Abstract
Context:
The immune response in autoimmune thyroid disease has been shown to occur primarily within the thyroid gland in which the most abundant antigens can be found. A variety of capture molecules are known to be expressed by thyroid epithelial cells and serve to attract and help retain an intrathyroidal immune infiltrate.
Objective:
To explore the entire repertoire of expressed genes in human thyroid tissue, we have deep sequenced the transcriptome (referred to as mRNA-Seq).
Design and Patients:
We applied mRNA-Seq to thyroid tissue from nine patients with Graves' disease subjected to total thyroidectomy and compared the data with 12 samples of normal thyroid tissue obtained from patients having a thyroid nodule removed. The expression for each gene was calculated from the sequencing data by taking the median of the coverage across the length of the gene. The expression levels were quantile normalized and a gene signature was derived from these.
Results:
On comparison of expression levels in tissues derived from Graves' patients and controls, there was clear evidence for overexpression of the antigen presentation pathway consisting of HLA and associated genes. We also found a robust disease signature and discovered active innate and adaptive immune signaling networks.
Conclusions:
These data reveal an active immune defense system in Graves' disease, which involves novel molecular mechanisms in its pathogenesis and development.
Autoimmune thyroid disease (AITD) shows a broad clinical spectrum, ranging from the immune-mediated thyroid damage of Hashimoto's thyroiditis to signal transduction-induced Graves' disease (1, 2). The immune response in AITD occurs predominantly within the thyroid gland itself, the site of major thyroid antigen expression (3, 4). A variety of immune cell capture molecules are expressed on the surface of thyroid epithelial cells, which attract and help maintain an active intrathyroidal immune infiltrate. Although much insight has been gained concerning the genetic susceptibility to AITD (5, 6), the molecular mechanisms initiating and leading to these diseases remain unclear.
Deep sequencing technologies provide a novel and powerful tool for gaining new insights into the complexity of gene expression and gene regulation, especially in such diverse cell populations as found in human tissues. mRNA sequencing (mRNA-Seq), the application of deep sequencing to transcriptome analysis, has proven to be useful in quantitative measurement of gene expression. The aims of the present study were to harness mRNA-Seq to examine the gene expression repertoire in thyroid tissue from patients with Graves' disease when compared with normal thyroid tissue. We hypothesized that such a repertoire would reveal a robust Graves' disease fingerprint and demonstrate the signaling pathways preferentially active in the thyroid gland of patients with Graves' disease.
Materials and Methods
Subjects
We obtained nine thyroid tissues from Graves' disease patients and 12 normal thyroid tissues via the Mount Sinai Biorepository. All Graves' disease patients had been subjected to antithyroid drug therapy before surgery for various lengths of time. These studies were approved by the Institutional Review Board of the Icahn School of Medicine at Mount Sinai.
mRNA sequencing
Total RNA was isolated from samples using Ambion's RNA isolation and purification kit. PolyA-tailed mRNA was selected using beads with oligodeoxythymidine and then fragmented. cDNAs were created using random hexamers and ligated with bar-coded adaptors compatible with Illumina's HiSeq 2000 sequencing. Single-end, 100-nucleotide-long reads were sequenced at the Genomics Core of the Icahn School of Medicine at Mount Sinai.
Analysis of mRNA-Seq data
Custom-built software was used to map the reads to the human genome (hg19) and estimate the coverage of each gene (7, 8). Briefly, the reads were split into three 32-bp pairs after trimming 2 nt at each end, and the parts were mapped to the genome using a suffix-array based approach. The median of coverage across the transcript was used as an estimate of gene expression. The expression values were quantile normalized, and ratios were calculated by comparing the mean of the samples from Graves' patients against the mean of samples from controls. We also used the coverage to cluster the data and determine outliers, which were excluded from further analyses.
For mRNA-Seq data, the distribution of expression values was plotted to identify the peak in the distribution, which helps estimate the noise in the system. The values were regularized by adding the noise to each gene's expression level before the ratios were calculated. This ensures that genes with low expression do not contribute to the list of genes with large fold changes so that the signature genes can be chosen from the significantly higher and differentially expressed genes (P < .05). The Panther program (http://www.pantherdb.org) was used to illustrate the top 100 gene expression profiles in normal and Graves' disease thyroids. The heat map was generated by custom software.
Pathway analysis
We used the Ingenuity Pathway Analysis system (http://www.ingenuity.com) and MetaCore software (http://www.thomsonreuters.com/metacore) to analyze and compare the pathways in normal and Graves' disease thyroid with a cutoff value of P < .05 and fold change of 2.3. A Fisher's exact test was used to calculate the P value for the probability that a pathway was significantly enriched in input genes when compared with the genome database. The P value was then used to rank the pathways/networks.
Results
Gene expression profiling in Graves' disease and normal thyroid tissue
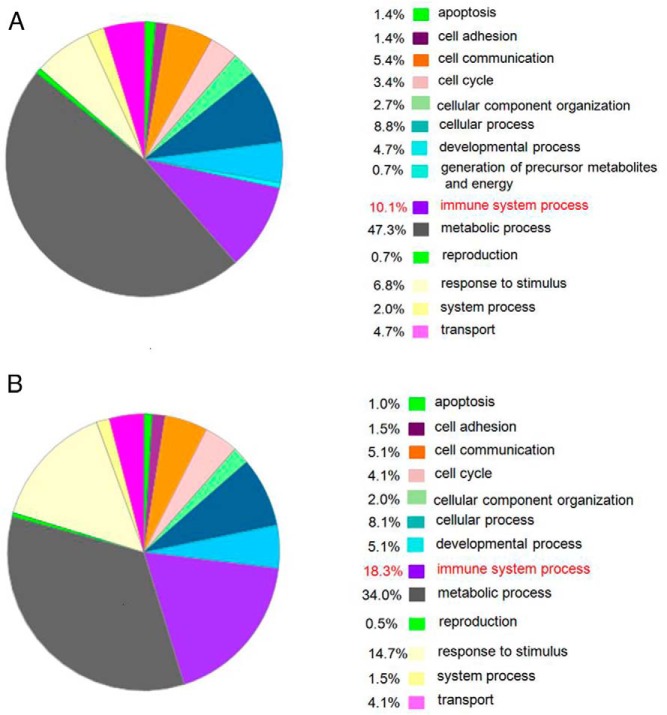
We used the top 100 most highly expressed genes from normal and Graves' disease thyroid for further analysis, and their gene expression is illustrated in Figure 1A. The pie on the left panel shows the percent of genes involved in the different cellular processes described. Genes in the immune system process made up 10.1% of the expressed genes in the normal tissues, suggesting that such control tissue likely contained immune cells from retained blood, inflammatory infiltrates secondary to nodules or other disease processes, or the natural retention of immune cells within the normal thyroid (bystander immune cells). Analysis of these data most likely showed the continuous low level of immune cell activity that is essential to health. However, immune gene expression increased to approximately 18.3% in the Graves' disease thyroid tissues (Figure 1B), indicating the expected immune hyperactivity of Graves' disease. Of particular import was the observation that B cell genes were most active and formed part of the top 100 genes. Signal responsive genes were elevated from 6.8% in normal to 14.7% in Graves' disease (Figure 1, A and B), most likely secondary to the increased immune response. As a result, metabolic processing genes were reduced from 47.3% in normal to 34.0% in Graves' disease (Figure 1, A and B) secondary to the increased immune related and signaling activity.
Figure 1.
A, Genes expressed in normal thyroid tissue. B, Genes expressed in thyroid tissue from patients with Graves' disease.
Highly expressed genes in Graves' disease
We next selected the 15 most up-regulated genes in Graves' disease thyroids (Table 1). The fold changes varied from 3.4 to 9.7. Table 1 groups these Graves' disease-overexpressed genes as six human leukocyte antigen (HLA) genes, four cytokine- and chemokine-related genes and regulators, four growth- and synthesis-related genes, and one gene encoding an uncharacterized protein. We suggest that these genes represent a Graves' disease signature.
Table 1.
A Graves' Disease Signature
| Function | Gene | P Value | Fold Change |
|---|---|---|---|
| Cytokine- and chemokine-related genes | CCL19 | .027 | 9.7 |
| SOCS3 | .003 | 5.3 | |
| CCL2 | .034 | 5.2 | |
| GADD45A | .047 | 3.6 | |
| Growth- and synthesis-related genes | SLC5A5 (NIS) | .02 | 6.9 |
| NR4A1 | .005 | 6.2 | |
| KLF6 | .017 | 5.6 | |
| FOSB | .022 | 4.9 | |
| HLA | HLA I-C | .003 | 5.2 |
| HLA II-DRB1 | .03 | 4.0 | |
| DRB6 | .042 | 3.5 | |
| DPA1 | .018 | 3.5 | |
| DRA | .003 | 3.4 | |
| CD74 | .005 | 3.4 | |
| Uncharacterized protein | C11orf96 | .041 | 4.5 |
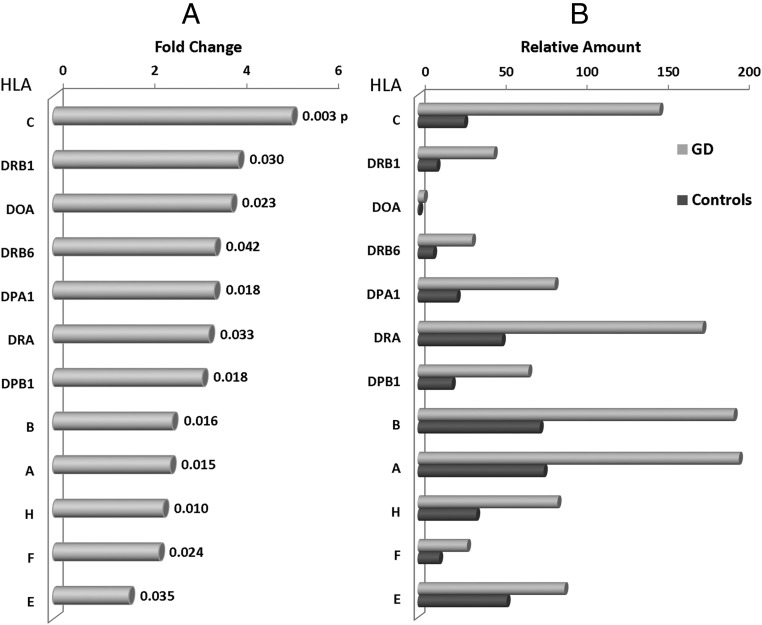
HLA gene expression
HLA genes encode cell-surface antigen-presenting proteins and have many other functions in humans. As expected, they were significantly overexpressed in thyroid tissues from patients with Graves' disease. Figure 2, A and B, show the changes in expression of some of the HLA genes in normal and Graves' disease thyroids. HLA-A, HLA-B, HLA-C, and DRA appeared to be overexpressed in Graves' disease tissue. HLA-C was the most overexpressed, suggesting that HLA class I molecules are intimately involved in the disease pathology. Because HLA class I molecules are involved in the presentation of endogenous antigens to the immune system, including those derived from viruses, such overexpression may reflect key environmental triggers for AITD. Similarly, HLA-H is involved in the presentation of foreign antigen to the immune system again implying environmental (virus or bacterial) antigens are involved in Graves' disease.
Figure 2.
A, Fold change in HLA genes in Graves' disease. B, Relative amount of HLA in normal and Graves' disease.
Cytokine gene expression
Of note, both CCL19 and CCL2 were overexpressed in Graves' tissue. CCL19 plays a role in normal lymphocyte recirculation and homing and is important in trafficking of T cells in the thymus and in T cell and B cell migration to secondary lymphoid organs. CCL19 binds to chemokine (C-C motif) receptor (CCR)-7 and has similar actions to chemokine (C-C motif) ligand (CCL)-21. We have previously reported that CCR7 deficiency in nucleotide-binding oligomerization domain receptors (NOD) mice leads to thyroiditis (9). Therefore, CCL19 may also play an important role in human AITD. CCL2 displays chemotactic activity for monocytes and basophils and has been implicated in the pathogenesis of diseases characterized by monocytic infiltrates including psoriasis and rheumatoid arthritis. CCL2 binds to chemokine receptors CCR2 and CCR4 and its overexpression suggests that it may also have a major role in intrathyroidal monocyte attraction.
Thyroid-related gene expression
Supplemental Table 1 shows the changes in expression of all the thyroid-related genes and transcription factor genes. Tg and TPO were highly expressed in both groups, but the SLC5A5 (NIS) gene had the highest fold change (6.9) and was the only thyroid-related gene that was overexpressed in this series of samples. This gene is also expressed in breast, stomach, and salivary glands, so it is not thyroid specific (10). However, it is responsible for the uptake of iodine into thyroid tissues, which is then incorporated into T3 and T4. These Graves' thyroids were from patients treated with antithyroid drugs, which inhibit iodide organification and may be partly responsible for this increased expression. However, most genes involved in thyroid hormone synthesis pathway were not highly expressed secondary to the antithyroid drug treatment.
Network analyses
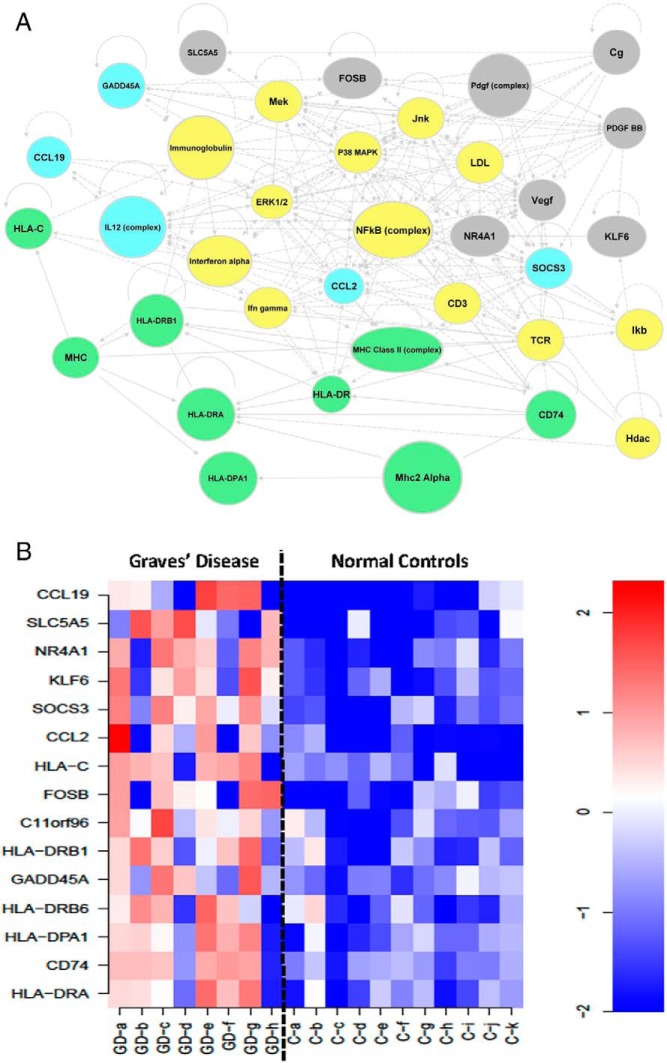
Using Ingenuity software, we developed a view of the active gene networks associated with the Graves' disease signature genes (Figure 3A). This revealed that the nuclear factor-kappa light polypeptide gene enhancer in B-cells (NF-κB) complex may act as a major regulator mostly linking and interacting with many of these pathways. Another approach to such an analysis is by a heat map from the Graves' disease signature (Figure 3B), which demonstrates the distinct difference in gene expression profiles between normal and Graves' disease thyroid tissue.
Figure 3.
A, Network built from Graves' disease signature. HLA-related genes are labeled in green; cytokine- and chemokine-related genes and regulators are in blue; growth- and synthesis-related genes are in gray; and multiple functional genes and regulators are in yellow. B, Heat map generated from the Graves' disease signature. Red indicates increased expression and blue indicates decreased expression. The expression levels of the individual signature genes are shown for individual Graves' disease (GD) samples and control samples.
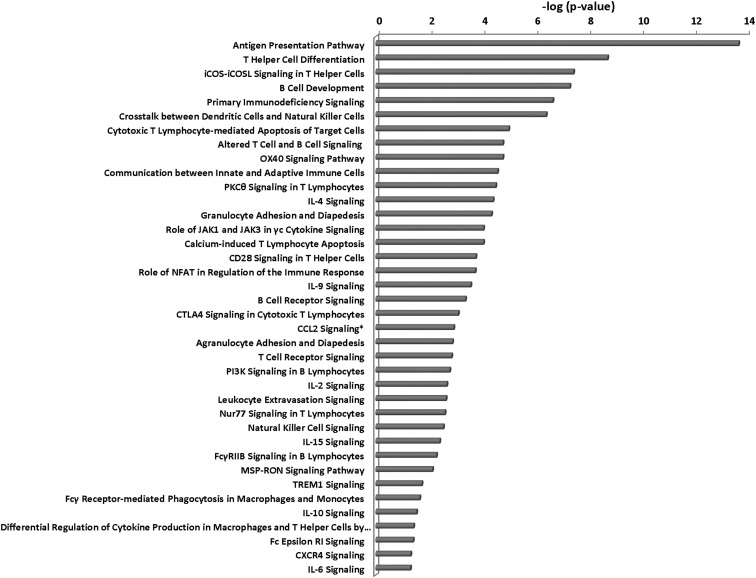
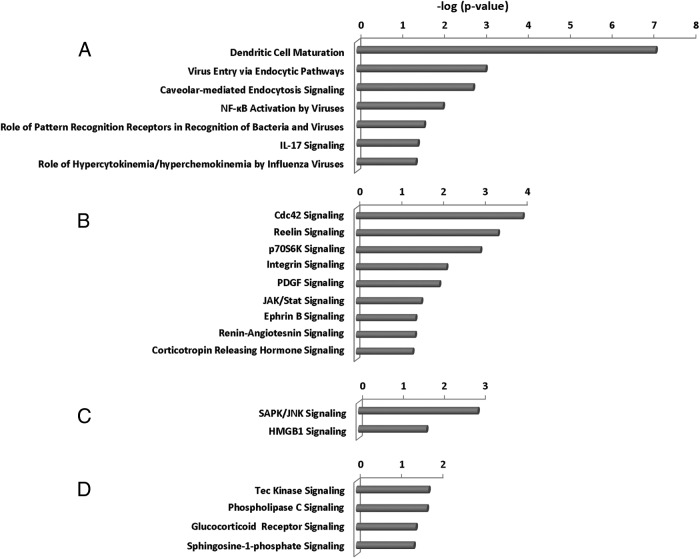
From the pathway analyses, we found five major categories of overactive pathways in Graves' disease: immune responses (Figure 4), pathogen influenced, thyroid growth, stress responses, and intracellular and second-messenger signaling pathways (Figure 5). The immune response pathways were most dominant, with evidence of both innate and adaptive immunity being active, as were cytokine and chemokine regulatory pathways and evidence of apoptosis and inflammation. Of all this activity, the antigen presentation pathway was the most significantly increased in Graves' disease. Antigen recognition appears to be ongoing in Graves' disease, with evidence of T cell receptor (TCR) complex and B cell receptor (BCR) complex (adaptive immunity) gene expression. In addition, evidence for the use of recently discovered pathogen-associated molecular patterns (PAMPs) and endogenous danger/damage-associated molecular patterns (innate immunity) were present. At the same time, Graves' tissue cells exhibited stress responses, especially those involving the stress-activated protein kinase/c-Jun N-terminal kinase signaling and high mobility group protein B1 signaling.
Figure 4.
Immune response pathways in Graves' disease. *, CCL2 pathway was obtained from MetaCore software.
Figure 5.
A, Pathogen-influenced pathways. B, Thyroid growth pathways. C, Stress response pathways. D, Intracellular and second-messenger signaling pathways in Graves' disease.
Innate immune system gene expression in Graves' disease
The innate immune system defends the host from infections in a nonspecific manner. Innate immune systems provide immediate defense against infection. Supplemental Table 2A lists some significantly differentially expressed genes from the innate immune system found in the Graves' disease thyroid, indicating the high degree of activity in the innate-immune system.
Adaptive immune system gene expression in Graves' disease
The adaptive immune system is composed of highly specialized T cells and B cells and other systemic cells and processes that eliminate or prevent pathogen growth and have often been associated with autoimmune disease such as AITD in addition to the HLA genes discussed earlier. Supplemental Table 2B lists some significantly differentially expressed genes from the adaptive immune system in the Graves' disease thyroid, confirming the expected high activity of this system.
Discussion
Graves' disease is a well-characterized autoimmune disease with a large number of known susceptibility genes (5, 6, 11), involvement of both T cells (12, 13) and B cells (14), which results in the production of autoantibodies to the TSH receptor (TSHR). The stimulating varieties of TSHR antibodies act as TSH agonists, causing excessive secretion of thyroid hormones. The intrathyroidal inflammatory infiltrate of Graves' disease tends to be heterogenous and less intense than in Hashimoto's thyroiditis, although such an impression has been obtained from thyroid specimens taken from patients treated with antithyroid drugs, which are known to act as local immune suppressants and can reduce the immune infiltrate (15). Applying the technical power of RNA-Seq to the examination of thyroid tissues from patients with Graves' disease allowed us a view of simultaneous activity within the tissue with the caveat that all the patients had been pretreated with antithyroid drugs. Despite this caveat, much of what we expected to find was indeed revealed, and in addition, strong evidence for the role of the innate immune system was obtained.
Although we had no way of knowing the source of the background immune gene activity in the pathologically normal control tissue, it was clear that an extensive autoimmune reaction was ongoing within the Graves' thyroid glands. In view of extensive earlier studies, it was not surprising to see strong evidence of the adaptive immune system with many HLA class II genes overexpressed and evidence for ongoing antigen presentation. Hence, a variety of genes related to B cell function and T cell activity were active. For example, CD79A and CD79B cooperate in the initiation of the signal transduction cascade activated by binding of antigen to the BCR, which leads to internalization of the complex, trafficking to late endosomes, and antigen presentation. CD19 encodes a cell surface molecule that assembles with the antigen receptor of B lymphocytes to decrease the threshold for antigen receptor-dependent stimulation. The CD8A gene encodes the CD8 α-chain. The CD8 antigen is a cell surface glycoprotein found on most cytotoxic T lymphocytes that mediates efficient cell-cell interactions within the immune system. The CD8 antigen acts as a coreceptor with the T cell receptor on T lymphocytes to recognize antigens displayed by an antigen-presenting cell in the context of HLA class I molecules. The finding of the differentially expressed CD8 gene in Graves' disease thyroid suggests that it may play an important role in T cell immunity and HLA class I function.
The extent of the HLA class I gene expression was also emphasized by a variety of additional related genes that were overexpressed in the Graves' tissue. For example, TLR9 encodes a nucleotide-sensing toll-like receptor (TLR), which is activated by unmethylated cytidine-phosphate-guanosine dinucleotides. It acts via MYD88 and TRAF6 (TNF receptor-associated factor-6, E3 ubiquitin protein ligase), leading to NF-κB activation, cytokine secretion, and the inflammatory response. TLRs play a fundamental role in pathogen recognition and activation of innate immunity. NLRC5, also called nucleotide-binding oligomerization domain receptors (NOD)-like receptor C5, encodes a member of the caspase recruitment domain-containing NOD-like receptor family. This gene is probably a regulator of the NF-κB and type I interferon signaling pathways and may also regulate the type II interferon signaling pathway. Moreover, it plays a role in homeostatic control of innate immunity and in antiviral defense mechanisms.
NLRP2 is a component of the inflammasome and whose function is activation of proinflammatory caspases. CD1C encodes a member of the CD1 family of transmembrane glycoproteins, which are structurally related to the major histocompatibility complex proteins and form heterodimers with β2-microglobulin. The CD1 proteins mediate the presentation of primarily lipid and glycolipid antigens of self or microbial origin to T cells. Complement component C3 plays a central role in the activation of the complement system. Its activation is required for both classical and alternative complement activation pathways. Pathogens coated with fragments of the complement protein C3 are not only opsonized for phagocytosis but also bind more strongly to B cells that have bound the pathogen through their BCR. This synergistic effect enables antibody production to occur at much lower doses of antigen. Therefore, such adaptive immunity may not be possible without the assistance of the mechanisms of innate immunity.
The detection of enhanced gene expression and the pathways analysis in the thyroid glands from Graves' disease patients emphasizes that the innate and adaptive immune systems interact and provide further evidence for environmental factors influencing the autoimmune response (Supplemental Figure 1). Environmental factors, infectious organisms, and spontaneous cell death or apoptosis can induce pathogen-associated molecular patterns and damage-associated molecular patterns. Such influences may affect only thyroid cells endowed with susceptible genetic and epigenetic backgrounds, leading to the release of thyroid-specific autoantigens. These autoantigens, such as the TSHR, thyroglobulin (Tg), and thyroid peroxidase (TPO) along with nucleic-acid fragments will activate and stimulate circulating B cells through their BCR, pattern-recognition receptors such as TLRs (eg, TLR 9), NOD-like receptors (eg, NLRC5), inflammasomes (eg, NLRP2), and complement proteins (eg, C3), resulting in the release of autoantibodies (anti-TSHR, anti-TG, and anti-TPO) from the plasma B cells.
In another view, viruses, bacteria, or other initiators may be taken up by macrophages and neutrophils via phagocytosis and endocytosis. After processing and displaying autoantigens containing protein and nucleic acid fragments (either thyroid antigens or mimics that cross-react with them) or presenting lipid antigen on the cell surface, they work as antigen presenting cells (APCs). Such a process also involves cytokines and chemokines (eg, IL-6, CCL2, and CCL19) that also are found to be overexpressed in the Graves' tissues. Meanwhile, the cytokines secreted from macrophages activate and recruit dendritic cells, which also take up autoantigens from thyrocytes and act as APCs. Dendritic cells and IL-17 play a central role in linking and integrating the innate and adaptive immune responses.
It would appear therefore that APCs can support three different routes to AITD. The first one is APCs with HLA-II molecules interact with CD4+ T helper (Th) cells through their TCRs and CD28 for antigen recognition. After activation and proliferation, Th cells develop into Th2 cells, which interact with B cells through their TCRs and HLA-II. This process also involves cytokine (eg, IL-2, IL-4, IL-10, and IL-17) production and regulation. After B cell activation, autoantibodies (anti-TSHR, anti-Tg, and anti-TPO) are produced from plasma B cells. The resulting autoantibodies lead to thyrocyte growth and T3/T4 release.
The second is APCs with HLA-I molecules interact with CD8+ Th cells through their TCRs. After antigen recognition, functional cytotoxic T cells and natural killer cells are activated to aid cell destruction and also release cytokines or chemokines. The resulting apoptotic cells are seen in Graves' thyroids (16, 17), and in addition, certain neutral TSHR-antibodies may also induce apoptosis (18).
Third, APCs with CD1C molecules interact with CD8+ Th cells through their TCRs. After recognition of lipid antigen, natural killer cells are activated to aid cell destruction.
In conclusion, deep sequencing offers important insights into the immunopathology of autoimmune thyroid disease, provides an overview or landscape of intrathyroidal gene expression and facilitates the discovery of novel disease-related genes, signature, and pathways. These data provide new directions for further studies and may lead to potential therapeutic targets for drug discovery with more efficient treatment for patients with Graves's disease. The data raise the possibility that innate and adaptive immunity respectively reflect the acute and chronic phases of classical Graves' disease.
Acknowledgments
We thank all of our colleagues including Yaron Tomer, Risheng Ma, Ramkumarie Baliram, and Rejwan Ali for their advice.
This work was supported in part by Grants DK052464 and DK69713 from the National Institutes of Health-National Institute of Diabetes and Digestive and Kidney Diseases, the David Owen Segal Endowment, the James J. Peters Veterans Affairs Medical Center, and the Veterans Affairs Merit Award program.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- AITD
- autoimmune thyroid disease
- APC
- antigen presenting cell
- BCR
- B cell receptor
- CCL
- chemokine (C-C motif) ligand
- CCR
- chemokine (C-C motif) receptor
- HLA
- human leukocyte antigen
- mRNA-Seq
- mRNA sequencing
- NF-κB
- nuclear factor of kappa light polypeptide gene enhancer in B-cells
- NOD
- nucleotide-binding oligomerization domain receptors
- PAMP
- pathogen-associated molecular pattern
- TCR
- T cell receptor
- Tg
- thyroglobulin
- Th
- T helper
- TLR
- toll-like receptor
- TPO
- thyroid peroxidase
- TSHR
- TSH receptor.
References
- 1. Davies TF, Amino N. A new classification for human autoimmune thyroid disease. Thyroid. 1993;3:331–333 [DOI] [PubMed] [Google Scholar]
- 2. Orgiazzi J. The spectrum of autoimmune thyroid diseases (AITD). Ann Med Interne (Paris). 1999;150:294–300 [PubMed] [Google Scholar]
- 3. Davies TF, Martin A, Concepcion ES, Graves P, Cohen L, Ben-Nun A. Evidence of limited variability of antigen receptors on intrathyroidal T cells in autoimmune thyroid disease. N Engl J Med. 1991;325:238–244 [DOI] [PubMed] [Google Scholar]
- 4. Weetman AP. Cellular immune responses in autoimmune thyroid disease. Clin Endocrinol (Oxf). 2004;61:405–413 [DOI] [PubMed] [Google Scholar]
- 5. Tomer Y. Genetic susceptibility to autoimmune thyroid disease: past, present, and future. Thyroid. 2010;20:715–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Davies TF, Latif R, Yin X. New genetic insights from autoimmune thyroid disease. J Thyroid Res. 2012;2012:623852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Olson AJ, Brennecke J, Aravin AA, Hannon GJ, Sachidanandam R. Analysis of large-scale sequencing of small RNAs. Pac Symp Biocomput 2008;126–136 [DOI] [PubMed] [Google Scholar]
- 8. Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747 [DOI] [PubMed] [Google Scholar]
- 9. Martin AP, Marinkovic T, Canasto-Chibuque C, et al. CCR7 deficiency in NOD mice leads to thyroiditis and primary hypothyroidism. J Immunol. 2009;183:3073–3080 [DOI] [PubMed] [Google Scholar]
- 10. Bruno R, Giannasio P, Ronga G, et al. Sodium iodide symporter expression and radioiodine distribution in extrathyroidal tissues. J Endocrinol Invest. 2004;27:1010–1014 [DOI] [PubMed] [Google Scholar]
- 11. Simmonds MJ. GWAS in autoimmune thyroid disease: redefining our understanding of pathogenesis. Nat Rev Endocrinol. 2013;9:277–287 [DOI] [PubMed] [Google Scholar]
- 12. Morshed SA, Latif R, Davies TF. Delineating the autoimmune mechanisms in Graves' disease. Immunol Res. 2012;54:191–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Martin A, Barbesino G, Davies TF. T-cell receptors and autoimmune thyroid disease—signposts for T-cell-antigen driven diseases. Int Rev Immunol. 1999;18:111–140 [DOI] [PubMed] [Google Scholar]
- 14. Brown J, Dorrington KJ, Ensor J, Smith BR, Munro DS. Proc R Soc Med. 1968;61:1301–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weiss I, Davies TF. Inhibition of immunoglobulin-secreting cells by antithyroid drugs. J Clin Endocrinol Metab. 1981;53:1223–1228 [DOI] [PubMed] [Google Scholar]
- 16. Stassi G, De Maria R. Autoimmune thyroid disease: new models of cell death in autoimmunity. Nat Rev Immunol. 2002;2:195–204 [DOI] [PubMed] [Google Scholar]
- 17. Morshed SA, Ma R, Latif R, Davies TF. How one TSH receptor antibody induces thyrocyte proliferation while another induces apoptosis. J Autoimmun. 2013;47C:17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morshed SA, Ando T, Latif R, Davies TF. Neutral antibodies to the TSH receptor are present in Graves' disease and regulate selective signaling cascades. Endocrinology. 2010;151:5537–5549 [DOI] [PMC free article] [PubMed] [Google Scholar]