Abstract
Bacteriophages have been used as natural biocontrol and therapeutic agents, but also as biotechnological tools for bacterial engineering. We showed recently that the transducing bacteriophage ϕMAM1 is a ViI-like phage and a member of the new genus, ‘Viunalikevirus'. Here, we show that four additional ViI-like phages and three new environmentally isolated viunalikeviruses, all infecting plant and human pathogens, are very efficient generalised transducers capable of transducing chromosomal markers at frequencies of up to 10−4 transductants per plaque-forming unit. We also demonstrate the interstrain transduction of plasmids and chromosomal markers, including genes involved in anabolism, genes for virulence and genes encoding secondary metabolites involved in biocontrol. We propose that all viunalikeviruses are likely to perform efficient horizontal gene transfer. Viunalikeviruses therefore represent useful agents for functional genomics and bacterial engineering, and for chemical and synthetic biology studies, but could be viewed as inappropriate choices for phage therapy.
Combined morphological, genomic and phylogenetic analyses have recently led to the proposed creation of a new phage genus, ‘Viunalikevirus', within the Myoviridae family (Adriaenssens et al., 2012a). The first member of this proposed genus, Salmonella phage ViI, was isolated in the 1930s (Craigie and Yen, 1938) and multiple viunalikeviruses have been sequenced and characterised since 2010 (Pickard et al., 2010; Anany et al., 2011; Hooton et al., 2011; Kutter et al., 2011; Matilla and Salmond, 2012; Park et al., 2012; Adriaenssens et al., 2012a, 2012b; Hsu et al., 2013; Luna et al., 2013; Shahrbabak et al., 2013). Viunalikeviruses are characterised as virulent (lytic) phages showing similar genome size, extensive DNA homology, strong gene synteny and a complex adsorption apparatus, which uses tail spike proteins as host-recognition determinants (Adriaenssens et al., 2012a).
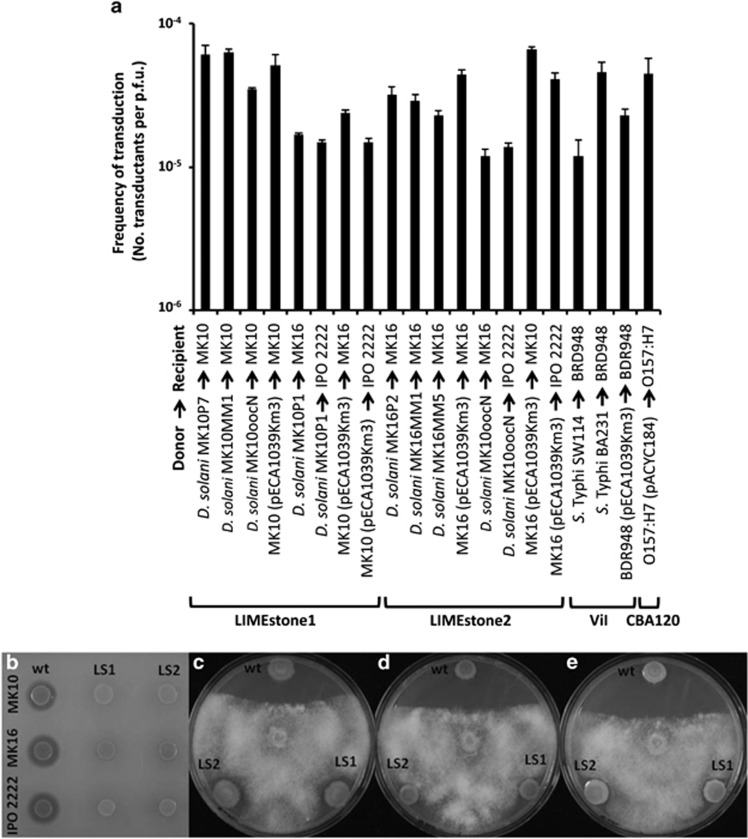
We recently isolated the ViI-like phage, ϕMAM1, that infects several environmental and clinical isolates belonging to Serratia and Kluyvera genera (Matilla and Salmond, 2012). During the characterisation of ϕMAM1, we showed that it mediates highly efficient generalised transduction (Matilla and Salmond, submitted for publication). These observations were consistent with a previous report, that the Salmonella phage ViI was also capable of transduction (Cerquetti and Hooke, 1993) and we have confirmed that phage ViI can transduce chromosomal markers and plasmids at frequencies of up to 4.6 × 10−5 transductants per plaque-forming unit (p.f.u.; Figure 1a; Supplementary Table 1).
Figure 1.
Transduction capabilities of viunalikeviruses. (a) Transduction frequencies of LIMEstone1, LIMEstone2, ViI and CBA120 phages. The graph also shows transduction efficiencies of LIMEstone phages within and between Dickeya solani strains. Transduction efficiency was defined as the number of transductants obtained per p.f.u. In all cases, error bars represent the standard deviations (n=3). (b) Skimmed milk agar plates showing protease production in the wild-type (wt) Dickeya solani strains MK10, MK16 and IPO 2222. LIMEstone1- (LS1) and LIMEstone2- (LS2) mediated transduction of the spp::Km marker from the protease negative mutant strain MK10P1 to the wild-type strains MK10, MK16 and IPO 2222 result in a protease-negative phenotype. (c–e) LIMEstone-mediated transduction of the oocN::Km marker from the oocydin A-negative mutant strain MK10oocN to the wild-type strains MK10 (c), MK16 (d) and IPO 2222 (e) results in an oocydin A-negative phenotype and, consequently, in the generation of strains defective in their antimicrobial activity against the plant pathogenic oomycete, Pythium ultimum. The anti-oomycete assays were performed as described previously (Matilla et al., 2012).
Most generalised transducers utilise a headful packing strategy where phage terminases recognise specific sequences (pac sites) in the DNA and perform cycles of packing that result in mature phage particles (Fineran et al., 2009a). Indeed, phage terminases with reduced specificity for pac sequences may lead to the evolution of efficient transducing phages (Schmeiger, 1972). Based on the high similarity between the terminases of ϕMAM1, ViI and those of other previously sequenced viunalikeviruses, we hypothesised that all of these ViI-like phages should be capable of transduction in their respective bacterial hosts. To test this hypothesis, we investigated three additional viunalikeviruses, Escherichia coli phage CBA120 (Kutter et al., 2011), and Dickeya phages LIMEstone1 and LIMEstone2 (Adriaenssens et al., 2012b). All the bacteriophages, bacterial strains, plasmids and primers used in this study are listed in the Supplementary Tables 2 and 3. Experimental procedures are presented as Supplementary Material.
The LIMEstone phages specifically infect some strains of the emerging plant pathogen, Dickeya solani (Adriaenssens et al., 2012b), and here we showed that they also infect the recently sequenced D. solani strains MK10, MK16 and IPO 2222. As predicted, we confirmed that the LIMEstone phages effected efficient transduction of various auxotrophic markers between Dickeya solani strains (Figure 1a; Supplementary Table 4). To our knowledge, only one Dickeya transducing phage, ϕEC2, has been isolated previously (Resibois et al., 1984). Additional mutant strains were constructed and the generalised nature of the transduction was confirmed by transfer of multiple chromosomal markers, including mutations in the gene cluster encoding biosynthesis of the anti-oomycete haterumalide, oocydin A (Matilla et al., 2012) and in the locus for synthesis and secretion of protease virulence factors. Transduction frequency was higher at an multiplicity of infection (m.o.i.) of 0.1 and 0.01 with efficiencies of up to 10−4 transductants per p.f.u. (Figure 1a; Supplementary Tables 4 and 5).
We also demonstrated transduction of a kanamycin resistance-marked plasmid pECA1039-Km3 between strains MK10, MK16 and IPO 2222 at frequencies of up to 8.6 × 10−5 (Supplementary Table 4). Plasmid pECA1039 (originally isolated from the phytopathogen, Pectobacterium atrosepticum) encodes a bifunctional type III Toxin-Antitoxin (TA) system, ToxIN, with abortive infection capacity. Although ToxIN aborts infection of various enterobacteria by diverse phages (Fineran et al., 2009b) it did not protect against infection by the tested viunalikeviruses, ϕMAM1, ViI, CBA120, LIMEstone1 or LIMEstone2 (not shown). Furthermore, another type III TA system, TenpIN, from the insect pathogen, Photorhabdus luminescens (Blower et al., 2012), failed to protect against any of the five ViI-like phages (not shown).
In addition, we also tested the transduction capacity of the E. coli phage, CBA120, and confirmed transduction of plasmid-borne antibiotic resistances at a frequency of up to 10−4 transductants per p.f.u. (Figure 1a; Supplementary Table 6).
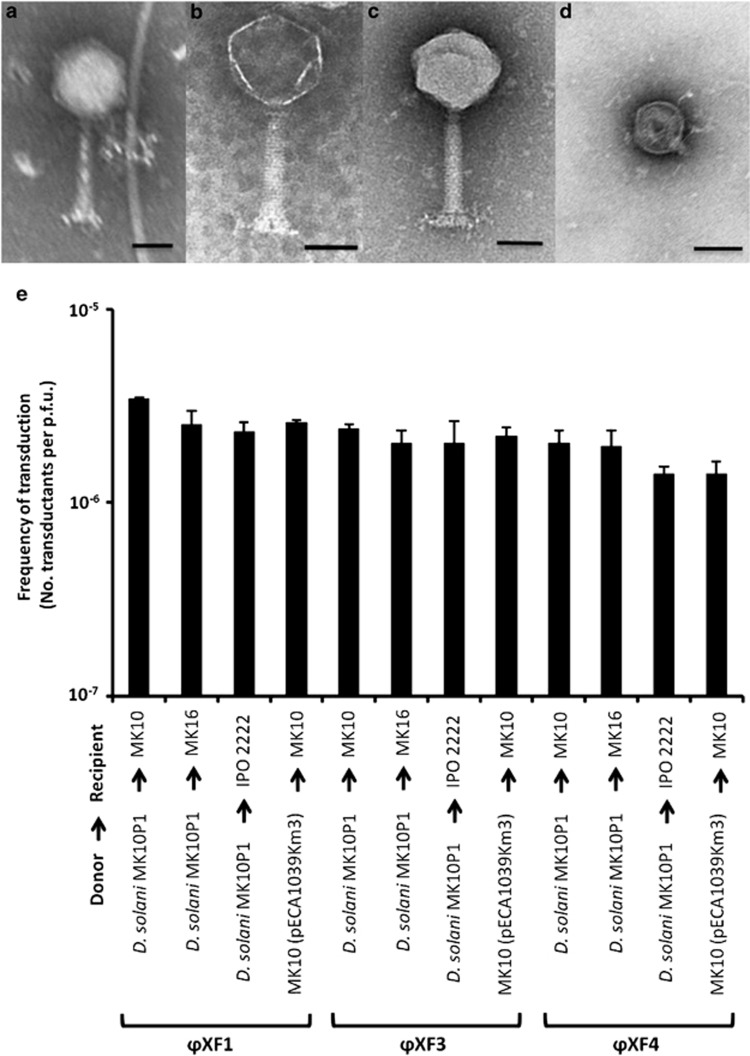
We decided to test our hypothesis that the viunalikeviruses may all be generalised transducers by first isolating new viunalikeviruses from the environment. From treated sewage effluent, we isolated three new bacteriophages infecting Dickeya solani, ϕXF1, ϕXF3 and ϕXF4, as defined initially by their very characteristic ViI-like morphology in electron microscopy (Figures 2a–c). As predicted, all of these new phages were able to transduce chromosomal markers and plasmids at frequencies of up to 3 × 10−6 transductants per p.f.u. (Figure 2e; Supplementary Table 7). Sequencing of structural and non-structural protein-encoding genes of ϕXF1, ϕXF3 and ϕXF4 showed high nucleotide homology (between 80% and 100%) with the corresponding orthologs in LIMEstone1 (Supplementary Figure 1), indicating that these virgin environmental isolates also clade within the Viunalikevirus genus.
Figure 2.
Environmental isolation and characterisation of new viunalikeviruses with generalised transduction functionality. Transmission electron micrographs of phages ϕXF1 (a), ϕXF3 (b), ϕXF4 (c) and ϕXF28 (d) are shown. As an internal control, ϕXF28 was an example of a new lytic phage isolated from the same environment but showing no transduction capabilities. Bars, 50 nm. (e) Transduction frequencies of the new viunalikeviruses ϕXF1, ϕXF3 and ϕXF4. Transduction experiments were performed using 109 cells with ϕXF1, ϕXF3, ϕXF4 at an m.o.i. of 0.01. Transduction efficiency was defined as the number of transductants obtained per p.f.u. Error bars represent the standard deviations (n=3).
Although we did not have access to other ViI-like Salmonella phages SFP10 (Park et al., 2012), ϕSH19 (Hooton et al., 2011) and Marshall (Luna et al., 2013), Escherichia phage PhaxI (Shahrbabak et al., 2013), Shigella phage ϕSboM-AG3 (Anany et al., 2011) and Klebsiella phage 0507-KN2-1 (Hsu et al., 2013), our results allow us to predict that all of these phages will mediate generalised transduction. Importantly, these phages would be expected to contribute to the horizontal gene transfer of virulence factors and antimicrobial-resistance determinants in diverse environments.
Viunalikeviruses do not seem to be limited to the enterobacteria as bacteriophages showing ViI-like morphology have been isolated in Acinetobacter (Ackermann et al., 1994), Bordetella (Adriaenssens et al., 2012b) and Sinorhizobium (Werquin et al., 1988). Furthermore, another ViI-like morphotype phage (ϕM12 of Sinorhizobium meliloti) has also been shown to be an efficient transducer (Finan et al., 1984). Taken together, these results suggest that, even in the absence of strongly predictive comparative genomic detail, a characteristically discrete ViI-like morphology in electron microscopy may be sufficient to identify new phages as strong candidates for possession of generalised transduction capacity.
The emergence and dissemination of antibiotic-resistant pathogens coupled with low discovery rates for new antimicrobials, plus increasing legal constraints on the use of chemical pesticides, have (re)focussed attention on the potential use of bacteriophages for ‘natural biocontrol' of human, animal and plant pathogens. Several viunalikeviruses have been proposed as candidate therapeutic agents for the control of bacterial infections (Anany et al., 2011; Hooton et al., 2011; Park et al., 2012; Hsu et al., 2013; Shahrbabak et al., 2013) and the LIMEstone phages have been used in successful field trials for biocontrol of D. solani infections (Adriaenssens et al., 2012b). However, their efficient transduction capacities could provide a route for dissemination of virulence factors, such as proteases (Marits et al., 1999). In fact, we have demonstrated the interstrain transduction of plasmids and oocydin A, auxotrophy and protease markers between three different D. solani strains, at high frequencies (Figures 1 and 2; Supplementary Tables 4 and 7). Also, the irregular distribution of the oocydin A gene cluster within the Dickeya genus and the fact that its genomic context varies between strains raises the possibility of phage-mediated horizontal gene transfer between bacterial strains. These results emphasize strongly that when considering the genomics of phages for ‘phage therapy' the absence of genes readily defined as playing roles in lysogeny or bacterial virulence may be insufficient to inspire confidence that use of a particular therapeutic phage presents no risk–particularly among the high efficiency-transducing viunalikeviruses.
Concluding remarks
Our results predict that all viunalikeviruses are likely to be capable of highly efficient horizontal gene transfer between their cognate bacterial hosts. This capacity could be exploited for use in fundamental research in bacterial functional genomics, and biotechnologically, for genetic engineering in chemical biology and synthetic biology applications. However, phages that show efficient horizontal gene transfer capacity could present biosafety implications for manipulation of bacterial pathogens. Obviously, the transduction capabilities of viunalikeviruses should encourage a cautious reconsideration of their appropriateness for phage therapy in human, animal or plant pathology.
Acknowledgments
This research was supported by the EU Marie-Curie Intra-European Fellowship for Career Development (FP7-PEOPLE-2011-IEF) grant number 298003. The Salmond laboratory is supported by funding through the Biotechnology and Biological Sciences Research Council (UK). Work with plant pathogens was carried out under DEFRA licence No. 50864/197900/1. We thank Rob Lavigne, Ian Toth, Elizabeth Kutter, Derek Pickard, Nicholas Thomson and Gordon Dougan for the very generous donation of phages and bacterial strains, and for helpful discussions.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Ackermann HW, Brochu G, Emadi Konjin HP. Classification of Acinetobacter phages. Arch Virol. 1994;135:345–354. doi: 10.1007/BF01310019. [DOI] [PubMed] [Google Scholar]
- Adriaenssens EM, Ackermann HW, Anany H, Blasdel B, Connerton IF, Goulding D, et al. A suggested new bacteriophage genus: ‘Viunalikevirus'. Arch Virol. 2012;157:2035–2046. doi: 10.1007/s00705-012-1360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adriaenssens EM, Van Vaerenbergh J, Vandenheuvel D, Dunon V, Ceyssens PJ, De Proft M, et al. T4-related bacteriophage LIMEstone isolates for the control of soft rot on potato caused by 'Dickeya solani'. PLoS One. 2012;7:e33227. doi: 10.1371/journal.pone.0033227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anany H, Lingohr E, Villegas A, Ackermann HW, She YM, Griffiths M, et al. A Shigella boydii bacteriophage which resembles Salmonella phage ViI. Virol J. 2011;8:242. doi: 10.1186/1743-422X-8-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower TR, Short FL, Rao F, Mizuguchi K, Pei XY, Fineran PC, et al. Identification and classification of bacterial Type III toxin-antitoxin systems encoded in chromosomal and plasmid genomes. Nucleic Acids Res. 2012;40:6158–6173. doi: 10.1093/nar/gks231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerquetti MC, Hooke AM. Vi I typing phage for generalized transduction of Salmonella typhi. J Bacteriol. 1993;175:5294–5296. doi: 10.1128/jb.175.16.5294-5296.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigie J, Yen CH. The demonstration of types of B. typhosus by means of preparations of type II Vi phage. I. Principles and technique. Can Public Health J. 1938;29:448–463. [Google Scholar]
- Finan TM, Hartwieg E, LeMieux K, Bergman K, Walker GC, Signer ER. General transduction in Rhizobium meliloti. J Bacteriol. 1984;159:120–124. doi: 10.1128/jb.159.1.120-124.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineran PC, Petty NK, Salmond GPC.2009Transduction: host DNA transfer by bacteriophagesIn Schaechter M, (ed)The Encyclopedia of Microbiology3rd ednElsevier: Oxford; 666–679. [Google Scholar]
- Fineran PC, Blower TR, Foulds IJ, Humphreys DP, Lilley KS, Salmond GP. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc Natl Acad Sci USA. 2009;106:894–899. doi: 10.1073/pnas.0808832106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooton SP, Timms AR, Rowsell J, Wilson R, Connerton IF. Salmonella Typhimurium-specific bacteriophage PhiSH19 and the origins of species specificity in the Vi01-like phage family. Virol J. 2011;8:498. doi: 10.1186/1743-422X-8-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CR, Lin TL, Pan YJ, Hsieh PF, Wang JT. Isolation of a bacteriophage specific for a new capsular type of Klebsiella pneumoniae and characterization of its polysaccharide depolymerase. PLoS One. 2013;8:e70092. doi: 10.1371/journal.pone.0070092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutter EM, Skutt-Kakaria K, Blasdel B, El-Shibiny A, Castano A, Bryan D, et al. Characterization of a ViI-like phage specific to Escherichia coli O157:H7. Virol J. 2011;8:430. doi: 10.1186/1743-422X-8-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna AJ, Wood TL, Chamakura KR, Kuty Everett GF. Complete genome of Salmonella enterica serovar Enteritidis myophage Marshall. Genome Announc. 2013;1:e00867–13. doi: 10.1128/genomeA.00867-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marits R, Kõiv V, Laasik E, Mäe A. Isolation of an extracellular protease gene of Erwinia carotovora subsp. carotovora strain SCC3193 by transposon mutagenesis and the role of protease in phytopathogenicity. Microbiology. 1999;145:1959–1966. doi: 10.1099/13500872-145-8-1959. [DOI] [PubMed] [Google Scholar]
- Matilla MA, Salmond GP. Complete genome sequence of Serratia plymuthica bacteriophage ϕMAM1. J Virol. 2012;86:13872–13873. doi: 10.1128/JVI.02702-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla MA, Stöckmann H, Leeper FJ, Salmond GP. Bacterial biosynthetic gene clusters encoding the anti-cancer haterumalide class of molecules: biogenesis of the broad spectrum antifungal and anti-oomycete compound, oocydin A. J. Biol Chem. 2012;287:39125–39138. doi: 10.1074/jbc.M112.401026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Lee JH, Shin H, Kim M, Choi J, Kang DH, et al. Characterization and comparative genomic analysis of a novel bacteriophage, SFP10, simultaneously inhibiting both Salmonella enterica and Escherichia coli O157:H7. Appl Environ Microbiol. 2012;78:58–69. doi: 10.1128/AEM.06231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard D, Toribio AL, Petty NK, van Tonder A, Yu L, Goulding D, et al. A conserved acetyl esterase domain targets diverse bacteriophages to the Vi capsular receptor of Salmonella enterica serovar Typhi. J Bacteriol. 2010;192:5746–5754. doi: 10.1128/JB.00659-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resibois A, Colet M, Faelen M, Schoonejans E, Toussaint A. phiEC2, a new generalized transducing phage of Erwinia chrysanthemi. Virology. 1984;137:102–112. doi: 10.1016/0042-6822(84)90013-8. [DOI] [PubMed] [Google Scholar]
- Schmieger H. Phage P22 mutants with increased or decreased transduction activities. Mol Gen Genet. 1972;119:75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- Shahrbabak SS, Khodabandehlou Z, Shahverdi AR, Skurnik M, Ackermann HW, Varjosalo M, et al. Isolation, characterization and complete genome sequence of PhaxI: a phage of Escherichia coli O157: H7. Microbiology. 2013;159:1629–1638. doi: 10.1099/mic.0.063776-0. [DOI] [PubMed] [Google Scholar]
- Werquin M, Ackermann HW, Levesque RC. A study of bacteriophages of Rhizobium meliloti. Appl Environ Microbiol. 1988;54:188–196. doi: 10.1128/aem.54.1.188-196.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.