantly better in patients with miR-194 high expressing tumors than in those with low expressing tumors (P<0.01, Fig. 6B), indicating that miR-194 expression was significantly associated with patient survival in CRC. These data confirmed that the expression change of miR-194 occurred at a relative early stage during CRC carcinogenesis. Moreover, the expression patterns of miR-194 in tissue samples could be represented by stool sample when used as a diagnostic biomarker, which makes the clinical utility simple and non-invasive.
Figure 6.
(A) Statistical analysis of miR-194 in 90 CRC and their corresponding adjacent normal mucosa (N-C). Data are shown as mean ± SD, n=180. (B) Survival analysis showed that CRC patients with low miR-194 expression had poorer survival than the patients with high expression, HR=0.438; 95% CI, 0.231-0.828, P<0.01. (C) Univariable and Multivariable analysis were performed in the 90 CRC cases of Renji dataset.
Table 1.
Clinicopathological characteristics of miR-194 expression in patients with CRC.
| Characteristics | Total | MiR-194 expression | P- value | |||
|---|---|---|---|---|---|---|
| High | % | Low | % | |||
| Age | 90 | 67.7± 1.7 | 70.0± 1.6 | 0.456 | ||
| Gender | ||||||
| Male | 47 | 22 | 46.8 | 25 | 53.2 | |
| Female | 43 | 25 | 58.1 | 28 | 41.9 | 0.282 |
| Location | ||||||
| Proximal colon | 43 | 23 | 53.5 | 20 | 46.5 | |
| Distal colon | 47 | 24 | 51.3 | 23 | 48.7 | 0.818 |
| Tumor size (cm3) | ||||||
| <30 | 33 | 25 | 69.4 | 11 | 30.6 | |
| ≥30 | 57 | 21 | 40.7 | 32 | 59.3 | 0.006* |
| T stage | ||||||
| T1+T2 | 11 | 5 | 45.5 | 6 | 55.5 | |
| T3+T4 | 79 | 42 | 53.8 | 37 | 47.4 | 0.195 |
| Lymph node metastasis | ||||||
| Absent | 54 | 34 | 63.0 | 20 | 37.0 | |
| Present | 36 | 13 | 36.1 | 23 | 63.9 | 0.012* |
| Distant metastasis | ||||||
| M0 | 87 | 47 | 54.0 | 40 | 46.0 | |
| M1a | 3 | 0 | 0.0 | 3 | 100.0 | 0.3345 |
| Histological grade | ||||||
| I, I-II, II, | 54 | 26 | 48.1 | 28 | 51.9 | |
| II-III, III | 36 | 21 | 58.3 | 15 | 41.7 | 0.343 |
Table 2.
Univariate Cox regression analysis of potential prognostic factors for colorectal cancer patients factors.
| Variable | HR (95%CI) | P-value |
|---|---|---|
| Age | 1.716(0.875-3.366) | 0.146 |
| Gender | ||
| Female | 1 | |
| Male | 0.933(0.505-1.725) | 0.826 |
| Tumor size (cm3) | ||
| <30 | 1 | |
| ≥30 | 1.744(0.912-3.332) | 0.092 |
| T Stage | ||
| T1+T2 | 1 | |
| T3+T4 | 1.262(0.487-3.168) | 0.650 |
| Lymph node metastasis | ||
| Absent | 1 | |
| Present | 3.278(1.738-6.181) | <0.000* |
| Distant metastasis | ||
| Absent | 1 | |
| Present | 2.604(0.802-8.457) | 0.111 |
| Histological grade | ||
| I, I-II, II | 1 | |
| II-III, III | 1.080(0.572-2.040) | 0.812 |
| MiR-194 expression | ||
| High expression | 1 | |
| Low expression | 2.349(1.242-4.442) | 0.009* |
Abbreviations: CI, confidence interval; HR, hazard ratio.
Table 3.
Multivariate Cox regression analysis of potential prognostic factors for colorectal cancer patients factors.
| Variable | HR (95%CI) | P-value |
|---|---|---|
| Age | 1.691(0.850-3.362) | 0.134 |
| Gender | ||
| Female | 1 | |
| Male | 1.262(0.657-2.421) | 0.485 |
| Tumor size (cm3) | ||
| <30 | 1 | |
| ≥30 | 1.446(0.722-2.883) | 0.298 |
| T Stage | ||
| T1+T2 | 1 | |
| T3+T4 | 1.443(0.346-6.014) | 0.614 |
| Lymph node metastasis | ||
| Absent | 1 | |
| Present | 2.946(1.487-5.835) | 0.002* |
| Distant metastasis | ||
| Absent | 1 | |
| Present | 2.595(0.417-16.147) | 0.317 |
| Histological grade | ||
| I, I-II, II | 1 | |
| II-III, III | 1.321(0.686-2.563) | 0.404 |
| MiR-194 expression | ||
| High expression | 1 | |
| Low expression | 1.608(0.808-3.202) | 0.176 |
Abbreviations: CI, confidence interval; HR, hazard ratio.
Discussion
Previous study suggests that miR-194 is significantly deregulated in CRC patients, as well as in three CRC cell lines 17. The other study on the small intestine finds that miR-194 is highly induced during intestinal epithelial maturation 23. However, another study reveals that elimination of miR-194 could reactivate thrombospondin-1 (TSP-1), impair CRC angiogenesis and reduce xenografts growth 18, which were contrast to the former two studies. Here, we systematically explored the potential biological function of miR-194 in CRC. Our in vitro data suggested that overexpression of miR-194 inhibited cell proliferation, induced G0/G1 phase or S phase arrest in HCT116 and SW1116 cells separately, increased apoptosis of HCT116, and deregulated cells invasion and migration in both cells. Furthermore, overexpression of miR-194 suppressed CRC xenograft growth, while loss of miR-194 in APCMin/+ mice colons promoted de novo tumor formation. Collectively, all above data confirmed the tumor-suppressive role of miR-194 in CRC.
HCT116 cell line, which has highly invasive and metastasis ability, was used to construct an orthotopic mice model for research of the in vivo invasion of human colon cancer cells previously 24. However, it's very difficult to establish an orthotopic mice model for primary colon cancer and it is not easy to implant the human CRC tissues into the colon of the mice. Furthermore, most of the mice will die very early because of the colorectal infection after the surgery of CRC tissues implantation. More studies were using HCT116 cells to provide information in regards to tumor growth and metastasis in xenograft mouse model later 25-27. The mouse model of HCT116 CRC subcutaneous metastasis is easier to establish and has been verified by different research groups 25-27, including our group. In addition, HCT116 cells injected subcutaneously into mice are more likely to metastasize to the lungs and it's easier to detect the lung metastasis by H&E staining. Therefore, we used the mouse model of HCT116 CRC subcutaneous metastasis in BALB/c nude mice to detect whether miR-194 participates in the CRC cell invasion in vivo.
(PI3K)/AKT signaling pathway which plays a key role in cell proliferation, survival, migration, and invasion in various cancers, including colorectal cancer 28. The previous studies have revealed that AKT2 is the predominant isoform involved in CRC carcinogenesis 29, AKT2 inhibition suppresses CRC cell growths in vitro and in vivo30, and also plays a key role in CRC metastasis 31. Significantly, our study identified that miR-194 and AKT2 were inversely correlated: Luciferase assays suggested that miR-194 decreased the transcriptional activity of PDK1, AKT2 and XIAP; overexpression of miR-194 in CRC cells decreased endogenous PDK1, AKT2, pAKT2 and XIAP expression levels. As the functional assay showed that introduction of miR-194 mimics significantly blocked cell proliferation and induced apoptosis. Through the literatures we found that PDK1 may activate AKT2 phosphorylation 32and the anti-apoptotic protein XIAP can be activated by AKT233. Our data indicate that miR-194 may target PDK1/AKT2/XIAP pathway to block cell proliferation and induce CRC cell apoptosis. Furthermore, upregualtion of miR-194 in human CRC xenografts led to reduced PDK1, AKT2, pAKT2 and XIAP expressions and block of miR-194 in APCMin/+ mice intestinal tissues increased AKT2, pAKT2 protein levels, suggesting that miR-194 was a significant tumor suppressor through suppressing AKT2 pathway which might be a major downstream effecter of miR-194 in its target network. In addition, we found that miR-194 may co-downregulate PDK1, AKT2 and XIAP genes in AKT pathway to decrease cell proliferation and increase apoptosis of CRC cells. The data is consistent with other reports that miR-210 disturbs mitosis through targeting multi-genes involved in mitotic progression, which may contribute to its inhibitory role on tumor growth 34.
Recent studies have indicated that P53 activation induces a significant increase of miR-194 expression in growth arrest and apoptosis 16, 18 In addition, other researchers have demonstrated that AKT2 can hyperphosphorylated murine double minute 2 (Mdm2) which promoted P53 degradation and attenuated its Ser-15 phosphorylation35-36. Furthermore, our data demonstrated that miR-194 directly suppressed the activation of AKT2. A possible double-negative AKT2/P53 feedback loop may exist, in which AKT2 and P53 were not only having opposing functions, but also reciprocally control one another by regulating expression of miR-194 to keep balance. Moreover, Mdm2 has been validated to be a direct target of miR-194 in multiple myeloma 16. Collectively, such a reciprocal loop could attenuate the expression of AKT2, Mdm2 and protect the activation of P53 by regulating miR-194 expression, which may account for the mechanism of miR-194 inhibiting CRC carcinogenesis.
To figure out the mechanism of miR-194-mediated blocking of CRC cell migration and invasion, we looked through the literatures and found that Wnt/β-catenin signal pathway is very important in cancer cell progression, including CRC 21. Generally, Wnt signaling activation is the stabilization of β-catenin in the cytoplasm, resulting in an increased translocation of β-catenin to the nucleus. Nuclear β-catenin forms a complex with the TCF/LEF transcription factor leading to activating of Wnt target gene expression 37. The Glycogen synthase kinase 3-β (GSK-3β) may inhibit the translation of β-catenin in Wnt/β-catenin pathway by phosphorylation and degradation of β-catenin 38, and the GSK-3β is negatively regulated by AKT2 20. The real time PCR data showed that introduction of miR-194 mimics significantly decreased the expression of β-catenin. However, we cannot find any seed region of miR-194 in the 3'UTR region of β-catenin. The data indicates that miR-194 may inhibit the expression of β-catenin via inhibiting of the AKT pathway. The cell-cell adhesion molecule E-cadherin sequesters β-catenin to the cell membrane and the Wnt/β-catenin pathway can be activated through accumulation of β-catenin in the cytoplasm and its translocation to the nucleus 21. In our study, we found that introduction of the miR-194 mimics dramatically decreased the invasion ability of CRC cells. Furthermore, we found that introduction of miR-194 mimics significantly increased the expression of β-catenin and the connection of β-catenin/E-cadherin on the cell membrane of CRC cells, which indicating that miR-194 may inhibit the invasion of CRC cells via blocking the Wnt/β-catenin pathway.
The expression of miR-194 may be regulated by p53 16 and it has been reported that the transcription factors, HNF-1α and Tcf1 may regulate miR-194 expression in intestinal epithelial cell line 39 and adult liver cells 40. However, it has not been reported whether miR-194 can be regulated by epigenetic modification. In this study, we first examined whether the expression of miR-194 can be regulated by DNA methylation and histone modification. The lower expression of miR-194 could not be rescued by 5'-AZA treatment (a specific inhibitor of DNA methylation) in CRC cells (data not shown), suggesting that the expression of miR-194 may not be regulated by DNA methylation. However, we found that Trichostatin A (TSA, a member of Histone deacetylase inhibitors) treatment significantly increased miR-194 expression in CRC cells (Additional File 1: Supplementary Fig. 3A), indicating that the expression of miR-194 may be regulated by histone modification. As EZH2 may regulate histone H3K27 methylation and plays an important role in CRC progression, we detected the expression of miR-194 after knockdown of EZH2 in CRC and gastric cancer cells. From the Supplementary Fig.3B, we found that knockdown of EZH2 significantly increased miR-194 expression both in CRC and gastric cancer cells, suggesting that EZH2 may play an important role in the regulation of miR-194 expression.
It has been well established that colorectal cancers have an altered miRNA expression profile 8, 41-42, and several studies have already assessed the feasibility of miRNA as a biomarker for CRC screening 43-44. Here, we examined whether miR-194 could be a candidate biomarker for CRC. Our clinical data showed that expression of miR-194 in tissues and stool samples was gradually and significantly decreased from N-N to CRC via CRA; indicating that downregulation of miR-194 may be an early event in colorectal carcinogenesis. Moreover, the association of miR-194 with the CRA, which had a greater risk of CRC, further confirmed the potential of miR-194 as a promising prognostic biomarker. Furthermore, expression level of AKT2, pAKT2 in human tissues was gradually enhanced from N-N to CRC via CRA, which is consistent with the previous report 29 and kept an inverse correlation with miR-194. Moreover, the ROC curves analysis revealed that miR-194 might be a significant parameter to discriminate between normal and abnormal tissues and stool samples. Most importantly, our data revealed that miR-194 expression having strongly positive correlation between stool and FFPE specimens, and stool/tissue miR-194 has practical sensitivity and specificity for discriminating between normal and CRC tissues, so stool sample may replace tissue specimens to be a more simple and practical diagnostic biomarker for early detection of CRC. Moreover, the clinicopathologic features analysis and patients survival data suggested that miR-194 was significantly correlated with poor survival, suggesting that miR-194 maybe a potential prognostic biomarker for CRC. Taken together, these results indicated that miR-194 had the potential to be a significant diagnostic and prognostic biomarker for CRC.
Overall, our findings revealed a potential inhibitory role of miR-194 in CRC tumorigenesis via inhibition of AKT2 pathway in vitro and in vivo. Our clinical data confirmed miR-194 downregulation in human tissues and stool samples from patients with adenomas and CRC. Clinicopathologic features data demonstrated that low miR-194 expression was associated with poor survival and occurrence of CRC. Based on all data, it was inspiring to speculate that miR-194 could be a significant diagnostic and prognostic biomarker for CRC.
Supplementary Material
Supplementary Table 1, Supplementary Figures 1-3.
Acknowledgments
This work was supported by the grant from the National Natural Science Foundation (No. 91129724), National Natural Science Foundation (No. 31271366) and Shanghai Rising-Star Program (No: 12QA1402000) and The Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning(No. 201268), National Natural Science Foundation (No. 81372267), and Key Lab Promotion Grant of Shenzhen (ZDSY20120619141025668).
Abbreviations
- CRC
colorectal cancer
- CRA
colorectal adenomas
- miRNA
mature microRNAs
- MRE
microRNA response element
- 3'-UTRs
3' untranslated regions
- ISH
In situ hybridization
- IHC
Immunohistochemistry
- ROC
receiver operating characteristic.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Mandel JS, Bond JH, Church TR, Snover DC, Bradley GM, Schuman LM. et al. Reducing mortality from colorectal cancer by screening for fecal occult blood. Minnesota Colon Cancer Control Study. N Engl J Med. 1993;328:1365–71. doi: 10.1056/NEJM199305133281901. doi:10.1056/NEJM199305133281901. [DOI] [PubMed] [Google Scholar]
- 3.Yamashita K, Watanabe M. Clinical significance of tumor markers and an emerging perspective on colorectal cancer. Cancer Sci. 2009;100:195–9. doi: 10.1111/j.1349-7006.2008.01022.x. doi:10.1111/j.1349-7006.2008.01022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pepe MS, Etzioni R, Feng Z, Potter JD, Thompson ML, Thornquist M. et al. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst. 2001;93:1054–61. doi: 10.1093/jnci/93.14.1054. [DOI] [PubMed] [Google Scholar]
- 5.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–85. doi: 10.1038/nrm1644. doi:10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 6.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–24. doi: 10.1101/gad.1399806. doi:10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 7.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. doi:10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 8.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D. et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. doi:10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 9.Lanza G, Ferracin M, Gafa R, Veronese A, Spizzo R, Pichiorri F. et al. mRNA/microRNA gene expression profile in microsatellite unstable colorectal cancer. Mol Cancer. 2007;6:54.. doi: 10.1186/1476-4598-6-54. doi:10.1186/1476-4598-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N. et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–36. doi: 10.1001/jama.299.4.425. doi:10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Padi SK, Zhang Q, Rustum YM, Morrison C, Guo B. MicroRNA-627 mediates the epigenetic mechanisms of vitamin D to suppress proliferation of human colorectal cancer cells and growth of xenograft tumors in mice. Gastroenterology. 2013;145:437–46. doi: 10.1053/j.gastro.2013.04.012. doi:10.1053/j.gastro.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen MB, Wei MX, Han JY, Wu XY, Li C, Wang J. et al. MicroRNA-451 regulates AMPK/mTORC1 signaling and fascin1 expression in HT-29 colorectal cancer. Cell Signal. 2014;26:102–9. doi: 10.1016/j.cellsig.2013.07.017. doi:10.1016/j.cellsig.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 13.Meng Z, Fu X, Chen X, Zeng S, Tian Y, Jove R. et al. miR-194 is a marker of hepatic epithelial cells and suppresses metastasis of liver cancer cells in mice. Hepatology. 2010;52:2148–57. doi: 10.1002/hep.23915. doi:10.1002/hep.23915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu X, Liu T, Fang O, Leach LJ, Hu X, Luo Z. miR-194 suppresses metastasis of non-small cell lung cancer through regulating expression of BMP1 and p27. Oncogene. 2013 doi: 10.1038/onc.2013.108. doi:10.1038/onc.2013.108. [DOI] [PubMed] [Google Scholar]
- 15.Song Y, Zhao F, Wang Z, Liu Z, Chiang Y, Xu Y. et al. Inverse association between miR-194 expression and tumor invasion in gastric cancer. Ann Surg Oncol. 2012;19(Suppl 3):S509–17. doi: 10.1245/s10434-011-1999-2. doi:10.1245/s10434-011-1999-2. [DOI] [PubMed] [Google Scholar]
- 16.Pichiorri F, Suh SS, Rocci A, De Luca L, Taccioli C, Santhanam R. et al. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell. 2010;18:367–81. doi: 10.1016/j.ccr.2010.09.005. doi:10.1016/j.ccr.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Chiang Y, Song Y, Wang Z, Liu Z, Gao P, Liang J. et al. microRNA-192, -194 and -215 are frequently downregulated in colorectal cancer. Exp Ther Med. 2012;3:560–6. doi: 10.3892/etm.2011.436. doi:10.3892/etm.2011.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sundaram P, Hultine S, Smith LM, Dews M, Fox JL, Biyashev D. et al. p53-responsive miR-194 inhibits thrombospondin-1 and promotes angiogenesis in colon cancers. Cancer Res. 2011;71:7490–501. doi: 10.1158/0008-5472.CAN-11-1124. doi:10.1158/0008-5472.CAN-11-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiong H, Hong J, Du W, Lin YW, Ren LL, Wang YC. et al. Roles of STAT3 and ZEB1 proteins in E-cadherin down-regulation and human colorectal cancer epithelial-mesenchymal transition. J Biol Chem. 2012;287:5819–32. doi: 10.1074/jbc.M111.295964. doi:M111.295964 [pii] 10.1074/jbc.M111.295964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christian SL, Sims PV, Gold MR. The B cell antigen receptor regulates the transcriptional activator beta-catenin via protein kinase C-mediated inhibition of glycogen synthase kinase-3. J Immunol. 2002;169:758–69. doi: 10.4049/jimmunol.169.2.758. [DOI] [PubMed] [Google Scholar]
- 21.Fodde R, Brabletz T. Wnt/beta-catenin signaling in cancer stemness and malignant behavior. Curr Opin Cell Biol. 2007;19:150–8. doi: 10.1016/j.ceb.2007.02.007. doi:S0955-0674(07)00021-X [pii] 10.1016/j.ceb.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 22.Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–4. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 23.Hino K, Tsuchiya K, Fukao T, Kiga K, Okamoto R, Kanai T. et al. Inducible expression of microRNA-194 is regulated by HNF-1alpha during intestinal epithelial cell differentiation. RNA. 2008;14:1433–42. doi: 10.1261/rna.810208. doi:10.1261/rna.810208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajput A, Dominguez San Martin I, Rose R, Beko A, Levea C, Sharratt E. et al. Characterization of HCT116 human colon cancer cells in an orthotopic model. J Surg Res. 2008;147:276–81. doi: 10.1016/j.jss.2007.04.021. doi:S0022-4804(07)00278-8 [pii] 10.1016/j.jss.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 25.Monnier Y, Farmer P, Bieler G, Imaizumi N, Sengstag T, Alghisi GC. et al. CYR61 and alphaVbeta5 integrin cooperate to promote invasion and metastasis of tumors growing in preirradiated stroma. Cancer Res. 2008;68:7323–31. doi: 10.1158/0008-5472.CAN-08-0841. doi:68/18/7323 [pii] 10.1158/0008-5472.CAN-08-0841. [DOI] [PubMed] [Google Scholar]
- 26.Kundu ST, Gosavi P, Khapare N, Patel R, Hosing AS, Maru GB. et al. Plakophilin3 downregulation leads to a decrease in cell adhesion and promotes metastasis. Int J Cancer. 2008;123:2303–14. doi: 10.1002/ijc.23797. doi:10.1002/ijc.23797. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H, Hao Y, Yang J, Zhou Y, Li J, Yin S. et al. Genome-wide functional screening of miR-23b as a pleiotropic modulator suppressing cancer metastasis. Nat Commun. 2011;2:554.. doi: 10.1038/ncomms1555. doi:ncomms1555 [pii] 10.1038/ncomms1555. [DOI] [PubMed] [Google Scholar]
- 28.Rychahou PG, Jackson LN, Silva SR, Rajaraman S, Evers BM. Targeted molecular therapy of the PI3K pathway: therapeutic significance of PI3K subunit targeting in colorectal carcinoma. Ann Surg. 2006;243:833–42. doi: 10.1097/01.sla.0000220040.66012.a9. discussion 43-4. doi:10.1097/01.sla.0000220040.66012.a9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roy HK, Olusola BF, Clemens DL, Karolski WJ, Ratashak A, Lynch HT. et al. AKT proto-oncogene overexpression is an early event during sporadic colon carcinogenesis. Carcinogenesis. 2002;23:201–5. doi: 10.1093/carcin/23.1.201. [DOI] [PubMed] [Google Scholar]
- 30.Ericson K, Gan C, Cheong I, Rago C, Samuels Y, Velculescu VE. et al. Genetic inactivation of AKT1, AKT2, and PDPK1 in human colorectal cancer cells clarifies their roles in tumor growth regulation. Proc Natl Acad Sci U S A. 2010;107:2598–603. doi: 10.1073/pnas.0914018107. doi:10.1073/pnas.0914018107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rychahou PG, Kang J, Gulhati P, Doan HQ, Chen LA, Xiao SY. et al. Akt2 overexpression plays a critical role in the establishment of colorectal cancer metastasis. Proc Natl Acad Sci U S A. 2008;105:20315–20. doi: 10.1073/pnas.0810715105. doi:10.1073/pnas.0810715105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hemmings BA, Restuccia DF. PI3K-PKB/Akt pathway. Cold Spring Harb Perspect Biol. 2012;4:a011189.. doi: 10.1101/cshperspect.a011189. doi:4/9/a011189 [pii] 10.1101/cshperspect.a011189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dan HC, Sun M, Kaneko S, Feldman RI, Nicosia SV, Wang HG. et al. Akt phosphorylation and stabilization of X-linked inhibitor of apoptosis protein (XIAP) J Biol Chem. 2004;279:5405–12. doi: 10.1074/jbc.M312044200. doi:10.1074/jbc.M312044200 M312044200 [pii] [DOI] [PubMed] [Google Scholar]
- 34.He J, Wu J, Xu N, Xie W, Li M, Li J. et al. MiR-210 disturbs mitotic progression through regulating a group of mitosis-related genes. Nucleic Acids Res. 2013;41:498–508. doi: 10.1093/nar/gks995. doi:gks995 [pii] 10.1093/nar/gks995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L, Sun S, Zhou J, Liu J, Lv JH, Yu XQ. et al. Knockdown of Akt1 promotes Akt2 upregulation and resistance to oxidative-stress-induced apoptosis through control of multiple signaling pathways. Antioxid Redox Signal. 2011;15:1–17. doi: 10.1089/ars.2010.3560. doi:10.1089/ars.2010.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moro L, Arbini AA, Yao JL, di Sant'Agnese PA, Marra E, Greco M. Mitochondrial DNA depletion in prostate epithelial cells promotes anoikis resistance and invasion through activation of PI3K/Akt2. Cell Death Differ. 2009;16:571–83. doi: 10.1038/cdd.2008.178. doi:10.1038/cdd.2008.178. [DOI] [PubMed] [Google Scholar]
- 37.Behrens J, von Kries JP, Kuhl M, Bruhn L, Wedlich D, Grosschedl R. et al. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–42. doi: 10.1038/382638a0. doi:10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 38.Mills CN, Nowsheen S, Bonner JA, Yang ES. Emerging roles of glycogen synthase kinase 3 in the treatment of brain tumors. Front Mol Neurosci. 2011;4:47.. doi: 10.3389/fnmol.2011.00047. doi:10.3389/fnmol.2011.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hino K, Fukao T, Watanabe M. Regulatory interaction of HNF1-alpha to microRNA-194 gene during intestinal epithelial cell differentiation. Nucleic Acids Symp Ser (Oxf) 2007:415–6. doi: 10.1093/nass/nrm208. doi:51/1/415 [pii] 10.1093/nass/nrm208. [DOI] [PubMed] [Google Scholar]
- 40.Krutzfeldt J, Rosch N, Hausser J, Manoharan M, Zavolan M, Stoffel M. MicroRNA-194 is a target of transcription factor 1 (Tcf1, HNF1alpha) in adult liver and controls expression of frizzled-6. Hepatology. 2012;55:98–107. doi: 10.1002/hep.24658. doi:10.1002/hep.24658. [DOI] [PubMed] [Google Scholar]
- 41.Bandres E, Cubedo E, Agirre X, Malumbres R, Zarate R, Ramirez N. et al. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29.. doi: 10.1186/1476-4598-5-29. doi:10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA Jr, Sjoblom T. et al. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103:3687–92. doi: 10.1073/pnas.0511155103. doi:10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahmed FE, Jeffries CD, Vos PW, Flake G, Nuovo GJ, Sinar DR. et al. Diagnostic microRNA markers for screening sporadic human colon cancer and active ulcerative colitis in stool and tissue. Cancer Genomics Proteomics. 2009;6:281–95. [PubMed] [Google Scholar]
- 44.Link A, Balaguer F, Shen Y, Nagasaka T, Lozano JJ, Boland CR. et al. Fecal MicroRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol Biomarkers Prev. 2010;19:1766–74. doi: 10.1158/1055-9965.EPI-10-0027. doi:10.1158/1055-9965.EPI-10-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1, Supplementary Figures 1-3.