Abstract
Functionally important proteins at the interface of cell and soil are of potentially low abundance when compared with commonly recovered intracellular proteins. A novel approach was developed and used to extract the metaexoproteome, the subset of proteins found outside the cell, in the context of a soil enriched with the nitrogen-containing recalcitrant polymer chitin. The majority of proteins recovered was of bacterial origin and localized to the outer membrane or extracellular milieu. A wide variety of transporter proteins were identified, particularly those associated with amino-acid and phosphate uptake. The metaexoproteome extract retained chitinolytic activity and we were successful in detecting Nocardiopsis-like chitinases that correlated with the glycoside hydrolase family 18 (GH18) chi gene data and metataxonomic analysis. Nocardiopsis-like chitinases appeared to be solely responsible for chitinolytic activity in soil. This is the first study to detect and sequence bacterial exoenzymes with proven activity in the soil enzyme pool.
Metaproteomics is an emerging technique for directly assessing cellular function and interactions within an environment. In complex environments such as soil, there is a vast dynamic range of microbial species abundance and protein expression levels. Data acquisition is biased towards high-abundance proteins, for example, chaperonins, ribosomal proteins, elongation factors and ATP synthases (Benndorf et al., 2007; Dill et al., 2010). Removal of these intracellular proteins could allow access to functionally important low-abundance proteins in the soil enzyme pool and at the interface of cell and soil, the soil metaexoproteome.
Chitin provides one of the dominant sources of organic nitrogen in soil (Gooday, 1990) and chitinases are implicated in its mineralization in a wide range of contexts (Rhazi et al., 2000; Muzzarelli, 2011), especially in nitrogen-poor soils (Olander and Vitousek, 2000). The molecular diversity of chitinases in soil microbial communities has been studied (Williamson et al., 2000; Metcalfe et al., 2002; Hjort et al., 2010) but very few have focused on the functional contributions of members of the chitinolytic bacterial community. We report here the first attempt to recover and analyse extracellular proteins in soil adopting a novel approach to extract the metaexoproteome. Our data indicate that one actinobacterial group was disproportionately responsible for chitin breakdown.
Soil was sampled from an island off the north coast of Cuba known for its high biodiversity and wide range of chitinolytic bacteria (Williamson et al., 2000; Williamson, 2001). Microcosms were constructed and amended with 1% crude crab shell (α-chitin) or squid pen (β-chitin) to enrich the microbial community, an unamended control was included for the 16S rRNA gene metataxonomic analysis (Supplementary Method S1). Community DNA was extracted and sequenced on a 454 GS FLX instrument with titanium reagents (Roche, Basel, Switzerland) using eubacterial primers Gray28F and Gray519R (Dowd et al., 2008) and GH18 Group A chi primers GASQF and GASQR (Williamson et al., 2000); the data were analysed with the bioinformatics package QIIME (Caporaso et al., 2010) (Supplementary Method S2). The metaexoproteome extraction is a modification of Masciandaro et al. (2008). In brief, 100 g soil was gently agitated with a K2SO4-based extraction solution (1:3 w/v) then the solid fraction and cells removed by centrifugation and filter sterilization before dilution (3:1 v/v) with 18.2 MΩ cm water and dialysis overnight. The retentate was concentrated to a final volume of ∼1 ml by ultrafiltration and using a centrifugal concentrator for direct loading onto a one-dimensional SDS–polyacrylamide gel electrophoresis gel. Gel-dependent nanoflow liquid chromatography-tandem MS (nanoLC-MS/MS) analysis was performed and the resultant Micromass peak list files interrogated with the NCBInr database using the MASCOT search engine (Matrix Science, London, UK). The full list of proteins was filtered to remove the few eukaryotic proteins and hits with <2 significant unique peptides (Supplementary Methods S3 and S4).
To successfully target the metaexoproteome, cell integrity must be maintained. Minimal cell lysis during the extraction was demonstrated experimentally by spiking soil with Escherichia coli overexpressing His-tagged phosphoribosyl isomerase A in the cytoplasm and attempting to detect the His-tag in the extract by western blot (Supplementary Method S5), as no protein was detected we believe the method did not lyse cells. The majority of 52 recovered proteins were Gram-negative in origin and attributed to the extracellular fraction or outer membrane (Supplementary Tables S1 and S2). Across both amendments, 73% of proteins were predicted to have a signal peptide (Nielsen et al., 1997), 13% to have transmembrane helices (Sonnhammer et al., 1998; Krogh et al., 2001), 17% to be TRAP transporters and 52% to be ABC transporters. These features are suggestive of export or being membrane bound and indicate that the metaexoproteome is representing the functional interface between cell and environment. In vitro secretomes commonly feature a similar range of TRAP and ABC transporters in addition to selected extracellular enzymes depending on the enrichment (Adav et al., 2010; Christie-Oleza and Armengaud, 2010; Christie-Oleza et al., 2012). The only extracellular enzymes identified were chitinases.
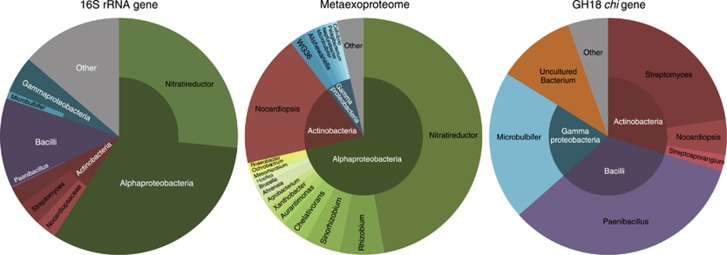
Recovered proteins were affiliated with three phyla, Proteobacteria, Actinobacteria and Bacteroidetes, this correlated well with the 16S rRNA gene data set (Figure 1). Only two genera dominated the metaexoproteome, both in terms of number of proteins recovered and protein abundance measured by emPAI (Ishihama et al., 2005), the actinomycete Nocardiopsis and the rhizobiale Nitratireductor. Approximately 17% of the identified proteins were matched to Nocardiopsis. The family Nocardiopsaceae was undetected in the unamended 16S rRNA gene data set but was one of the few actinobacterial groups to increase in abundance with α-chitin amendment, accounting for 3.7% of the community.
Figure 1.
A visual summary of the assigned bacterial community structure, recovered metaexoproteome community and GH18 chi gene taxonomic matches for the combined α- and β-chitin-amended soil. For clarity, low-abundance taxa have been grouped under ‘Other' and for the GH18 chi gene pie chart Stenotrophomonas, Amycolatopsis and Verrucosispora are not labelled as each account for <0.06% of their respective class segment.
The majority of proteins were related to the transport and metabolism of amino acids, carbohydrates and inorganic ions, namely phosphate and phosphonate. Two GH18 chitinases were identified by peptides from within their catalytic domains, ChiA from Nocardiopsis lucentensis and N. dassonvillei (Supplementary Table S1). Corresponding Nocardiopsis chiA-like sequences were identified in the GH18 chi gene pyrosequencing data set (Figure 1). Nocardiopsis chitinases have been shown to have chitinolytic activity against α- and β-chitin (Tsujibo et al., 2003) and to be capable of fast and complete degradation of crystalline chitin in liquid media (Sorokin et al., 2012).
A fluorogenic chitinase assay (Sigma-Aldrich, St Louis, MO, USA) was performed on the extracts from α-chitin-amended microcosm soil and metaexoproteome (Supplementary Method S6). Both extracts showed activity against the monomeric substrate but the metaexoproteome extract had proportionally higher activity against the more representative di-NAG and tri-NAG substrates. It is probable that the chitinase activity detected in the metaexoproteome extract is attributable to the Nocardiopsis chiA-like chitinases detected in the sequenced aliquot of the extract and represents the first example of an active exoenzyme extracted, assayed and sequenced from a soil.
The efficiency of mass spectrometry via in-gel digestion would preclude recovery of low-abundance peptides. Nocardiopsis-like proteins must therefore contribute disproportionately to the functional activity of the soil and thus the degradation of chitin. This is in marked contrast to the prevalence data for 16S rRNA gene analysis and GH18 chi gene analysis. Despite numerous attempts it was not possible to cultivate Nocardiopsis-like strains directly from the soil.
Acknowledgments
We gratefully acknowledge financial support by the Biotechnology and biological Sciences Research Council partnering award to EMHW and the EU METAEXPLORE project (KBBE-222625). ASJ-R was in receipt of a Natural Environment Research Council studentship and travel award.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Adav SS, Ng CS, Arulmani M, Sze SK. Quantitative iTRAQ secretome analysis of cellulolytic Thermobifida fusca. J Proteome Res. 2010;9:3016–3024. doi: 10.1021/pr901174z. [DOI] [PubMed] [Google Scholar]
- Benndorf D, Balcke GU, Harms H, von Bergen M. Functional metaproteome analysis of protein extracts from contaminated soil and groundwater. ISME J. 2007;1:224–234. doi: 10.1038/ismej.2007.39. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie-Oleza JA, Armengaud J. In-depth analysis of exoproteomes from marine bacteria by shotgun liquid chromatography-tandem mass spectrometry: the Ruegeria pomeroyi DSS-3 case-study. Mar Drugs. 2010;8:2223–2239. doi: 10.3390/md8082223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie-Oleza JA, Piña-Villalonga JM, Bosch R, Nogales B, Armengaud J.2012Comparative proteogenomics of twelve Roseobacter exoproteomes reveals different adaptive strategies among these marine bacteria Mol Cell Proteomics 11M111.013110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill BD, Young JC, Carey PA, VerBerkmoes NC.2010Metaproteomics: techniques and applications Environmental Molecular MicrobiologyLiu W-T, Jansson JK (eds)Caister Academic Press: Norfolk, UK; 37–61. [Google Scholar]
- Dowd SE, Sun Y, Secor PR, Rhoads DD, Wolcott BM, James GA, et al. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008;8:43. doi: 10.1186/1471-2180-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooday GW. The ecology of chitin degradation. Adv Microb Ecol. 1990;11:387–430. [Google Scholar]
- Hjort K, Bergström M, Adesina MF, Jansson JK, Smalla K, Sjöling S. Chitinase genes revealed and compared in bacterial isolates, DNA extracts and a metagenomic library from a phytopathogen-suppressive soil. Fems Micriobiology Ecol. 2010;71:197–207. doi: 10.1111/j.1574-6941.2009.00801.x. [DOI] [PubMed] [Google Scholar]
- Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, et al. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Masciandaro G, Macci C, Doni S, Maserti BE, LA Calvo-Bado, Ceccanti B, et al. Comparison of extraction methods for recovery of extracellular beta-glucosidase in two different forest soils. Soil Biol Biochem. 2008;40:2156–2161. [Google Scholar]
- Metcalfe AC, Krsek M, Gooday GW, Prosser JI, Wellington EMH. Molecular analysis of a bacterial chitinolytic community in an upland pasture. Appl Environ Microbiol. 2002;68:5042–5050. doi: 10.1128/AEM.68.10.5042-5050.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzzarelli RAA.2011Chitin nanostructures in living organisms ChitinGupta NS. (ed).Vol. 34Springer: Dordrecht, The Netherlands; 1–34. [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Olander LP, Vitousek PM. Regulation of soil phosphatase and chitinase activity by N and P availability. Biogeochemistry. 2000;49:175–191. [Google Scholar]
- Rhazi M, Desbrières J, Tolaimate A, Alagui A, Vottero P. Investigation of different natural sources of chitin: influence of the source and deacetylation process on the physicochemical characteristics of chitosan. Polym Int. 2000;49:337–344. [Google Scholar]
- Sonnhammer ELL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- Sorokin DY, Tourova TP, Sukhacheva MV, Mardanov AV, Ravin NV. Bacterial chitin utilisation at extremely haloalkaline conditions. Extremophiles. 2012;16:883–894. doi: 10.1007/s00792-012-0484-6. [DOI] [PubMed] [Google Scholar]
- Tsujibo H, Kubota T, Yamamoto M, Miyamoto K, Inamori Y. Characterization of chitinase genes from an alkaliphilic actinomycete, Nocardiopsis prasina OPC-131. Appl Environ Microbiol. 2003;69:894–900. doi: 10.1128/AEM.69.2.894-900.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson N.2001Molecular detection of chitinolytic actinomycete communities in the Cayo Blanco soilsPhD Thesis, University of Warwick, UK.
- Williamson N, Brian P, Wellington EMH. Molecular detection of bacterial and streptomycete chitinases in the environment. Antonie Van Leeuwenhoek Int J Gen Mol Microbiol. 2000;78:315–321. doi: 10.1023/a:1010225909148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.