Abstract
RNA editing can lead to amino acid substitutions in protein sequences, alternative pre-mRNA splicing and changes in gene expression levels. The exact in vivo modes of interaction of the RNA editing enzymes with their targets are not well understood. Alterations in RNA editing have been linked to various human disorders and the improved understanding of the editing mechanism and specificity can explain the phenotypes that result from mis-regulation of RNA editing. Unbiased high-throughput methods of detection of RNA editing events genome-wide in human cells are necessary for the task of deciphering the RNA editing regulatory code. With the rapidly falling cost of genome re-sequencing, the future method of choice for the detection of RNA editing events will be whole-genome gDNA and cDNA sequencing. We describe a detailed procedure for the computational identification of RNA editing targets using the data from the deep sequencing of DNA and RNA from the peripheral blood mononuclear cells of a human individual with severe hemophilia A who is resistant to HIV infection. Interestingly, we find that mRNAs of the cyclin-dependent kinase CDK13 and the DNA repair enzyme NEIL1 undergo extensive A→ I RNA editing that lead to amino acid substitutions in protein sequences.
Keywords: RNA editing, Single nucleotide variants, High-throughput sequencing, Bioinformatics, Human immunodeficiency virus infection
1. Introduction
RNA editing is the post-transcriptional alteration of RNA sequences through the insertion, deletion or modification of nucleotides, excluding changes due to processes such as RNA splicing and polyadenylation (1). Such alterations in RNA sequences can bring about amino acid substitutions in protein sequences, alternative pre-mRNA splicing and changes in gene expression levels (2). In higher eukaryotes, the most prevalent type of RNA editing is mediated by adenosine deaminase acting on RNA (ADAR) enzymes that convert adenosines to inosines (A→ I editing) in double-stranded RNA substrates (3). The three major types of A→ I editing targets are: protein-coding pre-mRNAs, repetitive elements such as Alu repeats located in exons or introns, and microRNA precursors (3). The exact in vivo modes of interaction of the RNA editing enzymes with their targets are unknown, but the base-paired RNA structures are believed to guide the enzymes to edit a single nucleotide with high specificity and efficiency (2). Alterations in RNA editing have been linked to various human disorders and the improved understanding of the editing mechanism and specificity can explain the phenotypic features that result from mis-regulation of RNA editing (4). Unbiased high-throughput methods of detection of RNA editing events genome-wide in normal and abnormal human cells are necessary for the task of deciphering the RNA editing regulatory code. Recent rapid developments in massively parallel DNA sequencing technologies (5, 6) have allowed the identification of RNA editing targets in human cells at 36,000 genomic loci by a targeted sequencing (7). With the rapidly falling cost of genome re-sequencing (8), the future method of choice for the detection of RNA editing events will be whole-genome genomic DNA (gDNA) and complementary DNA (cDNA) sequencing. Herein, we describe a detailed procedure for the computational identification of RNA editing targets using a dataset of raw sequence reads from the deep sequencing of DNA and RNA from one human individual.
2. Materials
2.1. Deep sequencing dataset
We obtained cDNA and gDNA raw sequencing data from the study published by Cirulli et al. (9). Let us briefly describe the samples from this work. The DNA and RNA were extracted from peripheral blood mononuclear cells (PBMCs) from an individual with severe hemophilia A, who is resistant to HIV infection. The DNA was prepared for sequencing according to Illumina’s gDNA sample prep kit protocol. The total RNA was prepared according to the Illumina RNA-Seq protocol that involved globin reduction and polyA enrichment. For alternative RNA preparation procedures, see Note 1.
The paired-end reads for the gDNA and cDNA libraries are each around 75 bp long. There are 1,450 million and 280 million reads in gDNA and cDNA libraries, respectively.
The raw sequencing data is in standard Sanger FASTQ format. A FASTQ file uses four lines per nucleotide sequence. An example entry for a single sequence in a FASTQ file is shown below:
@SRR037167.9742210 CCCGACGTTACATCATCTGCCCCGTTGTATGCAACA +SRR037167.9742210 6-66+06(63+&0(&666+66(-6&+1(03&(.)&)
For each entry in a FASTQ file, the first line begins with a “@” character and is followed by a sequence identifier and an optional description. The second line is the raw sequence. The third line begins with a “+” character and is optionally followed by a description. The fourth line encodes Phred quality scores for the sequence from the second line, and contains the same number of symbols as letters in the sequence. Phred quality score Q of a base call is defined as Q = −10 log10 p, where p is the probability that the base call is incorrect. The Phred quality score Q encoded by an ASCII of the character x is given by Q = ord(x) − 33, where ord(x) is the decimal integer ASCII value corresponding to x. For example, the symbol “&” corresponds to the Phred score Q = ord(&) − 33 = 38 − 33 = 5.
2.2. Computational hardware
The handling and processing of large datasets generated from deep sequencing experiments is most convenient on Linux and Unix-based computers. The storage of the raw sequence dataset described in Subheading 2.1 alone requires more than 300 GB of disk space. There are several additional large processed files such as alignment files that need to be stored. In principle, the Unix piping utilities can be used to deal with compressed datasets to save some disk space at the expense of CPU time. Nevertheless, we recommend a computer with at least 2TB of free disk space in order to comfortably work with the large datasets. It is also desirable that the computer has multiple processors/cores and at least 20GB of RAM so that several computationally intensive jobs can be run concurrently.
2.3. Computational software
Bowtie is an ultrafast, memory-efficient alignment program for aligning short DNA sequence reads to large genomes (10). For the human genome, Bowtie aligns more than 15 million 75 bp reads per CPU hour. The pre-compiled executable of Bowtie and pre-built index of human hg18 genome can be downloaded from (http://bowtie-bio.sourceforge.net).
SAMtools is a library and software package for parsing and manipulating alignments in the SAM/BAM formats (11). The source code for SAMtools is available from (http://samtools.sourceforge.net/). SAMtools needs to be compiled using GNU C compiler (http://gcc.gnu.org/).
Picard comprises Java-based command-line utilities that manipulate SAM files. It is available from (http://picard.sourceforge.net/). We use Picard to remove PCR amplification bias in alignment data.
3. Methods
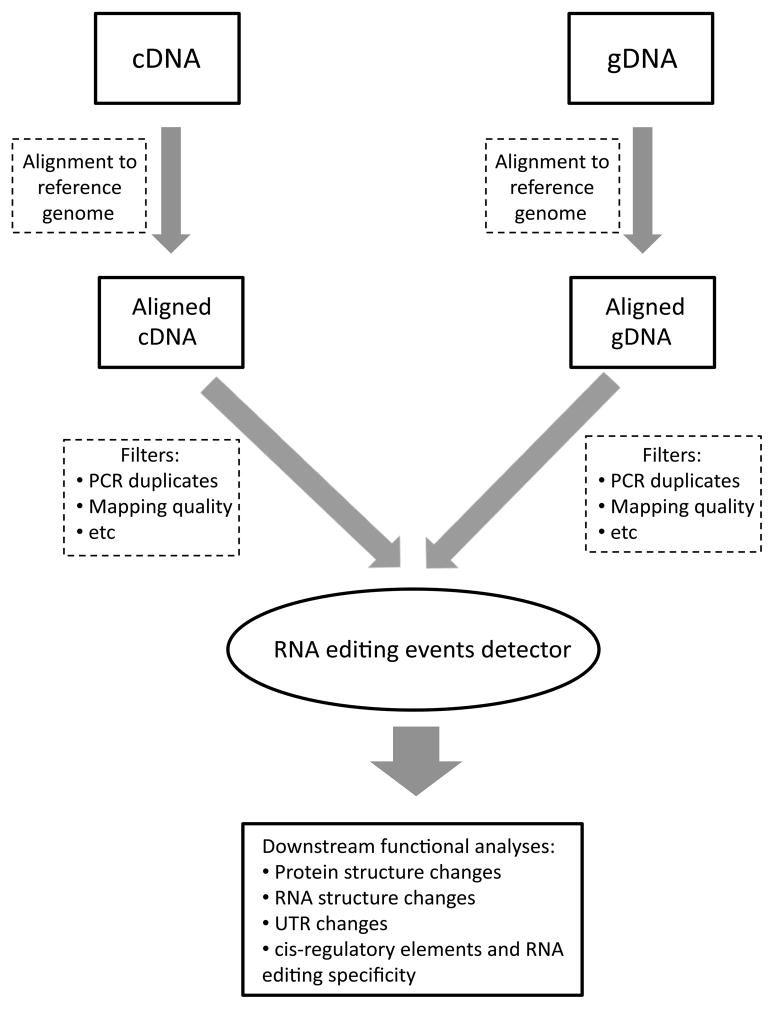
The RNA editing analysis is conceptually simple as illustrated in Fig. 1. The cDNA and gDNA sequencing reads are aligned to a reference genome. The aligned reads are then passed into various filters. The aligned and filtered cDNA and gDNA reads are then fed into the RNA editing events detector which outputs a list of candidate editing sites in the genome. The putative editing sites can then be analyzed for their functional consequences such as protein structure/function changes due to non-synonymous base substitutions, changes in RNA structure and UTR. The identification of RNA editing sites genome-wide and in multiple cell types facilitates the decoding of the RNA editing regulatory code that involves a complex interaction of cis-regulatory sequences, base-paired RNA structures and trans-acting elements.
Figure 1.
3.1. Sequence alignment
The Sequence Alignment/Map (SAM) format is a common alignment format that supports all sequence types (11). It is designed to scale to alignment sets of 1011 or more base pairs, which is typical for the deep sequencing of one human individual. Starting from the FASTQ file of raw reads data, we can align reads to hg18 reference human genome using Bowtie as follows:
bowtie hg18 -S -p 4 -n 2 -l 30 –e 70 –k 1 --best cDNA.fastq cDNA.sam
In the above command, the “-S” option sets the output alignment format to SAM. The “-p 4” allows the speed-up of the alignment process by using 4 processors/cores in parallel. The specification “-k 1 --best” guarantees that the best valid alignment per read will be reported. The validity of an alignment is specified as “-n 2 -l 30 –e 70”. The latter specification means that (a) Alignments may have no more than 2 mismatches (“-n” option) in the first 30 bases (“-l” option) on the high-quality end of the read, (b) the sum of Phred quality values at all mismatched positions may not exceed 70 (“-e” option). See Note 2 for an alignment method more appropriate for cDNA sequences.
3.2. Post-processing alignments
3.2.1. Compressing alignment files and indexing
The Binary Alignment/Map (BAM) format is the binary representation of SAM and keeps the same information as SAM (11). The BAM alignment file for the gDNA data from (9) requires around 90 GB disk space which is equivalent to a compression rate of approximately 1.0 byte per input base. The command to generate BAM file from gDNA SAM file is:
samtools view -bS -o gDNA.bam gDNA.sam
The BAM alignment file should be sorted by coordinate for an efficient data processing and to avoid loading extra alignments into memory. A BAM file can be sorted as follows:
samtools sort gDNA.bam gDNA_sorted
The position sorted BAM file “gDNA_sorted.bam” can now be indexed to achieve fast random retrieval of alignments overlapping a specific genomic region as follows:
samtools index gDNA_sorted.bam
Let us also index the FASTA file “hg18.fa” of the reference human genome sequence using the command:
samtools faidx hg18.fa
We can now view alignments at any genomic location using a text-based viewer using “tview” option as follows:
samtools tview gDNA_sorted.bam hg18.fa
3.2.2. Duplicate reads filtering
In order to remove possible PCR amplification artifacts, it is reasonable to retain only one or a few reads that align to the same genomic position (12). We used Picard’s “MarkDuplicates” function to properly remove duplicate reads. If multiple reads align to the same genomic location, Picard retains a single read with the best sequence quality. The following command removes duplicate reads from the sorted BAM file “cDNA_sorted.bam” and returns the sorted BAM file “cDNA_sorted_rmdup_picard.bam”.
java -Xmx4g -jar MarkDuplicates.jar INPUT=cDNA_sorted.bam \ OUTPUT=cDNA_sorted_rmdup_picard.bam REMOVE_DUPLICATES=true METRICS_FILE=MF.txt \ AS=true
In the above command, the –Xmx4g option sets the maximum java heap size to 4GB.
3.2.3. Pileup format and variant calling
The Pileup format describes the base-pair information at each chromosomal position. This format facilitates SNP/indel calling. An example pileup format file is shown below:
chr1 2012 T 6 ,C,... CBBAAC chr1 2586 T 8 CcGGCCC. #BA##A;? chr1 8745 A 5 c,... ##AB? chr1 8754 T 7 ,,...,c ;9BA;## chr1 8769 T 10 ,,...,,,.c ;3AAA#/:B# chr1 8772 t 11 ,,...,,,.,c 8>BB?#1;B#@ chr1 8773 c 12 ,,...,,,.,t, A?BB9;=8@#6#
Here each line consists of a chromosome, a 1-based coordinate, a reference base, the number of reads covering the site, the read bases and the base qualities. At the read base column, a dot stands for a match to the reference base on the forward strand, a comma for a match on the reverse strand, “ACGTN” for a mismatch on the forward strand and “acgtn” for a mismatch on the reverse strand. The first line in the above example shows that (a) there are six sequence reads that cover position chr1:2,012 in the genome, (b) the reference nucleotide at this position is T, (c) five reads have matching nucleotides at this position with two of these reads aligning to the reverse strand of the genome and three to the forward strand, and (d) the remaining read aligns to the forward strand with the mismatch nucleotide C at this position. More details on the pileup format can be found in SAMtools manual. Given a FASTA file “hg18.fa” of human genome sequence and sorted BAM file “gDNA.bam” of aligned gDNA sequences, a huge pileup file “gDNA_pileup.txt” for all genomic locations covered by at least one sequencing read is generated as follows:
samtools pileup -f hg18.fa gDNA.bam > gDNA_pileup.txt
If option -c is applied to SAMtools, the IUPAC consensus base, Phred-scaled consensus quality, SNP quality and root mean square mapping quality of the reads covering the site will be inserted between the “reference base” and the “number of reads covering the site” columns as in the following example:
chr1 95543 t K 36 36 60 14 ..GG,.G...G.,. B@@BB@BBBABCAC chr1 98160 a C 38 39 60 5 .cccc #>;;# chr1 98173 t C 50 51 60 9 ccccCccac B?@4/B9#.
3.2.4. Efficient data processing with UNIX pipes
Whenever possible, generation of huge files should be avoided. UNIX pipes should be used for streaming the output of one program into the input of another program so that disk space usage is minimized. For example, after consensus base calling, we want to filter out sites with very high read coverage because such sites are error prone. We can use SAMtools to set maximum read depth using the “–D” option. We then apply a filter to keep only those sites that have mapping quality equal or greater than 20. SAMtools works well with UNIX pipes. Let “cDNA.bam” and “gDNA.bam” be duplicate-reads filtered sorted BAM files for cDNA and gDNA alignments, respectively. The variant calling and the two filters can be combined using pipes as follows:
samtools pileup -f hg18.fa –c gDNA.bam | samtools.pl varFilter -D100 | awk '$6>=20' > gDNA_variants.txt
Similarly, instead of generating a full pileup file from cDNA alignment data, we can use pipes to keep only those sites that have read coverage of at least five and have at least one read with nucleotide mismatch as follows:
samtools pileup -f hg18.fa cDNA.bam | perl variant_site.pl > cDNA_pileup.txt
Here the script “variant_site.pl” is given by the following simple Perl code:
while(<>){
@row = split /\t/;
if ($row[4] =~ /[ACGTacgt]/){
if ($row[3] >= 5){
print $_;
}
}
}
3.3. Probabilistic framework for detecting RNA editing sites
3.3.1. Theory
At a given single-nucleotide position in the diploid human genome let the genotype be X1/X2. The genotype can be heterozygous (X1 ≠ X2) or homozygous (X1 = X2). We look for the evidence of RNA editing only at homozygous sites in gDNA because such sites constitute an overwhelming majority of sites in the genome and because it is somewhat complicated to do RNA editing analysis of the heterozygous sites. So, let the genotype at the homozygous locus x be XgDNA/XgDNA. Let the nucleotide at the position x in the reference hg18 human genome be Xhg18. There are two possibilities: XgDNA = Xhg18 or XgDNA ≠ Xhg18. We only consider homozygous loci in gDNA where XgDNA = Xhg18 since such loci constitute the overwhelming majority of homozygous loci in the human genome. See Note 3 for the XgDNA ≠ Xhg18 case.
Let the homozygous site x with the genotype XgDNA/XgDNA be located in a genomic region that is transcribed. In the absence of RNA editing the cDNA will have nucleotide XcDNA = XgDNA at position x. In general, there will be two species of cDNA: a fraction f of cDNA will be unedited and have XcDNA = XgDNA and a fraction 1− f of cDNA will be edited and have XcDNA ≠ XgDNA at position x. For the sake of simplicity, we only consider the most prevalent type of RNA editing: A→ I editing. Inosine is interpreted as guanosine by the translational machinery, and therefore, A→ I editing is functionally equivalent to an A→ G conversion.
If the sequencing error rate were identically zero, the likelihood of observing n(A) of A nucleotides and n(G) of G nucleotides at the position x (the conditional probability of observed data given the un-edited fraction f of RNA species), would be given by the binomial probability P(D | f) = f n(A) (1 − f)n(G). The maximum likelihood estimate (MLE) of f is given by fML =n(A)/[n(A)+n(G)].
In reality the sequencing error rate is non-zero and the probability of base error, Phred probability, needs to be taken into consideration. Let D be the observed sequence data, which is generated by sampling RNA species and noisy sequencing measurements. If the maximum likelihood of data assuming non-zero fraction of edited RNA species, maxf P(D | f), is much greater than the likelihood of the data assuming no RNA editing, P(D | f =1), we have a strong evidence for an RNA editing event (7).
If the base error probabilities are small, P(D | f ≠ 1) can still be approximated by the binomial distribution mentioned above. Otherwise one can proceed as follows. As a prerequisite, the reader is referred to Li et al. (13) for an introduction to a probabilistic theory of base error rates and variant calling. In the absence of any sequencing errors there is still a variability in the number of observed A’s and G’s due to the sampling noise. Let us denote by R the unobserved ‘sequencing-error-free’ data. P(D | f) can then be expanded as follows: P(D | f) = ΣR P(D | R) P(R | f). The conditional probability P(D | R) can be computed using Phred base error probabilities as in (13). The conditional probability P(R | f) describes the sampling noise and is given by f n(A) (1 − f)n(G), where n(A) and n(G) are numbers of A’s and G’s in the data R.
The probability P(D | f =1) can be computed using Phred base error probabilities as follows. Let Sa and Sg be two sets of cDNA reads that contain reads with called bases ‘A’ and ‘G’ at a homozygous A/A genomic locus x, respectively. Since it is assumed that there is no RNA editing at the locus x, the base call ‘G’ should be treated as a base calling error. If we denote by p the base error probabilities, we have P(D | f =1) = (Πm ∈Sg pm) (Πk ∈Sa (1−pk)). The base error probability is related to Phred base quality score as Q = −10 log10 p.
3.3.2. Implementation
We now have almost everything at hand for detecting RNA editing sites. The pileup file “cDNA_pileup.txt” from Subheading 3.2.4 contains around 5.8 million sites that are covered by at least five cDNA reads and at least one read has single-nucleotide mismatch with hg18 reference human genome. As explained in Subheading 3.3.1 we restrict our analysis to homozygous gDNA loci that match the hg18 genome. In Subheading 3.2.4 we obtained “gDNA_variants.txt” file that contains homozygous and heterozygous sites in gDNA that have mismatches with the hg18 genome. We thus removed all these gDNA variant sites from the file “cDNA_pileup.txt”. This procedure filtered out around 38,000 sites from the latter file. From the resulting list we also removed sites that have gDNA reads coverage less than 10 because such sites can represent false negatives in gDNA variant discovery. The coverage of gDNA reads at a list of genomic sites can be computed using the “pileup -l” option in SAMtools. We then retrieved genomic coordinates of hg18 exons from Ensembl database (www.ensembl.org) and retained only the putative RNA editing sites in exons. The resulting filtered “cDNA_pileup_filtered.txt” file contains all necessary information for the calculation of likelihood ratios using the theory from Subheading 3.3.1.
Note that ASCII integer values of characters can be computed using the “ord” function in Perl. One additional thing to remember for an efficient computation of probabilities is that products of base error probabilities correspond to the sum of Phred quality values.
We set the cutoff for the log likelihood ratio to be 4: log10 [maxf P(D | f)/P(D | f = 1)] ≥ 4, and identified 7,955 A→ G editing sites in human exons.
3.4. Functional analysis of RNA editing sites
We obtained genomic coordinates of 5′ and 3′ UTR regions from the Ensembl database. 413 A→ G editing sites are located in 5′UTR regions whereas 1,813 are in 3′UTR regions. We recommend BEDTools (14), a suite of utilities to work with BED format files, to identify editing sites that overlap genomic features such as UTR regions.
For the purposes of identifying non-synonymous sites, miscellaneous information about hg18 transcripts such as transcription start and end sites, coding start and end sites, genomic strand, exon start and end sites, and exon frames was retrieved from the UCSC genome Table Browser (http://genome.ucsc.edu). We identified 1,860 non-synonymous A→ G editing sites. See Note 4 for a functional test of these sites.
To further narrow down the list of putative non-synonymous editing sites to a very high-confidence list, we selected sites that fulfill the following criteria: (a) log likelihood ratio ≥ 20, (b) the number of reads with “G” at the putative editing site is at least 10 i.e. n(G) ≥10 and (c) the editing level, defined as 100 n(G)/[n(A)+n(G)], is at least 20%. There are 161 editing sites that fulfill these criteria. Interestingly, A→ G editing at chr6: 32,822,103 in the HLA-DQA2 gene, which is known to interact with a number of HIV proteins (15), results in the amino acid substitution Q → R. A closer inspection reveals, however, that the putative RNA editing site in HLA-DQA2 is likely a false positive. The sequence around the putative RNA editing site in the HLA-DQA2 gene is identical to the sequence surrounding an A/G single nucleotide polymorphism at chr6:32,718,473 in the HLA-DQA1 gene.
The results of analysis of high-throughput data should always be carefully checked. Only after various confounding factors are excluded as possible explanation of the results, we can assume biological validity of conclusions. In the context identification of RNA editing sites in mature microRNAs, it was noted that RNA sequences obtained from deep sequencing experiments could be inadvertently mapped to incorrect locations (16). Such cross-mapping of sequencing reads can lead to overrepresented mismatches at specific locations between the genome sequence and the RNA sequence, giving the appearance of RNA editing. The putative editing sites located in genes belonging to multi-gene families are more likely to be false positives. In order to reduce the number of cross-mapping events, the “uniqueness” of mapped reads can be controlled using the “-m” option in the Bowtie alignment program.
Interestingly, we found that 72% of CDK13 mRNAs undergo A→ I editing at chr7:39,957,073 which results in the Q103R amino acid substitution in the protein product. CDK13, cyclin-dependent kinase 13, is known to interact with HIV-1 trans-activator Tat protein and regulate viral mRNA splicing (17). CDK13 is also a known target of RNA editing in the brain (18). Intriguingly, we observed that 84% of mRNAs of NEIL1, an enzyme involved in base-excision repair of oxidative DNA damage (19), undergo A→ I editing at chr15:73,433,139 which results in the K242R amino-acid substitution in the protein product. A very recent study (20) showed that the edited and the genome-encoded forms of NEIL1 have very distinct enzymatic properties, thus demonstrating the functional importance of RNA editing of NEIL1.
Acknowledgments
I am grateful to Liz Cirulli and David Goldstein for providing the raw sequence data from their study (9). This work was supported by the Division of Intramural Research Program of the NIH, National Heart, Lung, and Blood Institute.
Footnotes
We used cDNA sequencing data from the polyA-enriched RNA sample. Since non-coding RNAs such as microRNAs and intronic RNAs are removed by the polyA enrichment procedure, no information about RNA editing of these RNA classes can be obtained. Specialized protocols should be used for the isolation and sequencing of specific classes of RNA as was, for example, done for the microRNAs in the study of Morin et al. (21).
For the sake of simplicity, we used Bowtie for aligning cDNA data and restricted our analysis to RNA editing sites in exons. Splice sites were thus excluded from the analysis. To analyze RNA editing in the vicinity of splice sites, an algorithm tailored for aligning RNA sequencing data, such as TopHat (22), should be used instead.
As discussed in Subheading 3.3.1, for the sake of simplicity, we restricted RNA editing analysis to those homozygous sites in the sample genome that match the reference hg18 human genome. The variant homozygous sites can be analyzed as follows. We first extract the locations of homozygous variants from the file “gDNA_variants.txt” where homozygous sites correspond to lines with IUPAC symbols “ACGT” in the fourth column. Let us name the resulting space-separated two-column file as “pos.txt”. We then pileup at these locations as:
samtools pileup -f hg18.fa -l pos.txt cDNA.bam | perl variant_site.pl > cDNA_pileup.txt
Non-synonymous base substitutions that result due to RNA editing may be computationally tested for functional importance using tools such as PolyPhen (23).
References
- 1.Gott JM, Emeson RB. Functions and mechanisms of RNA editing. Annu Rev Genet. 2000;34:499–531. doi: 10.1146/annurev.genet.34.1.499. [DOI] [PubMed] [Google Scholar]
- 2.Farajollahi S, Maas S. Molecular diversity through RNA editing: a balancing act. Trends Genet. 2010;26(5):221–30. doi: 10.1016/j.tig.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishikura K. Functions and Regulation of RNA Editing by ADAR Deaminases. Annu Rev Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maas S, Kawahara Y, Tamburro KM, Nishikura K. A-to-I RNA editing and human disease. RNA Biol. 2006;3(1):1–9. doi: 10.4161/rna.3.1.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11(1):31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 6.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li JB, Levanon EY, Yoon JK, Aach J, Xie B, Leproust E, et al. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324(5931):1210–3. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 8.The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cirulli ET, Singh A, Shianna KV, Ge D, Smith JP, Maia JM, et al. Screening the human exome: a comparison of whole genome and whole transcriptome sequencing. Genome Biol. 2010;11(5):R57. doi: 10.1186/gb-2010-11-5-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. (Software available at http://bowtie-bio.sourceforge.net) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. Genome Project Data Processing Subgroup (2009) The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 1000;25:2078–9. doi: 10.1093/bioinformatics/btp352. (Software available at http://samtools.sourceforge.net/) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chepelev I, Wei G, Tang Q, Zhao K. Detection of single nucleotide variations in expressed exons of the human genome using RNA-Seq. Nucleic Acids Res. 2009;37(16):e106. doi: 10.1093/nar/gkp507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18(11):1851–8. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu W, Sanders-Beer BE, Katz KS, Maglott DR, Pruitt KD, Ptak RG. Human immunodeficiency virus type 1, human protein interaction database at NCBI. Nucleic Acids Research. 2009;37:D417–22. doi: 10.1093/nar/gkn708. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Hoon MJ, Taft RJ, Hashimoto T, Kanamori-Katayama M, Kawaji H, Kawano M, et al. Cross-mapping and the identification of editing sites in mature microRNAs in high-throughput sequencing libraries. Genome Res. 2010;20(2):257–64. doi: 10.1101/gr.095273.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berro R, Pedati C, Kehn-Hall K, Wu W, Klase Z, Even Y, et al. CDK13, a new potential human immunodeficiency virus type 1 inhibitory factor regulating viral mRNA splicing. J Virol. 2008;82(14):7155–66. doi: 10.1128/JVI.02543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiran A, Baranov PV. DARNED: a DAtabase of RNa EDiting in humans. Bioinformatics. 2010;26(14):1772–6. doi: 10.1093/bioinformatics/btq285. [DOI] [PubMed] [Google Scholar]
- 19.David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447(7147):941–50. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeo J, Goodman RA, Schirle NT, David SS, Beal PA. RNA editing changes the lesion specificity for the DNA repair enzyme NEIL1. Proc Natl Acad Sci U S A. 2010;107(48):20715–9. doi: 10.1073/pnas.1009231107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morin RD, O’Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18(4):610–21. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30(17):3894–900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]