Abstract
A replicate long-term experiment was conducted using anaerobic digestion (AD) as a model process to determine the relative role of niche and neutral theory on microbial community assembly, and to link community dynamics to system performance. AD is performed by a complex network of microorganisms and process stability relies entirely on the synergistic interactions between populations belonging to different functional guilds. In this study, three independent replicate anaerobic digesters were seeded with the same diverse inoculum, supplied with a model substrate, α-cellulose, and operated for 362 days at a 10-day hydraulic residence time under mesophilic conditions. Selective pressure imposed by the operational conditions and model substrate caused large reproducible changes in community composition including an overall decrease in richness in the first month of operation, followed by synchronised population dynamics that correlated with changes in reactor performance. This included the synchronised emergence and decline of distinct Ruminococcus phylotypes at day 148, and emergence of a Clostridium and Methanosaeta phylotype at day 178, when performance became stable in all reactors. These data suggest that many dynamic functional niches are predictably filled by phylogenetically coherent populations over long time scales. Neutral theory would predict that a complex community with a high degree of recognised functional redundancy would lead to stochastic changes in populations and community divergence over time. We conclude that deterministic processes may play a larger role in microbial community dynamics than currently appreciated, and under controlled conditions it may be possible to reliably predict community structural and functional changes over time.
Keywords: anaerobic digestion, deterministic, neutral theory, niche specialisation, synchronised dynamics
Introduction
A number of ecological theories have been used to explain observed patterns in microbial community assembly and dynamics (Sloan et al., 2006; Falk et al., 2009; Ofiteru et al., 2010; Wang et al., 2013). Historically, community composition is thought to be governed by deterministic factors such as interspecies interactions (for example, competition, syntrophy and predation) and niche differentiation (Pholchan et al., 2013; Zhou et al., 2013). According to this traditional niche-based theory, there is a strong relationship between taxon traits and the environment. More recently, an alternative theory that disregards competition between populations, neutral theory, has emerged that only considers stochastic processes such as birth, death, colonisation, immigration, speciation and dispersal limitations (Sloan et al., 2006; Ofiteru et al., 2010; Harpole, 2012). Recent studies suggest that both deterministic and stochastic processes play a role in structuring microbial communities (Ofiteru et al., 2010; Caruso et al., 2011); however, because of the complexity of many natural ecosystems, and associated lack of controlled conditions and replicated experimental design, the mechanisms and factors that affect microbial diversity and community assembly remain poorly understood (Zhou et al., 2013).
Engineered systems offer a controlled environment in which to study complex microbial communities, and together with modern culture-independent techniques that provide an objective view of community composition, bioreactors are gaining popularity as environments for testing ecological theories (Falk et al., 2009; Zhou et al., 2013). There is also a growing appreciation of understanding microbial ecology to improve the efficiency and robustness of engineered systems (Briones and Raskin, 2003; Rittmann et al., 2006; Gentile et al., 2007; Pholchan et al., 2013).
Anaerobic digestion (AD) is a microbially mediated technology for the degradation and stabilisation of organic matter, resulting in the production of energy-rich compounds such as alcohols, volatile fatty acids (VFAs) and methane. This process involves four sequential steps: hydrolysis, fermentation (acidogenesis), acetogenesis (dehydrogenation) and methanogenesis (acetoclastic or hydrogenotrophic), that depend on the synergistic interactions of microorganisms, forming a complex metabolic network. During hydrolysis and fermentation, a diverse range of bacteria degrade complex polymers (carbohydrates, lipids and proteins), yielding soluble organic molecules (sugars, fatty acids and amino acids) that are fermented into short-chain VFAs, alcohols, CO2, H2 and other by-products. Some of these fermentation products (acetate, CO2 and H2) can directly be used as substrates for methanogenesis, whereas others are first oxidised to CO2 and H2 by acetogenic bacteria that typically form a syntrophic relationship with hydrogenotrophic methanogens (Schlüter et al., 2008; Amani et al., 2010).
Culture-independent molecular techniques have been used to characterise AD-associated microbial communities under a range of process configurations, conditions, feedstocks and using different inocula (Jaenicke et al., 2010; Werner et al., 2010; Nelson et al., 2011; Pervin et al., 2013). These studies have provided substantial insight into the methanogenic populations (Lee et al., 2008b; Steinberg and Regan, 2011), but the interactions between and within functional guilds and their link to reactor performance remain poorly understood. Functional redundancy is thought to be at the core of stable reactor performance, as it ensures the presence of a reservoir of populations able to perform the same ecological function (Briones and Raskin, 2003; Rittmann et al., 2006; Allison and Martiny, 2008; Falk et al., 2009). However, few studies have examined the relationship between community dynamics and functional stability in replicated lab-scale bioreactors, and the existing studies provide conflicting results (Fernandez et al., 1999; LaPara et al., 2001; McGuinness et al., 2006; Gentile et al., 2007). This lack of consensus is likely because of a combination of variables, including community complexity, reactor design and operation (Falk et al., 2009).
In this study, a replicate time series experiment was performed using AD as a model process to determine the relative role of deterministic and stochastic processes on microbial community assembly and dynamics. The replicate reactors were seeded with the same diverse inoculum and were supplied with a sterile model substrate (α-cellulose). The microbial community composition was monitored over time and correlated with reactor performance parameters to develop a better understanding of the relationship between community dynamics and functional stability. We demonstrate that population dynamics in these complex communities are largely synchronised over long periods, suggesting that deterministic rather than stochastic processes drive population succession when environmental conditions are normalised.
Materials and methods
Inoculum
Three replicate anaerobic digesters were set up in parallel and seeded with the same diverse inoculum (20% v/v) that consisted of a mixture of samples (equal volatile suspended solids ratios) taken from eight different anaerobic locations: six well-functioning engineered systems (three mesophilic ADs, thermophilic AD, Upflow Anaerobic Sludge Bioreactor and anaerobic lagoon), and two natural environments (rumen and lake sediment) (Supplementary Figure S1 and Supplementary Table S1).
Reactor set-up and operation
The replicate anaerobic digesters (2 l working volume) were run as semi-continuous completely mixed reactors with a hydraulic and sludge retention time of 10 days. This is the minimum retention time needed to prevent washout of slow-growing methanogens (Amani et al., 2010) and allows us to highlight differences in hydrolysis rate by running the process at an active point on the first order operating curve (Jensen et al., 2009; based on a cellulose hydrolysis rate of 0.2 per day; Jensen et al., 2011). The triplicate reactors were designated AD1, AD2 and AD3. The temperature was held at 37 °C (±1 °C) and a pH of 7 was maintained by adding 1 M NaOH solution. A sterile model substrate was used as a feedstock to reduce the substrate complexity and minimise influence of microorganisms otherwise entering the system through a nonsterile feed. α-Cellulose (Sigma Aldrich, Castle Hill, NSW, Australia) was selected because it is the purest and most polymerised form of cellulose. The sterile medium consisted of 3 g l−1 Na2HPO4, 1 g l−1 NH4Cl, 0.5 g l−1 NaCl, 0.2465 g l−1 MgSO4.7 H2O, 1.5 g l−1 KH2PO4, 14.7 mg l−1 CaCl2, 2.6 g l−1 NaHCO3, 0.5 g l−1 C3H7NO2S, 0.25 g l−1 Na2S.9 H2O and 1 ml of trace solution containing 1.5 g l−1 FeSO4.7 H2O, 0.15 g l−1 H3BO3, 0.03 g l−1 CuSO4.5 H2O, 0.18 g l−1 KI, 0.12 g l−1 MnCl2.4 H2O, 0.06 g l−1 Na2Mo4.2 H2O, 0.12 g l−1 ZnSO4.7 H2O, 0.15 g l−1 CoCl2.6 H2O, 10 g l−1 EDTA and 23 mg l−1 NiCl2.6 H2O (Rabaey et al., 2005). The medium was sparged with N2 and then autoclaved at 121 °C for 60 min for oxygen removal and sterilisation, respectively. The pH was adjusted to ∼7.2 by addition of HCl (37 vol%). The reactors were fed semi-continuously with α-cellulose (5 g cellulose per l medium) four times daily at 6 hour intervals. During these feed events, ∼50 ml of feed was pumped in the systems and an equal amount of reactor sludge was wasted simultaneously using multi-head peristaltic pumps (John Morris Scientific, Brisbane, QLD, Australia). This resulted in an organic loading rate of 0.5 g α-cellulose per l reactor volume per day. During the start-up phase, feeding and wasting was switched off from day 12 to day 24 (until a sufficient decrease in VFA concentration was observed) in order to minimise washout of slow-growing microorganisms and to allow the biomass in the reactors to increase. After day 24, the reactors were again fed and wasted semi-continuously.
Digester performance monitoring
Biogas production was measured continuously from each digester using tipping bucket gas metres, and logged daily. Gas composition (CH4, CO2, H2) was determined by gas chromatography with thermal conductivity detector (GC-TCD) (Gopalan et al., 2013). Slurry samples were collected from each reactor twice per week and analysed for total chemical oxygen demand (COD), soluble COD (sCOD) and VFA concentration. Sample preparation and analyses were performed as described previously (Ge et al., 2011) and according to standard methods (APHA, 2005). The extent of cellulose solubilisation was calculated using the total COD concentration in the feed and the particulate COD (pCOD) concentration in the reactor as a proxy for the residual cellulose concentration.
Sample fixation and fluorescence in situ hybridisation
Samples for fluorescence in situ hybridisation (FISH) were collected on a weekly basis. These samples were immediately fixed in 4% paraformaldehyde for 4 h and washed twice with 1% phosphate-buffered saline solution before being stored at −20 °C in a 50:50 mixture of 1% phosphate-buffered saline and 100% ethanol (Amann et al., 1995). Cells were hybridised with universal bacterial and archaeal probes (Amann et al., 1995) in combination with population-specific probes targeting Bacteroidales, Clostridiales and Fibrobacterales populations (Table 1). FISH preparations were visualised using a Zeiss LSM512 confocal laser scanning microscope (Zeiss, Oberkochen, Germany). Brightness, contrast and registration were modified using Adobe Photoshop 6.0 (Sydney, NSW, Australia).
Table 1. Oligonucleotide probes used in fluorescence in situ hybridisation (FISH), their sequences, formamide percentage (FA) and specificity.
| Probe | Sequence (5′–3′) | FA | Specificity | Reference | |
|---|---|---|---|---|---|
| ARC951 | FITC | GTGCTCCCCCGCCAATTCCT | 20 | Archaea | Stahl and Amann, 1991 |
| EUB338 | Cy5 | GCTGCCTCCCGTAGGAGT | 20 | Bacteria | |
| EUB338+ | Cy5 | GCWGCCACCCGTAGGTGT | 20 | Daims et al., 1999 | |
| Bac1080 | Cy3 | GCACTTAAGCCGACACCT | 20 | Bacteroidales | Kong et al., 2012 |
| CF319a | Cy3 | TGGTCCGTGTCTCAGTAC | 20 | ||
| Clo549 | Cy3 | CAATCATTCCGGACAACG | 30 | Clostridiales | Kong et al., 2012 |
| Ace731 | Cy3 | TACTGTCCAGATAGCCGC | 30 | ||
| Rum831 | Cy3 | GGTCAGTCCCCCCACA | 30 | ||
| Fibr225 | FITC | AATCGGACGCAAGCTCATCCC | 20 | Fibrobacterales | Kong et al., 2012 |
DNA extraction and 16S rRNA gene amplicon sequencing
Samples for DNA extraction were taken twice per week, snap-frozen in liquid nitrogen and stored at −80 °C. DNA extractions were performed using FastDNA Spin kits for Soil (MP Biomedicals Australasia, Seven Hills, NSW, Australia) according to the manufacturer's instructions. DNA quality was assessed using gel electrophoresis (1% agarose) and DNA concentrations were measured using Quant-iT dsDNA BR Assay kits and a Qubit fluorometer (Life Technologies, Mulgrave, VIC, Australia).
Genomic DNA was extracted from samples taken from each reactor at 14 time points (days 27, 70, 88, 95, 113, 130, 148, 178, 209, 243, 276, 305, 325 and 362) and 16S rRNA genes were amplified as described previously (Cayford et al., 2012; Dennis et al., 2013). Briefly, PCR reactions (50 μl) were prepared with 20 ng of template DNA, 5 μl 10 × buffer, 1 μl dNTP mix (10 mM each), 4 μl 25 mM MgCl, 1 μl forward primer (10 mM), 1 μl reverse primer (10 mM), 0.2 μl Taq polymerase and 1.5 μl BSA (Life Technologies) and 1 μl (10 mM) of each of the universal primers targeting the V6-V8 region of the bacterial and archaeal 16S rRNA gene (Engelbrektson et al., 2010): 926F (5′-AAACTYAAAKGAATTGRCGG-3′) and 1392R (5′-ACGGGCGGTGTGTRC-3′) modified on the 5′ end to contain 454 sequencing adaptor sequences. The reverse primer also contained a 5–6 base sample-specific barcode sequence. The PCR program included one cycle at 95 °C for 3 min, followed by 30 cycles at 95 °C for 30 s, 55 °C for 30 s and 75 °C for 30 s, and then a final extension at 74 °C for 10 min. Post amplification, amplicons were pooled and sequenced using the Roche 454 GS-FLX Titanium platform (Roche Diagnostics, Castle Hill, NSW, Australia) at the Australian Centre for Ecogenomics. Sequences were submitted to the National Centre for Biotechnology Information (NCBI) Short Read Archive with the following accession numbers: SRR1175890, SRR1175892, SRR1175894 and SRR1175897.
Amplicon sequences were quality filtered, trimmed to 250 base pairs and dereplicated using the QIIME pipeline (Caporaso et al., 2010). Chimeric sequences were removed using UCHIME (Edgar et al., 2011) and Acacia (Bragg et al., 2012) was used to correct for homopolymer errors. Sequences were clustered at 97% identity using CD-Hit OTU (Wu et al., 2011) and cluster representatives were selected. Sequences were also clustered at 100% identity with uclust (Edgar, 2010) to compare the community profiles at a higher phylogenetic resolution. BLASTn (Altschul et al., 1990) was used to assign a GreenGenes taxonomy (DeSantis et al., 2006) to each cluster representative. The 97% operational taxonomic unit (OTU) data set was normalised to 1100 sequences per sample and the 100% OTU data set was normalised to 2400 sequences to allow comparison of diversity without bias from unequal sampling effort. The libraries were repeatedly subsampled and the number of OTUs observed at equal number of sequences between samples (richness) and Simpson's diversity indices (evenness) were calculated. A table with the OTUs corresponding to distinct populations and their taxonomic assignments (to the lowest possible level of classification) in each sample was generated.
Real-time quantitative PCR
Total bacterial and archaeal biomass was estimated using real-time PCR for the three reactors at the same 14 time points subjected to 16S rRNA gene amplicon sequencing. The primers 1406F (5′-GYACWCACCGCCCGT-3′) and 1525R (5′-AAGGAGGTGWTCCARCC-3′) were used to amplify bacterial and archaeal 16S rRNA genes. For inhibition control, the rpsL F (5′-GTAAAGTATGCCGTGTTCGT-3′) and rpsL R (5′-AGCCTGCTTACGGTCTTTA-3′) primer set was used that is specific for Escherichia coli DH10B rRNA. Two dilutions (1/100, 1/1000) were made of the microbial template DNA and run in parallel with an inhibition control test using Escherichia coli DH10B genomic DNA. The PCR reaction was set up using 5 μl 2 × SYBR Green/AmpliTaq Gold DNA Polymerase mix (Life Technologies), 4 μl template DNA and 1 μM primer mix (0.4 μM 1406F/1525R, 0.2 μM rpsL F/R) and each sample was run in triplicate. The PCR program included one cycle at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. A melt curve was produced by running one cycle at 95 °C for 2 min and a final cycle at 60 °C for 15 s. The cycle threshold (Ct) values were recorded and analysed using ABI SDS 2.4.1 software.
Statistical analyses
All statistical analyses were performed in R Studio (version 2.15.0) using the R CRAN packages: vegan (Oksanen et al., 2012), RColowBrewer (Neuwirth, 2011) and shapes (Dryden, 2013). A heatmap showing relative abundances was generated based on the OTU table. Hellinger-transformed OTU abundances (Legendre and Gallagher, 2001), richness, evenness and performance parameters were compared between reactors over time using Tukey's Honestly Significant Differences tests. Differences in community composition were further explored and visualised using complete linkage hierarchal clustering and principle component analysis (PCA). Procrustes plots and metrics were calculated to compare the PCA results for each replicate. Correlations between microbial community composition and performance parameters were calculated using environmental parameter fitting.
Results
Anaerobic digester performance
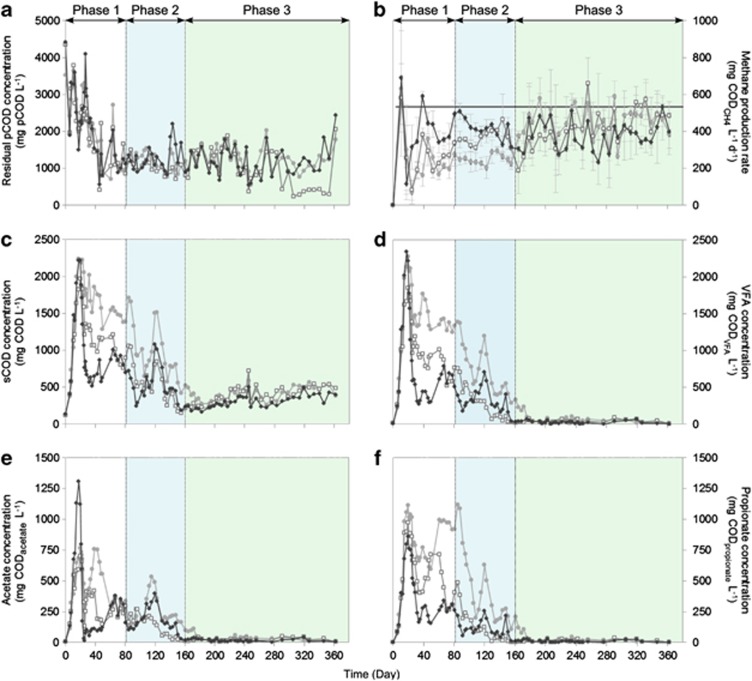
Three replicate anaerobic digesters were seeded with the same inoculum mixture and operated for 362 days with α-cellulose as the sole carbon and energy source. Based on the pCOD, sCOD and VFA profiles, three performance phases could be identified: phase 1—start-up (days 0–81), phase 2—stable hydrolysis (days 82–160) and phase 3—steady-state digestion performance (days 161–362) (Figure 1).
Figure 1.
Reactor performance parameters over time for reactors AD1 (○ grey), AD2 (□ light grey) and AD3 (◊ black). Three distinct phases can be identified (phase 1—start-up, phase 2—stable hydrolysis and phase 3—steady state) showing differences in pCOD concentration (a), methane production rate (b), sCOD concentration (c), total VFA concentration (d), acetate concentration (e) and propionate concentration (f). Methane production was measured daily and average methane production rates were calculated on a weekly basis (error bars represent s.d.).
After an initial variable start-up phase (phase 1), the residual pCOD profiles stabilised in the three reactors (Figure 1a and Table 2), indicating stable hydrolysis (phase 2). During this phase, the cellulose degradation efficiency was 78±5% and did not differ between reactors (P>0.984). Phase 2 was characterised by variable sCOD and VFA profiles (Figures 1c and d) which indicated instability in fermentation, acetogenesis and/or methanogenesis. Also, the sCOD concentrations, VFA profiles and methane production rate of AD1 differed significantly (P<0.02, P<0.005 and P<0.001, respectively) from those of AD2 and AD3 (Figures 1b and f and Table 2). VFA concentrations decreased with time and ranged between 290 and 1390 mg CODVFA per l for AD1, between 25 and 760 mg CODVFA per l for AD2 and between 40 and 710 mg CODVFA per l for AD3. Total VFA concentrations were tightly linked to the sCOD concentration, indicating that the majority of soluble products (on average 50–75%) were fermented to VFAs. The main VFA constituents were acetate and propionate, contributing ∼92% (0.8:1 acetate to propionate respectively) of the total VFAs for AD1, ∼90% (1.8:1) for AD2 and ∼91% (1.8:1) for AD3 (Figures 1e and f).
Table 2. Operational performance parameters.
| Reactor | Cellulose degradation | pCOD | sCOD | VFA | CH4 production rate | |||||
|---|---|---|---|---|---|---|---|---|---|---|
|
(%) |
(mg COD per l) |
(mg COD per l) |
(mg CODVFA
per l) |
(mg CODCH4
per l per day) |
||||||
| Phase 2 | Phase 3 | Phase 2 | Phase 3 | Phase 2 | Phase 3 | Phase 2 | Phase 3 | Phase 2 | Phase 3 | |
| AD1 | 77±5 | 76±7 | 1200±240 | 1300±310 | 990±370 | 410±90 | 750±340 | 70±70 | 250±48 | 450±132 |
| AD2 | 80±4 | 81±10 | 1100±190 | 1000±510 | 550±260 | 390±120 | 290±220 | 40±20 | 360±83 | 420±137 |
| AD3 | 76±7 | 78±8 | 1300±350 | 1200±400 | 520±230 | 310±80 | 320±160 | 30±20 | 410±68 | 380±124 |
Abbreviations: AD, anaerobic digestion; pCOD, particulate chemical oxygen demand; sCOD, soluble chemical oxygen demand; VFA, volatile fatty acids.
The average values of all performance parameters are given for AD1, AD2 and AD3 during stable hydrolysis and steady-state performance (average values and s.d. for phases 2 and 3): cellulose degradation, pCOD, sCOD and VFA concentrations and methane production rate.
During phase 3, the three reactors converged on a performance level with no significant differences in pCOD (P=1.00), sCOD (P>0.92) and VFA concentrations (P=1.00), or methane production rates (P>0.75). The cellulose degradation efficiency was 78±8% during phase 3. Residual VFA concentrations further decreased during phase 3 and ranged between 8 and 240 mg CODVFA per l for AD1, between 5 and 90 mg CODVFA per l for AD2 and between 2 and 80 mg CODVFA per l for AD3 (Figure 1d). The methane production rate in AD1 increased significantly compared to phase 2 (P<0.001) and is consistent with improved conversion of VFA to methane in this reactor. Methane production also increased for AD2 and decreased for AD3 (Table 2), but this change was not significant (P>0.65). During all phases, the CH4 content of the biogas was 63% (±5%) on a volume basis and no H2 was detected.
Microbial community dynamics related to performance in the ADs
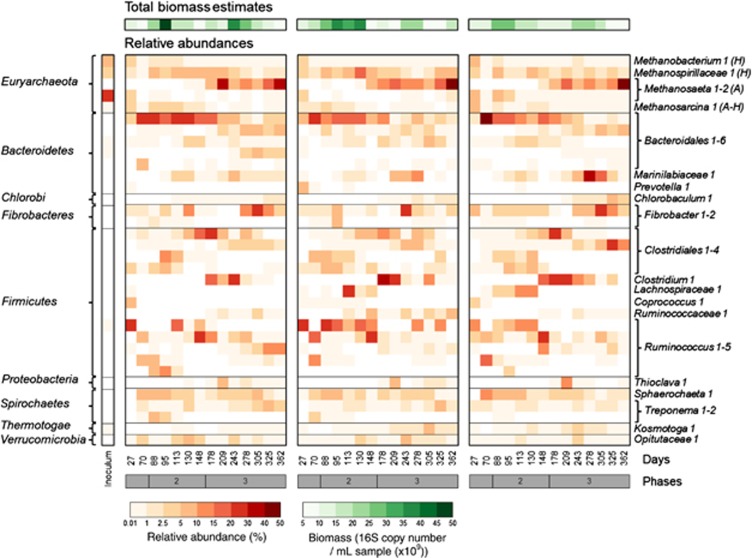
The composition (97% OTUs) and total biomass of the microbial communities associated with the replicate ADs were characterised at 14 time points in order to monitor the development and succession of populations, and to correlate the community dynamics to reactor performance parameters. The inoculum mixture was dominated by methanogens (53% relative abundance) belonging to the genera Methanobacterium and Methanosaeta. The most abundant bacterial populations in the inoculum belonged to the phyla Bacteroidetes, Proteobacteria, OP11 and WS6. Upon introduction into the anaerobic digesters, the microbial community rapidly changed (Figure 2 and Supplementary Figures S2 and S3) and decreased in richness (Table 3 and Supplementary Figure S4). Complete linkage hierarchal clustering and PCA indicated that the composition of microbial communities in the digesters differed between the three phases of reactor operation. The microbial community composition in all digesters was highly dynamic over time (Figures 2 and 3 and Supplementary Figure S5) and there was a significant difference in composition between phases 2 and 3 (P<0.001). Although minor differences between the reactor communities could be observed at individual time points, the communities were significantly similar within phases (P=0.81). Similarly, there was no significant difference in the richness and evenness of the microbial communities between reactors within a phase (Table 3, P=0.94 and P=0.80, respectively). Procrustes analysis on the PCAs from each replicate confirmed a high correlation (>0.8, P=0.001) between the community compositions (Supplementary Figure S6 and Supplementary Table S2). Minor differences in community composition between reactors are likely explained by operational variability and the relatively long time between sampling events (∼3 residence times). The estimated biomass in the reactors fluctuated over time between 6.0 × 109 and 4.9 × 1010 cells per ml of reactor sample, and trends in biomass concentration tracked one another in the three reactors (Figure 2).
Figure 2.
Microbial community composition and synchronised dynamics. This heatmap shows the total biomass estimates and relative abundance of the most dominant microbial populations (>5% relative abundance in at least one of the samples) in the three reactors (AD1, AD2 and AD3) at 14 time points. Different populations were associated with the three performance phases and changes in membership occurred in parallel in the three reactors over time. Taxonomy was determined at the phylum level (left column) and at the lowest possible assignment (right column). H: hydrogenotrophic methanogens; A: acetoclastic methanogens. Darker colour intensity indicates a higher total biomass estimate (green) or relative abundance (red).
Table 3. Richness and evenness of the microbial communities.
| Name | Richness (observed OTUs) | Evenness (Simpson's diversity index) |
|---|---|---|
| Inoculum | 201 | 0.92 |
| AD1 | 89±16 | 0.89±0.03 |
| AD2 | 93±15 | 0.89±0.06 |
| AD3 | 93±16 | 0.89±0.06 |
Abbreviations: AD, anaerobic digestion; OTU, operational taxonomic unit.
Richness (observed OTUs) and evenness (Simpsons's diversity index) are given for the inoculum mixture and reactors (AD1, AD2 and AD3 averaged over 14 time points) after normalisation of the data to 1100 sequences. Error margins represent s.d. values.
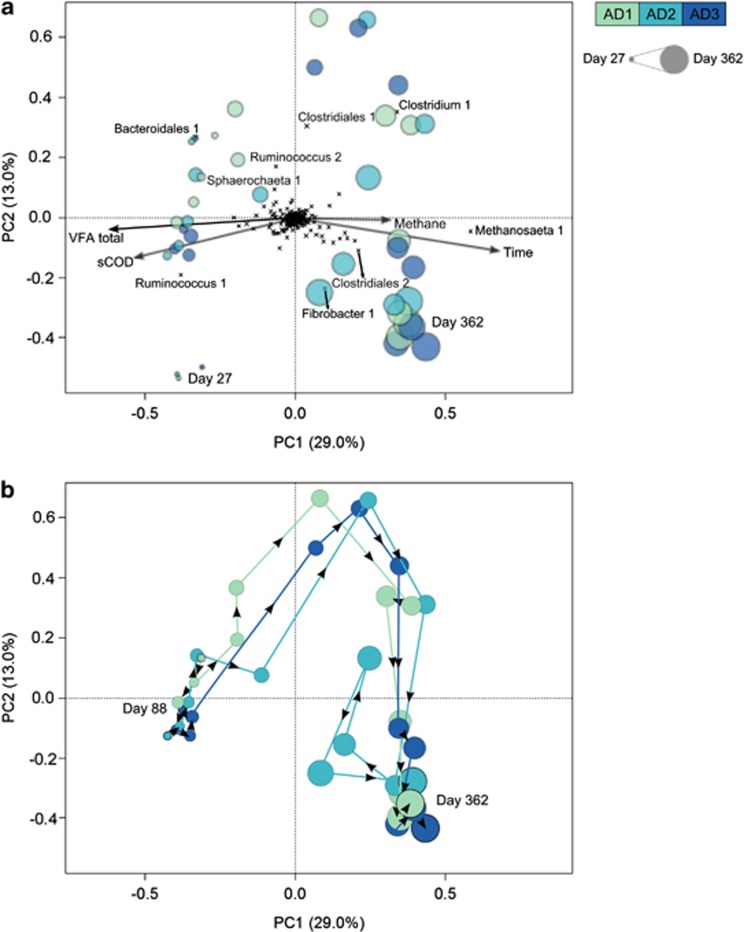
The most abundant populations in all reactors (average relative abundance >4%) belonged to the bacterial orders Bacteroidales, Clostridiales, Fibrobacterales and Sphaerochaetales, and the archaeal orders Methanomicrobiales and Methanosarcinales. Despite being independent systems, the relative abundance profiles of these populations were similar in the three reactors and tracked one another over time (Figures 2 and 3b and Supplementary Figure 5S). These synchronised dynamics were also observed when the community was analysed with increased resolution (100% OTUs) (Supplementary Figure S7). Synchronised shifts in population relative abundance were especially prominent at the transition between phases 2 and 3 (days 148–178), when reactor performance reached steady state. Different members of the order Clostridiales were present at high relative abundance (35±14%) in the three digesters at all time points and correlated with higher sCOD and VFA concentrations, and methane production rates (Figure 3a and Supplementary Figure S8). During phase 2, the microbial communities dominated by Bacteroidales, Ruminococcus (a member of the order Clostridiales) and Sphaerochaetales populations were significantly correlated with higher sCOD and VFA concentrations (P=0.001; Figure 3a and Supplementary Figure S8a). The relative abundance of these populations decreased between day 88 (phase 2) and day 362 (phase 3) from 25±4.9% to 13±6.4% for Bacteroidales, 19±9.5% to 8±6.4% for Ruminococcus and 9±3.0% to 1±0.9% for Sphaerochaetales. The relative abundance of a Fibrobacter population peaked between days 243 and 276 in all reactors, and was inversely correlated to higher pCOD concentrations. The relative abundance of methanogens increased over time from 14±6.2% at day 88 (phase 2) to 53±11.0% at day 362 (phase 3). This coincided with a shift in dominance from a Methanomicrobiales to a Methanosarcinales population around day 178, and was significantly correlated to higher methane production rates (Figures 2 and 3a; P=0.026). Populations belonging to the orders Desulfobacterales, Desulfovibrionales and Syntrophobacterales were typically found in low abundance (0–1.1%), but their relative abundance increased between phases 2 and 3.
Figure 3.
Microbial community dynamics linked to reactor performance. The PCA shows the microbial community composition at the OTU level (Hellinger transformed) for the three reactors (AD1, AD2 and AD3) at 14 time points. (a) Each sample is symbolised by a single circle coloured by reactor. The size of the circle increases with time. Individual OTUs are represented as black crosses and the taxonomy of those contributing most to the variability between community profiles is provided. Environmental parameter fitting was performed to correlate reactor performance parameters to the community composition and significant correlations are presented by the arrows. (b) Tracking of the microbial community composition in the three reactors over time, starting from day 88. Each line represents the trajectory in the PCA plot (based on community structure) of each reactor over time.
Spatial distribution of bacterial and archaeal community members
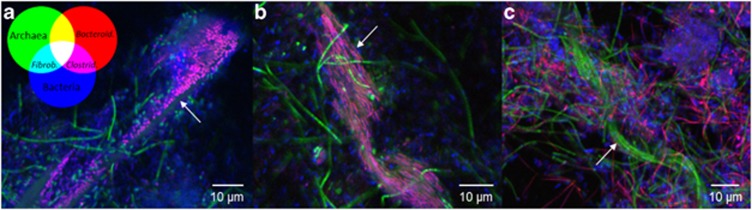
FISH probes broadly targeting bacteria and archaea were used in combination with specific probes to determine the morphology and spatial distribution of phylogenetic groups identified in the communities through 16S rRNA gene amplicon sequencing (Table 1 and Figure 4). The Bacteroidales-specific probes (Bac1080 and CF319a) revealed two morphotypes, straight rods (length 2–4 μm) and filaments (length up to 10 μm), that were more abundant in the planktonic phase, but were occasionally found attached to α-cellulose particles. Clostridiales- (Clo549 and Ace731), Ruminococcus- (Rum831) and Fibrobacter-specific probes (Fibr225) revealed large rods (3–5 μm), chain-forming cocci (diameter <1 μm) and short rods (1–2 μm), respectively, that were predominantly attached to cellulose particles (Figure 4). In most cases, α-cellulose particles appeared to be colonised by multiple populations. Archaeal cells (ARC951) were mainly seen in the planktonic phase and did not appear to be associated with cellulose. Three archaeal morphotypes could be distinguished, including cocci (single cells with a diameter of 1–2 μm), clusters of cocci (diameter 3–7 μm) and rods (3–10 μm). Rod-shaped archaeal cells also formed large filaments (15–100 μm) or were arranged as a chain within a common sheath (up to ∼100 μm; Figure 4).
Figure 4.
Morphology and spatial distribution of dominant populations. FISH micrographs are shown of the microbial community from AD3 on (a) day 88, (b) day 178 and (c) day 362. Universal probes targeting archaea (ARC951, green) and bacteria (EUB338 and EUB338+, blue) were used in combination with population-specific probes targeting Bacteroidales (Bac1080 and CF319a, red), Clostridiales (Clo549 and Ace731, magenta) and Fibrobacterales (Fibr225, cyan). The arrows point to possibly hydrolytic populations attached to cellulose particles (a, b), and clustering of archaeal rod-shaped cells into a sheathed filament (c).
Discussion
Community composition and dynamics linked to reactor performance
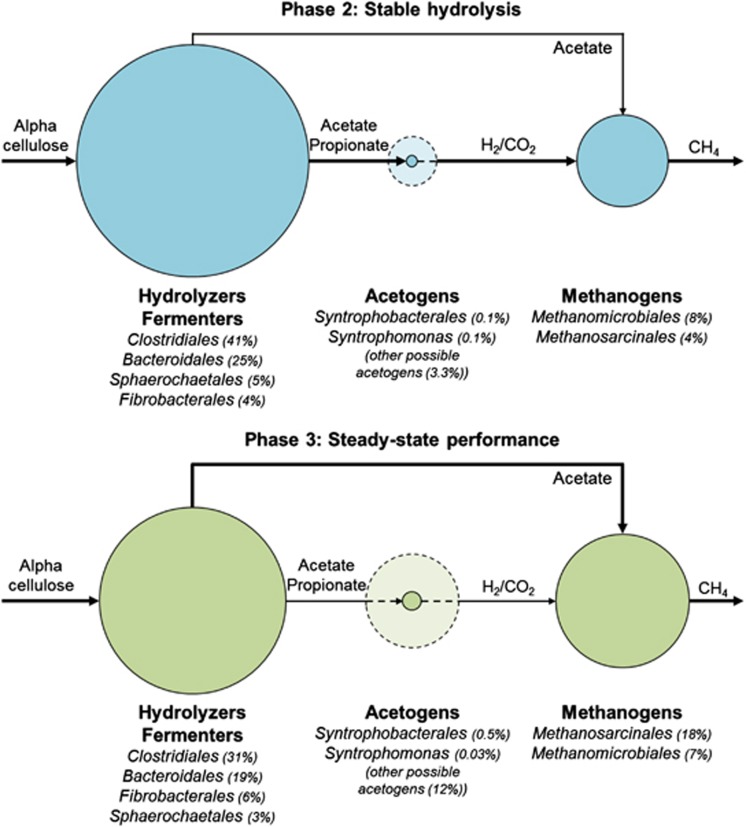
In this study, triplicate anaerobic digesters were run for 362 days under identical operating conditions with the same complex inoculum and model cellulosic feedstock. A diverse community was present in each reactor with multiple populations capable of hydrolysing cellulose, fermenting soluble intermediates and producing methane (Figure 5). By monitoring the microbial community dynamics and performance parameters in replicate digesters, strong correlations between composition and function were observed.
Figure 5.
Schematic of the proposed populations involved in the different steps of anaerobic digestion. The circle size represents the average relative abundance of the populations during phase 2 (blue) and phase 3 (green). During steady-state performance, the functional guilds are more evenly distributed. The dotted circles represent populations belonging to the genus Clostridium and the orders Desulfovibrionales, Desulfuromonadales and Synergistales that may be capable of syntrophic metabolism (McDonald et al., 2008). The arrows indicate the flow of carbon through the system.
The reactor communities rapidly shifted away from the inoculum community profile and converged to a highly similar composition under the selective pressures imposed by the operating conditions (Figure 2 and Supplementary Figure S2). The hydraulic residence time was shorter than that of the source environments of the inocula that initially caused partial washout of slow-growing microorganisms including methanogens and resulted in reactor communities dominated by bacteria. The dominant bacterial phylotypes were present at very low abundance or were below detection in the inoculum, suggesting that the shorter hydraulic residence time, mesophilic temperature and simple cellulosic substrate created specific niches different to the native inoculum environments. As anticipated, these selective pressures not only influenced the community composition but also led to a decrease in phylotype richness (Table 3).
The microbial communities in this experiment were dominated by bacterial populations belonging to the orders Bacteroidales, Clostridiales, Fibrobacterales and Sphaerochaetales (Figure 2). The most abundant archaeal populations belonged to the orders Methanomicrobiales and Methanosarcinales that are often found separately or together as the dominant methanogens in anaerobic digesters (Ariesyada et al., 2007; Jaenicke et al., 2010; Nelson et al., 2011). Recently, characterisation of the microbial community composition of 21 full-scale ADs highlighted dominant populations belonging to the phyla Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Chloroflexi and Spirochaetes (Sundberg et al., 2013). Similarly, a meta-analysis of all publicly available 16S rRNA gene sequences from AD-associated microbial communities supplied a variety of feedstocks showed that many dominant populations belong to the phyla Chloroflexi and Proteobacteria (Nelson et al., 2011). Several representatives of these phyla were detected in the inoculum (>5%), but they were only present at low relative abundance in the reactors (<1%). This is likely related to the use of α-cellulose as model feedstock in this study that reduces the number of metabolic niches that can be occupied by hydrolytic and fermentative microorganisms. Consistent with this finding, it has previously been observed that the type of substrate strongly influences the composition of AD microbial communities (Merlino et al., 2012; Regueiro et al., 2012; Sundberg et al., 2013; Ziganshin et al., 2013).
Hydrolysis
Populations belonging to the order Bacteroidales and the genus Ruminococcus were likely the initial main cellulose degraders, and contributed to stable hydrolysis during phase 2. As the relative abundance of these populations decreased, members of the genus Clostridium increased in all reactors, followed by a shift in dominance to a population belonging to the genus Fibrobacter. Members of each of these orders/genera have previously been implicated in cellulose hydrolysis (Bayer et al., 2004; Jacob-Lopes et al., 2009; Magnuson et al., 2009; Nelson et al., 2011; Sundberg et al., 2013). FISH confirmed that these populations were attached to or in close proximity to cellulose particles (Figure 4), strengthening the likelihood that these microorganisms were major cellulose hydrolysers. As some of these populations were also present in the planktonic phase, it is hypothesised that different mechanisms of cellulose degradation may be used, that is, via extracellular enzymes or by a cell-attached cellulosome complex which has previously been observed for Clostridium cellulovorans (Matano et al., 1994). Several Bacteroidales and Ruminococcus populations were significantly correlated to higher VFA concentrations (Figure 3a), consistent with their potential role in cellulose hydrolysis yielding soluble substrates for VFA production.
Fermentation
Members of the orders Bacteroidales, Clostridiales and Sphaerochaetales are known as saccharolytic chemoorganotrophic heterotrophs (Magnuson et al., 2009; Jaenicke et al., 2010; Sundberg et al., 2013). Our results suggest that these populations are the dominant fermenters in the digesters, supported by a significant correlation with higher VFA concentrations and high relative abundance of these populations in the planktonic phase. Clostridiales populations were also significantly correlated with higher methane production (Figure 3). Although members of this order are not capable of producing methane, they do have diverse metabolic capabilities that yield substrates for methanogenesis, including cellulose degradation, polysaccharide fermentation to VFAs, alcohols and hydrogen, homoacetogenesis and syntrophic acetate oxidation (Stieb and Schink, 1985; Schnurer et al., 1996; Lee et al., 2008a; Sundberg et al., 2013). Our results suggest a possible syntrophic interaction between specific Clostridiales and methanogenic populations as they both increased in relative abundance during steady-state performance, although additional functional data are required to further support this hypothesis. During phase 3, VFA concentrations decreased (Figure 1) as the relative abundance of populations capable of utilising VFAs increased, such as potentially syntrophic acetogens belonging to the order Syntrophobacterales that are known to oxidise acetate and propionate to H2 and CO2 (Muller et al., 2010; Muller et al., 2013).
Methanogenesis
In contrast to hydrolytic and fermentative populations that have relatively high growth rates (Griffin et al., 1997), methanogens tend to respond poorly to a change in operational conditions and short hydraulic residence time. Stress and partial washout of methanogenic populations led to an imbalance between functional guilds during start-up, resulting in increased VFA concentrations and a corresponding drop in pH (∼6). The latter may have further inhibited methanogenesis as optimal pH for methane production varies between pH 7 and 8 (Weiland, 2010). Methanogenesis remained the rate-limiting step during phase 2, as indicated by high residual VFA concentrations (Figure 1). During this phase, the most abundant methanogenic population belonged to the hydrogenotrophic family Methanospirillaceae. Members of the genus Methanosarcina were the second most dominant methanogen, likely explained by their low acetate substrate affinity and preference for high acetate concentrations (>60 mg l−1; Liu and Whitman, 2008) comparable to the reactors in phase 2 (average >200 mg l−1; (Figures 1e and 2). FISH confirmed the presence of this population as the characteristic tetrad clusters. During phase 3, the methanogens increased in relative abundance and this was correlated with higher methane production and lower VFA concentrations (Figures 1 and 2). Members of the genus Methanosaeta thrive at low acetate concentrations (as low as 0.3–1.2 mg l−1; Liu and Whitman, 2008) because of their high acetate substrate affinity, potentially explaining why they became the most dominant methanogenic population when acetate concentrations were low during phase 3 (average <40 mg l−1; Figures 1e and 2). Methanosaeta cells have previously been observed together in a common sheath (Kamagata and Mikami, 1991, Ma et al., 2006) and FISH confirmed clustering of these filaments in the reactors during steady state. The change in dominant methanogen likely corresponded to a shift in primary methanogenic pathway in the reactors from hydrogenotrophic to acetoclastic methanogenesis. The increase in abundance of Methanosaeta correlated with changes in the bacterial community composition and transition to steady-state performance; however, the causality of this relationship is not yet fully understood. Interestingly, the decrease in the relative abundance of populations known to be capable of hydrogenotrophic methanogenesis coincided with an increased abundance of potentially syntrophic acetogens. As H2 was not detected, other H2-scavenging microorganisms may have functioned as sinks within the reactors. Consistent with this finding, the relative abundance of sulphate-reducing bacteria, belonging to the orders Desulfobacterales and Desulfovibrionales, increased during phase 3. These bacteria can use the available acetate and H2 (Griffin et al., 1997); however, the low levels of sulphate present in the medium may have restricted their abundance and ability to out-compete the methanogens (Dar et al., 2008).
The increase in functional stability during phase 3 was correlated with a major shift in community composition on all functional levels and an increased abundance of possible syntrophic acetogens and methanogens (Figure 5), suggesting strong microbial interactions. Despite this functional stability, the communities did not increase in similarity over time (Supplementary Figure S9) and remained dynamic (Figure 2 and Supplementary Figure S10). It has been hypothesised that processes and interactions that promote functional stability result from greater functional redundancy and niche complementation by ensuring a reservoir of species using parallel pathways for substrate conversion (Briones and Raskin, 2003). Based on the composition of the community in the reactors over time, it is clear that many phylogenetically diverse populations are capable of fulfilling the same functional guild within these systems (Figure 5).
Deterministic processes drive microbial community structure in replicate anaerobic digesters
A major goal of this study was to test the relative influence of deterministic and stochastic processes on community assembly and dynamics over the course of a year using a highly controlled replicated model system. A complex inoculum of eight natural and engineered anoxic environments was used to provide a biologically diverse starting point from which to monitor convergence or divergence of communities between reactors over time.
The composition and diversity of the microbial communities were statistically similar within the three distinct performance phases. The communities were dynamic and changes in relative abundance of many phylotypes were synchronised between reactors over long time scales (Figures 2 and 3). Although the microbial communities in the replicates showed minor differences at individual time points, the communities followed equivalent trajectories in terms of performance and composition (Figure 3b). Given that the reactors were closed systems run under identical conditions, the largely synchronised phylotype dynamics indicate that reproducible changes in synergistic and competitive interactions within the community are occurring under the controlled conditions. Neutral theory, as a null hypothesis, predicts random drift of populations under identical conditions because it assumes that all individuals are ecologically identical (Harpole, 2012). In this study, we endeavoured to replicate the bioreactor operating conditions as closely as possible to identify bona fide random drift. Our results suggest that multiple populations capable of fulfilling the same functional niche do not randomly become dominant in the system; instead, their abundance appears to be highly controlled by deterministic processes such as microbial interactions, substrate availability and operational conditions. For example, many hydrogenotrophic methanogens were present in the inoculum that could have independently come to dominance in the parallel reactors over the course of the experiment. Instead, population synchronisation was observed among this functionally redundant guild (Figure 2 and Supplementary Figure S5). Our results reject the neutral theory null hypothesis and suggest that niche differences between populations have a critical influence on community assembly and dynamics.
Previous studies have observed reproducible temporal community dynamics in replicate denitrifying reactors (McGuinness et al., 2006), ammonia-oxidising membrane bioreactors (Falk et al., 2009) and activated sludge bioreactors (Valentin-Vargas et al., 2012), suggesting that these systems are not driven by stochastic processes, but are highly reproducible and predictable. In contrast, Zhou et al. (2013) concluded that stochastic assembly of communities dominated in replicate microbial electrolysis cell reactors, invoking neutral theory as the underlying model. The authors suggest that initial stochastic colonisation of the anodes was critical to random community drift and allowed considerable site-to-site variation in community composition. It was likely that deterministic processes influenced subsequent dynamics as there was a strong link between community structure and function (Zhou et al., 2013). The population dynamics in waste water treatment plants have also been studied and were consistent with neutral theory which the authors attributed to the open design and continuous influx of microorganisms and protozoa (Ofiteru et al., 2010). Our results suggest that microbial community assembly and dynamics in highly controlled anaerobic digesters is primarily driven by deterministic rather than stochastic processes. This observation also indicates the importance of high-resolution monitoring of microbial communities over long time periods to determine the relative influence of niche and neutral theory on population dynamics.
Conclusion
In this study, we identified multiple phylogenetically diverse populations associated with each of the main steps in AD (hydrolysis, fermentation and methanogenesis) that appear to have at least some functional redundancy. However, community dynamics were strongly linked with reactor performance and numerous populations were synchronised over long time periods, suggesting niche specialisation. This rejects the null hypothesis of neutral theory and highlights the importance of deterministic factors such as operational conditions, substrate availability and interspecies interactions. It should be noted that 16S amplicon sequencing only allows community member resolution down to approximately genus (97% OTU)/species (100% OTU) level, and it is possible therefore that undetected stochastic changes occur at the strain level. Recovery of population genomes from metagenomic data (Tyson et al., 2004; Albertsen et al., 2013) will provide the resolution needed to confirm the major role for deterministic processes within these highly controlled, closed replicated systems.
Acknowledgments
This study was supported by the Commonwealth Scientific & Industrial Research Organisation (CSIRO) Flagship Cluster ‘Biotechnological solutions to Australia's transport, energy and greenhouse gas challenges'. IV acknowledges support by the University of Queensland International Scholarship, and GWT is supported by an ARC Queen Elizabeth II fellowship (DP1093175). KR acknowledges support by the European Research Council (Starter Grant Electrotalk). We thank Fiona May at the Australian Centre for Ecogenomics for the 16S rRNA gene amplicon pyrosequencing, Adam Skarshewski for his assistance with the pyrotag data analysis and Nancy Lachner and Serene Low for their help with the RT-PCR. Beatrice Keller and Nathan Clayton from the ASL analysis lab are acknowledged for the VFA analyses. We also acknowledge Dr Michael Imelfort for his contribution to the project.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KA, Tyson GW, Nielsen PH. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol. 2013;31:533–538. doi: 10.1038/nbt.2579. [DOI] [PubMed] [Google Scholar]
- Allison SD, Martiny JBH. Resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci USA. 2008;105:11512–11519. doi: 10.1073/pnas.0801925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipma DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amani T, Nosrati M, Sreekrishnan TR. Anaerobic digestion from the viewpoint of microbiological, chemical and operational aspects - a review. Environ Rev. 2010;18:255–278. [Google Scholar]
- Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- APHA 2005Standard Methods for the Examination of Water and Wastewater21st edn.American Public Health Association: Washington DC [Google Scholar]
- Ariesyada HD, Ito T, Okabe S. Functional bacterial and archaeal community structures of major trophic groups in a full-scale anaerobic sludge digester. Water Res. 2007;41:1554–1568S. doi: 10.1016/j.watres.2006.12.036. [DOI] [PubMed] [Google Scholar]
- Bayer EA, Belaich J-P, Shoham Y, Lamed R. The cellulosome: multienzyme machines for degradation of plant cell wall polysaccharides. Annu Rev Microbiol. 2004;58:521–554. doi: 10.1146/annurev.micro.57.030502.091022. [DOI] [PubMed] [Google Scholar]
- Bragg L, Stone G, Imelfort M, Hugenholtz P, Tyson G. Fast, accurate error-correction of amplicon pyrosequences using Acacia. Nat Methods. 2012;9:425–426. doi: 10.1038/nmeth.1990. [DOI] [PubMed] [Google Scholar]
- Briones A, Raskin L. Diversity and dynamics of microbial communities in engineered environments and their implications for process stability. Curr Opin Biotechnol. 2003;14:270–276. doi: 10.1016/s0958-1669(03)00065-x. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso T, Chan Y, Lacap DC, Lau MCY, McKay CP, Pointing SB. Stochastic and deterministic processes interact in the assembly of desert microbial communities on a global scale. ISME J. 2011;5:1406–1413. doi: 10.1038/ismej.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayford BI, Dennis PG, Keller J, Tyson GW, Bond PL. High-throughput amplicon sequencing reveals distinct communities within a corroding concrete sewer system. Appl Environ Microbiol. 2012;78:7160–7162. doi: 10.1128/AEM.01582-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H, Bruhl A, Amann R, Schleifer KH, Wagner M. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol. 1999;22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- Dar SA, Kleerebezem R, Stams AJM, Kuenen JG, Muyzer G. Competition and coexistence of sulfate-reducing bacteria, acetogens and methanogens in a lab-scale anaerobic bioreactor as affected by changing substrate to sulfate ratio. Appl Microb Biotechnol. 2008;78:1045–1055. doi: 10.1007/s00253-008-1391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis PD, Guo K, Imelfort M, Jensen PD, Tyson GW, Rabaey K. Spatial uniformity of microbial diversity in a continuous bioelectrochemical system. Bioresour Technol. 2013;129:599–605. doi: 10.1016/j.biortech.2012.11.098. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden IL.2013. Shapes: Statistical Shape Analysis. R package version 1.1-9 http://CRAN.R-project.org/package=shapes .
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelbrektson A, Kunin V, Wrighton KC, Zvenigorodsky N, Chen F, Ochman H, et al. Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J. 2010;4:642–647. doi: 10.1038/ismej.2009.153. [DOI] [PubMed] [Google Scholar]
- Falk MW, Song K-G, NMatiasek MG, Wuertz S. Microbial community dynamics in replicate membrane bioreactors - natural reproducible fluctuations. Water Res. 2009;43:842–852. doi: 10.1016/j.watres.2008.11.021. [DOI] [PubMed] [Google Scholar]
- Fernandez A, Huang S, Xing J, Hickey R, Criddle C, Tiedje J. How stable is stable? Function versus community composition. Appl Environ Microbiol. 1999;65:3697–3704. doi: 10.1128/aem.65.8.3697-3704.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Jensen PD, Batstone DJ. Temperature phased anaerobic digestion increases apparent hydrolysis rate for waste activated sludge. Water Res. 2011;45:1597–1606. doi: 10.1016/j.watres.2010.11.042. [DOI] [PubMed] [Google Scholar]
- Gentile ME, Nyman JL, Criddle CS. Correlation of patterns of denitrification instability in replicated bioreactor communities with shifts in the relative abundance and denitrification patterns of specific populations. ISME J. 2007;1:714–728. doi: 10.1038/ismej.2007.87. [DOI] [PubMed] [Google Scholar]
- Gopalan P, Jensen PD, Batstone DJ. Anaerobic digestion of swine effluent: impact of production stages. Biomass Bioenergy. 2013;48:121–129. [Google Scholar]
- Griffin ME, McMahon KD, Mackie RI, Raskin L. Methanogenic population dynamics during start-up of anaerobic digesters treating municipal solid waste and biosolids. Biotechnol Bioeng. 1997;57:342–355. doi: 10.1002/(sici)1097-0290(19980205)57:3<342::aid-bit11>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Harpole WS. Neutral theory of species diversity. Nat Educ Knowledge. 2012;3:60. [Google Scholar]
- Jacob-Lopes E, Scoparo CHG, Lacerda L, Franco TT. Effect of light cycles (night/day) on CO2 fixation and biomass production by microalgae in photobioreactors. Chem Eng Proc. 2009;48:306–310. [Google Scholar]
- Jaenicke S, Ander C, Bekel T, Bisdorf R, Dröge M, Gartemann K-H, et al. Comparative and joint analysis of two metagenomic datasets from a biogs fermenter obtained by 454-pyrosequencing. PLoS One. 2010;6:1–15. doi: 10.1371/journal.pone.0014519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PD, Hardin MT, Clarke WP. Effect of biomass concentration and inoculum source in the rate of anaerobic cellulose solubilization. Biores Technol. 2009;2009:5219–5225. doi: 10.1016/j.biortech.2009.05.018. [DOI] [PubMed] [Google Scholar]
- Jensen PD, Ge H, Batstone DJ. Assessing the role of biochemical methane potential tests in determining anaerobic degradability rate and extent. Water Sci Technol. 2011;64:880–886. doi: 10.2166/wst.2011.662. [DOI] [PubMed] [Google Scholar]
- Kamagata Y, Mikami E. Isolation and characterization of a novel thermophilic Methanosaeta strain. Int J Syst Bacteriol. 1991;41:191–196. doi: 10.1099/00207713-42-3-463. [DOI] [PubMed] [Google Scholar]
- Kong Y, Xia Y, Seviour R, He M, McAllister T, Forster R. In situ identification of carboxymethyl cellulose–digesting bacteria in the rumen of cattle fed alfalfa or triticale. FEMS Microbiol Ecol. 2012;80:159–167. doi: 10.1111/j.1574-6941.2011.01295.x. [DOI] [PubMed] [Google Scholar]
- LaPara TM, Nakatsu CH, Pantea LM, Alleman JE. Stability of the bacterial communities supported by a seven-stage biological process treating pharmaceutical wastewater as revealed by PCR-DGGE. Water Res. 2001;36:638–646S. doi: 10.1016/s0043-1354(01)00277-9. [DOI] [PubMed] [Google Scholar]
- Lee C, Kim J, Shin SG, Hwang S. Monitoring bacterial and archaeal community shifts in a mesophilic anaerobic batch reactor treating a high-strength organic wastewater. FEMS Microbiol Ecol. 2008;65:544–554. doi: 10.1111/j.1574-6941.2008.00530.x. [DOI] [PubMed] [Google Scholar]
- Lee C, Kim JY, Hwang K, O'Flaherty V, Hwang S. Quantitative analysis of methanogenic community dynamics in three anaerobic batch digesters treating different wastewaters. Water Res. 2008;43:157–165. doi: 10.1016/j.watres.2008.09.032. [DOI] [PubMed] [Google Scholar]
- Legendre P, Gallagher ED. Ecologically meaningful transformations for ordination of species data. Oecologia. 2001;129:271–280. doi: 10.1007/s004420100716. [DOI] [PubMed] [Google Scholar]
- Liu Y, Whitman WB. Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann NY Acad Sci. 2008;1125:171–189. doi: 10.1196/annals.1419.019. [DOI] [PubMed] [Google Scholar]
- Ma K, Liu X, Dong X. Methanosaeta harundinacea sp. nov., a novel acetate-scavenging methanogen isolated from a UASB reactor. Int J Syst Evol Microbiol. 2006;56:127–131. doi: 10.1099/ijs.0.63887-0. [DOI] [PubMed] [Google Scholar]
- Magnuson A, Anderlund M, Johansson O, Lindblad P, Lomoth R, Polivka T, et al. Biomimetic and microbial approaches to solar fuel generation. Acc Chem Res. 2009;42:1899–1909. doi: 10.1021/ar900127h. [DOI] [PubMed] [Google Scholar]
- Matano Y, Park JS, Goldstein MA, Hoi RH. Cellulose promotes extracellular assembly of Clostridium cellulovorans cellulosomes. J Bacteriol. 1994;176:6952–6956. doi: 10.1128/jb.176.22.6952-6956.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald IR, Bodrossy L, Chen Y, Murrell JC. Molecular ecology techniques for the study of aerobic methanotrophs. Appl Environ Microbiol. 2008;74:1305–1315. doi: 10.1128/AEM.02233-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuinness LM, Salganik M, Vega L, Pickering KD, Kerkhof LJ. Replicability of bacterial communities in denitrifying bioreactors as measured by PCR/T-RFLP analysis. Environ Sci Technol. 2006;40:509–515. doi: 10.1021/es050900l. [DOI] [PubMed] [Google Scholar]
- Merlino G, Rizzi A, Villa F, Sorlini C, Brambilla M, Navarotto P, et al. Shifts of microbial community structure during anaerobic digestion of agro-industrial energy crops and food industry byproducts. J Chem Technol Biotechnol. 2012;87:1302–1311. [Google Scholar]
- Muller B, Sun L, Schnurer A. First insight into the syntrophic acetate-oxidizing bacteria - a genetic study. Microbiologyopen. 2013;2:35–53. doi: 10.1002/mbo3.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller N, Worm P, Schink B, Stams AJM, Plugge CM. Syntrophic butyrate and propionate oxidation processes: from genomes to reaction mechanisms. Environ Microbiol Rep. 2010;2:489–499. doi: 10.1111/j.1758-2229.2010.00147.x. [DOI] [PubMed] [Google Scholar]
- Nelson MC, Morrison M, Yu Z. A meta-analysis of the microbial diversity observed in anaerobic digesters. Bioresour Technol. 2011;102:3730–3739. doi: 10.1016/j.biortech.2010.11.119. [DOI] [PubMed] [Google Scholar]
- Neuwirth E.2011. RColorBrewer: ColorBrewer palettes. R package version 1.0-5 http://CRAN.R-project.org/package=RColorBrewer .
- Ofiteru ID, Lunn M, Curtis TP, Wells GF, Criddle CS, Francis CA, et al. Combined niche and neutral effects in a microbial wastewater treatment community. Proc Natl Acad Sci USA. 2010;107:15345–15350. doi: 10.1073/pnas.1000604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet G, Kindt R, Legendre P, Minchin PR, O'Hara RB, et al. 2012Vegan: Community ecology packageR package version 2.0-1. . http://CRAN.R-project.org/package=vegan .
- Pervin HM, GDennis PG, Lim HJ, Tyson GW, Batstone DJ, Bond PL. Drivers of microbial community composition in mesophilic and thermophilic temperature-phased anaerobic digestion pre-treatment reactors. Water Res. 2013;47:7098–7108. doi: 10.1016/j.watres.2013.07.053. [DOI] [PubMed] [Google Scholar]
- Pholchan MK, Baptista JC, Davenport RJ, Sloan WT, Curtis TP. Microbial community assembly, theory and rare functions. Front Microbiol. 2013;4:1–9. doi: 10.3389/fmicb.2013.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabaey K, Ossieur W, Verhaege M, Verstraete W. Continuous microbial fuel cells convert carbohydrates to electricity. Water Sci Technol. 2005;52:515–523. [PubMed] [Google Scholar]
- Regueiro L, Veiga P, Figueroa M, Alonso-Gutierrez J, Stams AJM, Lema JM, et al. Relationship between microbial activity and microbial community structure in six full-scale anaerobic digesters. Microbiol Res. 2012;167:581–589. doi: 10.1016/j.micres.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Rittmann BE, Hausner M, Loffler F, Love NG, Muyzer G, Okabe S, et al. A vista for microbial ecology and environmental biotechnology. Environ Sci Technol. 2006;15:1096–1103. doi: 10.1021/es062631k. [DOI] [PubMed] [Google Scholar]
- Schlüter A, Bekel T, Diaz NN, Dondrup M, Eichenlaub R, Gartemann K-H, et al. The metagenome of a biogas-producing microbial community of a production-scale biogas plant fermenter analysed by the 454-pyrosequencing technology. J Biotechnol. 2008;136:77–90. doi: 10.1016/j.jbiotec.2008.05.008. [DOI] [PubMed] [Google Scholar]
- Schnurer A, Schink B, Svensson BH. Clostridium ultunense sp. nov., a mesophilic bacterium oxidizing acetate in syntrophic association with a hydrogenotrophic methanogenic bacterium. Int J Syst Bacteriol. 1996;46:1145–1152. doi: 10.1099/00207713-46-4-1145. [DOI] [PubMed] [Google Scholar]
- Sloan WT, Lunn M, Woodcock S, Head IM, Nee S, Curtis TP. Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ Microbiol. 2006;8:732–740. doi: 10.1111/j.1462-2920.2005.00956.x. [DOI] [PubMed] [Google Scholar]
- Stahl DA, Amann R.1991Development and application of nucleic acid probesIn: Stackebrandt E, Goodfellow M (eds) Nucleic Acid Techniques in Bacterial Systematics John Wiley & Sons Ltd.: Chichester, England; 205–249. [Google Scholar]
- Steinberg LM, Regan JM. Response of lab-scale methanogenic reactors inoculated from different sources to organic loading rate shocks. Bioresour Technol. 2011;102:8790–8798. doi: 10.1016/j.biortech.2011.07.017. [DOI] [PubMed] [Google Scholar]
- Stieb M, Schink B. Anaerobic oxidation of fatty acids by Clostridium bryantii sp. nov., a sporeforming, obligately syntrophic bacterium. Arch Microbiol. 1985;140:387–390. [Google Scholar]
- Sundberg C, Al-Soud WA, Larsson M, Alm E, Yekta SS, Scvensson BH, et al. 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol Ecol. 2013;85:612–626. doi: 10.1111/1574-6941.12148. [DOI] [PubMed] [Google Scholar]
- Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- Valentin-Vargas A, Toro-Labrador G, Massol-Deya AA. Bacterial community dynamics in full-scale activated sludge bioreactors: Operation and ecological factors driving community assembly and performance. PLoS One. 2012;7:1–12. doi: 10.1371/journal.pone.0042524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Shen J, Wu Y, Chen T, Soininen J, Stegen JC, et al. Phylogenetic beta diversity in bacterial assemblages across ecosystems: deterministic versus stochastic processes. ISME J. 2013;7:1310–1321. doi: 10.1038/ismej.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiland P. Biogas production: current state and perspectives. Appl Microbiol Biotechnol. 2010;85:849–860. doi: 10.1007/s00253-009-2246-7. [DOI] [PubMed] [Google Scholar]
- Werner JJ, Knights D, Garcia ML, Scalfone NB, Smith S, Yarasheski K, et al. Bacterial community structures are unique and resilient in full-scale bioenergy systems. Proc Natl Acad Sci USA. 2010;108:4158–4163. doi: 10.1073/pnas.1015676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Zhu Z, Fu L, Niu B, Li W. WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genomics. 2011;12:1–9. doi: 10.1186/1471-2164-12-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liu W, Deng Y, Jiang Y-H, ZXue K, He Z, et al. Stochastic assembly leads to alternative communities with distinct functions in a bioreactor microbial community. mBio. 2013;4:1–8. doi: 10.1128/mBio.00584-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziganshin AM, Liebetrau J, Proter J, Kleinsteuber S. Microbial community structure and dynamics during anaerobic digestion of various agricultural waste materials. Appl Microbiol Biotechnol. 2013;97:5161–5174. doi: 10.1007/s00253-013-4867-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.