Abstract
For right-censored data perhaps the most commonly used tests are weighted logrank tests, such as the logrank and Wilcoxon-type tests. In this paper we review several generalizations of those weighted logrank tests to interval-censored data and present an R package, interval, to implement many of them. The interval package depends on the perm package, also presented here, which performs exact and asymptotic linear permutation tests. The perm package performs many of the tests included in the already available coin package, and provides an independent validation of coin. We review analysis methods for interval-censored data, and we describe and show how to use the interval and perm packages.
Keywords: logrank test, Wilcoxon test, exact tests, network algorithm, R
1. Introduction
In their original paper introducing the logrank test, Peto and Peto (1972) outlined how to perform permutation-based logrank tests for interval-censored data. Later, Finkelstein (1986) studied the application of the logrank test to interval-censored data in detail using a grouped continuous model. These logrank tests are appropriate for comparing treatment groups when the response is time to an event and time may only be known to fall into an interval. An example is time to progression-free survival (see e.g., Freidlin, Korn, Hunsberger, Gray, Saxman, and Zujewski 2007), where patients are monitored intermittently and progression is known to have occurred only to within the time since the last visit.
A na?ve approach, if one has interval-censored data, is to simply impute the mid-point of the intervals and perform the usual right-censored weighted logrank tests. Law and Brookmeyer (1992) studied this naive approach (Law and Brookmeyer 1992, use the term ‘doubly censored observations’ to mean interval-censored observations), and showed through simulation that when the censoring mechanism is different between the two groups, the type I error of a nominal 0.05 test was in one case estimated to be as high as 0.19. Thus, there is clearly a need for the tests specifically designed for interval-censored data.
Despite the demonstrated need for these methods and despite the development of several methods for generalizing the logrank test to interval-censored data, these tests are rarely used in the medical literature. This may be due to lack of knowledge about the interval-censored methods, but it also may be due to the lack of widely available software to test interval-censored data nonparametrically.
In this paper we describe an R package, called interval, to perform weighted logrank tests (WLRT) (including a generalization of the Wilcoxon-Mann-Whitney test) for interval-censored data. The open source freeware R (R Development Core Team 2010) easily accommodates packages, and all R packages mentioned in this paper are available from the Comprehensive R Archive Network at http://CRAN.R-project.org/.
For each type of rank score (e.g., logrank-type or Wilcoxon-type) the interval package allows three methods for creating tests: (1) score tests, (2) permutation tests derived from the score statistics in the grouped continuous model (GCM), both described in Fay (1999a), and (3) multiple imputation tests as described in Huang, Lee, and Yu (2008). The p-values from the permutation tests may be calculated either asymptotically using a permutational central limit theorem or exactly using either complete enumeration, a network algorithm (for the two-sample case only), or Monte Carlo resampling. We know of no readily available software to perform these tests (or other different generalizations of the WLRTs to interval-censored data), except for the S-PLUS functions (written by the first author) upon which the interval package is based (Fay 1999b).
In Section 2 we give a brief background on the analysis of interval-censored data, with emphasis on nonparametric maximum likelihood estimation of the distribution and generalizations of the WLRTs for interval-censored data. We review different forms of the likelihood as well as the different methods for carrying out the WLR tests. This section reviews what is known about the asymptotic validity of these tests for different interval-censoring settings. With this background, we hope to give intuition on the methods for users primarily familiar with the usual right-censored logrank tests and also provide information to guide the applied researcher to make an appropriate choice for the test for their setting of interest.
The mathematical formulation of the WLRTs used in interval package are outlined in Section 3. Section 4 provides step-by-step instructions for how to use the interval package to perform data analysis. This application section demonstrates the use of two main functions of interval: icfit which provide nonparametric maximum likelihood estimators (NPMLEs) of the survival distribution and ictest which performs weighted logrank tests.
As explained in Section 2, all implementations of the WLRTs require first fitting an NPMLE of the overall survival distribution and, for some kinds of inferences, permutation methods may be used. Because of this latter fact, the interval depends on the perm package which performs exact and asymptotic linear permutation tests. Note that this package is mostly redundant because nearly all of the tests performed by perm are available in the package coin (Hothorn, Hornik, van de Wiel, and Zeileis 2006, 2008), and the exact tests are generally faster in coin. The redundancy provides independent software to check the results from perm. In Section 5 we go into detail about the design of the perm package and how it differs from the coin package. In Section 6 we detail the design of the interval package. The interval package uses an expectation-maximization (EM) style algorithm to find the NPMLE of the overall distribution where the initial estimator of the distribution may come from a function from another package. The default initial estimate uses a function whose main calculation engine uses the computeMLE function from MLEcens, an available package for bivariate interval-censored data that is also applicable for univariate interval-censored data (Maathuis 2010). Additionally, the Icens package may be used to calculate an initial distribution estimate, and we have designed the interval package to be able to input NPMLEs from the Icens package (Gentleman and Vandal 2010), using the class structure designed there. Further, we show how the wlr_trafo function of the interval package may be called seamlessly from within coin to perform exact permutation tests on interval-censored data possibly more quickly than using the default perm package.
2. Background on interval-censored data analysis methods
2.1. Censoring assumptions
Interval censored data arise when the response of interest is a time-to-event variable, but instead of the response time, we only observe whether or not the event has yet occurred at a series of assessment times. For example, suppose the event is time until first disease occurrence, and we can assume that this disease will not spontaneously be cured without treatment (e.g., HIV). The response data may be a series of assessment times on the individual corresponding to times when blood samples used for detecting the disease were taken, and at each blood sample we determine if the disease is detected or not. Typically, we do not keep information about all the assessment times, but only record the last disease-free assessment time and the first assessment time where the disease was detected. When all the assessment times (not just the two that we keep information about) are independent of the event time we say that the censoring is non-informative. In the example, the censoring is non-informative if the subjects are not more or less likely to get a blood sample taken if they have the disease. More difficult situations where the assessment times may depend on the event can cause informative censoring and are not discussed in this paper or handled by the software described here. See Zhang, Sun, Sun, and Finkelstein (2007) and the references therein for methods for informative interval-censoring.
2.2. Nonparametric estimation of survival distributions
With right-censored data often we will plot the NPMLE of the survival function which in that case is called the Kaplan-Meier or product-limit estimator (see Kalbfleisch and Prentice 1980). The Kaplan-Meier estimator is typically called “undefined” after the last observation if that observation is right-censored. This is because the NPMLE is not unique in that space, as changes in the survival distribution after that last censored observation do not effect the likelihood of the observed data.
A similar and much more common non-uniqueness problem occurs with interval-censored data. Consider a simple case where every subject is assessed at day 0, day 7, day 14, and so on, for every 7 days until the study ends at 20 weeks and suppose that at least one failure is observed every week. Then no information about the survival time between days 0 and 7 is available and if two survival functions match at days 0, 7, 14, etc. they will give the same likelihood value. For this case, we represent the class of NPMLEs of the survival function by the probability mass associated with the intervals (0, 7], (7, 14], …, (133, 140]. This class does not represent one unique survival function, but any survival function within the class will maximize the likelihood. The interval package represents the class of NPMLEs of the survival distribution by putting gray rectangles in the area where the function is not unique (see Section 4). This type of non-uniqueness is called representational non-uniqueness by Gentleman and Vandal (2002). For ease of exposition (and in line with most of the literature) we will call the class of NPMLEs of the distribution ‘the’ NPMLE of the distribution.
For studies with irregular assessment times between individuals, we also represent the NPMLE of the survival distribution as a vector of probability masses and a set of intervals on which the masses are distributed; however, the determination of that set of intervals is not as obvious. Turnbull (1976) showed that the NPMLE of the survival distribution is only unique up to a set of intervals, which may be called the innermost intervals (these intervals are known also as the Turnbull intervals, or the regions of the maximal cliques, see Gentleman and Vandal 2001). Turnbull (1976) showed how the NPMLE can then be found by the self-consistent algorithm, which is a special case of the E-M algorithm. Convergence of the E-M algorithm does not guarantee convergence to the global maximum (see e.g., Tanner 1996); however, Gentleman and Geyer (1994) showed that for interval-censored data, a self-consistent estimate (i.e., an estimate that results at convergence of the self-consistent algorithm) is the NPMLE if it meets certain conditions, the Kuhn-Tucker conditions. Gentleman and Geyer (1994) proposed a “polishing algorithm” whereby we provisionally set the estimated probability mass in some intervals to zero if they are below some bound, then check the Kuhn-Tucker conditions to make sure that those values are truly zeros at the NPMLE. If those conditions are not met then a small probability is added back on and the E-M iterations continue. Convergence may be defined when the maximum reduced gradient is less than some minimum error, and the Kuhn-Tucker conditions are met (see Gentleman and Geyer 1994). For univariate interval-censored data, once the innermost intervals (i.e., regions of the maximal cliques) are determined, the probability assigned to those intervals which maximizes the likelihood is unique (see e.g., Gentleman and Vandal 2002, Lemma 4). For bivariate interval-censored data, this uniqueness of probabilities may not hold and that situation is called mixture non-uniqueness (see Gentleman and Vandal 2002).
There are many other algorithms for calculating the NPMLE, and the Icens package provides many of the different algorithms described in Gentleman and Vandal (2001). The default calculation of the NPMLE in the interval uses the E-M algorithm with the polishing algorithm of Gentleman and Geyer (1994) after calculating an initial estimator of the NPMLE using the computeMLE function of the MLEcens package. Although MLEcens was designed for bivariate interval-censored data, once the data are reduced to the set of maximal cliques, the calculation is the same as for the univariate interval-censored case. The optimization step (going from the set of maximal cliques to the NPMLE) in MLEcens uses the support reduction algorithm of Groeneboom, Jongbloed, and Wellner (2008). The advantage of the initial estimator is speed, and for completeness in interval the Kuhn-Tucker conditions are checked.
2.3. Overview of weighted logrank tests
For right-censored data, the logrank test is a score test for the equality of survival distributions under the proportional hazards model, thus it is an efficient test when the proportional hazards assumption holds. There are several different versions of the logrank test that have been developed (see Kalbfleisch and Prentice 1980). In particular, the likelihood could be the marginal likelihood of the ranks, the partial likelihood, or the grouped continuous model. Further, the variance could be estimated by the Fisher’s information from the likelihood, by martingale methods (see Fleming and Harrington 1991) or by permutation methods. The differences between the several different versions of the logrank test are often not a focus of applied statisticians; however, in this paper since we are emphasizing validation of software, these slight differences need to be considered to avoid confusion and will be discussed in detail in later sections (see e.g., Callaert 2003).
In addition to the logrank test, which is a WLRT with constant weight of 1 (or approximately 1), an important WLRT is the one that generalizes the Wilcoxon-Mann-Whitney test. We will call these latter tests Wilcoxon-type tests, but they are known by other names (e.g., Prentice-Wilcoxon test, proportional odds model, Harrington-Fleming Gρ class with ρ = 1). Similar to the logrank test, the Wilcoxon-type tests also have been derived using different likelihoods and using different variances. The important point for the applied researcher is that the Wilcoxon-type tests emphasize early differences in distributions (when there are more people at risk) more than the later differences (when there are fewer people at risk), while the logrank test gives constant (or near constant) weights when the test is written in the weighted logrank form (see Equation 6), which implies comparatively more weight to later differences than the Wilcoxon-type test.
We now summarize the next two subsections which detail the extension of logrank tests to interval-censored data. Both likelihoods that may be applied to interval-censored data, the likelihood under the grouped continuous model (LGCM) and the marginal likelihood of the ranks (MLR), should give similar answers. The permutation form of the tests are generally preferred over the score test forms when using the LGCM, since permuting allows exact inference when the censoring is not related to the covariate (e.g., treatment), and the permutation results avoid theoretical problems of the score test (see below and Fay 1996). When the censoring is related to treatment and there are few inspection times compared to the number of subjects, the usual score test is recommended since it is asymptotically valid in this case. Now we give some more details on the different tests for interval-censored data.
2.4. Choosing the likelihood for WLR tests
There has not been a single obvious approach for creating a likelihood to use for interval-censored data. Self and Grosman (1986) used the marginal likelihood of the ranks (MLR). This has the advantage that we need not estimate the baseline distribution (or equivalently the baseline hazard). The disadvantage of the MLR is that it is difficult to calculate. Note that even in the right-censored case with ties, the likelihood is usually only approximated (see Kalbfleisch and Prentice 1980, pp. 74{75). Satten (1996) introduces a stochastic approximation to the MLR using Gibbs sampling for the proportional hazards model and it is generalized to proportional odds and other models by Gu, Sun, and Zuo (2005).
Finkelstein (1986) (see also Fay 1996, 1999a) used the likelihood under the grouped continuous model (LGCM). In the LGCM, we estimate a baseline distribution, which is a monotonic function estimated at each observation point, and the function’s value at each of those points is a nuisance parameter that must be estimated. Because there are so many nuisance parameters and the number of them may depend on the sample size, the standard likelihood-based tests (i.e., score test, Wald test, and likelihood ratio test) may not follow the usual theory (see Fay 1996). Note, however, that the permutation test formed from the scores of the LGCM is theoretically justified and is known to be a valid test when the censoring is unrelated to the covariate (see the following section). We discuss the computational issues of the LGCM in the next section. For the non-censored case, Pettitt (1984) studied the two likelihoods and showed that both likelihoods give asymptotically equivalent score tests as long as either the number of categories of response is fixed, or the number of categories does not increase too quickly compared to the total sample size. Pettitt concluded (see Pettitt 1984, section 5.1) that the score test for the MLR was more efficient (i.e., had greater power) than the score test for the LGCM; however, Pettitt did not consider the permutation form of the test using the LGCM.
Finally, when imputation methods are used then martingale methods may be used (see Huang et al. 2008, and below).
The interval package allows the user to choose between the LGCM and imputation/martingale likelihood methods through the score option within ictest, as will be demonstrated in section 4. The MLR is not supported within the interval package at this time.
2.5. Choosing the inferential method for WLR tests
Once the likelihood is chosen, and the associated efficient score (the first derivative of the loglikelihood with respect to the parameter of interest evaluated under the null, i.e., the U in Equation 5 below) is calculated, then the distribution of that score under the null must be estimated so that the p-value corresponding to the test statistic can be calculated. There are several methods for doing this, but the three most common are using asymptotic methods with the observed Fisher’s information, which is commonly known as the score test, using permutation methods, or using multiple imputation (Huang et al. 2008).
When the censoring mechanism is the same for all treatment groups, the permutation test is known to be valid for either the MRL or the LGCM. In this case of equal censoring, the score test is only known to be asymptotically valid using the MRL; using the LGCM we require the additional assumption that the number of observation times remains fixed as the sample size goes to infinity (see Fay 1996, for a discussion of this issue).
When there is unequal censoring then the theory for the permutation method is not formally met. Thus, we have previously suggested that with unequal censoring the score variance is better (see e.g., Fay 1996, p. 820 for the interval-censoring case). Further work needs to be done to explore unequal interval-censoring; however, we can get some assurance for the practical use of the permutation method from the special case of right-censored data, where it has been shown through simulation that the permutation method usually controls the type I error except in extreme cases of unequal censoring and very unbalanced sample sizes between groups (Heinze, Gnant, and Schemper 2003). Heinzeet al. (2003) also developed an imputation method that controlled the type I simulated error for all cases, and we discuss other related imputation methods applied to interval-censored data next.
Another strategy to create WLRT for interval-censored data is to impute right-censored data from the interval-censored data and then properly adjust the variance. Huanget al. (2008) improved on some earlier attempts at this variance adjustment after imputation. This appears to be a reasonable strategy, and provides an independent check on the other methods. Since this imputation method is closely related to the within-cluster resampling of Hoffman, Sen, and Weinberg (2001), we use ‘wsr’ (for within subject resampling) to label these methods in the interval package. On each imputation Huanget al. (2008) only considered the usual martingale derived variance (use method = “wsr.HLY” in ictest), while the interval package additionally allows for permutational variance (method = “wsr.pclt”) and Monte Carlo estimation within each imputation (method = “wsr.mc”).
2.6. Regression in interval-censored data
This section is provided for completeness, but these methods are not a part of the interval package.
For parametric methods, it is straightforward to form the likelihood for interval-censored data under the accelerated failure time model and standard likelihood based methods may be applied (see Equation 1). These methods are provided in the survival package using the survreg function (Therneau and Lumley 2009). For right-censored data a more common regression method is the semi-parametric Cox proportional hazards regression. In this model the baseline hazard function is completely nonparametric, but does not need to be estimated. The score test from this model is the logrank test. The generalization of the model to interval-censored data typically uses the marginal likelihood of the ranks (see Satten 1996 ; Goggins, Finkelstein, and Zaslavsky 1999). The only available software for doing these models of which we are aware is an S function (which calls a compiled C program requiring access to a SPARC based workstation) to perform a Monte-Carlo EM algorithm for proportional hazards models described in Gogginset al. (1999) (Goggins 2007). Another approach to semi-parametric modeling is to specifically estimate the non-parametric part of the model with a piecewise constant intensity model (see Farrington 1996 ; Carstensen 1996). This is the approach taken with the Icens function in the Epi package (Carstensen, Plummer, Laara, and et. al. 2010).
3. Mathematical formulation of the scores for the WLRT
In this section, we provide the general form of rank invariant score test on the grouped continuous model, and for each of the three main rank scores available within ictest: those from the logistic (Sun 1996), the group proportional hazards (Finkelstein 1986), and the generalized Wilcoxon Mann Whitney (Fay 1996) models. In the details that follow, we briefly describe the underlying survival model (or hazard model) and the mathematical form of the individual scores. Further details on the derivation of the tests are given in Fay (1996) and Fay (1999a). The other rank scores available in interval are also described briefly.
Suppose we have n subjects. For the ith subject, use the following notation:
xi is the time to event, Xi is the associated random variable.
Li is the largest observation time before the event is known to have occurred.
Ri is the smallest observation time at or after the event is known to have occurred. In other words, we know that xi ∈ (Li, Ri]. (Note that interval allows the endpoints of each interval to be either included or excluded using Lin and Rin options, but for ease of explanation we assume the pattern just described.) We allow Ri = ∞ to denote right-censoring.
zi is a k × 1 vector of covariates.
Let the ordered potential observation times be 0 = t0 < t1 < t2 < ··· < tm < tm+1 = ∞. Partition the sample space by creating (m + 1) intervals, with the jth interval denoted Ij ≡ (tj−1, tj]. For simplicity, we assume that Li, Ri ∈ {t0, …, tm+1}. Let
We write the general model of the survival for the ith individual as
where β is a k × 1 vector of treatment parameters, and γ is an m ×1 vector of nuisance parameters for the unknown survival function.
In the interval package, there are several different ways we can model . Most of these ways use a model closely related to the accelerated failure time (AFT) model. Thus, we begin by defining the AFT model, where the time to event for the ith subject, Xi, is modeled as
| (1) |
where μ and σ are location and scale parameters and the distribution of εi is known to be F. In the grouped continuous model, we replace the log transformation with g(·), an unknown monotonic transformation that absorbs the μ and σ parameters, to get:
| (2) |
where here also εi ~ F and F is some known distribution (e.g., logistic, normal). Then the model of the survival distribution at time t is
| (3) |
and in the grouped continuous model, g(·) is described at all the places where the likelihood may change (i.e., at t1, …, tm) by the vector of nuisance parameters, γ.
The grouped continuous likelihood for interval-censored data is
| (4) |
To form the score statistic we take the derivative of log(L) with respect to β and evaluate it at β = 0. The MLE of the nuisance parameters when β = 0 (in terms of the baseline survival) are the NPMLE of survival, Ŝ(tj), j = 1, …, m. For convenience, let Ŝ(t0) = 1 and Ŝ(tm+1) = 0, even though these values are known by assumption.
We can write the efficient score vector for the parameter β (see Fay 1996, 1999a) as
| (5) |
where Ŝ′(t) is the derivative with respect to β evaluated at β = 0 and at g(t) = F−1(1−Ŝ(t)), i.e., Ŝ′(t) = f[F−1{1 − Ŝ(t)}] and f and F−1 are the density and quantile functions of F respectively.
When zi is an k ×1 vector of indicators of k treatments, we can rewrite the ℓth row of U as
| (6) |
where
and represents the expected value under the null of the number of deaths in Ij for the ℓth treatment group, represents the expected value under the null of the total number of deaths in Ij, similarly and represent the expected number at risk.
We now give the values for ci (from Equation 5) and wi (from Equation 6) for some different survival models provided in ictest. Although not developed first, we present the model of Sun (1996) first because it is the generalization of the logrank test most commonly used for right-censored data. Sun (1996) modeled the odds of discrete hazards as proportional to (see Fay 1999a), leading to the more complicated survival function:
Here and in the other two models, S(tj|γ) is a estimator of survival that does not depend on the covariates zi, and S(tj|γ) is nonparametric because the γ is m ×1 and there are only m unique time points observed in the data. Denote its estimator S(t|γ̂) ≡ Ŝ(t), which is the NPMLE of the survival function of all the data ignoring covariates. Under the model of Sun (1996) we get,
| (7) |
where , and λℓ = {Ŝ(tℓ−1) − Ŝ(tℓ)}/Ŝ(tℓ−1), and
This model is called from the interval package by the option scores = “logrank1”.
The second model we consider was actually developed first, it is the grouped proportional hazards model introduced by Finkelstein (1986), where the survival function is modeled as . This comes from the model of Equation 2 when F is the extreme minimum value distribution. Under this grouped proportional hazards model, the ci values are:
| (8) |
and
Note that because this model makes a proportional hazards assumption, we call the resulting test a logrank test also and the model is called by scores = “logrank2” in the interval package. When Ŝ(tj−1)/Ŝ(tj) ≈ 1 then wj ≈ 1.
Next, consider the model proposed by Peto and Peto (1972) (see Fay 1996) giving the Wilcoxon-type test, where the odds are proportional to exp(−ziβ) so that the survival function is
and we get
and
This comes from the model of Equation 2 when F is the logistic distribution. Since in the absence of censoring the resulting test reduces to the Wilcoxon-Mann-Whitney test, we call this model from the interval package by the option scores = “wmw”.
Other scores are possible (but less often used) from the model of Equation 2 by choosing different distributions for F. The user may specify that F is normal with scores = “normal”, or may supply an arbitrary distribution by using scores = “general” and specifying the function, f {F−1(·)} (see Equation 5), using the dqfunc option.
For illustrative purposes, we now give the form of the three most often used scores in the special case of right-censoring. For this, we introduce new notation. Suppose that there are m* observed failure times, . In other words there are m* subjects for which xi = Ri is known, so that Li = limε→0 Ri −ε ≡ Ri −0. Let nj and dj be the number of subjects who are at risk or fail respectively at . Then the Kaplan-Meier survival estimate is (see e.g., Kalbfleisch and Prentice 1980)
In Table 1 we summarize the formulation of the scores for the three model (score) choices in interval, as described above, for ordinary right-censored data.
Table 1.
Scores for right-censored data.
| Test (Model) | Score (ci) for Observed failure at | Score (ci′) for Right-censored observation at | ||
|---|---|---|---|---|
| Logrank1 (Logistic, Sun) |
|
|
||
| Logrank2 (Group Proportional Hazards, Finkelstein) |
|
|
||
| Generalized WMW (Proportional Odds) |
|
|
4. Application
The calls to the interval package are designed to be in the same format as in the survival package. As noted in the previous section, the icfit and ictest functions will also work on right-censored data (see demo(“right.censored.examples”)).
We demonstrate the two main functions in interval, icfit and ictest, with the commonly used interval-censored breast cosmesis data set of Finkelstein and Wolfe (1985). The data are from a study of two groups of breast cancer patients, those treated with radiation therapy with chemotherapy (treatment = “RadChem”) and those treated with radiation therapy alone (treatment = “Rad”). The response is time (in months) until the appearance of breast retraction, and the data are interval-censored between the last clinic visit before the event was observed (left) and the first visit when the event was observed (right) (or Inf if the event was not observed). The following provides the first few observations of the data set.
R> library(“interval”) R> data(“bcos”, package = “interval”) R> head(bcos) left right treatment 1 45 Inf Rad 2 6 10 Rad 3 0 7 Rad 4 46 Inf Rad 5 46 Inf Rad 6 7 16 Rad
4.1. Survival estimation
First, we calculate the NPMLE for each treatment group in the breast cosmesis data separately.
R> fit1<-icfit(Surv(left, right, type = “interval2”)~treatment, data = bcos) R> summary(fit1) treatment=Rad: Interval Probability 1 (4,5] 0.0463 2 (6,7] 0.0334 3 (7,8] 0.0887 4 (11,12] 0.0708 5 (24,25] 0.0926 6 (33,34] 0.0818 7 (38,40] 0.1209 8 (46,48] 0.4656 treatment=RadChem: Interval Probability 1 (4,5] 0.0433 2 (5,8] 0.0433 3 (11,12] 0.0692 4 (16,17] 0.1454 5 (18,19] 0.1411 6 (19,20] 0.1157 7 (24,25] 0.0999 8 (30,31] 0.0709 9 (35,36] 0.1608 10 (44,48] 0.0552 11 (48,60] 0.0552
The Surv function is from the survival package, and the type = “interval2” treats the variables left and right as the left and right endpoints of the interval. The assumption is that the left interval is excluded but the right one is included, except that if both are equal then both are included, and values of left may be 0 and values of right may be Inf. One can change the inclusion/exclusion defaults by using the Lin and Rin options.
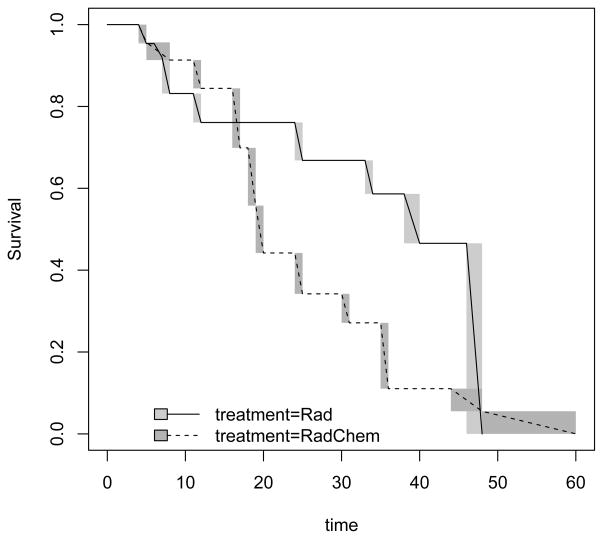
These results match those calculated from Icens, an available package for computing the NPMLE for censored and truncated survival data (see Gentleman and Vandal 2001 ). The summary function applied to an “icfit” object gives the intervals with positive probability for the NPMLE of the survival distribution function, i.e. where the estimated survival distribution drops; however, there are infinitely many functions which drop exactly the same increment within those intervals. The NPMLE is only unique outside of the intervals which are listed from the summary of the fit. For example, there are infinitely many survival functions for the treatment = “Rad” group, that have S(4) = 1 and S(5) = 1 − 0.0463 = 0.9537. Thus, as has been done in the Icens package, when plotting the NPMLEs we denote the areas with the indeterminate drops with grey rectangles. The function which linearly interpolates the survival within these indeterminate regions is also displayed on the graph. We plot the NPMLE for each treatment group using plot(fit1) to get Figure 4.1.
Figure 1.
Non-parametric maximum likelihood survival from breast cosmesis data.
4.2. Two-sample weighted logrank tests
There are five score tests available in ictest, which are selected by setting the scores argument to be one of “logrank1”, “logrank2”, “wmw”, “normal”, or “general”. As stated in Section 3, the two forms of the logrank scores are those associated with Finkelstein (1986) and those associated with Sun (1996). Although Finkelstein (1986) are perhaps more natural for interval-censored data, we make those of Sun (1996) the default (scores = “logrank1” or equivalently rho = 0) since these scores reduce to the usual logrank scores with right-censored data. The default method is the permutation test, and since the sample size is sufficiently large we automatically get the version based on the permutational central limit theorem:
R> icout<-ictest(Surv(left, right, type = “interval2”)~treatment, data = bcos)
R> icout
Asymptotic Logrank two-sample test (permutation form),
Sun’s scores
data: Surv(left, right, type = “interval2”) by treatment
Z = -2.6684, p-value = 0.007622
alternative hypothesis: survival distributions not equal
n Score Statistic*
treatment=Rad 46 -9.141846
treatment=RadChem 48 9.141846
* like Obs-Exp, positive implies earlier failures than expected
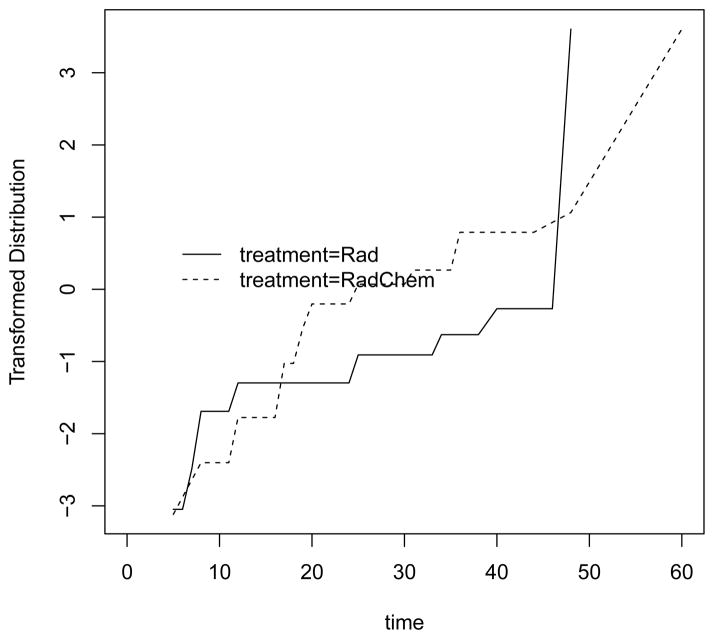
To pick which of the rank score methods is best, one may plot for each treatment group the NPMLE of the distribution transformed by the inverse of the error distribution from the grouped continuous model (see Fay 1996). In generalized linear models these transformations are known as links. For example, the proportional hazards is motivated by the extreme value error distribution whose inverse is the complementary log-log link, which is the default. In Figure 2 we plot this using the fit1 “icfit” object which contains the NPMLE for each treatment group using the code: plot(fit1, dtype = “link”). Other links besides the default complementary log-log link are possible: dlink = qlogis or dlink = qnorm for the fit of the proportional odds and normal model, respectively. Fay (1996) presents those plots for these data and none of the models looks clearly better than the others.
Figure 2.
Complementary log-log transformation of distribution from breast cosmesis data. If parallel then proportional hazards is a good model.
Because a major part of the calculation of the test statistic is estimating the NPMLE under the null hypothesis (i.e., for the pooled treatment groups), this NPMLE is saved as part of the output (icout$fit, in the above example) so that we can calculate this NPMLE once and reuse it for the calculation of the other two score tests. Here is code for the Finkelstein logrank formulation, which takes advantage of a precalculated NPMLE:
R> ictest(Surv(left, right, type = “interval2”)~treatment, data = bcos,
+ initfit = icout$fit, scores = “logrank2”)
Asymptotic Logrank two-sample test (permutation form),
Finkelstein’s scores
data: Surv(left, right, type = “interval2”) by treatment
Z = -2.6839, p-value = 0.007277
alternative hypothesis: survival distributions not equal
n Score Statistic*
treatment=Rad 46 -9.944182
treatment=RadChem 48 9.944182
* like Obs-Exp, positive implies earlier failures than expected
Notice how the two different logrank tests give very similar results, p close to 0.007 in both cases. We demonstrate the third score test, the generalization of the Wilcoxon-Mann-Whitney scores to interval-censored data, and also demonstrate the ictest function in default mode:
R> L<-with(bcos, left)
R> R<-with(bcos, right)
R> trt<-with(bcos, treatment)
R> ictest(L, R, trt, scores = “wmw”, initfit = icout$fit)
Asymptotic Wilcoxon two-sample test (permutation form)
data: {L,R} by trt
Z = -2.1672, p-value = 0.03022
alternative hypothesis: survival distributions not equal
n Score Statistic*
Rad 46 -5.656724
RadChem 48 5.656724
* like Obs-Exp, positive implies earlier failures than expected
4.3. K-sample and trend tests
We can perform k-sample tests using the ictest function. We create fake treatment assignments with four treatment groups for the individuals in the breast cosmesis data set to demonstrate.
R> fakeTrtGrps<-sample(letters[1:4], nrow(bcos), replace = TRUE)
R> ictest(L, R, fakeTrtGrps)
Asymptotic Logrank k-sample test (permutation form),
Sun’s scores
data: {L,R} by group
Chi Square = 1.3685, p-value = 0.7129
alternative hypothesis: survival distributions not equal
n Score Statistic*
d 27 -0.7520431
b 24 1.8821766
c 20 -2.8048270
a 23 1.6746934
* like Obs-Exp, positive implies earlier failures than expected
When scores = “wmw” and the responses are all non-overlapping intervals then this reduces to the Kruskal-Wallis test.
The function ictest performs a trend test when the covariate is numeric. The one-sided test with alternative = “less” rejects when the correlation between the generalized rank scores (e.g., WMW scores or logrank scores) and the covariate are small. Below, a continuous covariate z that is uncorrelated to the outcome is created for individuals in the cosmesis dataset to illustrate the trend test.
R> fakeZ<-rnorm(nrow(bcos))
R> ictest(L, R, fakeZ, alternative = “less”)
Asymptotic Logrank trend test(permutation form), Sun’s
scores
data: {L,R} by fakeZ
Z = 0.068, p-value = 0.5271
alternative hypothesis: higher fakeZ implies earlier event times
n Score Statistic*
[1,] 94 0.4421144
* postive so larger covariate values give earlier failures than expected
4.4. Exact permutation tests
We can also estimate the exact permutation p-value for any score choice in ictest using the exact argument. Here the logrank test using Sun (1996) scores is redone as an exact test:
R> ictest(Surv(left, right, type = “interval2”)~treatment, data = bcos, exact = TRUE, scores = “logrank1”)
Exact Logrank two-sample test (permutation form), Sun’s
scores
data: Surv(left, right, type = “interval2”) by treatment
p-value = 0.006
alternative hypothesis: survival distributions not equal
n Score Statistic*
treatment=Rad 46 -9.141846
treatment=RadChem 48 9.141846
* like Obs-Exp, positive implies earlier failures than expected
p-value estimated from 999 Monte Carlo replications
99 percent confidence interval on p-value:
0.0002072893 0.0184986927
The exact argument automatically chooses between an exact calculation by network algorithm or an approximation to the exact p-value by Monte Carlo through the methodRuleIC1 function. In this case the network algorithm was expected to take too long and the Monte Carlo approximation was used. If a more accurate approximation to the exact p-value is needed then more Monte Carlo simulations could be used and these are changed using the mcontrol option. Additionally, if icout is an “ictest” object created by the ictest function, then icout$scores will give the vector of rank scores, ci, which may be imputed into other software (e.g., StatXact) to create an exact permutation test. A more seamless interaction is possible with the coin package (see Section 5.3).
4.5. Other test options
All of the above are permutation based tests, but we may use other methods. Here are the results from the usual score test for interval-censored data:
R> ictest(Surv(left, right, type = “interval2”)~treatment, data = bcos, initfit = icout$fit, method = “scoretest”, scores = “logrank2”)
Asymptotic Logrank two-sample test (score form),
Finkelstein’s scores
data: Surv(left, right, type = “interval2”) by treatment
Chi Square = 7.8749, p-value = 0.005012
alternative hypothesis: survival distributions not equal
n Score Statistic*
treatment=Rad 46 -9.944182
treatment=RadChem 48 9.944182
* like Obs-Exp, positive implies earlier failures than expected
where in this case the nuisance parameters are defined after calculation of the NPMLE as described in Fay (1996). The results agree exactly with Fay (1996) and are similar to those in Finkelstein (1986). The very small differences may be due to differing convergence criteria in the NPMLE. The imputation method of Huang et al. (2008) may also be employed (note that scores = “logrank2”, “normal”, or “general” are not available for this method):
R> icoutHLY<-ictest(Surv(left, right, type = “interval2”)~treatment, data = bcos, initfit = icout$fit, method = “wsr.HLY”, mcontrol = mControl(nwsr = 99), scores = “logrank1”)
R> icoutHLY
Asymptotic Logrank 2-sample test(WSR HLY), Sun’s scores
data: Surv(left, right, type = “interval2”) by treatment
Chi Square = 7.1047, p-value = 0.007688
alternative hypothesis: survival distributions not equal
n Score Statistic*
treatment=Rad 46 -9.141846
treatment=RadChem 48 9.141846
* like Obs-Exp, positive implies earlier failures than expected
p-value estimated from Monte Carlo replications
These results agree with Huang et al. (2008) within the error to be expected from such an imputation method (Huang et al. 2008, had p = 0.0075).
For the breast cosmesis data, if we can assume that the assessment times are independent of treatment arm, then the assumptions needed for the permutation methods and the imputation methods are met. The assumptions for the theoretical use of the score function do not hold because the NPMLE is on the boundary of the parameter space since some masses were set to zero in the calculation of the NPMLE (although the ad hoc adjustment appears to give reasonable results). When some of these masses are set to zero then the anypzero element of the “icfit” object will be TRUE, as we see here:
R> icoutHLY$fit$anypzero [1] TRUE
If we cannot assume independence of assessment times and treatment arm then the exchange-ability assumption needed for the permutation method is not met, and the imputation method may be used. Further research is needed on the practical ramifications of the violation of any of these assumptions.
5. The perm package
The default method for inference in the interval package is the permutation test. The perm package presented here is a stand-alone R package that performs linear permutation tests. The tests that can be done in perm can also be done in the existing coin package; however, there are slight differences in the calculations and presentation, as outlined below.
5.1. Overview of methods
The perm package does linear permutation tests, meaning permutation tests where the test statistic is either of the form,
| (9) |
as in Equation 5, or of a quadratic version of T(y, x) (e.g., see k-sample tests below). We consider only the case where ci is a scalar and zi is either a scalar or a k × 1 vector (although more general cases are studied in Sen 1985; Hothorn et al. 2006). Following Sen (1985), we can write the mean and variance of T under the permutation distribution (i.e., permute indices of c1, …, cn and recalculate T, where there are n! different permutations with each equally likely) as,
where c̄ and z̄ are the sample means.
In the perm package, if zi is a scalar we define the one-sided p-value when alternative = “greater” as
where I(A) = 1 when A is true and 0 otherwise, Ti is the test statistic for the ith of n! possible permutations, and T0 is the observed value of T. When alternative = “less” then the p-value, say pL, is given as above except we reverse the direction of the comparison operator in the indicator function. Note that if you add or multiply by constants which do not change throughout all permutations then the one-sided p-values do not change. Thus, a permutation test on T can represent a test on the difference in means in the two-sample case, and can represent a test on the correlation when zi is numeric. When alternative = “two.sided”, then there are two options of how to calculate the p-value defined by the tsmethod option of the control variable. When tsmethod = “central”, the two-sided p-value is p2, twice the minimum one-sided p-value (i.e., p2 = min(1, 2 min(pL, pG))), and when tsmethod = “abs” then the two-sided p-value is
When zi is a k × 1 vector, we consider only the alternative = “two.sided” (in this case both tsmethod options give the same result), and the Wald-type test statistic Q = (T − U)⊤V−(T − U) is calculated, where V− is the generalized inverse of V.
To calculate the exact p-values, one may use complete enumeration, but this quickly becomes intractable and other algorithms are needed. One algorithm supplied is the network algorithm (see e.g., Agresti, Mehta, and Patel 1990). Monte Carlo approximations to the exact p-values may also be performed. Finally, the perm allows asymptotic methods such as the permutational central limit theorem (PCLT). Sen (1985) reviews the PCLT which shows that under the permutation distribution with standard regularity conditions on the ci and zi, V−1/2(T − U) is asymptotically approximately multivariate normal with mean 0 and variance the identity matrix.
Note that because of the way exact p-values are defined, minuscual differences in the way the computer treats ties between T0 and the Ti can lead to non-negligible differences in the calculated exact p-values for small samples. This is a problem for all permutation test software, but because of the generality of the perm package (i.e., the Ti can be any values) it can be a particularly difficult one. The solution taken by perm and by coin (Versions 1.0-8 or later) is to round so that differences between Ti and T0 that are close to zero become zero. In perm this is done with the digits option, which by default rounds the Ti to the nearest 12 digits. This can particularly be a problem for WLRTs, as is shown by the example in Section 6.3.
5.2. Design and implementation
The three main functions in perm perform the two-sample (permTS), k-sample (permKS), and trend (permTREND) tests. To demonstrate the package we will use the ChickWeight data set in the datasets package which is part of the base distribution of R. The data are from an experiment on chicks fed one of four diets. We use only the weight in grams of the chicks at day 21 (i.e., Time == 21) as the response. For the two-sample examples that follow, we use only the chicks given diets 3 and 4.
R> data(“ChickWeight”, package = “datasets”) R> head(ChickWeight) weight Time Chick Diet 1 42 0 1 1 2 51 2 1 1 3 59 4 1 1 4 64 6 1 1 5 76 8 1 1 6 93 10 1 1
The package uses the S3 methods, which allow object-oriented programming. The evaluation of any of the three main functions is determined by the class of the first object in the function call. This is similar to the calls to the analogous functions in the stats package. For example, as is possible using t.test or wilcox.test from the stats package, we can call the test using the formula structure, which automatically uses the permTS.formula function. The formula is weight~Diet where the ith element of weight represents ci and the ith element of Diet represents zi in Equation 9. We also use the data and subset variables to name the data set and pick out only the Day 21 weights of those chicks who got Diets 3 or 4.
R> permTS(weight~Diet, data = ChickWeight, subset = Diet %in% c(3, 4) & Time == 21)
Permutation Test using Asymptotic Approximation
data: weight by Diet
Z = 1.1412, p-value = 0.2538
alternative hypothesis: true mean Diet=3 - mean Diet=4 is not equal to 0
sample estimates:
mean Diet=3 - mean Diet=4
31.74444
Equivalently, we can define the responses explicitly and do the same test, with the default structure,
R> y3<-with(subset(ChickWeight, Time == 21 & Diet == 3), weight)
R> y4<-with(subset(ChickWeight, Time == 21 & Diet == 4), weight)
R> permTS(y3, y4)
Permutation Test using Asymptotic Approximation
data: y3 and y4
Z = 1.1412, p-value = 0.2538
alternative hypothesis: true mean y3 - mean y4 is not equal to 0
sample estimates:
mean y3 - mean y4
31.74444
The permTS uses a function (determined by the option methodRule) to automatically choose the method used in the permutation test. Since in this case the exact network method is not expected to give an quick answer, the default methodRule automatically chooses to use the PCLT. If the sample sizes are small enough, then exact methods are done automatically. For example, using only the first 5 chicks on each diet, then the methodRule function chooses the network algorithm:
R> permTS(y3[1:5], y4[1:5])
Exact Permutation Test (network algorithm)
data: y3[1:5] and y4[1:5]
p-value = 0.1825
alternative hypothesis: true mean y3[1:5] - mean y4[1:5] is not equal to 0
sample estimates:
mean y3[1:5] - mean y4[1:5]
60.6
Note that the network algorithm is written entirely in R code, so efficiency gains may be possible by translating portions of the code into C code (see e.g., Chambers 2008).
In addition to the pclt and exact.network methods, the two-sample test additionally has both a complete enumeration exact algorithm (method = “exact.ce”), which is useful for simulations involving a small number of observations in each group, and a Monte Carlo approximation to the exact p-value (method = “exact.mc”). The user may supply their own methodRule function, which must have three input values: the numeric vectors c = [c1, …, cn] and z = [z1, …, zn] (see Equation 9), and the logical variable exact given by the option of the same name. For the two-sample test the output of a methodRule function should be a character vector which is one of “pclt”, “exact.network”, “exact.ce”, or “exact.mc”. The logical variable exact causes the default methodRule for permTS to choose from among the three exact algorithms based on the estimated speed of the calculations (see help for methodRuleTS1).
All methods except “exact.mc” produce an “htest” object, which is a list with elements described in the help for permTS and printed according to the print method from the stats package. The “exact.mc” method creates an “mchtest” object, which additionally prints out confidence intervals on the p-value based on the Monte Carlo replications to help the users determine if the p-value would change much if the Monte Carlo procedure was repeated with a different random seed. Note that even if the confidence interval on the p-value is large, the given p-value from the “exact.mc” method (i.e., the quantity one plus the number of Monte Carlo replications that are equal to or more extreme than the observed test statistic divided by the quantity one plus the number of replications) is always a valid p-value (see e.g., Fay, Kim, and Hachey 2007, equation 5.3).
For the k-sample test the calls may also be made by formula structure,
R> permKS(weight~Diet, data = ChickWeight, subset = Time == 21)
K-Sample Asymptotic Permutation Test
data: weight by Diet
Chi Square = 11.1786, df = 3, p-value = 0.0108
or by explicit definition of the response (c) and group (z) vectors,
R> y<-ChickWeight[ChickWeight$Time == 21, “weight”]
R> g<-ChickWeight[ChickWeight$Time == 21, “Diet”]
R> permKS(y, g)
K-Sample Asymptotic Permutation Test
data: y and g
Chi Square = 11.1786, df = 3, p-value = 0.0108
The methodRule function works the same way as for permTS except a different default methodRule function is used since the exact.network method is not available for the k-sample test.
If we can assume for illustration that the diets are inherently ordered, then we may want to use the trend test. For the trend test (i.e., when zi is a scalar) the calls may also be made by formula structure (not shown), or the default,
R> permTREND(y, as.numeric(g))
Permutation Test using Asymptotic Approximation
data: y and COVARIATE
Z = 2.7879, p-value = 0.005305
alternative hypothesis: true correlation of y and COVARIATE is not equal to 0
sample estimates:
correlation of y and COVARIATE
0.4202893
Similar methodRule functions may be used (see help for permTREND).
Options for the algorithm methods are controlled by the variable control, and the function permControl allows changing of only a subset of the options. The output from permControl is a list of all the options. Most of the options pertain to the “exact.mc” method: nmc gives the number of Monte Carlo replications, p.conf.level gives the confidence level calculated on the p-value, seed gives the random number seed, and setSEED is a logical telling whether or not to set the seed. Other options are: digits adjusts the rounding of the test statistics to decide on which to call ties (see Section 6.3), tsmethod gives two options for the two-sided method for calculating the p-values (see Section 5.1 above). The last option is cm and is used in the following scenario. Suppose one wanted to do a simulation on data of the same size as the example. Then one could calculate the complete enumeration matrix once (using the chooseMatrix function), and then each simulation could reuse that matrix. This will save time as illustrated below on the ChickWeight data:
R> system.time(cm19c10<-chooseMatrix(length(y3)+length(y4), length(y3))) user system elapsed 1.95 0.00 1.95 R> system.time(PC<-permControl(cm = cm19c10)) user system elapsed 0.13 0.00 0.13 R> system.time(permTS(y3, y4, method = “exact.ce”, control = PC)) user system elapsed 0.03 0.00 0.03 R> system.time(permTS(y3, y4, method = “exact.network”)) user system elapsed 1.43 0.03 1.47
In a simulation, the first calculation only needs to be done once.
5.3. Comparison with coin package
This section compares the two permutation packages, coin (version 1.0-8) and perm (version 1.0-0.0).
The primary motivation for the creation of the perm package is for an independent, within R, validation of the coin package. All checks between coin and perm have shown no differences (see perm.coin.check.R in the test directory). In many ways, coin is the more comprehensive and general package of the two. For example, coin allows the following not supported in perm: user supplied transformations on the responses and covariates, other (nonlinear) test statistics, stratification, and multiple responses and covariates. Further, the exact algorithms in coin are faster than those in perm.
Here are some minor ways that perm is different from coin. First, the print method for coin prints a standardized Z statistic to show direction of the effect, while the print method for perm prints the difference in means and only additionally prints the Z statistic when the PCLT is used. Second, the perm package allows methodRule functions, as previously described, to automatically choose the calculation method. No similar feature is offered in coin. Third, when using the Monte Carlo approximation to the exact (method = “exact.mc” in perm and distribution = approximate() in coin), then perm prints confidence intervals automatically with the print method, while coin prints them only when using the pvalue function. Fourth, the perm package allows the “exact.ce” method, which does complete enumeration. If a simulation is desired for the two-sample test on a small sample size, then using the “exact.ce” method with the cm variable fixed by the control option (so that it does not need to be recalculated for each simulation) may give faster results than other algorithms. Fifth, the perm package allows two types of two-sided p-values (see permControl: option tsmethod = “central” (default) and tsmethod = “abs”), while the coin allows only one type of alternative (denoted tsmethod = “abs” in perm). We emphasize that these differences are minor and when the two packages do the same analysis, both are similar.
Further extensibility of the perm package may not be needed since many of the ways it may be expanded are covered by the coin package.
6. The interval package
We have already given some examples of how to use the interval package. In this section, we give more details on the structure of the package.
6.1. Design and implementation
The interval package uses S3 methods. The two main functions are the icfit function and the ictest function. Both functions allow a formula as well as a default implementation, and both implementation styles were presented in Section 4. Although the typical response for the formula will be of the “Surv” class, from the survival package, we also allow numeric responses and these are treated as exactly observed values.
The icfit function outputs an object of class “icfit” which is a list with the elements described in the help, and with most elements exactly as in the “icsurv” class of the Icens package. The “icfit” class is different from the “icsurv” class primarily because it allows the NPMLE of the distributions for multiple strata to be stored as one “icfit” object as is possible in the “surv” class of the survival package. For example, if the right hand side of the formula contains a factor object with k factors then the resulting “icfit” object will contain k separate NPMLEs, one for each factor. In that case the strata element will be a numeric vector of size k giving the number of elements in each strata, and the other objects (e.g., the vector of probability masses, pf) will be larger to include all the separate NPMLEs. The NPMLEs are separated by stratum when either the summary or plot methods are applied to the “icfit” objects.
The available methods for “icfit” objects are print, summary, “[“ and plot. The print method prints as a list except the ‘A’ matrix (described below) is not printed, only its dimensions are given. The summary and plot methods have been shown in Section 4 and they either print or plot on one graph the NPMLE for each stratum. The “[“ method allows picking out the ith stratum from an “icfit” object.
Here are some details on the calculations in icfit. The icfit function calls a separate functio n, Aintmap, that calculates an ‘A’ matrix and the ‘intmap’. The A matrix is an n × m matrix of zeros and ones with the ijth element being an indicator of whether the interval associated with the ith observation contains the jth interval of the intmap. The intmap is a matrix which gives left and right endpoints of the intervals associated with the columns of A, and the attributes of the intmap tell whether the endpoints are included or not as determined by the Lin and Rin options. The default is to exclude the left interval and include the right (i.e., (L, R]), except when either L = R (then the intervals are treated as exact, i.e., [R, R]) or R = ∞ which is not included. Differences in the inclusion of the endpoints can change the results (see Ng 2002). When the intervals of the intmap are regions of maximal cliques then the A matrix is the transpose of the incidence or clique matrix defined in Gentleman and Vandal (2002). The Aintmap is called internally by the icfit function, and the innermost intervals (i.e., regions of maximal cliques) are calculated to possibly reduce the number of columns of the A matrix.
Once an A matrix is calculated and reduced to represent only innermost intervals, the initial estimate of the survival distribution is needed for the E-M algorithm. The initfit option controls that initial estimate. An allowable option for initfit is to provide an initial NPMLE estimate as an object of either class “icfit” or “icsurv”. Another option for initfit is a character vector giving the name of a function to calculate an initial fit. This function must have as inputs any or all of five variables (L, R, Lin, Rin, and/or A), and must output a vector of probability masses estimating the distribution and optionally it may output the corresponding intmap. The default for initfit is initcomputeMLE, a function that calls computeMLE from the MLEcens package. Note that if the initfit function, such as the initcomputeMLE function, gives an error then the icfit ignores this calculation, gives a warning, and calculates a very simple initial distribution. In the control option of icfit, other values may be passed to the initfit function through the initfitOpts element of control, and initfitOpts must be a named list of options.
Once the initial distribution is calculated, it is used as the starting value in a modified E-M algorithm that allows ‘polishing’ elements to zero, then subsequently checking the Kuhn-Tucker conditions (see Gentleman and Geyer 1994). If the initial distribution is very close to the NPMLE, then convergence may happen on the first iteration. On the other hand, the initial distribution need not be very close to the NPMLE but convergence can still happen. If the initial distribution has some intervals set to zero that should not be, then the Kuhn-Tucker conditions will not be met and the message value of the resulting “icfit” object (e.g., fit$message) will state this fact.
We test in demo(“npmle”) that the NPMLE estimates from the Icens package match those from the icfit function. In that file we compare the NPMLE from the two packages for the cosmesis data. Additionally in the demo, we simulate 30 other data sets and show that the NPMLEs match for all the simulated data sets (data not shown).
The ictest function outputs an object of class “ictest” for which there is a print method, modeled after the print method for the “htest” class used in the stats package that comes with base R. Objects of class “ictest” are lists with many objects (see ictest help).
There are four choices for a predefined type of rank score: “logrank1” (scores described in Sun 1996), “logrank2” (scores described in Finkelstein 1986), “wmw” and “normal” (the WMW scores or normal scores described in Fay 1996). Additionally, the option “general” allows general scores for arbitrary error distributions on the grouped continuous model (see Fay 1996). To show the general scores we consider the logistic error distribution. We can show that these scores are equivalent to the Wilcoxon-type scores (within computation error):
R> icout<-ictest(Surv(left, right, type = “interval2”)~treatment, data = bcos, scores = “wmw”)
R> wmw.scores<-icout$scores
R> logistic.scores<-ictest(Surv(left, right, type = “interval2”)~treatment, data = bcos, icFIT = icout$fit. scores = “general”, dqfunc = function(x){ dlogis(qlogis(x))})$scores
R> max(abs(wmw.scores-logistic.scores))
[1] 8.881784e-16
There are many inferential methods available for ictest as described in Section 2.5 and the method may be either explicitly stated as a character vector or may be the result of a methodRule function. The methodRule function works similarly as in the perm package, except the input must be three objects: the vector of rank scores, the vector of group membership values, and exact, a logical value coming from the object of the same name in the input. Other methodRule functions may be created to automatically choose the method based on a function of these three objects, but the default is methodRuleIC1 (see help for that function for details). Note that permutation inferences are available for all types of rank scores, but other types of inferences are not available for all the scores; see Section 2.5 and the ictest help for details.
Here is an overview of the calculation functions used in ictest. First, unless icFIT is given, the NPMLE of the distribution for all individuals is calculated using the icfit function. Any options used with the icfit function may be passed from the ictest call by using the icontrol option. Using the resulting NPMLE from the icfit call, the rank scores are calculated using the wlr_trafo function.
Similar to the ictest function, wlr_trafo is an S3 function with a default method and one for “Surv” objects, but additionally there is a method for “data.frame” objects. In the “data.frame” method, there must be only one variable which has either a“Surv” or “numeric” class. The purpose of the “data.frame” method is to properly interact with the coin package (see Section 6.2 below). Once the rank scores are calculated, then other functions are called depending on the inferential method chosen: the icScoreTest function for the score test, the icWSR function for the imputation approach, and the functions from perm for the permutation approaches.
6.2. Interacting with the coin package
The coin package allows different transformations for the response variables and we can use the wlr_trafo function as a transformation function within coin.
R> library(“coin”)
R> independence_test(Surv(left, right, type = “interval2”)~treatment, data = bcos, ytrafo = wlr_trafo)
Asymptotic General Independence Test
data: Surv(left, right, type = “interval2”) by treatment (Rad, RadChem)
Z = -2.6684, p-value = 0.007622
alternative hypothesis: two.sided
This repeats the asymptotic results from the method = “pclt” of ictest. The usefulness of coin are the fast algorithms for exact permutation calculations. Even these fast methods are still intractable for the full breast cosmesis data set, but we show here how the method may be applied quickly to a subset of that data.
R> SUBSET<-c(1:5, 50:65)
R> independence_test(Surv(left, right, type = “interval2”)~treatment, data = bcos, subset = SUBSET, ytrafo = wlr_trafo, distribution = exact())
Exact General Independence Test
data: Surv(left, right, type = “interval2”) by treatment (Rad, RadChem)
Z = -1.0722, p-value = 0.2899
alternative hypothesis: two.sided
R> ictest(Surv(left, right, type = “interval2”)~treatment, data = bcos, subset = SUBSET, method = “exact.network”)
Exact Logrank two-sample test (permutation form), Sun’s
scores
data: Surv(left, right, type = “interval2”) by treatment
p-value = 0.2861
alternative hypothesis: survival distributions not equal
n Score Statistic*
treatment=Rad 5 -1.514936
treatment=RadChem 16 1.514936
* like Obs-Exp, positive implies earlier failures than expected
The p-values are different since, as discussed in Section 5.3, the default two-sided method is different for the coin package (corresponding to control option tsmethod = “abs” in perm) than the perm package (default uses tsmethod = “central”) and hence also the interval package. When we use tsmethod = “abs” then we reproduce the results from coin:
R> ictest(Surv(left, right, type = “interval2”)~treatment, data = bcos, subset = SUBSET, method = “exact.network”, mcontrol = mControl(tsmethod = “abs”))
Exact Logrank two-sample test (permutation form), Sun’s
scores
data: Surv(left, right, type = “interval2”) by treatment
p-value = 0.2899
alternative hypothesis: survival distributions not equal
n Score Statistic*
treatment=Rad 5 -1.514936
treatment=RadChem 16 1.514936
* like Obs-Exp, positive implies earlier failures than expected
Note that the algorithms in coin can be considerably faster. To show this consider a larger subset of the breast cosmesis data:
R> SUBSET2<-c(1:12, 47:58) R> system.time( + independence_test(Surv(left, right, type = “interval2”)~treatment, data = bcos, subset = SUBSET2, ytrafo = wlr_trafo, distribution = exact()) +) user system elapsed 0.03 0.00 0.03 R> system.time( + ictest(Surv(left, right, type = “interval2”)~treatment, data = bcos, subset = SUBSET2, method = “exact.network”, mcontrol = mControl(tsmethod = “abs”)) +) user system elapsed 0.83 0.00 0.83
6.3. On handling ties for exact permutation implementation
In implementing the exact version of permutation tests, the way ties are handled may change the resulting p-values by non-negligible amounts. In this section we detail a simple artificial example to show how the handling of ties is difficult, in terms of reliably reproducing results, and we show that the ictest function gives the correct results.
Consider the data set with:
R> L<-c(2, 5, 1, 1, 9, 8, 10) R> R<-c(3, 6, 7, 7, 12, 10, 13) R> group<-c(0, 0, 1, 1, 0, 1, 0) R> example1<-data.frame(L, R, group) R> example1 L R group 1 2 3 0 2 5 6 0 3 1 7 1 4 1 7 1 5 9 12 0 6 8 10 1 7 10 13 0
In this case we can calculate the NPMLE exactly and give it in Table 2.
Table 2.
NPMLE for example1.
| (L | R] | probability |
|---|---|---|
| 2 | 3 | 2/7 |
| 5 | 6 | 2/7 |
| 9 | 10 | 3/14 |
| 10 | 12 | 3/14 |
We calculate this NPMLE with the interval package as
R> summary(icfit(L, R), digits = 12) Interval Probability 1 (2,3] 0.285714285714 2 (5,6] 0.285714285714 3 (9,10] 0.214285714286 4 (10,12] 0.214285714286
which matches the exact to at least 12 digits:
R> print(3/14, digits = 12) [1] 0.214285714286
Usually the fit will not be this close, and the closeness of the fit is determined by the icfitControl list (see the help).
The problem relates to the numerical precision of the calculated rank scores and subsequent permutation p-value when there is a small number of permutations and ties in the test statistics with different permutations (for interval-censoring, possibly stemming from overlapping intervals). While not unique to interval-censored data, this combination of factors may be more common in this setting.
We can calculate the exact scores for the Sun method (Equation 7 and scores = “logrank1”) these are
These scores sum to zero (as do all such scores regardless of the model). There are unique permutations with equal probability. Note that the difference in means of the original scores, (with group=[0, 0, 1, 1, 0, 1, 0]), gives equivalent values to the permutation with group=[1, 1, 0, 0, 0, 1, 0] because the sum of the first and second scores equals the sum of the third and fourth scores. Thus, we have a tie in the permutation distribution. We need to make sure the computer treats it as a tie otherwise the p-value will be wrong. Dealing with ties in computer computations can be tricky (see R FAQ 7.31 Hornik 2010). To see the details, we completely enumerate all the sums of the scores in one group. We use the function chooseMatrix from perm to generate the full list of permutations of the original group variable. We print out only the first 9 of the 35 ordered test statistics, placing the difference in means in the 8th column, next to the permutation of the group allocation:
R> score1<-wlr_trafo(Surv(L, R, type = “interval2”))
R> cm<-chooseMatrix(7, 3)
R> T<- ((1-cm) %*% score1)/4 - (cm %*% score1)/3
R> cbind(cm, T)[order(T), ][1:9, ]
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8]
[1,] 1 0 1 1 0 0 0 -1.0166667
[2,] 1 1 1 0 0 0 0 -0.9000000
[3,] 1 1 0 1 0 0 0 -0.9000000
[4,] 0 1 1 1 0 0 0 -0.7833333
[5,] 1 0 1 0 0 1 0 -0.6083333
[6,] 1 0 0 1 0 1 0 -0.6083333
[7,] 1 1 0 0 0 1 0 -0.4916667
[8,] 0 0 1 1 0 1 0 -0.4916667
[9,] 0 1 1 0 0 1 0 -0.3750000
The seventh and eighth largest of the 35 test statistics are tied, and the eighth largest is equal to the original group assignment, so that the one sided p-value is 8/35 = 0.2286. The function ictest properly calculates this p-value. The way that perm can directly address the ties issue is to allow the user to specify numerical precision, i.e. rounding to the nearest permControl()$digits significant digits; and perm treats values of the permutation distribution that are tied for that many significant digits as true ties.
We have not shown that this method of breaking ties always works properly; however, it does work in all the cases we have tried. We would like to emphasize that this issue is only a problem with small sample sizes using exact permutation methods. Additionally, it is a problem with all permutation tests where the test statistics have a non-zero probability of creating a tie. Very small differences in the rank scores will only produce correspondingly small differences in the asymptotic approximation, so as the sample sizes get large and, as guaranteed by the permutational central limit theorem, the estimate becomes more accurate, the way ties are handled effects large sample results minimally.
Supplementary Material
Acknowledgments
We would like to thank the editors and anonymous reviewers for the Journal of Statistical Software for their valuable comments that have improved this paper and the packages.
Contributor Information
Michael P. Fay, National Institute of Allergy and Infectious Diseases
Pamela A. Shaw, National Institute of Allergy and Infectious Diseases
References
- Agresti A, Mehta C, Patel N. Exact Inference for Contingency Tables with Ordered Categories. Journal of the American Statistical Association. 1990;85:453–458. [Google Scholar]
- Callaert H. Comparing Statistical Software Packages: The Case of the Logrank Test in StatXact. American Statistician. 2003;57:214–217. [Google Scholar]
- Carstensen B. Regression models for Interval Censored Survival Data: Application to HIV Infection in Danish Homosexual Men. Statistics in Medicine. 1996;15:2177–2189. doi: 10.1002/(SICI)1097-0258(19961030)15:20<2177::AID-SIM345>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Carstensen B, Plummer M, Laara EMH, et al. Epi: A package for statistical analysis in epidemiology. R package version 1.1.14. 2010 URL http://CRAN.R-project.org/package=Epi.
- Chambers JM. Statistics and Computing. 1 Springer-Verlag; New York: 2008. Software for Data Analysis: Programming with R. [Google Scholar]
- Farrington C. Interval Censored Data: A Generalized Linear Modeling Approach. Statistics in Medicine. 1996;15:283–292. doi: 10.1002/(SICI)1097-0258(19960215)15:3<283::AID-SIM171>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Fay MP. Rank Invariant Tests for Interval Censored Data under the Grouped Continuous Model. Biometrics. 1996;52:811–822. [PubMed] [Google Scholar]
- Fay MP. Comparing Several Score Tests for Interval Censored Data. Statistics in Medicine. 1999a;18:273–285. doi: 10.1002/(sici)1097-0258(19990215)18:3<273::aid-sim19>3.0.co;2-7. Corr: 1999V18 p2681. [DOI] [PubMed] [Google Scholar]
- Fay MP. interval: S-PLUS functions for interval-censored data. S-PLUS functions version April 3, 1999. 1999b URL http://lib.stat.cmu.edu.
- Fay MP, Kim HJ, Hachey M. On Using Truncated Sequential Probability Ratio Test Boundaries for Monte Carlo Implementation of Hypothesis Tests. Journal of Computational and Graphical Statistics. 2007;16(4):946–967. doi: 10.1198/106186007X257025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein D, Wolfe R. A Semiparametric Model for Regression Analysis of Interval-Censored Failure Time Data. Biometrics. 1985;41:845–854. [PubMed] [Google Scholar]
- Finkelstein DM. A Proportional Hazards Model for Interval-Censored Failure Time Data. Biometrics. 1986;42:845–854. [PubMed] [Google Scholar]
- Fleming T, Harrington D. Counting Processes and Survival Analysis. John Wiley & Sons; New York: 1991. [Google Scholar]
- Freidlin B, Korn EL, Hunsberger S, Gray R, Saxman S, Zujewski JA. Proposal for the Use of Progression-Free Survival in Unblinded Randomized Trials. Journal of Clinical Oncology. 2007;25(15):2122–2126. doi: 10.1200/JCO.2006.09.6198. [DOI] [PubMed] [Google Scholar]
- Gentleman R, Geyer C. Maximum Likelihood for Interval Censored Data: Consistency and Computation. Biometrika. 1994;81:618–623. [Google Scholar]
- Gentleman R, Vandal A. Computational Algorithms for Censored-Data Problems Using Intersection Graphs. Journal of Compuational and Graphical Statistics. 2001;10:403– 421. [Google Scholar]
- Gentleman R, Vandal A. Nonparametric Estimation of the Bivariate CDF for Arbitrarily Censored Data. Canadian Journal of Statistics. 2002;30:557–571. [Google Scholar]
- Gentleman R, Vandal A. Icens: NPMLE for Censored and Truncated Data. R package version 1.20.0. 2010 URL http://CRAN.R-project.org/package=Icens.
- Goggins W. Fitting the Proportional Hazards Model for Interval Censored data. S program, calls SPARC object file. 2007 URL http://hedwig.mgh.harvard.edu/biostatistics/software.
- Goggins WB, Finkelstein DM, Zaslavsky AM. Applying the Cox Proportional Hazards Model When the Change Time of a Binary Time-Varying Covariate Is Interval Censored. Biometrics. 1999;55:445–451. doi: 10.1111/j.0006-341x.1999.00445.x. [DOI] [PubMed] [Google Scholar]
- Groeneboom P, Jongbloed G, Wellner J. The Support Reduction Algorithm for Computing Nonparametric Function Estimates in Mixture Models. Scandinavian Journal of Statistics. 2008;35:385–399. doi: 10.1111/j.1467-9469.2007.00588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Sun L, Zuo G. A Baseline-Free Procedure for Transformation Models Under Interval Censorship. Lifetime Data Analysis. 2005;11:473–488. doi: 10.1007/s10985-005-5235-x. [DOI] [PubMed] [Google Scholar]
- Heinze G, Gnant M, Schemper M. Exact Log-Rank Tests for Unequal Follow-Up. Biometrics. 2003;59:1151–1157. doi: 10.1111/j.0006-341x.2003.00132.x. [DOI] [PubMed] [Google Scholar]
- Hoffman EB, Sen PK, Weinberg CR. Within-Cluster Resampling. Biometrika. 2001;88:420–429. [Google Scholar]
- Hornik K. R FAQ: Frequently Asked Questions on R. R Foundation for Statistical Computing; Vienna, Austria: 2010. Version 2.11.2010-07-27. URL http://CRAN.R-project.org/doc/FAQ/R-FAQ.html. [Google Scholar]
- Hothorn T, Hornik K, van de Wiel MA, Zeileis A. A Lego System for Conditional Inference. The American Statistician. 2006;60(3):257–263. [Google Scholar]
- Hothorn T, Hornik K, van de Wiel MA, Zeileis A. Implementing a Class of Permutation Tests: The coin Package. Journal of Statistical Software. 2008;28(8):1–23. URL http://www.jstatsoft.org/v28/i08/ [Google Scholar]
- Huang J, Lee C, Yu Q. A Generalized Log-Rank Test for Interval-Censored Failure Time Data via Multiple Imputation. Statistics in Medicine. 2008;27:3217–3226. doi: 10.1002/sim.3211. [DOI] [PubMed] [Google Scholar]
- Kalbfleisch J, Prentice R. The Statistical Analysis of Failure Time Data. John Wiley & Sons; New York: 1980. [Google Scholar]
- Law C, Brookmeyer R. Effects of Mid-Point Imputation on the Analysis of Doubly Censored Data. Statistics in Medicine. 1992;11:1569–1578. doi: 10.1002/sim.4780111204. [DOI] [PubMed] [Google Scholar]
- Maathuis M. MLEcens: Computation of the MLE for Bivariate (Interval) Censored Data. R package version 0.1-3. 2010 URL http://CRAN.R-project.org/package=MLEcens.
- Ng M. A Modification of Peto’s Survival Curves for Interval-Censored Data. Biometrics. 2002;58:439–442. doi: 10.1111/j.0006-341x.2002.00439.x. [DOI] [PubMed] [Google Scholar]
- Peto R, Peto J. Asymptotically Efficient Rank Invariant Test Procedures. Journal of the Royal Statistical Society A. 1972;135:185–207. [Google Scholar]
- Pettitt A. Tied, Grouped Continuous and Ordered Categorical Data: A Comparison of Two Models. Biometrika. 1984;71:35–42. [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2010. URL http://www.R-project.org/ [Google Scholar]
- Satten GA. Rank-Based Inference in the Proportional Hazards Model for Interval Censored Data. Biometrika. 1996;83(2):355–370. [Google Scholar]
- Self SG, Grosman EA. Linear Rank Tests for Interval-Censored Data with Application to PCB Levels in Adipose Tissue of Transformer Repair Workers. Biometrics. 1986;42:521–530. [PubMed] [Google Scholar]
- Sen P. Permutational Central Limit Theorems. In: Kotz S, Johnson NL, editors. Encyclopedia of Statistics. Vol. 6. John Wiley & Sons; 1985. [Google Scholar]
- Sun J. A Non-Parametric Test for Interval-Censored Failure Time Data with Application to AIDS Studies. Statistics in Medicine. 1996;15:1387–1395. doi: 10.1002/(SICI)1097-0258(19960715)15:13<1387::AID-SIM268>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Tanner M. Tools for Statistical Inference. 3 Springer-Verlag; New York: 1996. [Google Scholar]
- Therneau T, Lumley T. survival: Survival Analysis Including Penalised Likelihood. R package version 2.35-8. 2009 URL http://CRAN.R-project.org/package=survival.
- Turnbull B. The Empirical Distribution Function with Arbitrarily Grouped, Censored and Truncated Data. Journal of the Royal Statistical Society B. 1976;38:290–295. [Google Scholar]
- Zhang Z, Sun L, Sun J, Finkelstein D. Regression Analysis of Failure Time Data with Informative Interval Censoring. Statistics in Medicine. 2007;26:2533–2546. doi: 10.1002/sim.2721. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.