Abstract
Mammalian embryonic stem (ES) cells and sperm exhibit unusual chromatin packaging that plays important roles in cellular function. Here, we extend a recently developed technique, based on deep paired-end sequencing of lightly digested chromatin, to assess footprints of nucleosomes and other DNA-binding proteins genome-wide in murine ES cells and sperm. In ES cells, we recover well-characterized features of chromatin such as promoter nucleosome depletion, and further identify widespread footprints of sequence-specific DNA-binding proteins such as CTCF, which we validate in knockdown studies. We document global differences in nuclease accessibility between ES cells and sperm, finding that the majority of histone retention in sperm preferentially occurs in large gene-poor genomic regions, with only a small subset of nucleosomes being retained over promoters of developmental regulators. Finally, we describe evidence that CTCF remains associated with the genome in mature sperm, where it could play a role in organizing the sperm genome.
INTRODUCTION
Eukaryotic genomes are packaged into a nucleoprotein complex known as chromatin, whose repeating subunit consists of ~147 bp of DNA wrapped around an octamer of histone proteins. Nucleosomes impact essentially all DNA-templated processes, and as a result there has been a great deal of interest over the past decade in characterizing nucleosome positions across the genomes of a variety of organisms (Jiang and Pugh, 2009; Radman-Livaja and Rando, 2010). In mammals, genome-wide maps have been reported for nucleosome positions in a number of cell types and tissues, including several immune cell types (Schones et al., 2008; Valouev et al., 2011), liver (Li et al., 2012), and embryonic stem (ES) cells, neural precursor cells, and embryonic fibroblasts (Li et al., 2012; Teif et al., 2012). These genome-wide maps reveal characteristic features that are conserved throughout eukaryotes, such as nucleosome depletion at promoters and other regulatory elements.
Several cell types exhibit unusual chromatin states that appear to be linked to biological function. ES cells, which are pluripotent, are characterized by “hyperdynamic” chromatin in which histone proteins exchange rapidly on and off of the genome (Meshorer et al., 2006). This has been proposed to contribute to a generally permissive chromatin state in which genes important for differentiation are accessible for rapid transcriptional activation. In contrast, mammalian sperm exhibit a highly unusual chromatin state that is vastly different from that of other cell types (Ooi and Henikoff, 2007); most of the histone proteins are lost during spermatogenesis, first replaced by transition proteins, and eventually replaced by small basic proteins termed protamines. However, not all histones are lost (murine sperm retain ~2% of their histones), and recent studies on histone retention in human and mouse sperm suggests that there is a bias for promoters of genes expressed early during development to be specifically packaged in histones (Arpanahi et al., 2009; Brykczynska et al., 2010; Erkek et al., 2013; Gardiner-Garden et al., 1998; Hammoud et al., 2009). These findings contrast with several lines of evidence suggesting that histone retention in sperm primarily occurs over repeat elements – small scale cloning of DNA released by nuclease digestion of sperm revealed primarily repeat elements such as LINE and SINE sequences (Pittoggi et al., 1999) and pericentric repeats (Govin et al., 2007), while immunostaining studies on mature sperm reveal colocalization of histone proteins with the repeat-enriched sperm chromocenter (Govin et al., 2007; van der Heijden et al., 2006). The discrepancies between these views of the sperm chromatin landscape remain unresolved.
In general, genome-wide nucleosome mapping relies on the characterization of the products of micrococcal nuclease (MNase) digestion of chromatin; MNase preferentially cleaves the linker DNA between nucleosomes, leaving nucleosomal DNA relatively intact as ~147bp footprints. Genome-wide characterization of MNase digestion products has proceeded rather rapidly from early studies using ~1 kb resolution microarrays, to higher resolution tiling microarrays, to the modern era of deep sequencing (Radman-Livaja and Rando, 2010). Most recently, the Kent and Henikoff groups reported a significant advance in chromatin mapping (Henikoff et al., 2011; Kent et al., 2011), by carrying out paired-end deep sequencing of an entire MNase digestion ladder (as opposed to using size-selected mononucleosomal DNA from such a ladder). These maps reveal not only nucleosome footprints of ~120–150bp (fragments shorter than 147 bp result from MNase “nibbling” on the ends of nucleosomal DNA), but also shorter (<80 bp) footprints of other DNA-binding proteins such as transcription factors.
Here, we use this method to analyze the chromatin structure of murine ES cells and sperm. We find that nucleosome positioning in ES cells is consistent with that of many other cell types, with broadly conserved features such as promoter nucleosome depletion that scales with transcription rate. More interestingly, we confirm that this protocol yields footprints of a wide variety of sequence-specific DNA-binding proteins, including pluripotency factors such as Oct4, as well as the genomic insulator CTCF. We confirm that short MNase footprints over CTCF binding sites correspond to bona fide CTCF binding events by showing that these footprints are lost upon CTCF knockdown in ES cells. Further, we show that sperm nuclease accessibility is broadly different than that of ES cells. In contrast to prior genome-wide studies, we demonstrate that the majority of nucleosomes in sperm are retained over gene-poor regions, and we independently confirm this result microscopically in mature sperm. Nonetheless, using a specific subset of MNase digestion conditions we are also able to recapitulate previous reports of histone retention at CpG-rich promoters, although we show this signal can only account for a minority of the total levels of histone retention in sperm. Lastly, we unexpectedly identify widespread CTCF footprints in sperm, suggesting that this factor may play a role in organizing the sperm genome for fertilization or for embryonic genome activation.
RESULTS
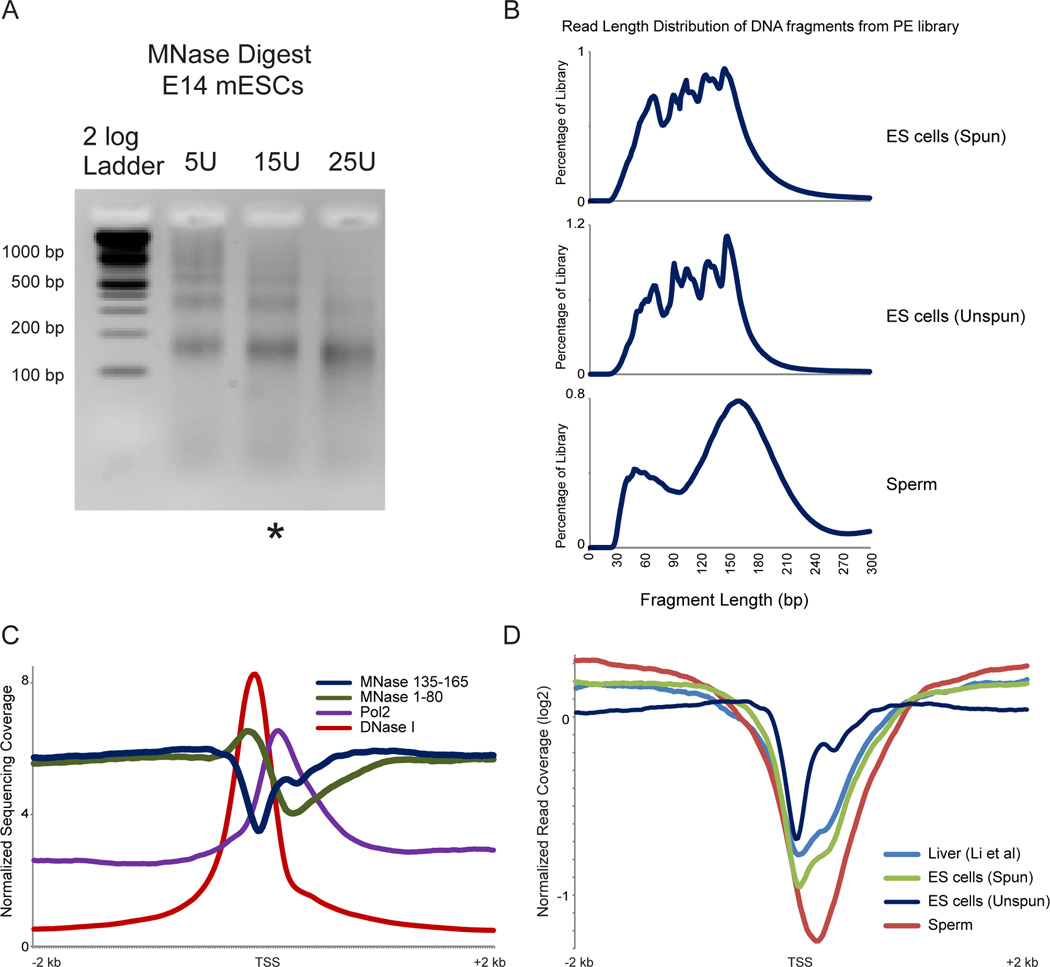
We set out to thoroughly characterize the chromatin structure in murine ES cells and sperm, using a protocol recently developed by the Henikoff group (Henikoff et al., 2011). As is common in nucleosome mapping studies, cells were crosslinked with formaldehyde, lysed, and subjected to MNase digestion. Purified DNA after MNase digestion from ES cells is visualized in Figure 1A, revealing the expected nucleosome ladder. Following digestion, protected genomic DNA was isolated, and the entire nucleosomal ladder was subject to paired-end deep sequencing. In this protocol there is no gel purification of mononucleosome-sized fragments, although several steps during sequencing library preparation select against DNA fragments longer than ~300 bp. After mapping sequencing reads back to the mouse genome, insert sizes were calculated for each fragment. Both libraries exhibited strong mononucleosome-sized protected fragments; 144 bp for ES cells and 157 bp for sperm (Figure 1B). For both libraries subnucleosomal size peaks are also present, with one predominant peak of ~60 bp in sperm (Montellier et al., 2013) and several smaller peaks in ES cells corresponding to 124, 104, 94, 68 bp. The successive 20bp reductions in peak size for ES cells likely represent the products of endonuclease cleavage at sharp bends in the nucleosomal DNA path where DNA interactions with the histone octamer are diminished (Crick and Klug, 1975; Noll and Kornberg, 1977).
Figure 1. Average promoter architecture in embryonic stem cells.
A. Gel electrophoresis of MNase digestion ladders for murine ES cells subject to EGFP knockdown. Characteristic nucleosomal laddering is evident in underdigested ES lanes. ES cell digestion patterns are shown here for unfractionated (unspun) samples – identical results are obtained for the supernatant of low-speed centrifugation (“spun” – not shown). * indicates the titration step used for paired-end deep sequencing library construction. B. Fragment size distribution for sequenced MNase digestion products. After deep sequencing of the indicated libraries, paired sequences were mapped back to the mouse genome and insert size was calculated from genomic distance between reads. ES “spun” indicates that MNase digestion was fractionated by centrifugation and library was constructed from supernatant material, whereas “unspun” library was constructed from the entire MNase digestion. C. Normalized deep sequencing data plotted relative to transcriptional start sites (TSS) of all mouse mm9 refGene transcripts for 1–80 bp MNase-protected fragments, 135–165bp fragments, DNaseI hypersensitivity (GSE40869), and RNAPII ChIP-Seq, as indicated. Y axis represents the average value, for all genes, in parts per million reads. D. Comparison between cell types, and comparison to previous ES cell data (Teif et al., 2012). 135–165 bp MNase footprints were selected for the indicated libraries, and occupancy was averaged for all TSS-aligned genes, shown as log2 of the enrichment relative to the genome-wide average read depth. Two key features are apparent here. First, sperm exhibit a strong nucleosome-depleted region on average. Second, the unspun ES cell library exhibits a relatively positioned +1 nucleosome on average, whereas the supernatant “spun” library exhibits the 3’-extended promoter NDR reported by Teif et al, indicating that 5’ nucleosomes are relatively insoluble under typical MNase digestion conditions.
We first focused on nucleosome-length (135–165 bp) MNase-protected fragments, expecting to recapitulate prior studies on ES cell chromatin structure (Li et al., 2012; Teif et al., 2012). Basic characteristics of nucleosome positioning observed here are consistent with prior observations in a multitude of species, including mammalian cell types (Radman-Livaja and Rando, 2010). Promoters typically exhibit a nucleosome-depleted region (NDR) which coincides with genomic hypersensitivity to DNase I, and both of these features are centered just upstream of both the annotated transcriptional start site (TSS) and the peak of RNA polymerase II (RNAPII) ChIP signal (Yildirim et al., 2011) (Figure 1C and see below). In a prior study on ES cell nucleosomes, Teif et al reported an unusual extension of promoter nucleosome depletion downstream of the TSS (Teif et al., 2012). We do not find such a 3’ NDR extension when deep sequencing unfractionated MNase-digested chromatin (Figure 1C), but this unusual NDR behavior was recapitulated in another dataset in which we centrifuged the MNase digestion and isolated relatively soluble chromatin prior to generating our deep sequencing library (Figures 1B and D). These results suggest that nucleosomes located at the extreme 5’ end of coding regions, such as the +1 nucleosome, may have reduced solubility due to their interactions with transcriptional machinery or other large protein complexes, consistent with prior studies on variability in nucleosomal solubility following nuclease digestion in flies (Henikoff et al., 2009). Our results using unfractionated MNase digests thus show that chromatin organization in ES cells is consistent with chromatin organization in other cell types and species, at least at the level of nucleosome positioning/occupancy.
Chromatin organization at the Transcriptional Start Site
In addition to nucleosome size DNA fragments (135–165 bp), we were also able to recover a wide range of digestion fragments including subnucleosomal-sized DNA fragments. In yeast, such MNase footprints (<80 bp) are particularly abundant over the binding locations of sequence-specific DNA-binding proteins such as transcription factors (TFs) (Henikoff et al., 2011; Kent et al., 2011). To extend these observations to a mammalian system, we averaged data for 1–80 bp fragments for all TSS-aligned genes. As shown in Figure 1C, these footprints were strongly enriched at the 5’ end of the nucleosome-depleted region (NDR), consistent with these footprints corresponding to transcription factors or the basal transcription machinery.
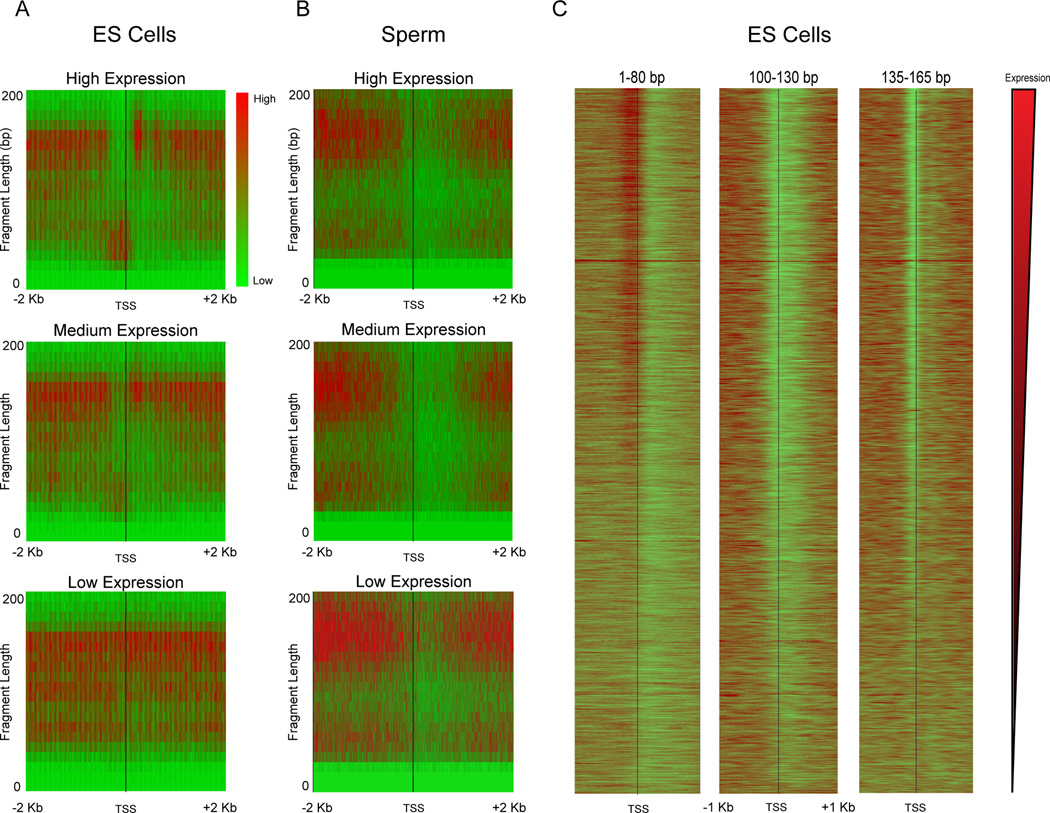
We further explored the connection between MNase footprints and promoter architecture using “V-plots” introduced in Henikoff et al (Henikoff et al., 2011). Briefly, in these plots sequencing reads are aligned by a specific genomic position (anchor); in this case, the annotated TSSs of all genes in the mouse genome. The x-axis represents the distance from the anchor along the genome, while the y-axis shows the sequencing fragment size. Enrichment or depletion of footprints is shown in red or green, respectively, in a heatmap. Figure 2A shows V-plots for TSS-aligned data in ES cells for three sets of genes grouped according to expression level. Examining nucleosome-size DNA fragments (size range 135–165 bp) reveals the expected behavior – nucleosomes are dramatically depleted from the promoters of highly-expressed genes, with modest nucleosome depletion occurring at moderately-expressed genes, and minimal nucleosome depletion at silent genes (Figure 2A). At highly-expressed genes, we observe a well-positioned +1 nucleosome with a slightly enlarged (~175 bp) footprint, potentially representing a nucleosome engaged either with RNAPII or with some component of the basal transcription machinery. In contrast, short DNA fragments (1–80 bp) reveal the converse behavior, with footprints being observed most strongly at promoters with the highest expression in ES cells. This is consistent with the notion that these footprints result from TFs or the basal transcription machinery, as previously suggested in flies (Teves and Henikoff, 2011). Finally, we note that intermediate size footprints (100–130 bp) are depleted surrounding the TSS in all three groups of genes. These footprints presumably result from MNase digestion of partially unwrapped or less stably bound nucleosomes, and show little correspondence with transcription rate.
Figure 2. Relationship between transcript abundance and MNase footprinting.
A–B. Enrichment of reads around TSSs is shown in red-green (enriched-depleted) heatmap for genes grouped according to mRNA abundance in E14 mES cells: High (top 5%), Mid (middle 5%), Low (bottom 5%). For each panel, x axis shows distance from the TSS, y axis shows size of the MNase footprint. Data for ES cell MNase digestion is shown in (A), data for mature sperm in (B). Note that sperm “expression levels” derive from mRNA abundance from round spermatids (Namekawa et al., 2006) as mature sperm are transcriptionally inactive. C. Gene-resolution heatmap of deep sequencing reads from ES cell MNase digestion, aligned by TSS for three prominent size classes: <80 bp, 100–130 bp, and 135–165 bp. Each row is a single gene, and rows are sorted from high to low mRNA abundance.
Next we compared these TSS plots in ES cells to sperm (Figure 2B). Most dramatically, in contrast to the strong TF signal at ES promoters, sperm promoters were strongly depleted of these subnucleosomal footprints. This lack of promoter TF signal is consistent with the global, or nearly-global, absence of active transcription in mature sperm (Kierszenbaum and Tres, 1975; Ward and Zalensky, 1996). Curiously, while no, or very little, transcription occurs in mouse sperm (expression values for sperm TSS plots were generated based on gene expression during the round spermatid stage), significant depletion of nucleosome-sized fragments remains evident at promoters (Figure 1D, Figure 2B). We speculate that this lack of apparent nucleosome occupancy at promoters could potentially be a remnant of transcriptional activity in round spermatids rather than the result of active repositioning or selective removal of histones during chromatin compaction in spermatogenesis (see also below). In addition to the lack of a strong signature of the basal transcription machinery (above), and the depletion of nucleosome footprints observed at promoters, we also noted a broadening of mononucleosomal footprint width in the sperm V-plots (Figure 2B, see also Figure 1B). The cause of this broadening is presently unclear.
The conclusions drawn from the averaged data were further confirmed on a gene-by-gene basis for ES cells. Figure 2C shows how the three size class fragments described above correlate with gene expression level – for each size class, all genes are ordered by ES cell mRNA abundance from high (top) to low (bottom). Together, our ES cell data validate our experimental approach, and extend our understanding of ES cell chromatin to include a broad atlas of DNA-binding proteins.
Sperm histone retention
Having demonstrated the utility of this protocol in ES cells, we next sought to understand the unusual chromatin state of mammalian sperm by comparing our sperm and ES cell datasets in more detail. First, it is notable that in our protocol MNase digestion of sperm results in a nucleosome ladder very similar to that observed for somatic cells (see below). While this was expected for ES cells, the ladder observed for sperm is intriguing given that only ~2% of histones are retained in mature mouse sperm. If sparse nucleosomes were randomly retained at 2% of genomic loci, then adjacent nucleosomes producing a dinucleosomal band would occur quite rarely; instead, the finding of a nucleosomal ladder indicates that nucleosome retention in sperm generally occurs in blocks of multiple adjacent nucleosomes.
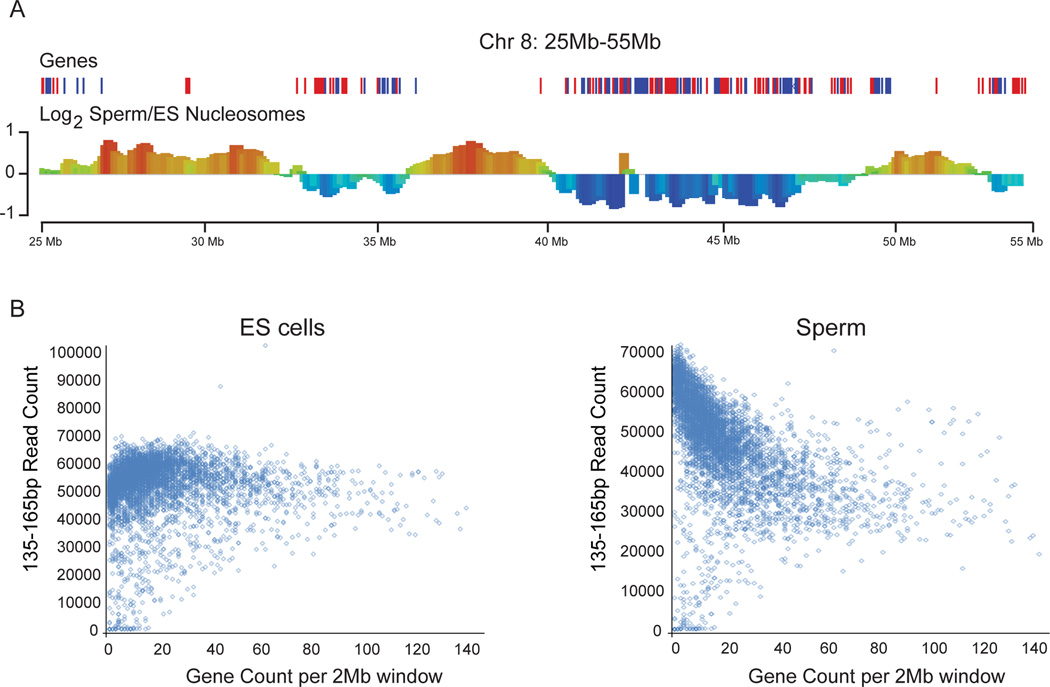
Turning to the deep sequencing data, we noted in initial surveys of genome browser tracks that nucleosome occupancy was fairly uniform across the genome in ES cells, but was substantially more variable at the megabase scale in murine sperm, with long blocks exhibiting ~2–3 fold variation in average nucleosome occupancy (Figure S1A–B). When comparing the enrichment of nucleosome size fragments (135–165bp) between mouse ES and sperm cells using a 2 Mb sliding window, we observed that differential nucleosome occupancy was strongly correlated with gene density (Figure 3A). Specifically, nucleosomes appear to be preferentially retained in gene-poor regions in sperm relative to ES cells. To quantitate this trend genome-wide, we binned the genome into 2 Mb bins and plotted total nucleosome-sized sequencing reads versus the number of genes within each window. As expected, ES cell chromatin exhibits uniform nucleosome occupancy across the genome, whereas in sperm the number of nucleosome-length fragments for a given window is strongly anti-correlated with the number of genes in that window (Figure 3B). Importantly, the low nucleosome occupancy over promoters (Figure 1D) and gene-rich regions (Figure 3B) inferred from nucleosome-sized MNase footprints was also observed using low-coverage anti-histone H3 ChIP-Seq (Figure S1C–D), validating the hypothesis that the ~150 bp MNase footprints (Figure 4A) represent bona fide histone retention in sperm.
Figure 3. Preferential histone retention occurs at gene deserts in sperm.
A. Sperm nucleosomes are retained in gene-poor regions compared to ES cell nucleosomes. Normalized mononucleosomal (135–165 bp) MNase footprints were averaged for 2 Mb bins, and the log2 of the relative enrichment for ES cells vs. sperm (y axis) is shown for a typical stretch of chromosome 8 (genomic coordinate on x axis). See also Figure S1. B. Scatterplot of gene density (x-axis) vs. number of mononucleosome-sized MNase sequencing reads (y-axis) for ES cells and sperm. The small number of points in the lower left corner depleted of sequencing reads correspond to 2 Mb bins comprised of largely unmappable or unannotated sequence. Low-depth ChIP-Seq using anti-H3 in sperm also reveals this anticorrelation between gene density and H3 signal (Figure S1D), strongly supporting the claim that mononucleosome-sized fragments released by MNase digestion of sperm indeed correspond to nucleosomes.
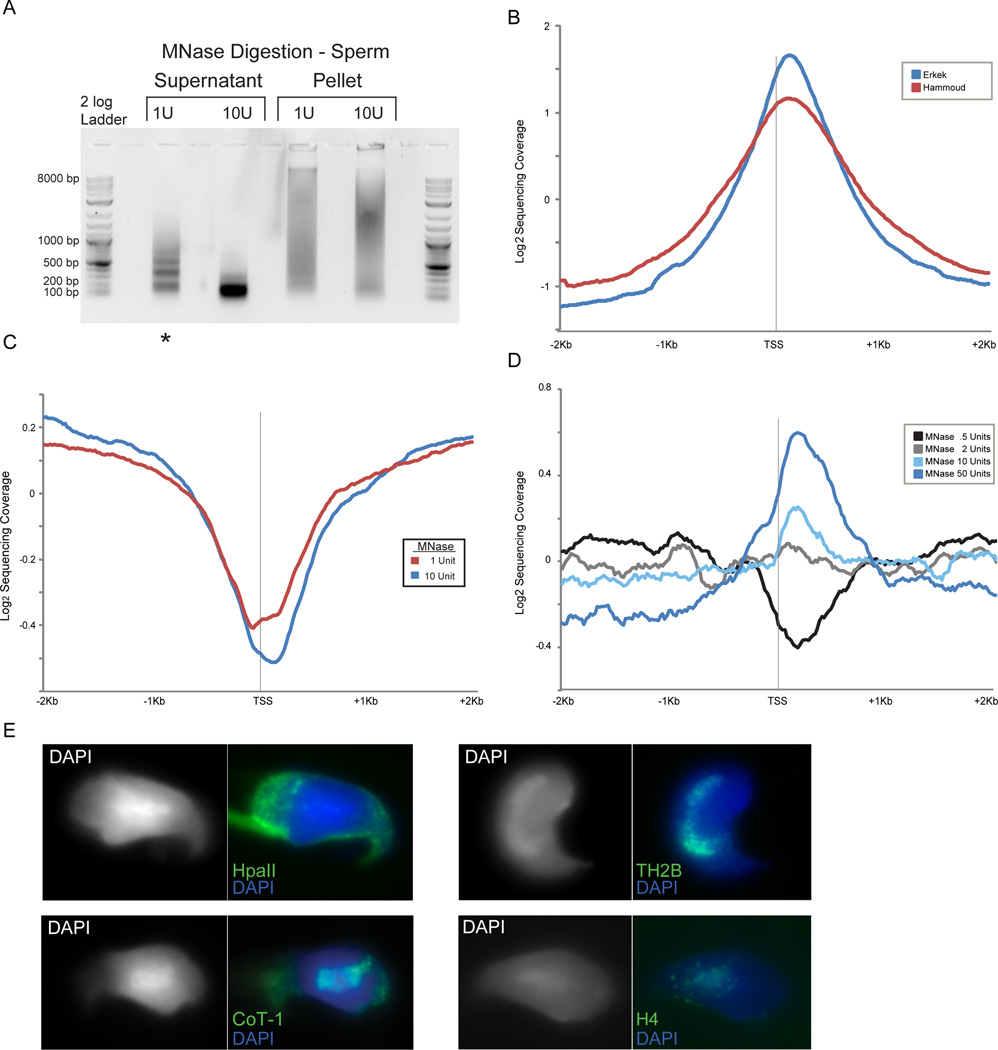
Figure 4. A small subset of nucleosomes are retained at promoters in sperm.
A. Gel electrophoresis of soluble vs. insoluble MNase digestion products from mature M. musculus sperm. Lane with asterisk designates level of digestion used for most analyses. Pellet fraction contains the majority of protamine protein, while supernatant carries the majority of histone proteins. B. Aggregation plot of data from Hammoud et al (H. sapiens) and Erkek et al datasets (M. musculus) aligned by TSS. C. Aggregation plot of MNase seq reads for 1 U and 10 U MNase digestion levels (digestions shown in (A)). These libraries were generated from the supernatant of the MNase digestion material. D. Aggregation plots of MNase-seq data for unspun, not size-selected, digestion series of formaldehyde-crosslinked M. musculus sperm. A peak of promoter nucleosome occupancy is revealed only after extensive MNase digestion (note that MNase levels are not directly comparable to those in (C) as fewer sperm were used in this digestion), and is more prominent when MNase digestions are not subject to centrifugation (Figures S2B–C). E. Representative images for fluorescent in situ hybridizations (FISH) with HpaII probe (poorly methylated CpG islands – see Methods) and Cot-1 probe (gene-poor heterochromatic regions), as well as immunofluorescence using antibodies to TH2B (testis specific H2B) and bulk H4, demonstrating that the majority of anti-histone staining overlaps the DAPI-rich chromocenter that also stains with Cot-1 probe.
Several previous studies examining histone retention in murine and human sperm reported strong retention of nucleosomes in the promoter regions of genes implicated in early development – these regions generally are also characterized by high CpG density and low DNA methylation (Erkek et al., 2013; Hammoud et al., 2009). This can be observed in our reanalysis of those published datasets, as TSS-aligned aggregation plots of read coverage confirms strong retention of promoter nucleosomes under the reported conditions (Figure 4B). In contrast, nucleosome-size MNase footprints in our dataset were generally depleted from promoters, including developmental promoters such as the Hox promoters (Figures 2B, 4C, and S1B). We speculated that these dramatic discrepancies might result from differences in the extent of micrococcal nuclease digestion – in yeast, several studies have reported a subset of so-called “fragile nucleosomes” that are particularly susceptible to extensive MNase digestion (Weiner et al.; Xi et al., 2011). Deep sequencing of mononucleosome-sized DNA released from sperm by varying levels of MNase digestion revealed that nucleosome depletion at promoters was also observed at relatively low MNase concentrations, arguing against promoter nucleosomes being fragile (Figure 4C). In contrast, after extensive MNase digestion of sperm we observed a ~150 bp footprint becoming enriched over promoters (Figure 4D), particularly over the “high CpG” class of promoters that are associated with developmental regulators (Figure S2A).
In addition to the extent of MNase digestion, our protocol differs from the Peters and Cairns protocols in that we digest formaldehyde-crosslinked sperm – we could not generate nucleosome-sized fragments from murine sperm in the absence of crosslinking – whereas both the Peters and Cairns groups carried out MNase digestions on uncrosslinked sperm that were first swelled in water/DTT prior to MNase digestion. Interestingly, we found that after centrifuging the sperm MNase digests to separate primarily histone-associated DNA (supernatant) from the more abundant protamine-associated genome in the pellet, promoter retention of ~150 bp footprints was somewhat stronger in the pellet than in the supernatant (Figures S2B–C). Thus, we conclude that the previously-reported histone retention at developmental promoters represents a specific subset of all histone retention in sperm in which promoter-bound nucleosomes are revealed only by extensive nuclease digestion – presumably “overdigestion” eventually degrades the more abundant nucleosomes found over gene deserts, revealing the more stable promoter nucleosomes – and may furthermore be partially lost during centrifugation due to formaldehyde crosslinking to some insoluble scaffold.
Our hypothesis that histones associated with developmental promoters represent a small subset of the nucleosomes retained in sperm is also supported by elementary arithmetical considerations – human sperm retain ~10% of the nucleosome complement of a somatic cell (Hammoud et al., 2009), yet even if histones were penetrantly retained in 100% of sperm across 1 kb of sequence at all ~30,000 promoters this would only account for 1% of the haploid genome. Nonetheless, we sought to independently assess our conclusion that most histone retention occurs in gene-poor regions in murine sperm (Figure 3B). Given the sensitivity to nuclease levels documented above for digestion-based assays, we used fluorescence in situ hybridization (FISH) and immunofluorescence as a completely independent assay for histone localization in mature mouse spermatozoa. Mammalian sperm contains a highly organized nucleus with a DAPI-dense heterochromatic center surrounded by a euchromatic periphery that exhibits more modest DAPI staining (Li et al., 2008; van der Heijden et al., 2006). To determine the location within the sperm nucleus of CpG-rich developmental promoters, we generated florescent probes from size-selected 50–500 bp DNA fragments of HpaII-digested genomic DNA. Cot-1 DNA probes were used to confirm the physical location of repetitive heterochromatic sequences. Signal from the HpaII probe is prominent in the nuclear periphery while Cot1 signal overlaps almost perfectly with the chromocenter, as expected (Figure 4E). We then used immunofluorescence to characterize the localization of histones H4 and TH2B, finding that the majority of histone staining occurs in the central DAPI-dense chromocenter, consistent with prior studies (Figure 4E) (Govin et al., 2007; Meyer-Ficca et al., 2013; van der Heijden et al., 2006). Taken together, our data confirm that a small subset of histone retention occurs at CpG-rich promoters, but demonstrate that the vast majority of histone retention in mammalian sperm occurs in gene deserts.
Short MNase-protected fragments coincide with DNA-binding protein footprints
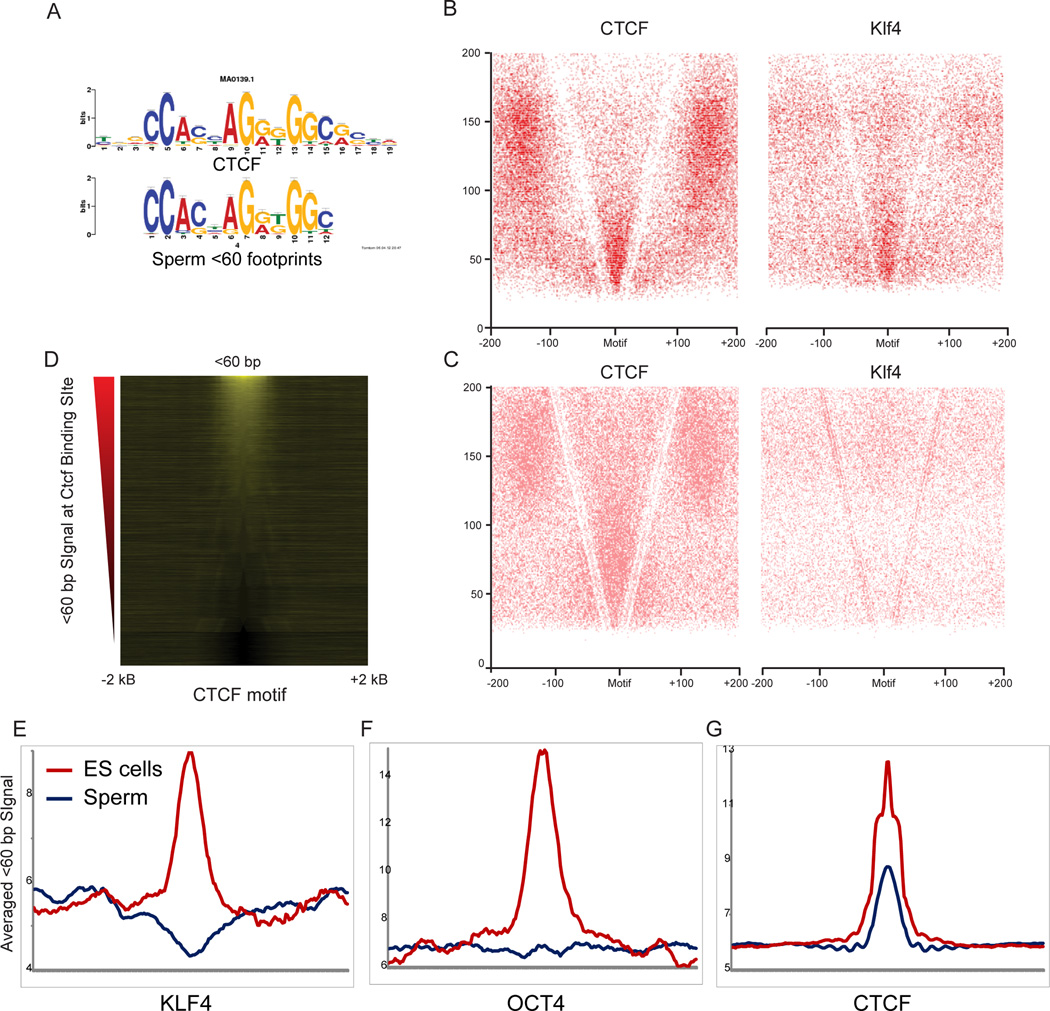
We finally turn to more detailed analysis of short MNase footprints in sperm and ES cells. As shown previously (Kent et al., 2011), short MNase-protected footprints can reveal binding sites for DNA-binding proteins such as transcription factors. In order to determine whether our data revealed the locations of specific DNA-binding proteins, we sought sequence motifs enriched in short MNase-protected fragments relative to the background of longer (nucleosome or dinucleosome) footprints (see Methods). In ES cells, significantly enriched motifs included sequence matches for a number of transcription factors that play key roles in the pluripotency transcriptional network such as Klf4, as well as a strong match to the binding site for key insulator and locus control regulator CTCF. Plotting short MNase footprints around in vivo binding sites for a variety of transcription factors reveals a range of footprinting behaviors for transcription factors (Figure S3). Despite the lack of active transcription in sperm, we were also able to identify a number of significantly enriched motifs among the short MNase footprints in sperm. Sperm footprints did not carry strong matches to the Klf4 binding site or most other pluripotency transcription factors, but instead were enriched for a subset of sequence motifs from the ES cell motif list such as Stat3, RelA, and, most notably, CTCF (Figures 5A and S3). In contrast, a number of TFs, including key pluripotency factors Oct4, Nanog, Sox2, and Klf4, were associated with strong 1–80 bp footprints specifically in ES cells, but not in sperm. Interestingly, we did not identify any motifs specifically protected in sperm but not in ES cells.
Figure 5. Enrichment of short MNase fragments over binding sites for sequence-specific TFs.
A. Short MNase footprints in sperm occur over CTCF binding sites. <60 bp MNase digestion products from sperm were searched for overrepresented sequence motifs, and the top motif hit is shown alongside the published sequence motif for CTCF. B–C. V-plots (as in Figure 2A, but showing all digestion products as points rather than enrichment/depletion) for ES cell (B) or sperm (C) MNase digestion products, anchored by CTCF and Klf4 binding sites. D. Short footprints in sperm are associated with a subset of the CTCF binding sites identified in ES cells. Heatmap shows enrichment of <60bp DNA fragments from sperm dataset, centered on CTCF motifs that are empirically CTCF-bound in ES cells (Chen et al., 2008). Sites are sorted from high to low signal intensity over 100 bp surrounding the binding site. E. Examples of short (<80 bp) footprints averaged over various TF motifs, reproduced from Figure S3.
To further explore MNase footprinting patterns around transcription factor binding sites, we focused on Klf4 and CTCF, both of which have previously been mapped in murine ES cells (Chen et al., 2008). Using matches to the relevant sequence motif located within previously identified ChIP-Seq peaks as anchors (n=15,657 for CTCF, n=1,813 for Klf4), we generated V-plots for ES cell MNase footprints (Figure 5B). As previously shown for yeast General Regulatory Factors such as Reb1 (Henikoff et al., 2011), three characteristics are apparent in these plots. First, strong enrichment in the 30–60 bp size range is observed centered on the TF motif, indicating presumed protection of DNA from MNase by the bound TF. Second, a V-shaped depletion of fragments results from MNase cleavage immediately adjacent to the TF. Finally, enriched 140–160 bp fragments located outside this V are consistent with well-positioned nucleosomes occurring flanking the TF binding sites, as previously reported for CTCF (Fu et al., 2008; Teif et al., 2012). Similar, albeit weaker, results were observed over Klf4 anchors.
In contrast to the extensive literature on ES cell transcription factor binding, little is known about the localization or even presence of sequence-specific DNA-binding factors in mature sperm. However, as noted above we identified a number of sequence motifs enriched among short MNase footprints in mature sperm, most notably the CTCF motif (Figure 5A). We repeated the V-plot analyses for CTCF and Klf4 binding sites for sperm MNase data (Figure 5C). Consistent with our motif searches, we find little evidence for Klf4 binding in mature sperm, but we confirm enrichment for <60bp fragments at CTCF binding sites (Figures 5C and E). The modest enrichment observed here relative to the ES cell peak is explained by the fact that only a subset (~40%) of the CTCF motifs that are bound in ES cells are associated with MNase footprints in mature sperm (Figure 5D).
Subnucleosomal MNase footprints represent bona fide CTCF binding events
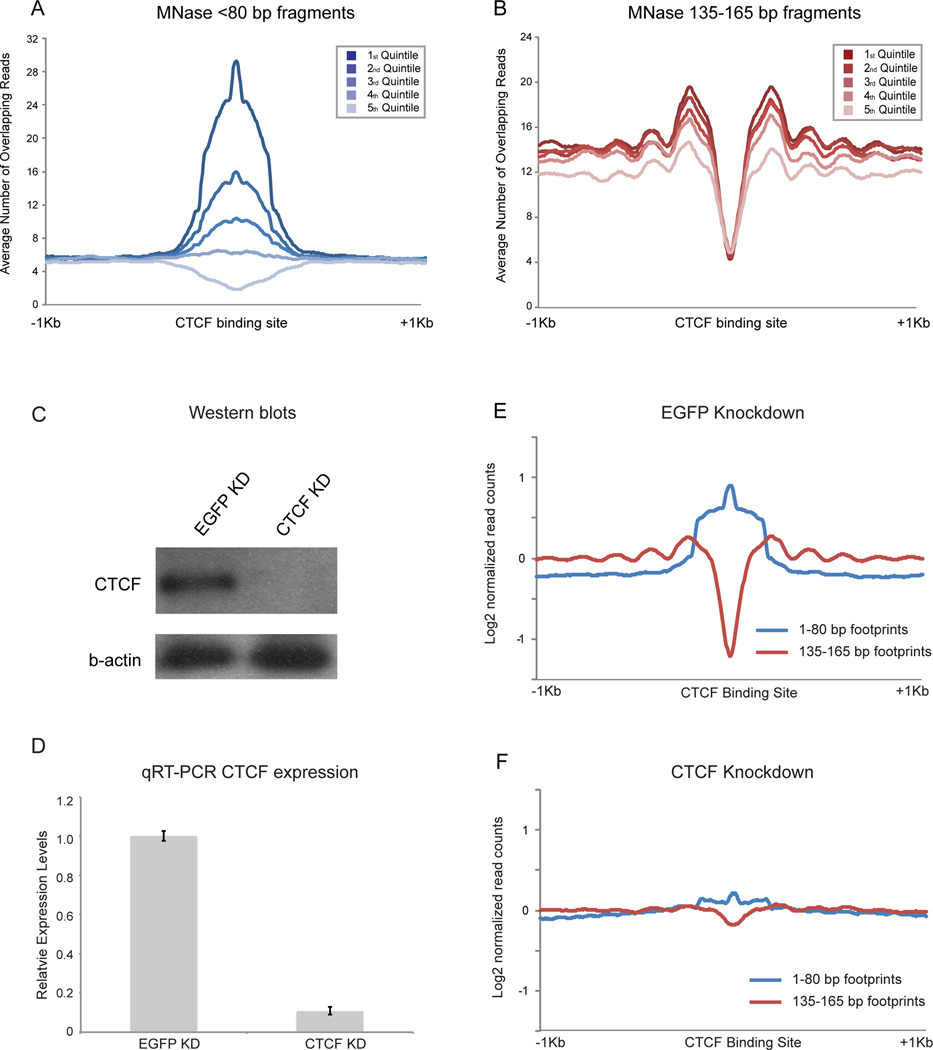
The presence of sequence motifs underlying short MNase footprints provides us with strong hypotheses regarding transcription factor binding events, which make several additional testable predictions. First, the extent of footprinting over CTCF motifs should correlate with in vivo occupancy levels as assayed by ChIP-Seq. Second, these footprints should be eliminated by CTCF knockdown (KD). To test these predictions, we carried out ChIP q-PCR for 6 loci predicted to be CTCF binding sites in ES cells, as well as 3 CTCF motifs only exhibiting MNase footprints in sperm. All 9 loci exhibited the expected CTCF binding in ES cells, and this binding was lost upon CTCF knockdown (KD) (Figure S4A). More globally, we made use of published CTCF ChIP-Seq localization data generated using ES cells (Chen et al., 2008). Enrichment of short MNase fragments over CTCF binding motifs was indeed correlated with in vivo CTCF occupancy, consistent with the idea that these short footprints correspond to CTCF protected-fragments (Figure 6A). Interestingly, nucleosome phasing surrounding CTCF binding sites also correlated with CTCF binding levels (Figure 6B), supporting the hypothesis that CTCF acts as a positioning barrier to establish nucleosome arrays (Radman-Livaja and Rando, 2010).
Figure 6. Nuclease-resistant footprints over CTCF motifs represent bona fide CTCF binding in ES cells.
A. Abundance of short MNase footprints over CTCF binding sites in ES cells correlates with CTCF ChIP-Seq enrichment. CTCF motifs are split into quintiles according to ChIP-seq signal (Chen et al., 2008). B. As in (A), but for mononucleosome-length footprints. Flanking nucleosomes are more strongly positioned when CTCF ChIP-seq signal is highest. C. Western blots of CTCF and EGFP esiRNA knockdown in mESCs, probed with anti-CTCF and β-actin antibodies. D. qRT-PCR of CTCF mRNA abundance. E. Aggregation plot for <80 bp and 135–165 bp digestion products in EGFP KD ES cells, aligned using CTCF motifs. F. Knockdown of CTCF in ES cells results in loss of <80bp footprint enrichment over CTCF motifs, with an associated increase in nucleosome occupancy over the CTCF motif and loss of surrounding nucleosome positioning. See also Figures S4–5.
Finally, we directly assessed whether short MNase footprints over CTCF binding sites resulted from bona fide CTCF binding (as opposed to MNase sequence biases, or binding by a factor other than CTCF such as its paralog CTCFL/BORIS) by repeating our MNase mapping experiment in ES cells following CTCF knockdown (KD). Overall we achieved a ~90% KD of CTCF as illustrated by both Western blotting and mRNA abundance (Figures 6C–D). Genome-wide, results for this experiment were strongly correlated with results from control or EGFP knockdown ES cells, with similar fragment size distributions, nucleosome depletion at promoters, and short transcription-correlated promoter footprints (not shown). However, as predicted, short fragments over CTCF binding sites were greatly diminished in the CTCF KD cells (Figure 6F, Figures S4B–C). The remaining short fragments could result either from residual CTCF following incomplete (~90%) knockdown, or from the CTCFL/BORIS protein that binds a similar sequence motif to CTCF (Jelinic et al., 2006). Nucleosome phasing was also lost surrounding CTCF binding motifs in the knockdown cells (Figure 6F), providing strong evidence that CTCF binding (rather than some aspect of DNA ”programming” of chromatin structure) is required for nucleosome positioning at these locations.
Finally, we asked whether MNase footprints in sperm were likely to result from CTCF itself or from CTCFL/BORIS, which binds to the same sequence and plays important roles in male reproduction (Jelinic et al., 2006; Loukinov et al., 2002). First, Western blotting confirms the presence of CTCF protein in mature sperm (Figure S4D), consistent with prior reports (Tang and Chen, 2006). Despite the presence of this protein in sperm, to date we have been unable to successfully recover ChIP material, likely due to the unique crosslinking and fragmentation challenges presented by the highly compact sperm nucleus. As an indirect approach to the question of the CTCF footprinting protein, we focused on CTCF-specific and BORIS-specific binding events recently described in ES cells (Sleutels et al., 2012) and asked whether these sites were occupied by short MNase DNA fragments from our sperm dataset. We find that <60bp MNase footprints over CTCF motifs in sperm preferentially occur over sites bound exclusively (in ES cells) to CTCF, while they are completely absent over known CTCFL-only binding sites (Figure S5). These results support our hypothesis that CTCF itself is associated with the genome in mature murine sperm.
DISCUSSION
By characterizing a wide size spectrum of MNase digestion products, we have profiled the chromatin architecture of two unusual mouse cell types, embryonic stem cells and sperm, illuminating nucleosome occupancy and transcription factor binding in each cell type, and providing strong evidence for the presence of bound transcription factors such as CTCF in mature sperm.
Embryonic Stem Cell chromatin
In ES cells, our analysis of mononucleosome-sized footprints confirms well-known aspects of chromatin biology, such as a strong relationship between promoter nucleosome depletion and transcription level. We therefore focused on analysis of features only appreciable in non-size-selected libraries. Extending our analysis to short MNase footprints reveals a strong correlation between these footprints at the 5’ ends of promoters and transcription rate, thus supporting the hypothesis (Teves and Henikoff, 2011) that these MNase footprints reflect the presence of the basal transcription machinery (among other DNA-binding proteins).
Short MNase footprints were enriched for sequence motifs associated with a subset of key pluripotency regulators, such as Klf4. Interestingly, we did not observe equally strong footprinting at experimentally validated binding sites for all transcription factors (Figure S3). This was previously observed in yeast studies, where an unusual subset of transcription factors (largely corresponding to the abundant “General Regulatory Factors”) showed far stronger footprinting than the majority of transcription factors (Henikoff et al., 2011). The absence of strong MNase footprinting at certain TF binding sites (such as for Esrrb in ES cells) could reflect weak crosslinking to DNA, low average occupancy, or inability of the DNA-bound TF to interfere with MNase digestion. It is interesting in this regard that the binding sites associated with strong footprints, such as CTCF and Klf4 binding sites, tended to be associated with relatively well-positioned flanking nucleosomes (Figure 5B). One potential explanation for this correlation would be that TF footprints are strongest for factors associated with particular chromatin remodeling complexes.
We tested the hypothesis that these short MNase footprints report on bona fide TF binding in vivo, finding that CTCF footprints exhibited strong correlation with CTCF ChIP-Seq data, and further showing that MNase footprints over CTCF binding sites are greatly reduced upon CTCF knockdown. Finally, we extended the prior observation that CTCF binding is associated with strong nucleosome phasing (Fu et al., 2008) by showing that the level of CTCF binding corresponds to the strength of nucleosome positioning, supporting the hypothesis that TFs can function as positioning barriers. Moreover, loss of CTCF binding upon knockdown leads to a loss of nucleosome phasing around this site. Taken together, these results lend further support to the hypothesis that TFs are capable of directing nucleosome positioning.
Analysis of sperm histone retention
The unusual packaging state of mammalian sperm, which retain ~2–15% of the histone load of somatic cells (Ooi and Henikoff, 2007), has been proposed to play roles in programming early embryonic gene expression (Brykczynska et al., 2010; Gardiner-Garden et al., 1998; Hammoud et al., 2009) and in epigenetic inheritance of cytosine methylation states (Nakamura et al., 2012). The specialized subset of genomic loci associated with retained histones in sperm are therefore of great interest.
In principle, retention of nucleosomes over 2% of the genome in sperm could occur stochastically (eg, each sperm cell retains nucleosomes over a different 2% of the genome), deterministically (all sperm retain nucleosomes over specific loci), or somewhere in between. Prior reports on sperm histone retention in humans and mice, using nuclease sensitivity as a key assay (Arpanahi et al., 2009; Brykczynska et al., 2010; Hammoud et al., 2009), came to somewhat different conclusions. Two studies isolated solubilized mononucleosome-sized fragments after MNase digestion of human sperm, finding enrichment over the promoters of key developmental regulators, with a very strong correlation with GC content (Brykczynska et al., 2010; Hammoud et al., 2009; Vavouri and Lehner, 2011). Somewhat different results were obtained via tiling microarray analysis of DNA released after light restriction enzyme or MNase digestion of human and mouse sperm, with gene and promoter-rich regions flanked by CTCF sites very broadly overrepresented in much longer (Megabase-scale) domains (Arpanahi et al., 2009; Saida et al., 2011).
Here, we confirm that nucleosome-sized fragments can be found over CpG-rich promoters, but find that these nucleosomes are unusual in that they are only released by extensive MNase digestion, and appear to be crosslinked by formaldehyde into a somewhat insoluble complex. Moreover, we show that these nucleosomes represent a small fraction of all nucleosomes retained in mature sperm – preferential histone retention over gene deserts is observed robustly across a range of MNase digestion levels, and fluorescence microscopy independently confirms that the majority of histone protein is localized in a distinct chromocenter occupied by repeat elements, rather than the CpG-rich fraction of the genome associated with developmental promoters (Figure 4E). It is currently unclear whether either of these classes of histone-bound loci serves any function in early embryonic processes (but see (Ihara et al., 2014)), and how these functions might differ between the broad blocks of nucleosomes occurring over repeat elements (eg gene deserts) and the more focal nucleosomes found over CpG islands. Interestingly, recent studies have shown that retention of H3K9me2-marked histones plays a role in protecting a fraction of the paternal genome from Tet3-dependent cytosine demethylation (Nakamura et al., 2012). Preliminary low-coverage anti-H3K9me2 ChIP-Seq shows preferential localization of this mark to gene-poor regions of the genome (not shown). Given that we find histone retention primarily occurring over gene deserts, our results suggest a model in which the sperm packaging state preferentially protects epigenetic silencing marks from removal at repeat elements. Advances in editing the epigenome should allow targeting histone eviction or covalent modifications at specific loci in gametes, to definitively determine the role for sperm histones in zygotic biology.
Finally, we provide evidence that a subset of CTCF binding sites are associated with strong MNase footprints in mature sperm. This suggests the hypothesis that mature sperm carry chromatin-associated CTCF at specific genomic locations. CTCF protein has previously been reported to be present in mature mouse sperm (Tang and Chen, 2006), although it was not shown to associate with the sperm genome. CTCF is intricately involved in epigenetic control of a number of well-studied imprinted genes (Phillips and Corces, 2009), raising the intriguing possibility that CTCF associated with the paternal genome plays a regulatory role in gene expression during early development. Future studies using conditional alleles of CTCF will be needed to assess if and how chromatin-bound CTCF protein in sperm influences gene expression upon zygotic genome activation, and whether inheritance of CTCF-bound sperm nuclei is essential for developmental gene regulation.
Methods
Tissue culture and sperm isolation
ES cell lines used in this study were mouse E14 lines cultured using standard conditions. Briefly, E14 ES cells were cultured on gelatin-coated dishes without feeder cells in standard media containing serum and LIF at 37°C with 5% CO2. ES cells were passed by washing with PBS, and dissociating with trypsin. Sperm were isolated from the caudal epididymis and vas deferens of 10 week old C57/6J mice by lacerating tissue in 1 mL of 37°C M2 medium (Sigma). Sperm were allowed to swim up for 1 hr, washed 1× in water, 1× PBS, crosslinked at 37°C with 1% formaldehyde for 15 min, quenched with 250mM glycine for 5 min and frozen in liquid nitrogen. The majority of figures were generated using data from control EGFP knockdown ES cultures, with the exception of Figures S3 and 4E, which used untreated ES cell data which had higher levels of short MNase footprints. Control and EGFP knockdown data yield nearly-identical results; EGFP knockdowns were used simply based on the higher sequencing depth for these libraries.
Sperm permeabilization
Frozen sperm were briefly thawed at 37°C, pelleted 4000g for 4 min at room temp (RT), and washed 1× in 1 mL PBS then pellet was resuspended in 1 mL Sperm Permeabilizion Buffer SPB (.025% Trypsin/EDTA, 10 mM DTT, 5 mM Tris pH 7.5, 5 mM NaCl, 1.5 mM MgCl2, .25% NP40, 1mM PMSF). Sperm are incubated in SPB for 5 min at 37°C. After 5 min, 10 µL of Fetal Bovine Serum was added to inactivate Trypsin and sample was incubated for additional 5 min at 37°C. Sperm are pellet 4000g for 4 min at room temp (RT), washed with PBS 2×, and resuspended in 500 µL PBS + 2 mM Cacl2 and incubated 5 min at 37°C.
RNAi-mediated knockdown
RNA interference using endoribonuclease III siRNAs (esiRNAs) was performed as previously described (Fazzio et al., 2008), transfecting E14 mouse ES cells using Lipofectamine2000. Briefly, templates for esiRNA production were produced using a two-step PCR procedure followed by in vitro transcription and dicing of double stranded RNA using RNase III. Knockdowns were performed for 48 hours to achieve effective KD.
MNase Digestion
Permeabilized ES and sperm nuclei were treated with 10 Units/106 cells and 1 Unit/108 cells of micrococcal nuclease (Worthington), respectively, for 5 min 37°C. Reaction was stopped with the addition of 10 mM EGTA on ice. For ES cells, nuclei were incubated for 4 hrs at 4°C with rotation, pelleted at 5000g for 5 min, and separated into supernatant and pellet (spun) or were treated the same without centrifugation and separation (unspun). After MNase digestion of sperm nuclei, digestion was centrifuged for 5000g for 5 min – the supernatant from this spin contained the vast majority of histone protein, whereas the vast majority of protamine protein was located in the pellet (not shown). All sperm data except Figures 4D and S2A, C were generated using the supernatant fraction – Figures 4D and S2A were generated using libraries constructed without any centrifugation step. For Figures S2B–C, libraries were prepared from sperm digested with a range of MNase concentrations 1, 5, 20, and 100 Units for 5 min @ 37°C, where digestion was stopped with EGTA on ice and incubated for 4 hrs at 4°C. Supernatant and Pellet fractions were separated by centrifugation at 5000g for 5 min, and the ~150 bp mononucleosome size DNA was used to generate deep sequencing libraries. In all cases, DNA was isolated by Phenol:Chloroform:Isoamyl Alcohol and EtOH precipitation.
Sperm ChIP-seq
Mouse sperm was isolated as described above. Chromatin isolated from 6 mice was used for the immunoprecipitation experiments. Crosslinked sperm was washed with 1× with PBS, resuspended in 600uL of 20mM Tris with 20% Glycerol and 10mM PMSF and subjected to bead-beating (.5mm glass beads). SDS was added to a final concentration of 1% and chromatin was sheared to ~300bp with Bioruptor. Samples were kept on ice at all stages. ChIP was performed overnight at 4°C with panH3 antibody abcam ab1791.
Deep sequencing
To characterize the entire range of MNase digestion products we generated paired-end libraries of DNA fragments from both ES and sperm digestions and performed sequencing using Illumina HiSeq technology. Libraries were prepared as described in Henikoff et al (Henikoff et al.). Reads were mapped to the mouse genome (mm9) using Bowtie2, and uniquely mapped reads were used for further analysis. For library insert size distributions see Figure 1B. For most figures, reads were normalized to parts per million reads. In Figures 4, 6, and S2, data are normalized to the average genome-wide coverage and represented as log2 of the fold-difference from the genomic average.
The majority of analyses were carried out using an ES cell (unspun) dataset comprising 388,389,043 total reads obtained from aggregating 2 biological replicates, ES cell (spun) with 384,462,519 reads, and a sperm dataset comprising 389,791,625 reads obtained from aggregating 3 biological replicate datasets. Data will be available via the Gene Expression Omnibus (GEO), Accession #GSE58101.
Motif Analysis
We first trimmed sequencing adaptors, considering reverse-complementarity of the paired-end reads, to recover reads sequenced from short DNA fragments. Then all reads were aligned to mouse genome (mm9) by Bowtie2 with default parameters (end-to-end alignment mode and random selection for multiple mappers). Only concordantly aligned read pairs were retained, and the length of the sequenced DNA fragments was calculated according to their mapped coordinates. Reads representing DNA fragments of mono-nucleosomal MNase (135–165 bp), subnucleosomal MNase (100–130 bp), and TF-sized (<80 bp) footprints were segregated for several analyses, as indicated. We performed peak-calling from TF-sized reads against other two groups (as background inputs) using MACS. We further selected the top 1000 most significant peaks and refined their borders by extending 20 bp both up- and down-stream of the peak summits, which are the points with highest read coverage within peaks. We then search for enriched motifs by MEME among the genomic sequences of the refined peaks. Enriched de novo motifs were compared to known motifs using TOMTOM with the motif frequency models from Jaspar.
FISH
Sperm were collected by swim up as previously described, and slides were prepared as in (van der Heijden et al., 2006). Sperm placed on coverslips were permeabilized in CSK buffer, 5% triton, and VRC (vanadyl ribonucleoside complex) for 3 min, then fixed in 4% Paraformaldehyde for 10 min, and stored in 70% EtOH. Antibodies used for immunohistochemistry were panH4 (Upstate Cat# 05-858 Lot# 27171) and TH2B (Abcam Cat# ab23913 Lot# 825930). Primary antibodies were detected with anti-rabbit 488 (Jackson ImmunoResearch). Slides were incubated with 1:250 primary antibody in 1%BSA, 1xPBS for 1 hour at 37°C, then washed, and immunodetected using 1:500 dilution of conjugated (Alexa 488 or Alexa 594, Invitrogen) secondary (anti-goat, mouse or rabbit) antibody, in 1xPBS with 1% BSA. HpaII probe was prepared by performing an overnight RE digest on purified mouse sperm DNA followed by gel purification of 50–500 bp DNA fragments, ligation of adapters and PCR amplification in the presence of Biotin-dCTP. Mouse CoT-1 DNA (Invitrogen) probe was nick-translated using biotin-11-dUTP (Roche). 200 ng HpaII biotinylated probe or 50 ng CoT-1 biotin nick translated probe was hybridized to fixed and denatured cells.
Supplementary Material
HIGHLIGHTS.
Genome-wide catalog of DNA-binding by nucleosomes and TFs in ES cells and sperm
Sperm histone retention primarily occurs in gene-poor regions
Nuclease overdigestion is required to reveal rare promoter nucleosomes in sperm
Insulator factor CTCF appears to be associated with the genome of mature sperm
Acknowledgements
We thank K. Ahmad and members of the Rando lab for insightful discussions and comments on the manuscript. OJR is supported in part by grant HD080224 from NICHD, and by the Harold and Leila Mathers Charitable Foundation. TGF is supported in part by grant HD072122 from NICHD and is a Pew Scholar in the Biomedical Sciences. ZW is supported by NSF grant DBI-0850008. BRC is supported by the T32HD007439 training grant. SJH is supported by the T32CA130807 training grant and by a Leukemia and Lymphoma postdoctoral fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arpanahi A, Brinkworth M, Iles D, Krawetz SA, Paradowska A, Platts AE, Saida M, Steger K, Tedder P, Miller D. Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences. Genome Res. 2009 doi: 10.1101/gr.094953.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, Beisel C, Schubeler D, Stadler MB, Peters AH. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nature structural & molecular biology. 2010;17:679–687. doi: 10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Crick FH, Klug A. Kinky helix. Nature. 1975;255:530–533. doi: 10.1038/255530a0. [DOI] [PubMed] [Google Scholar]
- Erkek S, Hisano M, Liang CY, Gill M, Murr R, Dieker J, Schubeler D, van der Vlag J, Stadler MB, Peters AH. Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nature structural & molecular biology. 2013;20:868–875. doi: 10.1038/nsmb.2599. [DOI] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sinha M, Peterson CL, Weng Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS genetics. 2008;4:e1000138. doi: 10.1371/journal.pgen.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M, Ballesteros M, Gordon M, Tam PP. Histone- and protamine-DNA association: conservation of different patterns within the beta-globin domain in human sperm. Mol Cell Biol. 1998;18:3350–3356. doi: 10.1128/mcb.18.6.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govin J, Escoffier E, Rousseaux S, Kuhn L, Ferro M, Thevenon J, Catena R, Davidson I, Garin J, Khochbin S, et al. Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis. The Journal of cell biology. 2007;176:283–294. doi: 10.1083/jcb.200604141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff JG, Belsky JA, Krassovsky K, MacAlpine DM, Henikoff S. Epigenome characterization at single base-pair resolution. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18318–18323. doi: 10.1073/pnas.1110731108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res. 2009;19:460–469. doi: 10.1101/gr.087619.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara M, Meyer-Ficca ML, Leu NA, Rao S, Li F, Gregory BD, Zalenskaya IA, Schultz RM, Meyer RG. Paternal poly (adp-ribose) metabolism modulates retention of inheritable sperm histones and early embryonic gene expression. PLoS genetics. 2014;10:e1004317. doi: 10.1371/journal.pgen.1004317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinic P, Stehle JC, Shaw P. The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol. 2006;4:e355. doi: 10.1371/journal.pbio.0040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent NA, Adams S, Moorhouse A, Paszkiewicz K. Chromatin particle spectrum analysis: a method for comparative chromatin structure analysis using paired-end mode next-generation DNA sequencing. Nucleic acids research. 2011;39:e26. doi: 10.1093/nar/gkq1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierszenbaum AL, Tres LL. Structural and transcriptional features of the mouse spermatid genome. The Journal of cell biology. 1975;65:258–270. doi: 10.1083/jcb.65.2.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Lalancette C, Miller D, Krawetz SA. Characterization of nucleohistone and nucleoprotamine components in the mature human sperm nucleus. Asian journal of andrology. 2008;10:535–541. doi: 10.1111/j.1745-7262.2008.00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Gadue P, Chen K, Jiao Y, Tuteja G, Schug J, Li W, Kaestner KH. Foxa2 and H2A.Z Mediate Nucleosome Depletion during Embryonic Stem Cell Differentiation. Cell. 2012;151:1608–1616. doi: 10.1016/j.cell.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loukinov DI, Pugacheva E, Vatolin S, Pack SD, Moon H, Chernukhin I, Mannan P, Larsson E, Kanduri C, Vostrov AA, et al. BORIS, a novel male germ-line-specific protein associated with epigenetic reprogramming events, shares the same 11-zinc-finger domain with CTCF, the insulator protein involved in reading imprinting marks in the soma. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:6806–6811. doi: 10.1073/pnas.092123699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Ficca ML, Lonchar JD, Ihara M, Bader JJ, Meyer RG. Alteration of poly(ADP-ribose) metabolism affects murine sperm nuclear architecture by impairing pericentric heterochromatin condensation. Chromosoma. 2013;122:319–335. doi: 10.1007/s00412-013-0416-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montellier E, Boussouar F, Rousseaux S, Zhang K, Buchou T, Fenaille F, Shiota H, Debernardi A, Hery P, Curtet S, et al. Chromatin-to-nucleoprotamine transition is controlled by the histone H2B variant TH2B. Genes & development. 2013;27:1680–1692. doi: 10.1101/gad.220095.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Liu YJ, Nakashima H, Umehara H, Inoue K, Matoba S, Tachibana M, Ogura A, Shinkai Y, Nakano T. PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature. 2012;486:415–419. doi: 10.1038/nature11093. [DOI] [PubMed] [Google Scholar]
- Namekawa SH, Park PJ, Zhang LF, Shima JE, McCarrey JR, Griswold MD, Lee JT. Postmeiotic sex chromatin in the male germline of mice. Current biology : CB. 2006;16:660–667. doi: 10.1016/j.cub.2006.01.066. [DOI] [PubMed] [Google Scholar]
- Noll M, Kornberg RD. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977;109:393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- Ooi SL, Henikoff S. Germline histone dynamics and epigenetics. Curr Opin Cell Biol. 2007;19:257–265. doi: 10.1016/j.ceb.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittoggi C, Renzi L, Zaccagnini G, Cimini D, Degrassi F, Giordano R, Magnano AR, Lorenzini R, Lavia P, Spadafora C. A fraction of mouse sperm chromatin is organized in nucleosomal hypersensitive domains enriched in retroposon DNA. Journal of cell science. 1999;112(Pt 20):3537–3548. doi: 10.1242/jcs.112.20.3537. [DOI] [PubMed] [Google Scholar]
- Radman-Livaja M, Rando OJ. Nucleosome positioning: How is it established, and why does it matter? Developmental biology. 2010 doi: 10.1016/j.ydbio.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saida M, Iles D, Elnefati A, Brinkworth M, Miller D. Key gene regulatory sequences with distinctive ontological signatures associate with differentially endonuclease-accessible mouse sperm chromatin. Reproduction. 2011;142:73–86. doi: 10.1530/REP-10-0536. [DOI] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels F, Soochit W, Bartkuhn M, Heath H, Dienstbach S, Bergmaier P, Franke V, Rosa-Garrido M, van de Nobelen S, Caesar L, et al. The male germ cell gene regulator CTCFL is functionally different from CTCF and binds CTCF-like consensus sites in a nucleosome composition-dependent manner. Epigenetics & chromatin. 2012;5:8. doi: 10.1186/1756-8935-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang JB, Chen YH. Identification of a tyrosine-phosphorylated CCCTC-binding nuclear factor in capacitated mouse spermatozoa. Proteomics. 2006;6:4800–4807. doi: 10.1002/pmic.200600256. [DOI] [PubMed] [Google Scholar]
- Teif VB, Vainshtein Y, Caudron-Herger M, Mallm JP, Marth C, Hofer T, Rippe K. Genome-wide nucleosome positioning during embryonic stem cell development. Nature structural & molecular biology. 2012;19:1185–1192. doi: 10.1038/nsmb.2419. [DOI] [PubMed] [Google Scholar]
- Teves SS, Henikoff S. Heat shock reduces stalled RNA polymerase II and nucleosome turnover genome-wide. Genes & development. 2011;25:2387–2397. doi: 10.1101/gad.177675.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–520. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden GW, Derijck AA, Ramos L, Giele M, van der Vlag J, de Boer P. Transmission of modified nucleosomes from the mouse male germline to the zygote and subsequent remodeling of paternal chromatin. Developmental biology. 2006;298:458–469. doi: 10.1016/j.ydbio.2006.06.051. [DOI] [PubMed] [Google Scholar]
- Vavouri T, Lehner B. Chromatin organization in sperm may be the major functional consequence of base composition variation in the human genome. PLoS genetics. 2011;7:e1002036. doi: 10.1371/journal.pgen.1002036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward WS, Zalensky AO. The unique, complex organization of the transcriptionally silent sperm chromatin. Crit Rev Eukaryot Gene Expr. 1996;6:139–147. doi: 10.1615/critreveukargeneexpr.v6.i2-3.30. [DOI] [PubMed] [Google Scholar]
- Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 2010 doi: 10.1101/gr.098509.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, Yao J, Chen R, Li W, He X. Nucleosome fragility reveals novel functional states of chromatin and poises genes for activation. Genome Res. 2011;21:718–724. doi: 10.1101/gr.117101.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Weng Z, Rando OJ, Fazzio TG. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.