Abstract
Despite microbes' key roles in driving biogeochemical cycles, the mechanism of microbe-mediated feedbacks to global changes remains elusive. Recently, soil transplant has been successfully established as a proxy to simulate climate changes, as the current trend of global warming coherently causes range shifts toward higher latitudes. Four years after southward soil transplant over large transects in China, we found that microbial functional diversity was increased, in addition to concurrent changes in microbial biomass, soil nutrient content and functional processes involved in the nitrogen cycle. However, soil transplant effects could be overridden by maize cropping, which was attributed to a negative interaction. Strikingly, abundances of nitrogen and carbon cycle genes were increased by these field experiments simulating global change, coinciding with higher soil nitrification potential and carbon dioxide (CO2) efflux. Further investigation revealed strong correlations between carbon cycle genes and CO2 efflux in bare soil but not cropped soil, and between nitrogen cycle genes and nitrification. These findings suggest that changes of soil carbon and nitrogen cycles by soil transplant and cropping were predictable by measuring microbial functional potentials, contributing to a better mechanistic understanding of these soil functional processes and suggesting a potential to incorporate microbial communities in greenhouse gas emission modeling.
Keywords: climate change, soil transplant, microbial community, biogeochemical cycle, GeoChip
Introduction
We are witnessing some of the most rapid changes in climate and land use practices in Earth's history. Nevertheless, their consequences remain elusive. Ecosystem feedbacks may either amplify or dampen the extent of global change, making them important targets for further investigation. Microbes constitute a major portion of the Earth's biosphere and have a key role in determining effluxes of greenhouse gases such as carbon dioxide (CO2), methane (CH4) and nitrous oxide (N2O), which are considered to be major feedback responses to global changes (Falkowski et al., 2008). Therefore, it is important to investigate soil microbial communities to accurately predict the future dynamics and consequences of global changes.
Assessing microbial communities in their native niches is difficult, partly because of their vast diversity, complex interaction, frequent genetic interexchange and lack of appropriate analysis tools (Singh et al., 2010). Moreover, a central issue in microbial ecology is how microbial community composition and functional potentials are linked to metabolism. Although a growing number of studies have demonstrated that microbial community composition has an essential role in biogeochemical cycles (Falkowski et al., 2008), the quantification of this linkage is neither consistent nor tractable. This knowledge gap has resulted in the disputable assumption of global climatic models that microbial composition is irrelevant to global change (Reed and Martiny, 2007) and has hampered the exploitation of microbes in mitigating greenhouse gas emission.
The use of soil transplant as a proxy to study the effects of global changes has been successfully demonstrated in both plant biology and microbiology (Balser and Firestone, 2005; Breeuwer et al., 2010; De Frenne et al., 2011; Lazzaro et al., 2011; Vanhala et al., 2011). It was reported that soil microbial community structure and community functions were altered when soil was transplanted into warmer regions to simulate global warming (Vanhala et al., 2011), which was consistent with a number of studies showing that warming altered microbial community (Petchey et al., 1999; Rinnan et al., 2007; Zhou et al., 2012). Nevertheless, it remains unclear how microbial functional potentials, representing the underlying mechanisms of microbial community metabolism, are altered.
To tackle this question, we initiated an integrated study in an agricultural ecosystem to investigate the microbial community via traditional molecular tools (phospholipid fatty acid (PLFA) and denaturing gradient gel electrophoresis (DGGE)) and a high-throughput metagenomics tool named GeoChip 3.0, which profiles microbial functional potentials by targeting a wide range of functional genes on a microarray (He et al., 2012). GeoChip 3.0 contains 27 812 probes in 292 gene families involved in microbe-mediated processes such as carbon (C), nitrogen (N), phosphorus (P) and sulfur (S) cycles, antibiotic resistance and organic pollutant remediation. With advantages in quantitative capability, robustness to contaminants and random sampling errors, GeoChip has been widely applied in analyzing samples from soil, seawater, landfill and microbial electrolysis cells (Lu et al., 2012; Zhou et al., 2012).
In this study, we analyze the interactive effects of southward soil transplant and maize cropping. We postulate that (i) southward soil transplant will result in an increase in microbial diversity, as community differentiation may be stimulated at warmer regions; (ii) changes of microbial functional potentials will affect processes such as nitrification and field CO2 efflux; and (iii) cropping will alter the transplant effects on microbial community structure and metabolism, introducing complexity and imposing a challenge to climate change research, as most of the current studies were based on vegetated soil (Balser and Firestone, 2005; Vanhala et al., 2011; Zhou et al., 2012). Our test of these hypotheses may provide valuable insights to understand how these simulated global changes affect microbial community and soil functional processes.
Materials and methods
Site description and soil sampling
In October 2005, a total of 18 (1.4 m × 1.2 m × 1.0 m (length × width × depth)) soil plots were set up at Hailun experimental station, Heilongjiang province (N site, E126°38′ and N47°26′). For each of these plots, the aboveground plant community was removed and soil was excavated. To ensure an intact soil matrix, soil was stratified with every 20 cm per layer during excavation. Six plots were mock transplanted but remained in-place to serve as control plots. Six other plots were transferred to Fengqiu, Henan province (C site, E114°24′ and N35°00′, transplanted samples were thereby designated as NC), and another six plots to Yingtan, Jiangxi Province (S site, E116°55′ and N28°15′, transplanted samples were thereby designated as NS). All of these plots were isolated from the surrounding environment by fencing with a 20-cm brick wall and with an underlayment of quartz sand. To test the vegetation effect, maize was planted each year, starting in 2006, to half of each site. Samples with maize cropping at these sites were designated as follows: Nm, NCm and NSm. For the noncropped plots, soils were kept to be bare by weed removal. The annual average temperature at N, NC and NS sites is 3.0, 13.9 and 17.6 °C, respectively. The annual precipitation at N, NC and NS sites is 530, 605 and 1795 mm, respectively. The soil type at the N, C and S site is Phaeozem with a pH of 6.5, Cambisol with a pH of 7.7 and Acrisol with a pH of 4.0, respectively.
In October 2005, 10 soil cores of 2 cm diameter were collected and composited from the surface soil (0–15 cm) at the N site (namely, 2005 N shown in Supplementary Table S1). In the late summer (August–September) of 2009, 10 soil cores of 2 cm diameter were collected and composited from the surface soil (0–15 cm) of each plot. As microbial biomass measurements by PLFA experiments were unsuccessful for the 2009 samples, soil was recollected in October, 2011 and used for PLFA experiments. Soil for DNA extraction was transported to the laboratory in liquid nitrogen and stored at −80 °C until analysis. Soil for geochemical analysis was transported on ice and stored at 4 °C.
Soil attribute and nitrification potential measurements
Soil pH was measured in a soil and water suspension (soil: water ratio of 2.5:1) with a pH meter (Mettler Toledo Instruments, Shanghai, China), and organic matter was measured by dichromate oxidation. The moisture content was measured by an oven-drying method (Lu, 1999). Total and available N were determined by Kjeldahl digestion and by the Illinois Soil Nitrogen Test (ISNT) diffusion method (Khan et al., 2001), respectively. Total P in soil was fused with sodium carbonate, and available P was extracted by sodium bicarbonate and both were determined by a molybdenum blue method (Olsen et al., 1954). Total and available potassium concentrations were determined with flame photometry FP6400A (CANY Precision Instrument Co., Ltd, Shanghai, China) after pretreatments of fusion with sodium hydroxide and extracted by ammonium acetate, respectively. Soil CO2 effluxes, which were the averages of July and August 2011, were measured by a Li 6400 Portable Photosynthesis System (LI-COR Inc., Lincoln, NE, USA). Annual precipitation, relative humidity and annual average temperature were recorded by the meteorological stations at three agricultural experiment stations.
Soil nitrification potential was determined based on a method described in a previous study (Smolders et al., 2001), with slight modifications. Briefly, a 31.5-mg (NH4)2SO4 solution was added to 70 g of soil (oven-dry weight equivalent sieved by 2 mm sieve), and additional water was added to adjust the final soil moisture content to 70% water-holding capacity. Soil was incubated in the dark at 25 °C for 14 days. On Days 0 and 14, soil NO3− was extracted with 1 M KCl extractant (soil: KCl extractant ratio of 1:5) for 2 h and then determined by a continuous flow analyzer (Bran+Luebbe GmbH Inc., Hamburg, Germany).
DGGE analyses
DGGE was performed to determine the composition of the soil microbial communities, using a method described previously with some modifications (Tong et al., 2007). Briefly, soil DNA was extracted using the FastDNA Spin Kit for Soil (MP Biomedicals, Santa Ana, CA, USA), and the V3 region of 16S ribosomal DNA was amplified using the primers F338 (5′-ACT CCT ACG GGA GGC AGC AG-3′) and R534 (5′-ATT ACC GCG GCT GCT GG-3′). The amplicons were electrophoresed using a CBS-DGGE 2000 system (C.B.S. Scientific Co. Inc., Del Mar, CA, USA) for 15 h at 60 °C, 70 V with a denaturing gradient ranging from 40 to 60% and then stained with SYBR green I dye (Cambrex Bioscience, Walkersville, MD, USA). Visualization and quantification were performed with the Gel Doc XR+ imaging System (Bio-Rad Laboratories Inc., Hercules, CA, USA) and Quantity One software (Bio-Rad Laboratories), respectively.
PLFA analyses
The PLFA content was measured for soil sampled in October, 2011. Extraction was performed using a modified Bligh-Dyer protocol, as described previously (Okabe et al., 2000). Briefly, 2 g of soil (dry weight) was incubated in a 50-ml flask with 15 ml of chloroform:methanol:phosphate (1:2:0.8 volume ratio) buffer. After shaking, centrifuging and filtering, the chloroform phase was collected. The lipid extract was then separated to phospholipids, glycolipids and neutral lipids by a silicic acid-bonded solid-phase extraction column. Subsequently, phospholipids were saponified and methylated to fatty-acid methyl esters and then measured using a Sherlock Microbial Identification System (MIDI Inc., Newark, DE, USA). PLFA peaks were identified by comparing retention times with known standards.
GeoChip analyses
Microbial genomic DNA for GeoChip analyses was extracted from 5 g of soil by a freeze-grinding method, as previously described (Zhou et al., 1996). Crude DNA was purified by agarose gel electrophoresis followed by successive extractions with phenol, chloroform and butanol. DNA quantity was measured by a PicoGreen method (Lu et al., 2012). DNA purity was determined by a Nanodrop (Nanodrop Inc., Wilmington, DE, USA) using ratios of A260/A230 and A260/A280.
DNA hybridization was performed with GeoChip 3.0. In brief, microarray slides were hybridized at 45 °C for 16 h with 50% formamide on a MAUI hybridization station (BioMicro, Salt Lake City, UT, USA) and then scanned with a ScanArray 5000 microarray analysis system (Packard BioChip Technologies, Billerica, MA, USA), as previously described (Yang et al., 2013, 2014).
The statistical analysis of GeoChip data
Processing of GeoChip data was described previously (Liu et al., 2014). Briefly, spots with a signal-to-noise ratio (SNR=(signal mean−background intensity)/background standard deviation) <2.0 or present only once in three replicates were removed. Subsequently, raw data were logarithmically transformed and then divided by the mean value of each slide. Further statistical analyses were carried out by the use of the vegan and mvpart packages in R software version 2.12.2 (R Development Core Team, R Foundation for Statistical Computing, Vienna, Austria) and included the following: (i) detrended correspondence analysis (Zhou et al., 2008); (ii) hierarchical cluster analysis of whole functional genes (Eisen et al., 1998); (iii) multiple regression tree analysis (De'Ath, 2002); (iv) β diversities of microbial community structure with Bray-Curtis index(Bray and Curtis, 1957); (v) canonical correspondence analysis (CCA) based on the variance inflation factors (VIF) with attributes with VIF over 20 removed (Ramette and Tiedje, 2007); (vi) partial CCA (variation partitioning analysis) (Borcard et al., 1992); and (vii) simple Mantel tests to determine the major environmental attributes shaping microbial structures (Smouse et al., 1986). Alpha diversities were calculated by Simpson index (Lande, 1996). To determine the significance of the differences, one-way Analysis of variance followed by the least significant difference test in SAS (version 6.1) (SAS Inc., Cary, NC, USA) (Wonnacott and Wonnacott, 1972) and two-tailed, unpaired t–tests by Microsoft Excel 2010 were performed. Correlations between environmental attributes and functional genes were conducted by Pearson's correlation and t-distribution tests for r- and P-values by Microsoft Excel 2010, respectively. We also conducted two-way analysis of variance in the PASW software version 18 (SPSS Inc., Chicago, IL, USA) by treating cropping and sites as two factors to determine the significance of interaction between transplant and maize cropping.
Results and discussion
The effects of soil transplant on soil attributes
The NC and NS sites are geographically distant from the in-place N site by ∼1700 and 2300 kilometers, respectively. These two sites were 10.9 and 14.6 °C higher in annual temperature and 75 and 1265 mm higher in annual precipitation than those at the N site, respectively (Table 1). Measurements of soil attributes showed that total N content remained unchanged at the NC site, but significantly (P<0.050) decreased from 2.13 g kg−1 at the N site to 1.79 g kg−1 at the NS site. Nitrate increased from 30.15 mg kg−1 at the N site to 50.07 mg kg−1 at the NC site, but decreased to 10.80 mg kg−1 at the NS site, which was indicative of disparate soil N cycles. Available N content was increased at the NC site from 161.72 to 181.53 mg kg−1, but remained unchanged at the NS site. By contrast, other soil geochemical attributes, including pH, organic matter, P, K, available P and K, remained largely unchanged at both NC and NS sites.
Table 1. Environmental attributes in plots without maize cropping.
| Environmental attributes | Na | NC | NS | C | S |
|---|---|---|---|---|---|
| Soil physical-chemical attributes | |||||
| pH | 6.26bcb | 5.97c | 6.50b | 7.70a | 5.97c |
| Organic matter (g kg−1) | 48.56a | 49.16a | 47.44a | 9.49b | 9.92b |
| Total N (g kg−1) | 2.13a | 2.20a | 1.79b | 0.61c | 0.64c |
| NO3− (mg kg−1) | 30.15bc | 50.07a | 10.80d | 32.88b | 15.30cd |
| NH4+ (mg kg−1) | 2.13bc | 2.21bc | 2.61ab | 1.70c | 2.98a |
| Available N (mg kg−1) | 161.72b | 181.53a | 167.23b | 41.81d | 52.81c |
| Total P (g kg−1) | 0.83a | 0.85a | 0.82a | 0.62b | 0.44c |
| Total K (g kg−1) | 18.54b | 19.17a | 18.83ab | 17.84c | 9.86d |
| Available P (mg kg−1) | 33.04a | 36.20a | 33.89a | 6.58c | 21.06b |
| Available K (mg kg−1) | 180.83a | 180.00a | 170.83ab | 95.83c | 131.25bc |
| Moisture content (%) | 14.98c | 11.63d | 29.42a | 5.91e | 21.62b |
| Soil functional process | |||||
| Nitrification potential (%) | 3.19c | 27.64a | 12.53b | 14.84b | 9.05bc |
| CO2 efflux (umolCO2 m−2 s−1) | 1.54c | 3.19ab | 2.68b | 3.42a | 1.02c |
| Climate parameters | |||||
| Annual temperature (°C) | 1.60c | 13.81b | 18.38a | 13.81b | 18.38a |
| Precipitation (mm) | 60.40c | 128.80b | 132.40a | 128.80b | 132.40a |
Abbreviations: K, potassium; N, nitrogen; P, phosphorus.
N: soil located in Hailun, Heilongjiang Province, Northern China; C: soil located in Fengqiu, Henan Province, Central China; S: soil located in Yingtan, Jiangxi Province, Southern China; NC: soil of Hailun transferred to Fengqiu; NS: soil of Hailun transferred to Yingtan.
Values in the table is the mean value of three replicates. Letters behind the values indicate significant differences (P<0.050) among treatments, as determined by one-way analysis of variance followed by least significant difference test in the SAS system version 6.1. If one treatment shares a letter with another treatment, the difference between these two treatments is insignificant (P>0.050).
As soil N cycle was the most sensitive to the disturbance of soil transplant, nitrification capacity was examined (Table 1). The results showed that soil nitrification, a process well known to be microbe-mediated, increased by three or eightfold at the transplanted sites. In addition, soil CO2 efflux, another process comprising autotrophic and heterotrophic respiration, was nearly doubled at the transplanted sites. Consistently, it has been well documented that soil nitrification and CO2 efflux are strongly affected by climate changes (Grundmann et al., 1995; Davidson et al., 1998).
The effects of soil transplant on microbial communities
To examine microbial responses to soil transplant, GeoChip 3.0 was used to examine microbial functional genes. The α diversities of microbial communities, calculated by Simpson index, increased from 1575 at the N site to 2034 and 2210 at the NC and NS sites, respectively (Supplementary Figure S1A). The β diversities between sites (N vs NC, N vs NS and NC vs NS sites) were significantly (P<0.030) different from those within sites (N, NC and NS sites) (Supplementary Figure S2A), suggesting that transplanted sites differed from the original N sites. Detrended correspondence analysis and hierarchical clustering analysis further verified this finding (Supplementary Figure S3).
Several experimental controls are essential for excluding possible artifacts introduced by soil transplant. To ensure that microbial communities assayed in the transplanted sites, which were collected in 2009, were derived from samples at the N site when transplant experiment started in 2005, NC and NS samples were compared with N samples collected in 2005 (2005 N). The results showed that 71.61±0.01% and 62.20±0.01% genes detected in the NC and NS samples, respectively, were found in 2005 N samples (data not shown). By contrast, 82.36±0.01% genes detected in N samples were found in 2005 N samples. These high percentages strongly supported the fact that microbial communities assayed in the transplanted sites resulted from initial samples at the N site. To exclude the possibility that transplanted soil was influenced by native soil over the adaption period of 4 years, transplanted samples were compared with native samples at the C or S site collected in 2009. Only 29.48±0.03% genes detected in NC samples were found in C samples, but 62.79±0.01% genes detected in NS samples were found in S samples. By contrast, 21.17±0.02% and 39.56±0.01% genes detected in 2005 N samples were found in the C and S samples, respectively. Thus, displacement of transplanted samples by native soil was unlikely for the C site. However, we could not exclude the possibility of soil displacement for the S site based on microbial community information. Thus, we examined its soil pH, which is usually stable over a long time course. Soil pH at the S site was 5.97 (Table 1). When soil was transplanted from the N site, pH was changed from 6.26 to 6.50, denying the possibility of soil displacement. Interestingly, our findings that (i) 71.61±0.01% and 62.20±0.01% genes detected in the NC and NS samples, respectively, were found in 2005 N samples, and that (ii) 29.48±0.03% genes detected in NC samples were found in C samples, but 62.79±0.01% genes detected in NS samples were found in S samples, raised a possibility that the rates of microbial adaptation occurred more rapidly at the warmer site. However, further studies are necessary to examine this possibility.
DGGE, a method to unveil coarse-scale microbial species richness and diversity, was subsequently performed. Higher α diversities were detected at the NC and NS sites (Supplementary Figure S1B), in accordance with GeoChip results, and suggested that both microbial phylogenetic and functional diversity were increased. These findings supported a framework that microbial diversity was positively correlated with temperature and negatively correlated with latitude (Fuhrman et al., 2008), but opposed to other large-scale studies showing that bacterial diversity was not correlated with temperature or latitude (Fierer and Jackson, 2006; Chu et al., 2010; Mi et al., 2012). This discrepancy might be attributed to the high level of soil heterogeneity in these large-scale studies, which is controlled in our or the study by Fuhrman et al. (2008). However, we cannot exclude the possibility that the change of microbial diversity might be a reflective, transient response in adapting to environmental disturbance after transplant.
Fungal, bacterial and total microbial biomass assessed by PLFA decreased by more than 50% at both NC and NS sites (Supplementary Figure S1C), in agreement with a recent study showing that climate warming decreased microbial biomass (Bradford et al., 2008). It is likely that there may be stress on soil biota to acclimate or adapt to a new environment. Home-site advantage, which predicts that indigenous species have an advantage to survive in their native niche in competing with foreign/invasive species, has been observed in plants and microbes (Montalvo and Ellstrand, 2001; Savolainen et al., 2007; Strickland et al., 2009).
The effects of soil transplant on selected functional genes
N cycle genes
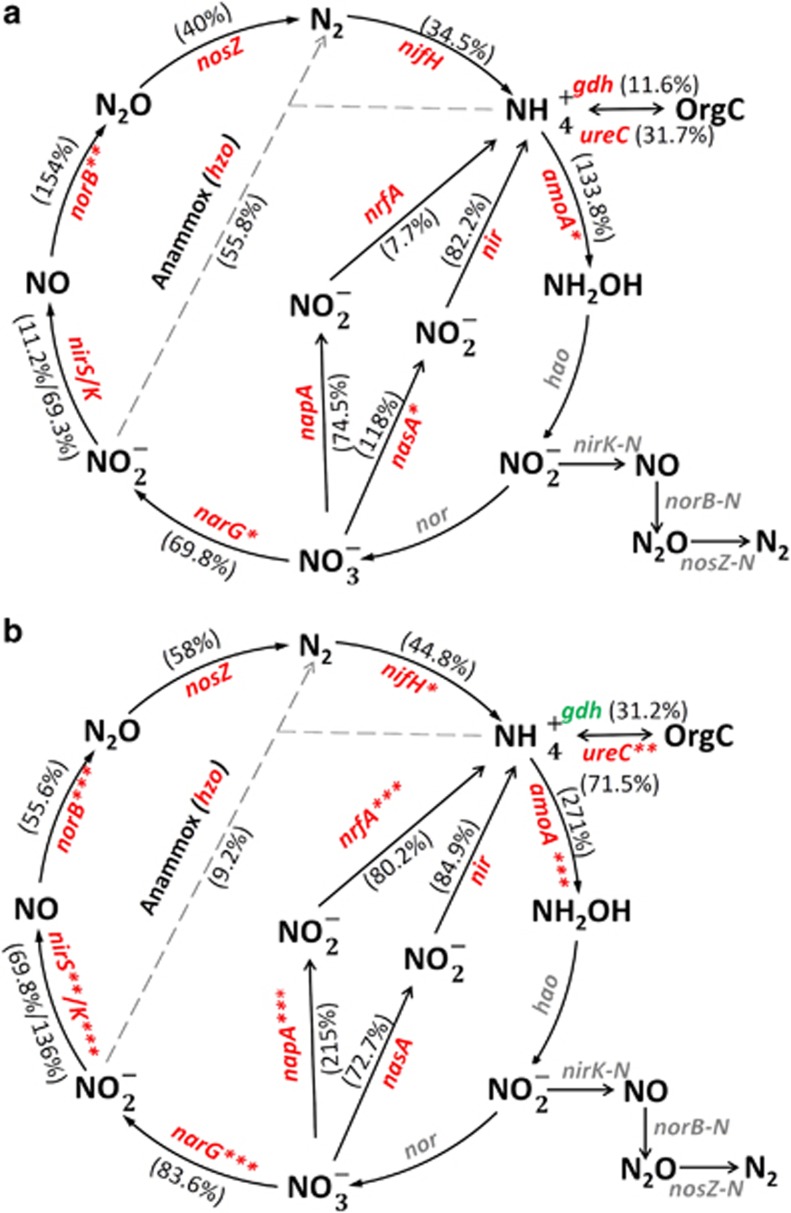
The total abundance of N cycle genes has been shown to respond to increased temperature (Szukics et al., 2010). Consistently, we found an increase in the relative abundance of microbial N cycle genes at the NS site, and to a lesser extent at the NC site (Figure 1). For example, the total abundance of amoA genes involved in nitrification was 2.7-fold higher at the NS site, whereas the increase in total abundances of narG, nirK/S, norB and nosZ genes involved in denitrification, hzo genes involved in anaerobic ammonium oxidation (anammox) and napA, nasA, nrfA and nir genes involved in assimilatory nitrogen reduction were less than 0.9-fold lower (compared with the 'home' site). Therefore, transplant elicited a range of impacts for different functional genes involved in the N cycle.
Figure 1.
Relative changes of gene abundances involved in the nitrogen cycle at (a) the NC site and (b) the NS site. The signal intensity of each gene was normalized by the mean value of all detected genes. The percentage in the bracket was the fold change (in positive or negative values) of signal intensity of each gene at the transplanted site compared to that at the N site. Significant fold changes are indicated by black font. *P<0.100, **P<0.050, ***P<0.010.
A group of 40 genes, predominantly genes involved in N fixation or denitrification, were detected only at the NC and NS sites but not at the N site (Supplementary Table S2). Most nitrogen fixation or denitrification genes were derived from unknown bacteria. A few exceptions were nifH genes derived from methanogenic archaea species Methanococcus maripaludis, Candidatus Methanoregula boonei and Methanosarcina barkeri. These archaea, with an optimum growth temperature of 37 °C, are known for nitrogen fixation activity (Leigh, 2000).
C cycle genes
Consistent with the increase of microbial functional diversity, an additional 114 and 109 C cycle genes, spanning a range of functions in C fixation, C degradation and methane cycles, were observed in the NC and NS samples, respectively. Among these, a number of genes were shared by NC and NS samples (Supplementary Table S3), including the C-fixing pcc gene derived from Deinococcus geothermalis, a radiation-resistant and slightly thermophilic species with an optimum growth temperature at 45–50 °C (Ferreira et al., 1997). Starch-degradation amyA genes derived from Shewanella amazonensis with an optimum growth temperature range around 35 °C (Venkateswaran et al., 1998) and chitin-degradation endochitinase genes derived from Bacillus thuringiensis were also shared.
P and S cycle genes
Genes present at both transplanted sites but not at the N site included 8 and 11 P and S cycle genes, respectively (Supplementary Table S4). Most P genes (ppk and ppx) were derived from Proteobacteria or Actinobacteria, two of the most abundant soil phyla (Janssen, 2006), whereas most S genes encoding sulfite reductase subunits (dsrA and dsrB) were derived from Proteobacteria or unknown bacteria.
Vegetation overrides soil transplant effects on microbial communities
We compared NC and NS samples with maize cropping (namely, NCm and NSm) against N samples with maize cropping (Nm). Total microbial biomass and bacterial biomass were decreased by transplant (Supplementary Figure S4C). Both microbial functional diversity and phylogenetic diversity remained unaltered by transplant (Supplementary Figures S4A and B). This was surprising, as soil transplant or maize cropping alone significantly increased microbial diversity (Supplementary Figures S1 and S4A). To explore this, we calculated the interactive effect of cropping and transplant on microbial diversity, using an established method (Klein et al., 2004). The results showed that the interactive effect of cropping and transplant was negative (Supplementary Figure S5).
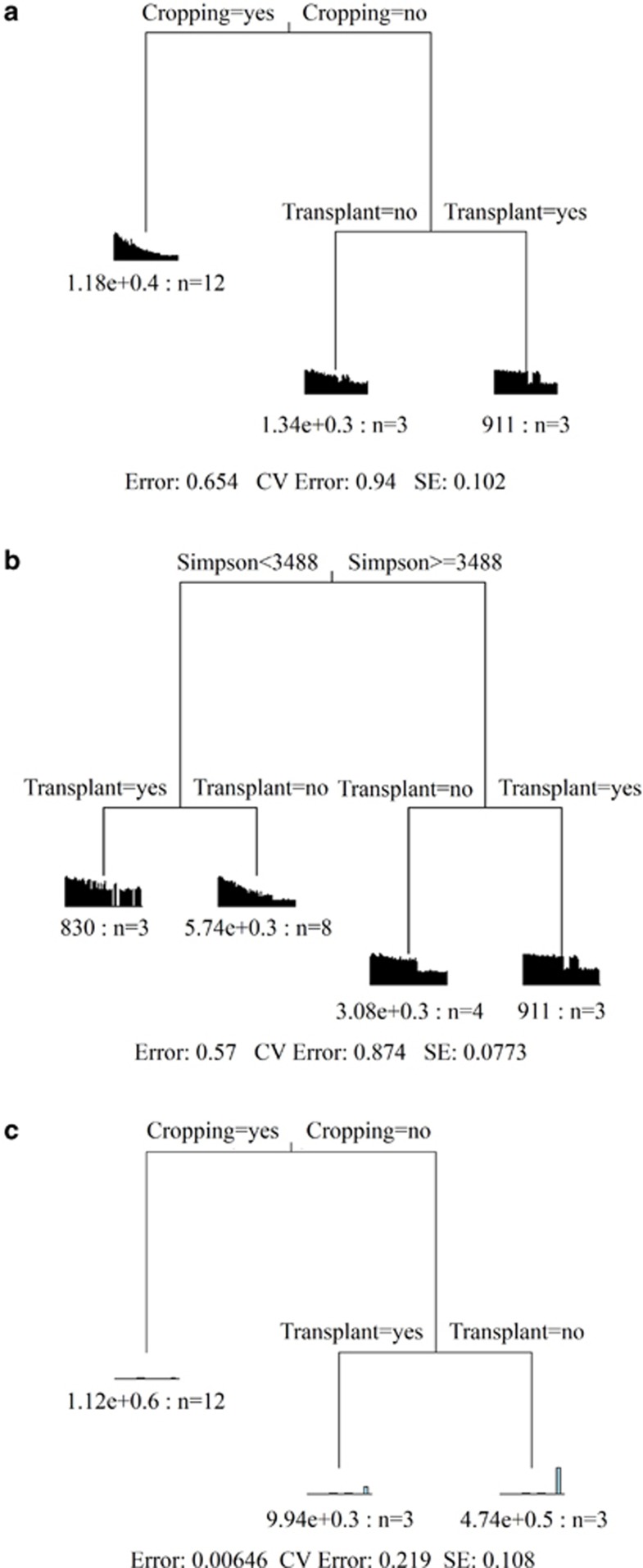
Multiple regression tree analysis, a statistical technique used to split the data into different levels and clusters based on environmental factors (De'Ath, 2002), was used to rank the relative importance of maize cropping and soil transplant effects on microbial community composition, showing that cropping overrode soil transplant in affecting microbial community structure, α diversity and soil attributes (Figure 2). This observation may raise concerns for many climate change studies, which are often based on soil with vegetation (Balser and Firestone, 2005; Vanhala et al., 2011; Zhou et al., 2012), and underscores the importance of disentangling these effects. Indeed, the effects of soil transplant on microbial community were distinct between soils with and without vegetation. Consistently, cropping has been shown to affect the structure, composition and functional activity of microbial communities through a number of intertwined mechanisms (He et al., 2010). Owing to the increased aboveground litter input to microbes, cropping has been shown to significantly increase microbial diversity (Zhang et al., 2005). Meanwhile, cropping directly affects the composition of root-associated microbial community or indirectly impacts the composition of root-associated microbial community by root exudates (Wardle et al., 2004).
Figure 2.
Multiple regression tree analysis of (a) microbial phylogenetic diversity, (b) microbial functional diversity and (c) soil variables for all samples comparing the relative importance of transplant and maize cropping.
Major environmental attributes shaping microbial community functional structure
To identify major environmental attributes shaping soil microbial communities, Mantel tests were performed with environmental attributes in all of the soil samples (Table 1 and Supplementary Table S5). Annual average temperature, precipitation, relative humidity, available N, ammonium (NH4+), total K, total P, available P, grain, maize biomass, CO2 efflux and soil nitrification capacity were significantly (P<0.050) linked to microbial community functional structures (Table 2), suggesting that climatic factors, soil properties and maize cropping were crucial for shaping the soil microbial community. By contrast, soil pH, considered a major attribute shaping microbial community (Fierer and Jackson, 2006), remained insignificant for our sites.
Table 2. The correlation between environmental attributes and microbial community functional structures by Mantel tests.
| Environmental attributes | r | P |
|---|---|---|
| pH | 0.08 | 0.215 |
| Organic matter | −0.11 | 0.788 |
| Moisture content | 0.11 | 0.144 |
| Relative humidity | 0.27 | 0.007a |
| Total N | 0.03 | 0.365 |
| Available N | 0.39 | 0.002 |
| NO3− | 0.00 | 0.455 |
| NH4+ | 0.27 | 0.013 |
| Total P | 0.24 | 0.038 |
| Available P | 0.34 | 0.003 |
| Total K | 0.20 | 0.050 |
| Available K | 0.17 | 0.108 |
| Precipitation | 0.21 | 0.029 |
| Annual temperature | 0.26 | 0.008 |
| Grain | 0.25 | 0.043 |
| Maize biomass | 0.37 | 0.009 |
| CO2 efflux | 0.58 | 0.001 |
| Nitrification potential | 0.35 | 0.002 |
Abbreviations: K, potassium; N, nitrogen; P, phosphorus.
Letters in bold are statistically significant (P<0.050).
Canonical correspondence analysis was performed to identify the major environmental attributes shaping the microbial community, resulting in a significant model (P<0.005) with ten environmental attributes that included moisture content, total N, available N, NH4+, NO3−, total P, available P, total K, maize biomass and ratio of precipitation to annual average temperature (P/T)(Supplementary Figure S6A). Among these, P/T, NH4+, available N and total P appeared to be major attributes contributing to variations of microbial functional structure.
As our results indicated that climatic factors, maize cropping, soil nitrogen components and total P were important for shaping the microbial community structure, variance partition analysis was performed to reveal both independent and interactive effects of these attributes on the microbial community. A total of 65.6% of the microbial community variation was explained by these attributes (Supplementary Figure S6B), which contributed significantly and relatively independently to the changes in microbial community structure. The interactive contribution of climate factors and soil geochemical components was 7.0%, suggesting that climate factors could affect microbial community functional structure through changes in the soil geochemical composition.
The influence of selected environmental attributes on microbial genes
Ammonium
A total of 73 functional genes were significantly (r=−0.56∼0.83, P<0.010) correlated with NH4+ (Supplementary Table S6). Among these, there were a number of pcc, nifH and narG genes, suggesting that C fixation, N fixation and denitrification were linked to NH4+ content. In addition, NH4+ was also correlated with a sensitive phylogenetic gene marker gyrB (Wang et al., 2007) derived from a number of Proteobacteria species. Some of these microbes have been well documented in NH4+ metabolism. For example, Burkholderia species are important N2-fixing microbes. Klebsiella pneumoniae can transform N2 to NO3− and assimilate NO3− and NO2− to NH4+ (Lin et al., 1994).
pH
pH was shown to be a major factor in shaping soil microbial community at the global scale (Fierer and Jackson, 2006) and to affect certain genes such as those involved in nitrification (Bru et al., 2010). Pearson's correlation tests indicated that a total of 65 genes spanning a number of categories such as the C cycle, N cycle, antibiotic and metal resistance were significantly (r=−0.62∼0.84, P<0.010) correlated with pH (Supplementary Table S7). Among these were B lactamase genes derived from Mycobacterium marinum, Rhodospirillum rubrum and several uncultured bacteria, and CopA genes derived from Archaeoglobus fulgidus, Methanococcus aeolicus, Methanocorpusculum labreanum and Sagittula stellate. For the latter, a recent DNA fingerprinting experiment showed that CopA genes were strongly influenced by pH (Lejon et al., 2007). pH has also been shown to affect both ammonia-oxidizing archaea and ammonia-oxidizing bacteria (Bru et al., 2010). Therefore, the correlation between pH and amoA-ammonia-oxidizing archaea/amoA-ammonia-oxidizing bacteria ratios was examined. A strong negative correlation (r=−0.68, P<0.010) (Supplementary Figure S7) was observed, demonstrating that pH had a role in affecting the relative abundance of ammonia-oxidizing archaea and ammonia-oxidizing bacteria.
Linking microbial genes to soil functional processes
Changes in microbial community composition affect soil functional processes and attributes. For example, in experimental microcosms inoculated with different microbial community composition, rates of litter decomposition substantially differed (Cavigelli and Robertson, 2000; Strickland et al., 2009), suggesting that microbial community composition determined soil functional processes and negated the assumption that C degradation was mediated equivalently by a wide array of microbes. However, it has also been shown that communities similar in taxonomic composition may be dissimilar in functional potentials (Strickland et al., 2009), suggesting that microbes belonging to similar taxonomic groups can have distinct functions. Further, it is possible that only a subset of microbes are key players in mediating processes. Thus, the overall microbial community composition can exhibit varying relationships with microbial community function. This observation makes it difficult to accurately predict how changes in the phylogenetic composition of microbial communities affect ecosystem functioning, underscoring the importance of using functional genes, rather than phylogenetic information, in modeling microbe-mediated functional processes such as greenhouse gas emission. Therefore, we analyzed the linkages between microbial genes and soil functional processes.
CO2 efflux
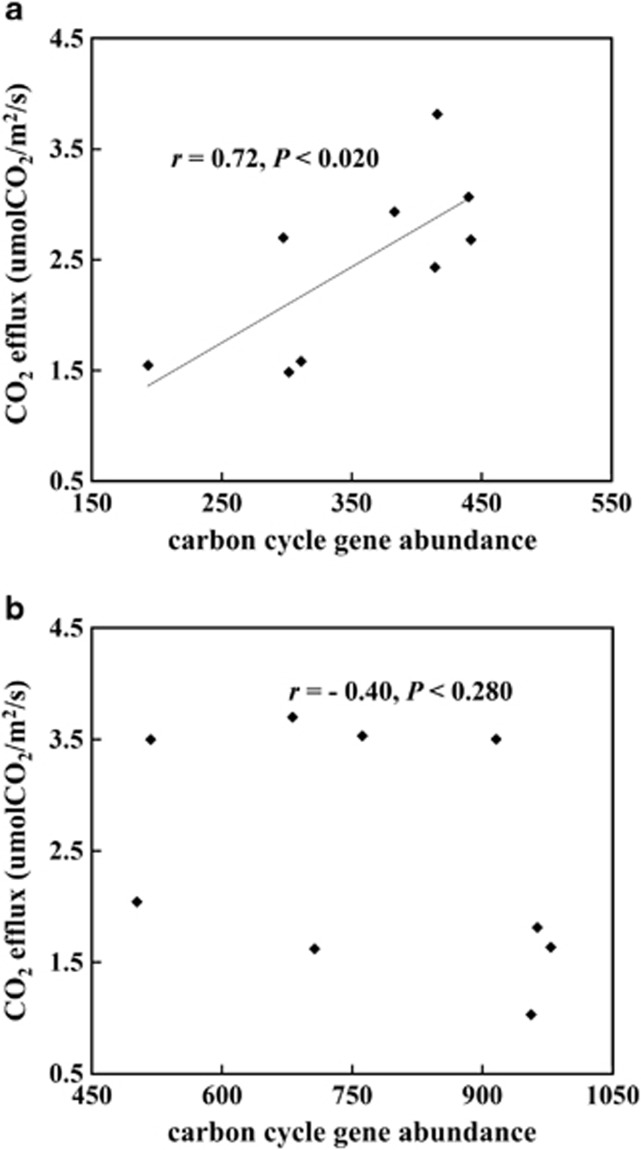
As soil CO2 efflux involves microbial respiration, the correlation between C cycle gene abundance and in situ CO2 efflux was examined. A strong positive correlation (r=0.72, P<0.020) was observed for bare soils but not cropped soils (r=−0.40, P<0.280) (Figure 3), suggesting that soil heterotrophic respiration, rather than autotrophic respiration, was linked to microbial C cycle genes. Ten C cycle genes were significantly (r>0.76, P<0.010) correlated with CO2 efflux (Supplementary Table S8), implying that they have a role in mediating CO2 efflux at these sites. Among these were C cycle genes derived from well-studied hemicellulose-degrading microbes such as Clostridia and Aspergillus species. The presence of Clostridia species, which are typically strict anaerobes, indicated that the surface soil contained anaerobic niches, which is somewhat expected, considering the highly heterogeneous nature of soil. In addition, CO2 efflux was positively correlated with a number of C fixation genes Rubisco, pcc and CODH derived from Proteobacteria and a few other bacteria, suggesting that C fixation was an important factor in affecting the volume of CO2 efflux.
Figure 3.
Correlations between in situ CO2 efflux and carbon cycle gene abundance in (a) bare soil samples and (b) maize-cropped soil samples. Statistic r and P was calculated by Pearson's correlation and t-distribution tests. Total abundance of all carbon cycle genes was used.
Nitrification capacity
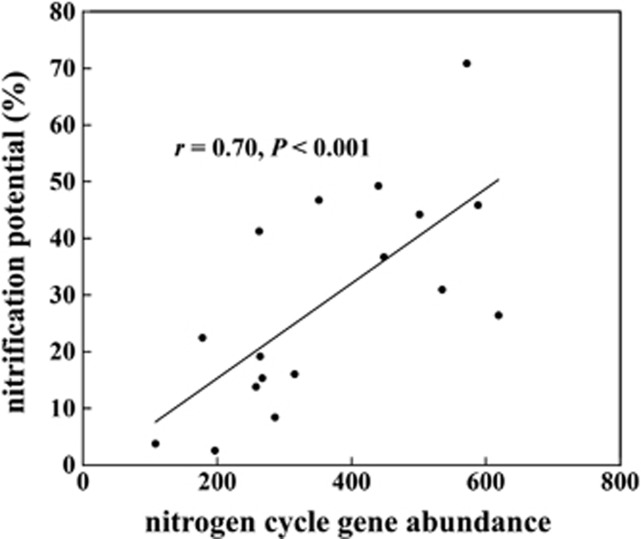
Similar to the observation with the CO2 efflux, our results showed that nitrification capacity was increased (Table 1). As soil nitrification is well known to be microbe-mediated (Grundmann et al., 1995; Lodhi et al., 2009), we examined the correlation between N gene abundance and nitrification in all of the soil samples. A strong positive correlation (r=0.70, P<0.001) was observed (Figure 4), suggesting that the increase of N cycle gene abundance in soils may partially explain the increase of the nitrification capacity in this study.
Figure 4.
Correlations between soil nitrification potential and nitrogen cycle gene abundance. Statistic r and P was calculated by Pearson's correlation and t-distribution tests. Total abundance of all nitrogen cycle genes was used.
A total of 26 N cycle genes were significantly (r>0.56, P<0.010) correlated with nitrification potential (Supplementary Table S9). Among these were genes involved in N fixation and denitrification such as a number of narG genes derived from Chromobacterium violaceum and uncultured bacteria. These results supported the coupling of nitrification to N fixation and denitrification (Wrage et al., 2001).
Understanding the mechanisms between microbial community composition and microbe-mediated processes is imperative, as current models of ecosystem functioning treat microbial communities as a static variable. Nevertheless, our results suggested that microbial communities should be taken into consideration for modeling. The correlation between microbial gene abundances and greenhouse gas emission raises a possibility that gene abundance can be used to indicate soil functional processes. Currently, it is commonly believed that microbial community DNA measures the metabolic potential, and only messenger RNA or protein represents functional activity. However, detection of soil messenger RNA from field samples imposes a number of challenges including interference from ribosomal RNA and transfer RNA, strict transport and storage requirements, rapid turnover and severe instability (Sessitsch et al., 2002), rendering messenger RNA profiling and quantification very difficult, if not intractable, in many environmental samples. By contrast, DNA profiling and quantification are substantially reliable compared with messenger RNA. We propose that changes in DNA can be used to estimate functional activity, because it indicates that the microbial population is active across the samples, thus providing a good alternative to short-lived RNAs in estimating functional activity. In support of this viewpoint, DNA abundance of nirS–nosZ genes was recently shown to correlate with N2O emission in soils, suggesting that these functional genes could serve as proxies for greenhouse gas (N2O) emissions (Morales et al., 2010).
Conclusion
Climate warming causes a shift in range toward higher latitudes across natural ecosystems (Parmesan and Yohe, 2003). Therefore, soil transplant provides a valuable strategy for examining ecosystem responses to changes in realistic climate regimes varying in factors such as temperature, precipitation and atmospheric nitrogen deposition. By examining the effects of soil transplant and cropping on microbial community, we found that changes of soil carbon and nitrogen cycles were predictable by measuring microbial functional potentials, which provided a better mechanistic understanding of soil functional processes. Nevertheless, it is important to mention the caveats imposed by the size and scope of this study. The field study was based on a relatively small number of samples and one time point sampling, which may not extrapolate to larger spatial or temporal scales. Bare soil was studied to remove the complication of plant community. Although it is present in agricultural environment, it was rare in the natural environment. To manifest transplant effects, soil was exposed to warmer regions beyond possible scenarios in the coming decades. Moreover, annual global temperature is likely to rise gradually over time, whereas transplant exposes soil to abrupt environmental changes. Therefore, this still needs to be investigated across different environments to identify linkages between microbial functional genes and soil processes, which suggests a potential to incorporate microbial communities into greenhouse gas modeling.
Acknowledgments
We thank Hailun, Fengqiu and Yingtan Research Station staff for sampling assistance, Christopher R Penton for manuscript editing and four anonymous reviewers and the editor for constructive comments and suggestions to improve this manuscript. This research was supported by grants to Yunfeng Yang from National Science Foundation of China (41171201) and National Key Basic Research Program of China (2013CB956601), to Bo Sun from National Basic Research Program of China (2011CB100506) and National Science Foundation of China (41271258), and to Jizhong Zhou from US National Science Foundation (EF-1065844).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Author contributions
This study was conceived and led by BS, JZ and YY; KX, FW and SB carried out GeoChip experiments and environmental measurements; MZ and SL performed the analytical work; and MZ and YY wrote the manuscript. All authors discussed the results and their implications and commented on the manuscript as it progressed.
Supplementary Material
References
- Balser TC, Firestone MK. Linking microbial community composition and soil processes in a California annual grassland and mixed-conifer forest. Biogeochemistry. 2005;73:395–415. [Google Scholar]
- Borcard D, Legendre P, Drapeau P. Partialling out the spatial component of ecological variation. Ecology. 1992;73:1045–1055. [Google Scholar]
- Bradford MA, Davies CA, Frey SD, Maddox TR, Melillo JM, Mohan JE, et al. Thermal adaptation of soil microbial respiration to elevated temperature. Ecol lett. 2008;11:1316–1327. doi: 10.1111/j.1461-0248.2008.01251.x. [DOI] [PubMed] [Google Scholar]
- Bray JR, Curtis JT. An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr. 1957;27:325–349. [Google Scholar]
- Breeuwer A, Heijmans MPD, Robroek BM, Berendse F. Field simulation of global change: transplanting Northern bog mesocosms Southward. Ecosystems. 2010;13:712–726. [Google Scholar]
- Bru D, Ramette A, Saby N, Dequiedt S, Ranjard L, Jolivet C, et al. Determinants of the distribution of nitrogen-cycling microbial communities at the landscape scale. ISME J. 2010;5:532–542. doi: 10.1038/ismej.2010.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavigelli MA, Robertson GP. The functional significance of denitrifier community composition in a terrestrial ecosystem. Ecology. 2000;81:1402–1414. [Google Scholar]
- Chu H, Fierer N, Lauber CL, Caporaso J, Knight R, Grogan P. Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ microbiol. 2010;12:2998–3006. doi: 10.1111/j.1462-2920.2010.02277.x. [DOI] [PubMed] [Google Scholar]
- Davidson E, Belk E, Boone RD. Soil water content and temperature as independent or confounded factors controlling soil respiration in a temperate mixed hardwood forest. Glob Chang Biol. 1998;4:217–227. [Google Scholar]
- De'Ath G. Multivariate regression trees: a new technique for modeling species-environment relationships. Ecology. 2002;83:1105–1117. [Google Scholar]
- De Frenne P, Brunet J, Shevtsova A, Kolb A, Graae BJ, Chabrerie O, et al. Temperature effects on forest herbs assessed by warming and transplant experiments along a latitudinal gradient. Glob Chang Biol. 2011;17:3240–3253. [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive Earth's biogeochemical cycles. Science. 2008;320:1034–1039. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]
- Ferreira AC, Nobre MF, Rainey FA, Silva MT, Wait R, Burghardt J, et al. Deinococcus geothermalis sp. nov. and Deinococcus murrayi sp. nov., two extremely radiation-resistant and slightly thermophilic species from hot springs. Int J syst bacteriol. 1997;47:939–947. doi: 10.1099/00207713-47-4-939. [DOI] [PubMed] [Google Scholar]
- Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA, Steele JA, Hewson I, Schwalbach MS, Brown MV, Green JL, et al. A latitudinal diversity gradient in planktonic marine bacteria. Proc Natl Acad Sci USA. 2008;105:7774–7778. doi: 10.1073/pnas.0803070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundmann G, Renault P, Rosso L, Bardin R. Differential effects of soil water content and temperature on nitrification and aeration. Soil Sci Soc Am J. 1995;59:1342–1349. [Google Scholar]
- He Z, Xu M, Deng Y, Kang S, Kellogg L, Wu L, et al. Metagenomic analysis reveals a marked divergence in the structure of belowground microbial communities at elevated CO2. Ecol lett. 2010;13:564–575. doi: 10.1111/j.1461-0248.2010.01453.x. [DOI] [PubMed] [Google Scholar]
- He Z, Deng Y, Zhou J. Development of functional gene microarrays for microbial community analysis. Curr Opin Biotechnol. 2012;23:49–55. doi: 10.1016/j.copbio.2011.11.001. [DOI] [PubMed] [Google Scholar]
- Janssen PH. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microb. 2006;72:1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan J, Wei JS, Ringnér M, Saal LH, Ladanyi M, Westermann F, et al. Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nature Med. 2001;7:673–679. doi: 10.1038/89044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein JA, Harte J, Zhao XQ. Experimental warming causes large and rapid species loss, dampened by simulated grazing, on the Tibetan Plateau. Ecol lett. 2004;7:1170–1179. [Google Scholar]
- Lande R. Statistics and partitioning of species diversity, and similarity among multiple communities. Oikos. 1996;76:5–13. [Google Scholar]
- Lazzaro A, Gauer A, Zeyer J. Field-scale transplantation experiment to investigate structures of soil bacterial communities at pioneering sites. Appl Environ Microb. 2011;77:8241–8248. doi: 10.1128/AEM.05778-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh JA. Nitrogen fixation in methanogens: the archaeal perspective. Curr Issues Mol Biol. 2000;2:125–131. [PubMed] [Google Scholar]
- Lejon DP, Nowak V, Bouko S, Pascault N, Mougel C, Martins JM, et al. Fingerprinting and diversity of bacterial copA genes in response to soil types, soil organic status and copper contamination. FEMS Microbiol Ecol. 2007;61:424–437. doi: 10.1111/j.1574-6941.2007.00365.x. [DOI] [PubMed] [Google Scholar]
- Lin JT, Goldman BS, Stewart V. The nasFEDCBA operon for nitrate and nitrite assimilation in Klebsiella pneumoniae M5al. J Bacteriol. 1994;176:2551–2559. doi: 10.1128/jb.176.9.2551-2559.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Wang F, Xue K, Sun B, Zhang Y, He Z, et al. 2014The interactive effects of soil transplant into colder regions and cropping on soil microbiology and biogeochemistry Environ Microbiole-pub ahead of print 15 January 2014; doi: 10.1111/1462-2920.12398 [DOI] [PubMed]
- Lodhi A, Arshad M, Azam F, Sajjad M, Ashraf M. Changes in mineral and mineralizable N of soil incubated at varying salinity, moisture and temperature regimes. Pak J Bot. 2009;41:967–980. [Google Scholar]
- Lu R. Soil Agricultural Chemical Analysis Methods. Agricultural Sci-Tech Press: Beijing: China; 1999. [Google Scholar]
- Lu Z, Deng Y, Van Nostrand JD, He Z, Voordeckers J, Zhou A, et al. Microbial gene functions enriched in the Deepwater Horizon deep-sea oil plume. ISME J. 2012;6:451–460. doi: 10.1038/ismej.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi L, Wang G, Jin J, Sui Y, Liu J, Liu X. Comparison of microbial community structures in four Black soils along a climatic gradient in northeast China. Can J Soil Sci. 2012;92:543–549. [Google Scholar]
- Montalvo AM, Ellstrand NC. Transplantation of the subshrub lotus scoparius: testing the home–site advantage hypothesis. Conserv Biol. 2001;14:1034–1045. [Google Scholar]
- Morales SE, Cosart T, Holben WE. Bacterial gene abundances as indicators of greenhouse gas emission in soils. ISME J. 2010;4:799–808. doi: 10.1038/ismej.2010.8. [DOI] [PubMed] [Google Scholar]
- Okabe A, Toyota K, Kimura M. Seasonal variations of phospholipid fatty acid composition in the floodwater of a Japanese paddy field under a long-term fertilizer trial. Soil Sci Plant Nutr. 2000;46:177–188. [Google Scholar]
- Olsen SR, Cole C, Watanabe FS, Dean L. Estimation of Available Phosphorus in Soils by Extraction with Sodium Bicarbonate. Vol. 939. USDA Press: Washington, DC; 1954. [Google Scholar]
- Parmesan C, Yohe G. A globally coherent fingerprint of climate change impacts across natural systems. Nature. 2003;421:37–42. doi: 10.1038/nature01286. [DOI] [PubMed] [Google Scholar]
- Petchey OL, McPhearson PT, Casey TM, Morin PJ. Environmental warming alters food-web structure and ecosystem function. Nature. 1999;402:69–72. [Google Scholar]
- Ramette A, Tiedje JM. Biogeography: an emerging cornerstone for understanding prokaryotic diversity, ecology, and evolution. Microb Ecol. 2007;53:197–207. doi: 10.1007/s00248-005-5010-2. [DOI] [PubMed] [Google Scholar]
- Reed HE, Martiny JBH. Testing the functional significance of microbial composition in natural communities. FEMS Microbiol Ecol. 2007;62:161–170. doi: 10.1111/j.1574-6941.2007.00386.x. [DOI] [PubMed] [Google Scholar]
- Rinnan R, Michelsen A, Bååth E, Jonasson S. Fifteen years of climate change manipulations alter soil microbial communities in a subarctic heath ecosystem. Glob Chang Biol. 2007;13:28–39. [Google Scholar]
- Savolainen O, Pyhäjärvi T, Knürr T. Gene flow and local adaptation in trees. Annu Rev Ecol Evol Syst. 2007;38:595–619. [Google Scholar]
- Sessitsch A, Gyamfi S, Stralis-Pavese N, Weilharter A, Pfeifer U. RNA isolation from soil for bacterial community and functional analysis: evaluation of different extraction and soil conservation protocols. J Microbiol Methods. 2002;51:171–179. doi: 10.1016/s0167-7012(02)00065-9. [DOI] [PubMed] [Google Scholar]
- Singh BK, Bardgett RD, Smith P, Reay DS. Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat Rev Microbiol. 2010;8:779–790. doi: 10.1038/nrmicro2439. [DOI] [PubMed] [Google Scholar]
- Smolders E, Brans K, Coppens F, Merckx R. Potential nitrification rate as a tool for screening toxicity in metal–contaminated soils. Environ Toxicol Chem. 2001;20:2469–2474. doi: 10.1897/1551-5028(2001)020<2469:pnraat>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Smouse PE, Long JC, Sokal RR. Multiple regression and correlation extensions of the Mantel test of matrix correspondence. Syst Zool. 1986;35:627–632. [Google Scholar]
- Strickland MS, Lauber C, Fierer N, Bradford MA. Testing the functional significance of microbial community composition. Ecology. 2009;90:441–451. doi: 10.1890/08-0296.1. [DOI] [PubMed] [Google Scholar]
- Szukics U, Abell GC, Hödl V, Mitter B, Sessitsch A, Hackl E, et al. Nitrifiers and denitrifiers respond rapidly to changed moisture and increasing temperature in a pristine forest soil. FEMS Microbiol Ecol. 2010;72:395–406. doi: 10.1111/j.1574-6941.2010.00853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Z, Bischoff M, Nies L, Applegate B, Turco RF. Impact of fullerene (C60) on a soil microbial community. Environ Sci Technol. 2007;41:2985–2991. doi: 10.1021/es061953l. [DOI] [PubMed] [Google Scholar]
- Vanhala P, Karhu K, Tuomi M, BjÖRklÖF K, Fritze H, HyvÄRinen H, et al. Transplantation of organic surface horizons of boreal soils into warmer regions alters microbiology but not the temperature sensitivity of decomposition. Glob Chang Biol. 2011;17:538–550. [Google Scholar]
- Venkateswaran K, Dollhopf ME, Aller R, Stackebrandt E, Nealson KH. Shewanella amazonensis sp. nov., a novel metal-reducing facultative anaerobe from Amazonian shelf muds. Int J Syst Bacteriol. 1998;48:965–972. doi: 10.1099/00207713-48-3-965. [DOI] [PubMed] [Google Scholar]
- Wang LT, Lee FL, Tai CJ, Kasai H. Comparison of gyrB gene sequences, 16S rRNA gene sequences and DNA–DNA hybridization in the Bacillus subtilis group. Int J Syst Evol Microbiol. 2007;57:1846–1850. doi: 10.1099/ijs.0.64685-0. [DOI] [PubMed] [Google Scholar]
- Wardle DA, Bardgett RD, Klironomos JN, Setala H, van der Putten WH, Wall DH. Ecological linkages between aboveground and belowground biota. Science. 2004;304:1629–1633. doi: 10.1126/science.1094875. [DOI] [PubMed] [Google Scholar]
- Wonnacott TH, Wonnacott RJ. Introductory Statistics. Vol. 19690. Wiley: New York; 1972. [Google Scholar]
- Wrage N, Velthof G, Van Beusichem M, Oenema O. Role of nitrifier denitrification in the production of nitrous oxide. Soil Biol Biochem. 2001;33:1723–1732. [Google Scholar]
- Yang Y, Gao Y, Wang S, Xu D, Yu H, Wu L, et al. The microbial gene diversity along an elevation gradient of the Tibetan grassland. ISME J. 2014;8:430–440. doi: 10.1038/ismej.2013.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wu L, Lin Q, Yuan M, Xu D, Yu H, et al. Responses of the functional structure of soil microbial community to livestock grazing in the Tibetan alpine grassland. Glob Chang Biol. 2013;19:637–648. doi: 10.1111/gcb.12065. [DOI] [PubMed] [Google Scholar]
- Zhang W, Parker KM, Luo Y, Wan S, Wallace LL, Hu S. Soil microbial responses to experimental warming and clipping in a tallgrass prairie. Glob Chang Biol. 2005;11:266–277. [Google Scholar]
- Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Kang S, Schadt CW, Garten CT. Spatial scaling of functional gene diversity across various microbial taxa. Proc Natl Acad Sci USA. 2008;105:7768. doi: 10.1073/pnas.0709016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Xue K, Xie J, Deng Y, Wu L, Cheng X, et al. Microbial mediation of carbon-cycle feedbacks to climate warming. Nature Clim Change. 2012;2:106–110. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.