Abstract
Two central steps for initiating eukaryotic DNA replication involve loading of the Mcm2–7 helicase onto double-stranded DNA and its activation by GINS–Cdc45. To better understand these events, we determined the structures of Mcm2–7 and the CMG complex by using single-particle electron microscopy. Mcm2–7 adopts two conformations—a lock-washer-shaped spiral state and a planar, gapped-ring form—in which Mcm2 and Mcm5 flank a breach in the helicase perimeter. GINS and Cdc45 bridge this gap, forming a topologically closed assembly with a large interior channel; nucleotide binding further seals off the discontinuity between Mcm2 and Mcm5, partitioning the channel into two smaller pores. Together, our data help explain how GINS and Cdc45 activate Mcm2–7, indicate that Mcm2–7 loading may be assisted by a natural predisposition of the hexamer to form open rings, and suggest a mechanism by which the CMG complex assists DNA strand separation.
A key step in DNA replication is the loading and activation of ring-shaped helicases, an event achieved by a variety of distinct strategies. In bacteria, the DnaC protein is thought to crack open and chaperone the hexameric helicase DnaB onto ssDNA at the nascent replication bubble1–3. By contrast, certain eukaryotic viruses possess helicases that autonomously self-assemble around duplex DNA to induce origin melting and form a competent replication fork4–7.
Eukaryotic cells use a multistep mechanism for helicase deposition and activation. First, the origin recognition complex (ORC), together with Cdc6 and Cdt1, loads a catalytically inactive double hexamer consisting of six minichromosome maintenance subunits (Mcm2–7) around duplex DNA8–10. Concomitant with entry into S phase, Cdc45 and the GINS tetramer associate with Mcm2–7 to form the 11-subunit CMG helicase11–13. Now activated, the CMG likely acts as the front end of the replisome progression complex (RPC), tracking 3′→5′ along the leading strand to provide single-stranded DNA (ssDNA) for DNA polymerases α, β and ε (refs. 14–17).
The six homologous Mcm subunits each contain an N-terminal DNA-interacting and assembly domain18,19 and a C-terminal motor domain belonging to the ATPases-associated-with-various-cellular-activities (AAA+) superfamily20. The subunit order of the Mcm2–7 assembly proceeds Mcm5 → Mcm3 → Mcm7 → Mcm4 → Mcm6 → Mcm2, with a comparatively weak interaction found to exist between Mcm5 and Mcm2 (refs. 21–23). Structural studies of orthologous archaeal Mcm systems have shown that the protein forms double-tiered, hexameric or heptameric rings with a large central pore that can engage duplex oligonucleotides18,24–27. The isolated N-terminal domain can assemble into a head-to-head double hexamer, which also possesses a channel sufficiently large to accommodate double-stranded DNA (dsDNA)18,28,29.
In contrast to its archaeal counterpart19,30,31, isolated Mcm2–7 is a rather weak helicase in vitro15,32. Activity is markedly improved upon incorporation into the CMG11,15. The GINS portion of this complex, which is composed of the four related polypeptides Psf1–3 and Sld5 (refs. 33,34), binds Mcm2–7, Cdc45 and ssDNA12,15,35. X-ray crystallo-graphic studies of GINS assemblies have shown that the complex forms a tightly compact and elongated structure36–38, although an EM report has suggested that an alternative, horseshoe-shaped configuration may also exist35. How GINS and Cdc45 associate with Mcm2–7 and each other, and how this subcomplex activates the latent Mcm2–7 helicase activity, are not known. Whether the Mcm2–7 ring can spontaneously open, or whether the ORC–Cdc6–Cdt1 complex must physically crack apart the helicase before loading, is also unresolved. Addressing these issues is necessary to understand the physical events needed to control replisome assembly and regulate cell proliferation.
Here, we use single-particle EM reconstructions to describe the architecture of Drosophila melanogaster Mcm2–7 and the full CMG complex. We find that on its own, Mcm2–7 forms open-ring hexamers with a discontinuity between Mcm2 and Mcm5, whereas GINS and Cdc45 both seal off this opening and reinforce a more planar helicase configuration. In the presence of a nonhydrolyzable ATP analog, a large central channel formed between the Mcm subunits and GINS–Cdc45 constricts into two topologically segregated smaller pores. Our findings provide a structural basis for activation of the eukaryotic replicative helicase and suggest a potential organization for separated strands during DNA unwinding.
RESULTS
A cracked-ring Mcm2–7 complex
The Mcm2–7 assembly was expressed and purified from cells infected with recombinant baculoviruses15. A glycerol gradient peak fraction, containing a stoichiometric complex of all six Mcm subunits, was used for negative-stain EM (Supplementary Fig. 1).
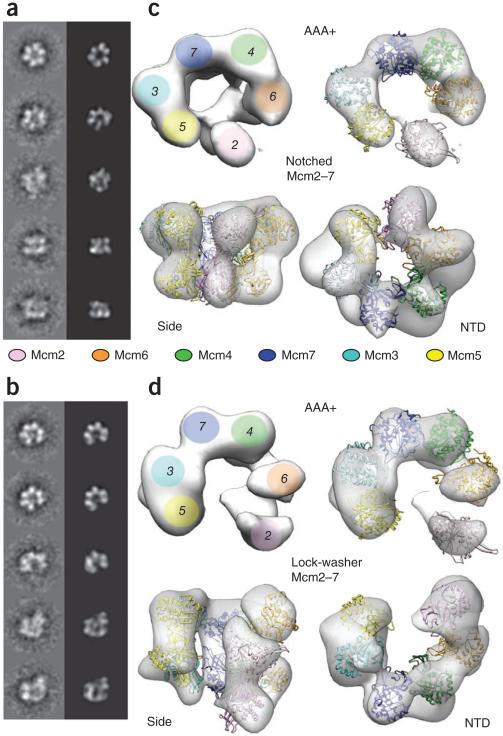
Two-dimensional image analysis on a set of 7,527 apo Mcm2–7 particles showed the presence of two distinct states: quasisymmetric rings with approximate six-fold symmetry, and asymmetric gapped particles (Fig. 1a,b). Analysis of the same complex purified in the presence of the ATP mimic ADP·BeF3 yielded a similar particle population distribution (based on a 53,000 particle dataset). Three-dimensional reconstruction and multimodel refinement39 allowed for the determination of two distinct structures of the Mcm2–7 assembly at 32 and 35 Å resolution (as measured by Fourier shell correlation (FSC), using the 0.5 criterion; Supplementary Fig. 1), one a planar, notched conformation and the other a right-handed spiral, lock-washer state (Fig. 1c,d and Supplementary Movie 1). Both structures show a symmetry mismatch, with a nearly six-fold–symmetric tier of elements sitting atop a more C-shaped set of lobes. Model fitting indicates that these regions correspond to the Mcm2–7 N-terminal domain and C-terminal AAA+ regions, respectively (see below). The lock-washer configuration is more heavily represented than the planar form in both states. At equilibrium, the two forms likely readily partition between each other.
Figure 1.
Mcm2–7 exists in two states. (a,b) Reference-free class averages and corresponding forward projections of notched-ring or lock-washer reconstructions. (c) 3D reconstruction of the notched ring viewed from the AAA+ face (full view, left; slab view, right), the side and the N-terminal face. Mcm homology models are fitted into the reconstruction. (d) 3D structure of the lock-washer ring viewed from the AAA+ (full or slab view), side and N-terminal faces, and fitted with homology models.
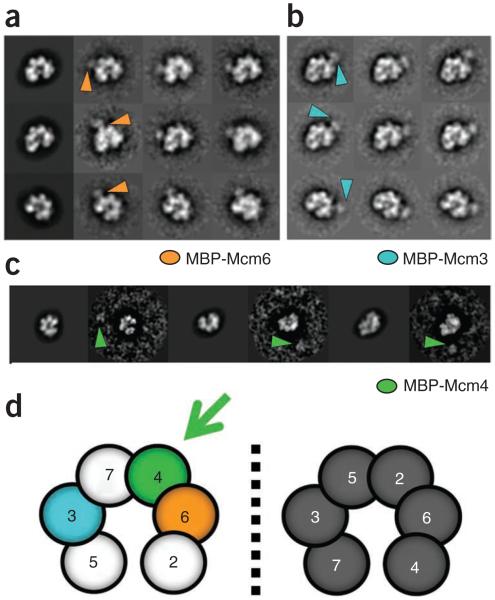
To determine the location of the gap observed in the complex, we collected EM data from ADP·BeF3-treated Mcm2–7 preparations containing an N-terminal MBP tag on Mcm3, Mcm6 or Mcm4, (Fig. 2a–c and Supplementary Fig. 2). We then did reference-free classification, followed by multi-reference alignment to forward projections of the three-dimensional lock-washer structure40. The tags on Mcm3 and Mcm6 clearly reveal that these subunits are both positioned one subunit away from the gap, on opposite sides of the complex. Initially, this arrangement was insufficient to identify the subunits flanking the gap, which conceivably could have corresponded to either subunits 2 and 5 or subunits 4 and 7 (Fig. 2d). The use of an MBP tag on Mcm4, however, resolved this issue, showing that although the tag projects out from the main volume of the complex, it always resides on the side of the Mcm ring distal from the gap (Fig. 2c and Supplementary Fig. 3). The opening in the complex thus occurs at the Mcm2–5 interface.
Figure 2.
Subunit mapping of Mcm2–7. (a) Projection per class average matching of the Mcm2–7 lock-washer state with an N-terminal MBP tag on Mcm6. Orange arrowheads mark additional density observed on the ring. (b) Class averages of Mcm2–7 containing MBP-tagged Mcm3. Orientations match those in a. Blue arrowheads mark additional density observed on the ring. (c) Class-average projection matching of Mcm2–7 containing MBP-tagged Mcm4. The tag appears detached from the ring density (green arrowheads), consistent with the presence of a 154-residue N-terminal tail on Mcm4. (d) Possible subunit arrangements for gapped Mcm2–7 complexes as defined by MBP-tagged Mcm3 and Mcm6. The correct configuration further defined by MBP-tagged Mcm4 is shown in color.
The observation of a gap provides direct visual evidence for a ‘gate’ that could be used to control the entry of nucleic acid segments into the central pore of the helicase. Notably, an opening at this position has been previously predicted, based on biochemical studies of yeast Mcm2–7 (refs. 23,32). Moreover, nonhydrolyzable ATP analogs can either prevent binding to, or trap Mcm2–7 on, circular ssDNA (depending on their relative order of addition), suggesting that the nucleotide can help seal a discontinuity in the Mcm ring. Consistent with this observation, we detected a shift in particle population distribution from the lock-washer configuration to the notched state upon treatment with ADP·BeF3. However, the change we observed is subtle (72% lock-washer versus 28% planar for apo, compared to 64.5% lock-washer versus 35.5% planar for ADP·BeF3), indicating that additional factors help tighten and activate the Mcm2–7 ring, at least in the Drosophila system.
GINS–Cdc45 bind the Mcm2–7 edge, closing the gate
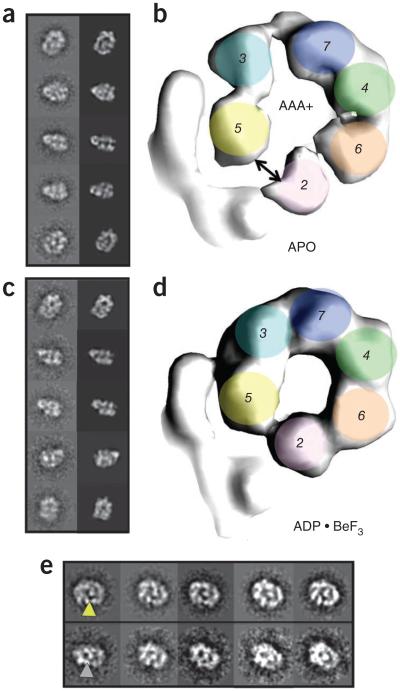
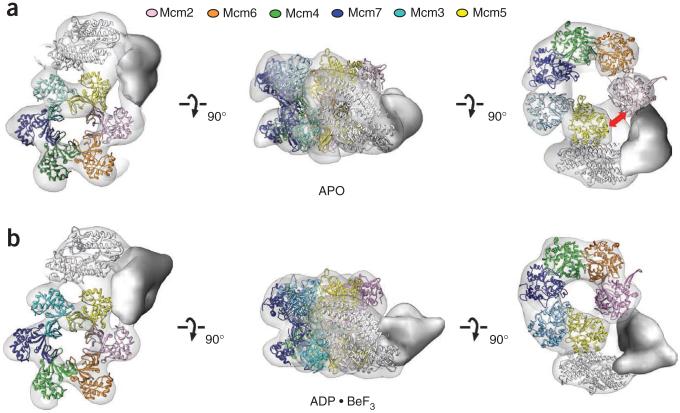
To investigate how GINS and Cdc45 engage and activate Mcm2–7, we next determined the structure of the full CMG complex. Protein expression and purification was done as previously reported15, and as with the Mcm2–7 complex, the peak fraction of a glycerol gradient was used for imaging by negative stain EM (Supplementary Figs. 3 and 4). Unlike the isolated Mcm2–7 assembly, two-dimensional class averages based on a dataset of 13,668 apo CMG particles showed exclusively a planar, hexameric Mcm2–7 ring. Moreover, additional density was visible as a protruding ‘handle’ on one side of the ring that appeared to wrap around several Mcm subunits (Fig. 3a). Free Mcm2–7 complexes were virtually absent from this dataset. Moreover, the CMG complexes used here (and below) all showed helicase activity, and activity profiles precisely followed protein concentration in fractions eluted from the sedimentation step. In all of the raw images and class averages collected for the CMG, only the single additional density element was observed appended to the hexameric ring. These findings establish our sample homogeneity, ruling out the possibility that mixtures of complexes might have confounded subsequent analyses and permitting the straightforward determination of the three-dimensional CMG structure to 28 Å resolution.
Figure 3.
CMG contains a notched, planar Mcm2–7 ring that is sealed upon nucleotide binding. (a) Class-average projection matching for apo CMG. (b) AAA+ view of apo CMG, showing a discontinuity between Mcm2–5. (c) Class-average projection matching for ADP·BeF3-bound CMG. (d) C-terminal AAA+ view of ADP·BeF3-bound CMG, showing a pinched-off gap between Mcm2–5. (e) Class averages of apo (top) or ADP·BeF3-bound CMG (bottom) in the same orientation, showing the presence (yellow arrowhead) or absence (gray arrowhead) of an Mcm2–5 gap.
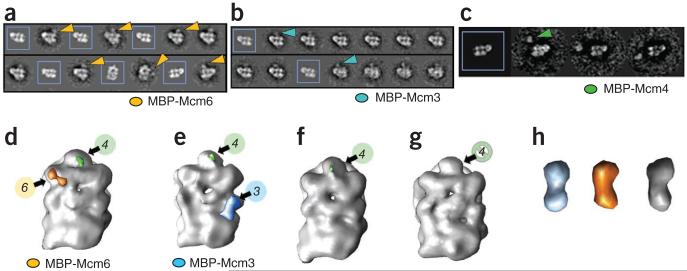
Overall, the configuration of the Mcm2–7 hexamer within the apo CMG assembly proved notably similar to the notched, planar configuration of the Mcm2–7 complex alone. This observation immediately suggested a relative orientation for the six Mcm subunits with respect to the new regions of density occupied by GINS and Cdc45 (Fig. 3 and Supplementary Fig. 2g). To further verify this assignment, we used two independent approaches to determine the register of Mcm2–7 in the CMG complex: (i) MBP tagging of the Mcm3, Mcm4 or Mcm6 N termini (combined with the deletion of the Mcm4 N-terminal tail, Fig. 4) and (ii) a rotational correlation function search of a hexameric, Mcm2–7 homology model (Supplementary Fig. 5). All methods defined a consistent order and placement of the six Mcm subunits in the central ring. The reconstructions also corroborated the existence of a gap between Mcm2 and Mcm5 in the apo CMG, albeit one that is considerably narrower than that seen in nucleotide-free Mcm2–7 alone (compare Figs. 1c and 3c). Additional density for what likely corresponds to the winged-helix domain that lies at the extreme C terminus of Mcm6 was further evident (Supplementary Fig. 6).
Figure 4.
Mcm-subunit mapping in the CMG. (a–c) Class-average projection matching for ADP·BeF3-bound CMG with (unboxed) or without (boxed) N-terminal MBP tags (arrowheads) on Mcm6 (a), Mcm3 (b) or Mcm4 (c). Mcm4 is distal from GINS and Cdc45 (see also Supplementary Fig. 5). (d–g) Unfiltered 3D reconstructions of ADP·BeF3-bound CMG with an N-terminal MBP tag on either Mcm6 (d) or Mcm3 (e) and compared to both untagged CMG (f) and a CMG complex with a 154-amino-acid deletion of the N-terminal Mcm4 tail (g). Extra density attributed to MBP on Mcm6 and Mcm3 is colored orange or light blue, respectively; only a portion of the Mcm4 tail is observable in the full-length constructs and is highlighted in green. (h) Density corresponding to MBP on Mcm3 (light blue) or Mcm6 (orange), cut out from the CMG density maps and compared to the density map calculated from the MBP crystal structure, PDB entry 3HPI51, filtered to 30 Å and showed at 5σ (gray).
We next imaged CMG purified in the presence of ADP·BeF3. Two-dimensional image analysis and three-dimensional structure determination based on a dataset of 25,594 particles (28-Å resolution) showed overall structural features similar to those of the apo CMG complex (Fig. 3c,d and Supplementary Fig. 7). However, inspection of the density map revealed that the C-terminal tier of AAA+ domains with the Mcm2–7 subcomplex is considerably more constricted, closing the Mcm2-Mcm5 gate and narrowing the diameter of the pore between the motor elements (as observed from both three-dimensional structures and end-on view reference-free class averages, Fig. 3 and Supplementary Movie 2). In addition, no density corresponding to the C terminus of Mcm6 could be detected, suggesting that this domain either became disordered or recessed into the body of the motor (Supplementary Fig. 6).
The interaction network of the CMG
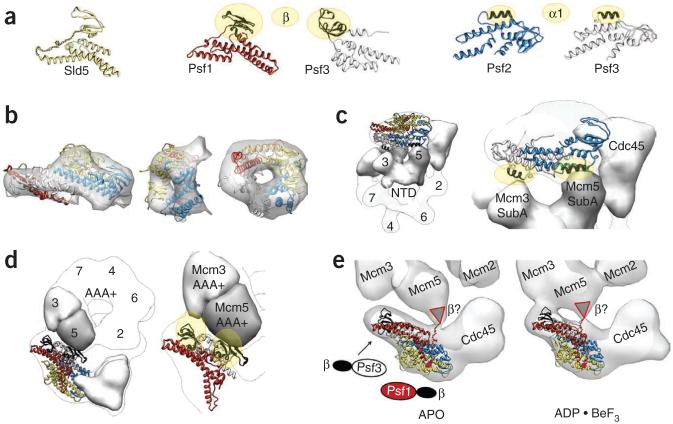
To gain further insights into CMG organization, we turned to antibody labeling and fitting using structural models of component subunit pieces. The atomic structure of the human GINS, which shares ~40% sequence identity on average with each of the four Drosophila subunits, was first used for docking into both the apo and ADP·BeF3 states of the CMG complex (Fig. 5 and Supplementary Fig. 5). This approach revealed a conclusive and consistent position for the four GINS subunits within the full assembly (Fig. 6 and Supplementary Fig. 5) and showed that GINS occupies slightly more than half of the prominent density feature that protrudes from the side of Mcm2–7. Docking also revealed that the GINS tetramer makes extensive contacts with the outer belt of the so-called ‘A-domains’ within the N-terminal tier of the Mcm2–7 ring (Figs. 5 and 6c). Confidence in docking of both the GINS complex (which measures 100 × 65 Å) and the N-MCM complex (120 × 59 Å) into our 28-Å-resolution maps is supported by cross-correlation analysis of multiple docking configurations, with the best solution scoring >10% higher than the second best (Supplementary Fig. 4). Notably, the volume of the remaining density (in other words, that left unfilled by GINS) equates to that expected for a ~70-kDa globular protein, a value in good agreement with the 66-kDa molecular weight predicted for Cdc45 (whose structure is not known). The location of Cdc45, which we further confirmed by imaging antibodies to Cdc45 (anti-Cdc45) bound to the CMG (Supplementary Fig. 8), indicates that Cdc45 serves as a bridge between GINS and the Mcm2–7 helicase core (Figs. 5 and 6c,d).
Figure 5.
Structure docking into CMG reconstructions. (a,b) From left to right: N-terminal, side and C-terminal (AAA+ domain) views of apo (a) or ADP·BeF3-bound CMG (b). The red double arrow in a indicates the location of a discontinuity in the AAA+ tier of Mcm2–7 in apo CMG. GINS is colored white; unoccupied density assigned to Cdc45 is solid gray.
Figure 6.
GINS contacts within the CMG. (a) Crystal structures of the four individual subunits of the GINS complex37. As Sld5 and Psf1 are close homologs, an orientation for the β-domain in Psf1 was generated using that seen in Sld5. The related β-domains of Psf1 and Psf3 are shown in black and highlighted with gold ovals. Psf2 and Psf3 share close structural similarity; a related helical element (α1) is shown in black and highlighted with gold ovals. (b) Three views of the crystal structure of GINS docked into the CMG density. (c) Zoomed-out (left) and close-up (right) views of GINS engaged with Cdc45 and the N-terminal domains of Mcm3 and Mcm5. (d) Zoomed-out (left) and close-up (right) views of GINS and its interactions with Cdc45 and the AAA+ domains of Mcm3 and Mcm5. (e) Nucleotide-triggered movements in the GINS-AAA+ domain interaction region appear to seal off the Mcm2–7 central channel.
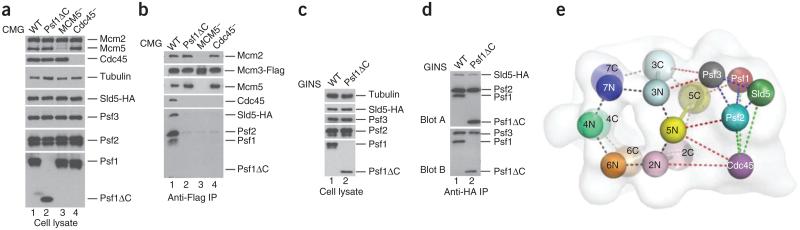
Other lines of evidence support these placements. Consistent with predictions by two independent studies15,41, our CMG reconstructions show that GINS and Cdc45 make extensive contacts with each other. In particular, Cdc45 appears to share a large interaction surface with the N-terminal α-helical domain of Psf2 (Fig. 6c), a finding in accord with yeast cell-lethality complementation experiments36 (Supplementary Fig. 8). Moreover, we observe that Cdc45 associates with the N-terminal domain of Mcm2, an observation buttressed by experiments in both the Drosophila15 and human systems41. We had shown previously using co-infections and pulldowns that only the isolated Mcm4 subunit was capable of associating with GINS15. This interaction prompted us to suggest that GINS might associate with Mcm4 in the context of the Mcm2–7 ring. However, the interaction revealed by these immunoprecipitation experiments was quite modest, and we had further noted that it was not possible to purify GINS– Mcm4 subcomplexes. Our present findings instead suggest that this interaction arose from the analysis of single Mcm subunits outside of the Mcm2–7 complex and is not relevant to the actual CMG structure. Indeed, when the isolated Mcm5 subunit is not provided from a co-infection with each of the remaining ten subunits, neither the GINS complex nor Cdc45 coprecipitates with Mcm3 (Fig. 7a,b). Similarly, when Cdc45 is withheld from co-infection with all other vectors, the GINS complex cannot be detected in the immunoprecipitation with tagged Mcm3 (Fig. 7a,b). We now reinterpret our prior results to suggest that Mcm4 has low affinity interactions with the GINS that are likely masked in the full Mcm2–7 complex, and the stability of the complete CMG is guaranteed only by the formation of a complicated and coordinated network of interactions involving subunits other than Mcm4. These complex multivalent interactions likely govern the stability of the CMG particle, explaining why attempts to purify stable pairwise or ternary subcomplexes have not been successful.
Figure 7.
CMG interactions. (a) Expression levels of individual proteins used for testing CMG formation. (b) Immunoprecipitation (IP) experiments testing CMG stability. 1, wild-type CMG; 2, CMG with a C-terminal truncation in Psf1; 3, CMG without MCM5; 4, CMG without Cdc45. Mcm3 immunoprecipitation yields the intact CMG complex, whereas deletion of the Psf1 β domain (Psf1ΔC, amino acids 1–139), or withholding Mcm5 or Cdc45, disrupts CMG formation. (c) Expression levels of individual proteins used for testing GINS formation. (d) IP experiments testing GINS stability. The C-terminal β-domain of Psf1 is dispensable for GINS formation. (e) Summary of interactions in the CMG. Subunits are demarcated by spheres, which have been placed into the 3D reconstruction obtained for the ADP·BeF3-bound complex (transparent surface). The N- and C-terminal regions of each Mcm subunit are highlighted separately and labeled 2N–7N and 2C–7C, respectively; other subunits are labeled by their full name. Dotted lines show noncovalent interactions observed in the complex and are colored as follows: black, intra-MCM; blue, intra-GINS; green, GINS with Cdc45; red, GINS and Cdc45 with Mcm2–7.
Together, our present assignments demonstrate that the GINS– Cdc45 subcomplex forms a handle that bridges the Mcm2–5 gap in the Mcm ring (Figs. 3c and 6e). In our model, we can see that GINS binds to the external face of the N-terminal A-domains of Mcm5 and Mcm3 through an intimate interaction with Psf2 and Psf3, likely through the structurally conserved α1 helices of both proteins (residues 68–81 human Psf2 and residues 94–102 of human Psf3) (Fig. 6a,c). In support of this assignment, helix α1 of Psf2 has been identified as essential in yeast cell-lethality complementation experiments36 (Supplementary Fig. 8). Psf2 and Psf3 also are closely related evolutionarily42, and many archaeal genomes contain a homologous ancestral GINS subunit (GINS23), which binds to the N-terminal domain of its corresponding MCM partner43.
Unexpectedly, our reconstructions—particularly the more compact ADP·BeF3-bound CMG structure—also show that GINS interacts with the AAA+ domain tier of Mcm2–7 (Figs. 5 and 6a,d,e). Our modeling suggests that this contact is mediated by the β-sheet domain present at the N terminus of Psf3 and the peripheral face of the Mcm3 ATPase region. When inspecting the CMG structures at slightly lower contour levels, additional connectivity can be observed between the central body of the GINS complex and the AAA+ domain of Mcm5 (Fig. 6e). The density for both of these connecting elements appears to move, depending on the nucleotide status of the CMG (Supplementary Movie 2), alternating between a somewhat looser (apo) or tighter (ADP·BeF3) configuration. This less substantial AAA+ contact may be mediated by the flexible C-terminal β-sheet domain of Psf1, which shows the same fold as the Mcm3-binding N-terminal domain of Psf3 (Figs. 5e and 6a,d) but has been reshuffled by a circular permutation event17.
Consistent with this idea, the C-terminal β-domain is not visible in the crystal structure of full-length, GINS-associated Psf1 (ref. 37), indicating that this element is highly flexible. Moreover, sequence comparisons indicate that Psf1 and Sld5 are closely related, and superposition of full-length Sld5 onto Psf1 suggests a location for the missing C terminus of Psf1 that would allow it interact with the AAA+ periphery of Mcm5. To test this assumption, we deleted the C-terminal β-sheet domain of Psf1 and asked if this construct might affect the formation of the CMG. Using co-immunoprecipitation, we found that this deleted version of Psf1 is expressed well and is capable of forming a complex with its other GINS partners but is unable to stably coassemble into the CMG (Fig. 7a–d). Others have similarly deleted this domain from human Psf1 (ref. 38) and found that, although able to associate into a GINS complex, the truncation mutant could not associate with chromatin or contribute to a replication complex. Together with our other biochemical and structural analyses, these findings reinforce the notion that CMG formation relies on an intricate network of interactions (Fig. 7e). Our model shows that the surface-exposed face of the N-terminal domain of Sld5 does not contact any partner in the CMG, despite observations indicating that the surface of this region is important for cell viability36 (Supplementary Fig. 8).
DISCUSSION
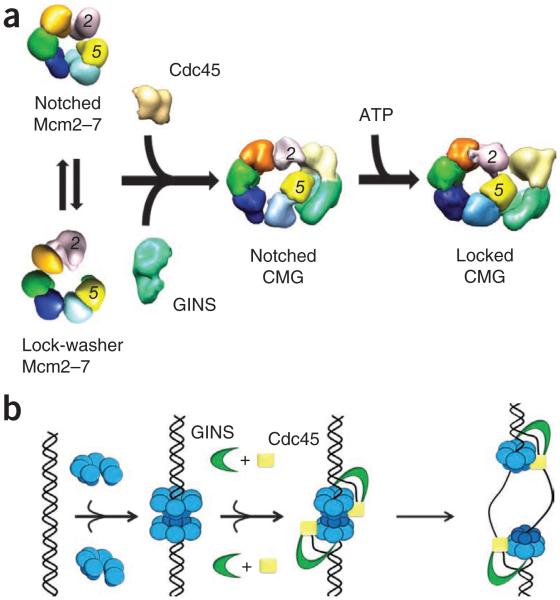
We have shown that the Mcm2–7 complex has a dynamic structure that alternates between planar, notched-ring and spiral, lock-washer conformations, and that there exists a persistent discontinuity at the interface between Mcm2 and Mcm5 (Fig. 8a). This gap is preserved in the nucleotide-free CMG, in which Mcm2–7 is restrained to its more planar configuration. Adoption of this state appears to be reinforced by the association of GINS and Cdc45 with the exterior of Mcms 2, 3 and 5. The binding of GINS–Cdc45 also generates a large, bilobed channel through the CMG complex that stretches from the center of the Mcm2–7 pore to the inner surface of the non-Mcm accessory subunits. However, upon binding an ATP analog, the CMG complex undergoes another conformational change, closing the Mcm2–5 gate, tightening the Mcm2–7 ring and enlarging the contact area of GINS to include the AAA+ domains of Mcm3 and Mcm5. This change bisects the single CMG channel into two topologically segregated conduits through the complex (Fig. 8a). The more intimate contacts we observe in the CMG help explain its enhanced ATPase activity compared to Mcm2–7, and together with the narrowing of the internal helicase channel, this may account for the tighter association of the complex with DNA15.
Figure 8.
Model for Mcm2–7 activation and function. (a) Free Mcm2–7 can exist in either an open, lock-washer or notched, planar configuration. Each form shows a discontinuity between Mcm2 and Mcm5. Binding of GINS–Cdc45 stabilizes the notched, planar Mcm2–7 state, whereas ATP binding promotes ring closure. (b) ORC, Cdc6 and Cdt1 load a preopened Mcm2–7 assembly onto dsDNA as an inactive double hexamer. GINS and Cdc45 bind to Mcm2–7, concomitant with an isomerization that creates or stabilizes melted DNA. The side channel formed by the GINS–Cdc45 subcomplex likely prevents DNA escape from Mcm2–7 and may help partition the lagging DNA strand from its complement following unwinding.
Another noteworthy facet of Mcm2–7 and CMG architecture revealed by our data is that the particles are highly dynamic. We emphasize this point because the CMG is known to be able to bind to and track along a single-stranded DNA circle to displace an annealed oligonucleotide in helicase assays11,15. Our data suggest that the assembly can ‘breathe’ between closed and open states to allow DNA access into the interior motor regions. Such behavior is not without precedent and has been reported for preformed SV40 T antigen and papilloma virus E1 hexamers as well44,45.
These findings have important ramifications for understanding the physical basis of Mcm loading, activation and function during replication in eukaryotes. Mcm2–7 is known to be loaded as a head-to-head double hexamer onto duplex DNA at origins of replication, before the switch into the S phase of the cell cycle9,10. Interestingly, single-particle electron microscopy suggests that the Mcm2–7 complex is closed in this configuration, as well as when bound to the Cdt1 loading cofactor9. These findings are not inconsistent with our present study: given the wealth of data establishing that Mcm2–7 forms a stable complex3,15,22,23, it has frequently been postulated that the ring must be breached at some point to facilitate its deposition on DNA. Our studies do suggest, however, that the eukaryotic replication initiation machinery need not necessarily expend chemical energy to crack apart a closed Mcm ring. Rather, ORC, Cdc6 and Cdt1 may take advantage of a predisposition of Mcm2–7 to form open-ring states to facilitate helicase loading. Our demonstration that the Mcm2–5 interface is indeed broken in the isolated Mcm2–7 complex is consistent with suggestions that DNA will be threaded into the motor through this breach32,46.
Once loaded, how does Mcm2–7 convert into a functional helicaseΔ Activation depends in part upon phosphorylation by the DDK and CDK kinases13, as well as the subsequent association of GINS and Cdc45. Our studies show that GINS–Cdc45 bridges the Mcm2–5 gap, an event that likely assists DNA capture by the CMG. However, nucleotide binding additionally alters the conformational state of the CMG by both strengthening the GINS–Mcm3–5 interaction and further closing the Mcm2–5 interface. This transition constricts the central Mcm2–7 pore and generates a second, external channel on the outer perimeter of the Mcm ring.
Our structural studies of the CMG do not concretely address how GINS and Cdc45, along with possibly other factors, may distort and/or disrupt double hexamers formed by ORC loading. Nevertheless, we have shown previously that, unlike isolated Mcm2–7, the CMG cannot bind duplex DNA9,10,15. Moreover, an off-rate for the CMG is not detectable in a competition experiment once the complex is bound to a fork15. These observations, coupled with the obvious need for the CMG to separate DNA strands during replication, indicate that some structural rearrangement likely occurs within the double hexamer on the path to forming a competent helicase. Although the physical disposition of the displaced DNA strands within the functioning CMG is still an open question15,35,47, the placement of GINS and Cdc45 to one side of Mcm2–7 argues strongly against the idea that these factors act as a type of plow to directly separate duplex DNA strands48,49. Instead, the structures and changes we see are consistent with a model in which the CMG directly promotes duplex opening and then segregates the melted nucleic acid segments from one another by partitioning them between the helicase pore and the GINS–Cdc45–Mcm235 vestibule (Fig. 8b). Following this event, the two helicases would dissociate and move away from the origin50, allowing the extruded strands to be engaged by RPA, Mcm10 and polymerase α-primase.
ONLINE METHODS
Baculovirus construction and protein production
Baculoviruses expressing single CMG subunits were generated using the Bac-to-Bac system from Invitrogen as previously described15. MBP-tagged versions of Mcm3, Mcm4 and Mcm6 were constructed by inserting the PCR-amplified MBP coding region from pMal-c2x plasmid (New England Biolabs) in front of the appropriate Mcm open reading frame. Both MBP-tagged and untagged MCM3 carried also an N-terminal flag sequence. ΔN-MCM4 lacks its first 154 amino acids. Mcm2–7 and CMG were expressed and purified as published previously (also see Supplementary Fig. 1)15.
Approximately 5 μg of CMG or MCM2–7 were loaded onto a 4 ml, 15–35% (v/v) glycerol gradient and run for 10–12 h on a Beckman L7 ultracentrifuge using an Sw60 rotor (42,000 r.p.m. at 4 °C). The gradient was fractionated (200 μl aliquots) and fractions analyzed by PAGE and silver staining to verify protein complex identity and stoichiometry. The peak fraction was immediately used for negative-stain EM sample preparation. ADP·BeF3-bound CMG was obtained by carrying out a glycerol gradient run, in which gradient buffers contained 1 mM ADP plus a fresh mix of 3 mM BeCl2 and 15 mM NaF.
Negative stain sample preparation
EM grids were prepared by depositing a thin layer of continuous carbon over a holey carbon layer on a 400-mesh copper grid (Electron Microscopy Sciences) using a custom-made carbon floating device. Four μl of Mcm2–7 or CMG complex (~30 ng) was applied onto a fresh, glow-discharged grid, and incubation time was varied from 30 s to 2 min to optimize particle density. The grid was laid on top of 75-μl drops of a freshly prepared 2% (w/v) uranyl formate solution and stirred gently for five subsequent 10-s staining steps. The CMG complex bound to an affinitypurified, anti-Cdc45 (ref. 11) was prepared by incubating the apo CMG (6 ng μl−1) and antibody (0.5ng μl−1) in a final volume of 10 μl for 1 min at room temperature, before applying to grids.
Electron microscopy data collection
Data were collected on a Tecnai 12 transmission EM (FEI) operating at 120 KeV and at a magnification of 30,000×, with a nominal defocus ranging from −0.8 to −0.3 μm. Images were acquired using a Gatan 1k × 1k CCD camera, resulting in a final pixel size of 5.72 Å. Low-dose conditions were used throughout this study, except for ΔN-Mcm4 CMG and all MBP-tagged complexes, for which normal-dose conditions were used.
Particle picking and dataset generation
Particle picking was done semiautomatically using Boxer (EMAN)52 or automatically using DoG picker53. A total of 7,527 particles were picked for apo Mcm2–7, 53,000 particles for ADP·BeF3-bound Mcm2–7, 4,983 particles for ADP·BeF3-bound N-MBP-Mcm3 Mcm2–7, 4,168 particles for ADP·BeF3-bound N-MBP-Mcm4 Mcm2–7, 15,838 particles for ADP·BeF3-bound N-MBP-Mcm6 Mcm2–7, 13,668 particles for apo CMG, 25,594 particles for ADP·BeF3-bound CMG, 7,599 particles for ADP·BeF3-bound N-MBP-Mcm3 CMG, 3,754 particles for ADP·BeF3-bound N-MBP-Mcm4 CMG, and 10,898 particles for ADP·BeF3-bound N-MBP-Mcm6 CMG. For the apo CMG–anti-Cdc45 complex, a dataset of 1,070 particles was picked interactively using Boxer. A 64 × 64-pixel particle window was used for all datasets, except for N-MBP-tagged Mcm4 complexes, for which a window of 100 × 100 pixels was used to account for the long N-terminal Mcm4 tail, which considerably increased the dimensions of the protein complex.
Two-dimensional image analysis
All datasets were band-pass filtered with a 200-Å high-pass cutoff and either a 20-Å low-pass cutoff (for initial classification and initial projection-matching refinement steps) or a 11.5-Å low-pass cutoff (for finer classification and final projection-matching refinement steps). Reference-free class averages were produced using multivariate statistical analysis (MSA) in Imagic40, with the following modification: after image centering, different in-plane orientations were explored for each particle by appending six copies of the same dataset, each rotated by 60°. Analysis of this enlarged and redundant dataset resulted in high-quality featured class averages, which virtually never contained rotated versions of the same raw image in the same class. Redundant class averages were removed after an additional multi-reference-alignment step. This approach allowed exhaustive MSA classification in one lone step. Overall, results were comparable to (if not better than) those obtained by traditional multi-reference-alignment-based approaches54, which require several rounds of refinement and are highly dependent on user intervention. On average, ~20 particles were included per class average.
To take into account the larger diameter of the antibody supercomplex, the CMG·anti-Cdc45 particle dataset was band-pass filtered with a 300-Å highpass cutoff and a 30-Å low-pass cutoff for initial classification procedures. This approach allowed averaging of (rather featureless) CMG complexes connected to an additional globular density. The generated classification file was used to average an equivalently transformed dataset of images that were band-pass filtered using a 200-Å high-pass cutoff and a 20-Å low-pass filtered cutoff.
Three-dimensional reconstruction
An initial structure of a split Mcm2–7 ring was determined by angular reconstitution and a C1-startup approach for de-novo 3D reconstruction based on common lines, using three selected top, tilted and side-view class averages as an initial anchor set40. Ten subsequent rounds of angular reconstitution refinement were done. The volume obtained was filtered to 120 Å and used for six rounds of projection-matching refinement using SPIDER55. At the same time, an initial model was generated after superposing the atomic coordinates of six full-length SsoMCM monomers (PDB entry: 3F9V55) onto a hexameric N-terminal domain of MthMCM (PDB entry: 1LTL18). This model was filtered to 120 Å and used as an initial reference in SPIDER for projection-matching refinement (using raw particles as input), resulting in a planar ring structure superposed to a structure presenting prominent out-of-plane elements. Subsequent multimodel refinement approaches (using the lockwasher and the planar structure filtered to 40 Å as starting references)39 allowed the separation of the split ring into a spiral, lock-washer–shaped particle and a planar notched-ring hexamer.
An initial structure of the CMG complex was first determined using traditional angular reconstitution approaches in Imagic40. Further refinement of the structure was done using projection matching, after filtering the starting model to 120 Å. A similar structure was obtained starting from a six-fold– symmetric, homo-oligomeric MCM ring reference filtered to 120 Å, providing confidence about the quality of our results. After obtaining a reconstruction of the ADP·BeF3-bound CMG complex, characterized by a nearly pseudo–six-fold–symmetric Mcm2–7 ring, we repeated projection-matching refinement of the apo CMG structure, using the nucleotide-treated sample as a starting model. The final apo structure again showed a markedly asymmetric character, further confirming its departure from any higher-order symmetry56. The same starting model also was used for projection matching refinement of the ADP·BeF3-bound CMG complex with an N-terminal MBP tag on Mcm3 or Mcm6, and of our CMG construct bearing a truncated N-terminal Mcm4 tail. Resolution of all maps was determined using Fourier Shell Correlation, with a 0.5 cut-off criterion, as implemented in SPIDER55.
EM map analysis, crystal structure docking and molecular modeling
The handedness of all electron density maps was assessed by three different approaches. First, independent docking of the atomic structures of the N-terminal MthMCM hexamer18 and the human GINS complex37 resulted in a clear preference for the same hand. Docking of a tetrameric SsoMCM complex (isolated from the six-fold–symmetric model hexamer)25 further confirmed the assigned hand for the planar, notched Mcm2–7 hexamer. An additional approach consisted of docking either hands of the notched Mcm2–7 ring into the apo CMG structure, or either hands of the spiral Mcm2–7 structure into the notched ring, and calculating correlation coefficients. This approach also allowed determination of the Mcm2–7 subunit register in various complexes. A third approach, exploiting either three-dimensional reconstruction or two-dimensional averaging, combined with multi-reference alignment of MBP-tagged complexes, allowed an independent and conclusive assessment of the hand of all maps and of the motor-subunit register in the context of the isolated Mcm2–7 or full CMG complexes.
All structure docking analyses were done using the ‘Fit in Map’ function of UCSF Chimera57. Cross-correlation coefficients were calculated after generating an electron density map from the atomic coordinates of interest (using the same sampling frequency as used for EM data collection) and fitting it into the map generated by single particle approaches. Homology models for the six Mcm2–7 subunits were generated using SWISS-MODEL58. Modeling of the Psf1 C-terminal domain was done based on the crystal structure of human GINS (PDB entry 2Q9Q37) by superposing the N-terminal α-domain of Sld5 into the atomic structure of Psf1, using Pymol (Delano Scientific) and EBI-SSM59. All two-dimensional class-average figures were generated using Imagic40 and all three-dimensional structure figures were generated using UCSF Chimera57. Movies and electron density map morphs were generated using UCSF Chimera57.
Immunoprecipitation experiments
Cell extracts were prepared and pulldown experiments were generally conducted as previously described15, except that 1ml of protein extract (from 2 × 107 infected Hi5 cells) was added to 15 μl of either anti-flag M2-agarose beads or anti-HA (HA-7) agarose beads (Sigma-Aldrich). Immunoprecipitation reactions were incubated in the presence of 50 μg ml−1 ethidium bromide. Polyclonal antibodies to CMG subunits and monoclonal antibodies have been described elsewhere11,15.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank A. Lyubimov and F. Bleichert for comments and help with the manuscript; and G. Lander, P. Grob, R. Hannah, R. Hall, M. Cianfrocco and C. Ciferri for technical help. This work was supported by a European Molecular Biology Organization long-term postdoctoral fellowship (to A.C.), a PhD fellowship from the Boehringer Ingelheim Fonds (to T.P.), the Human Frontier Science Program (RPG0039, to E.N.), the National Institute of General Medical Sciences (GM071747, to J.M.B.) and the National Cancer Institute (CA R37-30490, to M.R.B.). E.N. is a Howard Hughes Medical Institute investigator.
Footnotes
Accession codes. EM Data Bank: Three-dimensional EM maps of notched-ring Mcm2–7 and lock-washer Mcm2–7 have been deposited with accession codes EMD-1834 and EMD-1835. Three-dimensional EM maps of ADP·BeF3 CMG and apo CMG have been deposited with accession codes EMD-1832 and EMD-1833, respectively.
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Davey MJ, O’Donnell M. Replicative helicase loaders: ring breakers and ring makers. Curr. Biol. 2003;13:R594–R596. doi: 10.1016/s0960-9822(03)00523-2. [DOI] [PubMed] [Google Scholar]
- 2.Funnell BE, Baker TA, Kornberg A. In vitro assembly of a prepriming complex at the origin of the Escherichia coli chromosome. J. Biol. Chem. 1987;262:10327–10334. [PubMed] [Google Scholar]
- 3.Davey MJ, Fang L, McInerney P, Georgescu RE, O’Donnell M. The DnaC helicase loader is a dual ATP/ADP switch protein. EMBO J. 2002;21:3148–3159. doi: 10.1093/emboj/cdf308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valle M, Gruss C, Halmer L, Carazo JM, Donate LE. Large T-antigen double hexamers imaged at the simian virus 40 origin of replication. Mol. Cell. Biol. 2000;20:34–41. doi: 10.1128/mcb.20.1.34-41.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar A, et al. Model for T-antigen-dependent melting of the simian virus 40 core origin based on studies of the interaction of the beta-hairpin with DNA. J. Virol. 2007;81:4808–4818. doi: 10.1128/JVI.02451-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuck S, Stenlund A. Assembly of a double hexameric helicase. Mol. Cell. 2005;20:377–389. doi: 10.1016/j.molcel.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 7.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 8.Bowers JL, Randell JC, Chen S, Bell SP. ATP hydrolysis by ORC catalyzes reiterative Mcm2–7 assembly at a defined origin of replication. Mol. Cell. 2004;16:967–978. doi: 10.1016/j.molcel.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 9.Remus D, et al. Concerted loading of Mcm2–7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evrin C, et al. A double-hexameric MCM2–7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc. Natl. Acad. Sci. USA. 2009;106:20240–20245. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2–7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. USA. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gambus A, et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 13.Remus D, Diffley JF. Eukaryotic DNA replication control: lock and load, then fire. Curr. Opin. Cell Biol. 2009;21:771–777. doi: 10.1016/j.ceb.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Calzada A, Hodgson B, Kanemaki M, Bueno A, Labib K. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 2005;19:1905–1919. doi: 10.1101/gad.337205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2–7 helicase by association with Cdc45 and GINS proteins. Mol. Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC. Localization of MCM2–7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell. 2006;21:581–587. doi: 10.1016/j.molcel.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 17.MacNeill SA. Structure and function of the GINS complex, a key component of the eukaryotic replisome. Biochem. J. 2010;425:489–500. doi: 10.1042/BJ20091531. [DOI] [PubMed] [Google Scholar]
- 18.Fletcher RJ, et al. The structure and function of MCM from archaealM. thermoautotrophicum. Nat. Struct. Biol. 2003;10:160–167. doi: 10.1038/nsb893. [DOI] [PubMed] [Google Scholar]
- 19.McGeoch AT, Trakselis MA, Laskey RA, Bell SD. Organization of the archaeal MCM complex on DNA and implications for the helicase mechanism. Nat. Struct. Mol. Biol. 2005;12:756–762. doi: 10.1038/nsmb974. [DOI] [PubMed] [Google Scholar]
- 20.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 21.Davey MJ, Indiani C, O’Donnell M. Reconstitution of the Mcm2–7p heterohexamer, subunit arrangement, and ATP site architecture. J. Biol. Chem. 2003;278:4491–4499. doi: 10.1074/jbc.M210511200. [DOI] [PubMed] [Google Scholar]
- 22.Crevel G, Ivetic A, Ohno K, Yamaguchi M, Cotterill S. Nearest neighbour analysis of MCM protein complexes in Drosophila melanogaster. Nucleic Acids Res. 2001;29:4834–4842. doi: 10.1093/nar/29.23.4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bochman ML, Bell SP, Schwacha A. Subunit organization of Mcm2–7 and the unequal role of active sites in ATP hydrolysis and viability. Mol. Cell. Biol. 2008;28:5865–5873. doi: 10.1128/MCB.00161-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pape T, et al. Hexameric ring structure of the full-length archaeal MCM protein complex. EMBO Rep. 2003;4:1079–1083. doi: 10.1038/sj.embor.7400010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brewster AS, et al. Crystal structure of a near-full-length archaeal MCM: functional insights for an AAA+ hexameric helicase. Proc. Natl. Acad. Sci. USA. 2008;105:20191–20196. doi: 10.1073/pnas.0808037105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bae B, et al. Insights into the architecture of the replicative helicase from the structure of an archaeal MCM homolog. Structure. 2009;17:211–222. doi: 10.1016/j.str.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 27.Yu X, et al. The Methanobacterium thermoautotrophicum MCM protein can form heptameric rings. EMBO Rep. 2002;3:792–797. doi: 10.1093/embo-reports/kvf160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gómez-Llorente Y, Fletcher RJ, Chen XS, Carazo JM, San Martin C. Polymorphism and double hexamer structure in the archaeal minichromosome maintenance (MCM) helicase from Methanobacterium thermoautotrophicum. J. Biol. Chem. 2005;280:40909–40915. doi: 10.1074/jbc.M509760200. [DOI] [PubMed] [Google Scholar]
- 29.Costa A, et al. Structural basis of the Methanothermobacter thermautotrophicus MCM helicase activity. Nucleic Acids Res. 2006;34:5829–5838. doi: 10.1093/nar/gkl708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chong JP, Hayashi MK, Simon MN, Xu RM, Stillman B. A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase. Proc. Natl. Acad. Sci. USA. 2000;97:1530–1535. doi: 10.1073/pnas.030539597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelman Z, Lee JK, Hurwitz J. The single minichromosome maintenance protein of Methanobacterium thermoautotrophicum DeltaH contains DNA helicase activity. Proc. Natl. Acad. Sci. USA. 1999;96:14783–14788. doi: 10.1073/pnas.96.26.14783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bochman ML, Schwacha A. The Mcm2–7 complex has in vitro helicase activity. Mol. Cell. 2008;31:287–293. doi: 10.1016/j.molcel.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 33.Kanemaki M, Sanchez-Diaz A, Gambus A, Labib K. Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature. 2003;423:720–724. doi: 10.1038/nature01692. [DOI] [PubMed] [Google Scholar]
- 34.Takayama Y, et al. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev. 2003;17:1153–1165. doi: 10.1101/gad.1065903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boskovic J, et al. Molecular architecture of the human GINS complex. EMBO Rep. 2007;8:678–684. doi: 10.1038/sj.embor.7401002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi JM, Lim HS, Kim JJ, Song OK, Cho Y. Crystal structure of the human GINS complex. Genes Dev. 2007;21:1316–1321. doi: 10.1101/gad.1548107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang YP, Wang G, Bermudez V, Hurwitz J, Chen XS. Crystal structure of the GINS complex and functional insights into its role in DNA replication. Proc. Natl. Acad. Sci. USA. 2007;104:12685–12690. doi: 10.1073/pnas.0705558104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamada K, Kubota Y, Arata T, Shindo Y, Hanaoka F. Structure of the human GINS complex and its assembly and functional interface in replication initiation. Nat. Struct. Mol. Biol. 2007;14:388–396. doi: 10.1038/nsmb1231. [DOI] [PubMed] [Google Scholar]
- 39.Grob P, et al. Cryo-electron microscopy studies of human TFIID: conformational breathing in the integration of gene regulatory cues. Structure. 2006;14:511–520. doi: 10.1016/j.str.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 40.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 41.Im JS, et al. Assembly of the Cdc45-Mcm2–7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc. Natl. Acad. Sci. USA. 2009;106:15628–15632. doi: 10.1073/pnas.0908039106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Makarova KS, Wolf YI, Mekhedov SL, Mirkin BG, Koonin EV. Ancestral paralogs and pseudoparalogs and their role in the emergence of the eukaryotic cell. Nucleic Acids Res. 2005;33:4626–4638. doi: 10.1093/nar/gki775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marinsek N, et al. GINS, a central nexus in the archaeal DNA replication fork. EMBO Rep. 2006;7:539–545. doi: 10.1038/sj.embor.7400649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dean FB, Borowiec JA, Eki T, Hurwitz J. The simian virus 40 T antigen double hexamer assembles around the DNA at the replication origin. J. Biol. Chem. 1992;267:14129–14137. [PubMed] [Google Scholar]
- 45.Fouts ET, Yu X, Egelman EH, Botchan MR. Biochemical and electron microscopic image analysis of the hexameric E1 helicase. J. Biol. Chem. 1999;274:4447–4458. doi: 10.1074/jbc.274.7.4447. [DOI] [PubMed] [Google Scholar]
- 46.Bochman ML, Schwacha A. Differences in the single-stranded DNA binding activities of MCM2–7 and MCM467: MCM2 and MCM5 define a slow ATP-dependent step. J. Biol. Chem. 2007;282:33795–33804. doi: 10.1074/jbc.M703824200. [DOI] [PubMed] [Google Scholar]
- 47.Labib K, Gambus A. A key role for the GINS complex at DNA replication forks. Trends Cell Biol. 2007;17:271–278. doi: 10.1016/j.tcb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 48.Takahashi TS, Wigley DB, Walter JC. Pumps, paradoxes and ploughshares: mechanism of the MCM2–7 DNA helicase. Trends Biochem. Sci. 2005;30:437–444. doi: 10.1016/j.tibs.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 49.Takara TJ, Bell SP. Putting two heads together to unwind DNA. Cell. 2009;139:652–654. doi: 10.1016/j.cell.2009.10.037. [DOI] [PubMed] [Google Scholar]
- 50.Yardimci H, Loveland AB, Habuchi S, van Oijen AM, Walter JC. Uncoupling of sister replisomes during eukaryotic DNA replication. Mol. Cell. 2010;40:834–840. doi: 10.1016/j.molcel.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gould AD, Shilton BH. Studies of the maltose transport system reveal a mechanism for coupling ATP hydrolysis to substrate translocation without direct recognition of substrate. J. Biol. Chem. 2010;285:11290–11296. doi: 10.1074/jbc.M109.089078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high- resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 53.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 2009;166:205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Heel M, et al. Single-particle electron cryo-microscopy: towards atomic resolution. Q. Rev. Biophys. 2000;33:307–369. doi: 10.1017/s0033583500003644. [DOI] [PubMed] [Google Scholar]
- 55.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 56.Boekema EJ, Berden JA, van Heel MG. Structure of mitochondrial F1-ATPase studied by electron microscopy and image processing. Biochim. Biophys. Acta. 1986;851:353–360. doi: 10.1016/0005-2728(86)90071-x. [DOI] [PubMed] [Google Scholar]
- 57.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 58.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 59.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.