Abstract
Opinions diverge on whether mapping the synaptic connectivity of the brain is a good idea. Here we argue that albeit their limitations, such maps will reveal essential characteristics of neural circuits that would otherwise be inaccessible.
Neuroscience is in its heyday: large initiatives in Europe1 and the United States2 are getting under way. The annual neuroscience meetings, with their tens of thousands of attendees, feel like small cities. The number of neuroscience research papers published more than doubled over the last two decades, overtaking those from fields such as biochemistry, molecular biology and cell biology3. Given the number of momentous advances we hear about, surely must we not be on the threshold of knowing how the healthy brain works and how to fix it when it does not?
Alas, the brain remains a tough nut to crack. No other organ system is associated with as long a list of incurable diseases. Worse still, for many common nervous system illnesses there is not only no cure but no clear idea of what is wrong. Few psychiatric illnesses, learning disorders or even severe pain syndromes such as migraine have pathognomonic signs: no blood tests, radiographic or electrophysiological findings, or even brain biopsies enable diagnoses. This predicament is unlike that for other organ systems, where disease is nearly always associated with tissue pathological and/or biochemical signs. A patient may come for help because of pain in their belly, but the physician discovers the cellular or molecular cause of the pain before a treatment regimen begins. Why is this so different for the brain?
Those of us with an interest in a deeper understanding of the structure of the brain, not surprisingly, see the problem in structural terms. The nervous system is a physical tissue that is quite unlike other organ systems4. For one thing, it contains far more types of cells. The retina alone has more than 50 different kinds of cells5, whereas the liver has closer to five. Second, neurons, by virtue of their complicated geometry, connect (via synapses) to many more cellular partners than the limited associations of immediately adjacent cells in other organs. Third, the neuronal contacts give rise to directional circuits that have no analogs in other tissues. Fourth, the fine structure of these neural circuits is quite diverse and differs from the marked structural redundancy in other organs where the same multicellular motif (for example, the renal nephron) is iterated many times. Fifth, perhaps the most intriguing difference between neural tissue and other organs is that the cellular structure of neural tissue is a product of both genetic instruction and experience. Thus, the structure of each of our nervous systems is personalized by our own set of experiences.
The special structural features of the nervous system are likely the reason why it is more difficult to understand than other organs are. We contend, however, that a more complete rendering of neural circuit synaptic connectivity (that is, connectomics6) would go a long way to solving this problem. With this understanding, diseases that manifest as abnormalities of behavior, thought or learning, or as pain might become as clearly linked to underlying pathological structure, as is the case for diseases in other organ systems. Knowing what is wrong is a good first step to finding a solution.
For some proponents of connectomics, the potential clinical payoffs provide sufficient justification for such studies. But not everyone agrees. First, connectomics can require an industrialized effort that is akin to initiatives that allowed genomics to flourish. In light of present severe budget limitations, connectomics might be ill-advised. Moreover, some have argued that pursuing connectomics would be a waste of money, even if it were free. They have argued that anatomical maps fundamentally do not reveal how the brain works. Below we address some of these arguments.
Top ten arguments against connectomics
Number ten: circuit structure is different from circuit function
One argument put forward is that the nervous system’s macroscopic functions (that is, behaviors) are derived directly from the functional (that is, electrical) properties of the neurons rather than the anatomical connections between neurons. Hence it is the relationship between the firing patterns of action potentials of neurons and function that is the key to bridge the gap between the cells of the nervous system and behavior. The recent proposal of an initiative to get a ‘brain activity map’ reflects this view, as do many initiatives for Ca2+ imaging from ever larger numbers of nerve cells.
Although no one doubts that neural connections underlie signaling between nerve cells, the focus on neuronal function (action potentials) is based in part on the belief that the structural wiring details per se are insufficient to derive the firing patterns. Part of the problem is that the spiking properties of a neuron come not only from the electrical signals they receive from their presynaptic partners but also from the distribution and characteristics of their intrinsic voltage-gated channels. Although in principle connectomics can reveal all the presynaptic partners, distributions of ion channels would not be easily revealed by a connectional map. Furthermore, even if one knew the molecular identities and precise location of all the voltage-gated channels in every neuron (determined by very sophisticated immunological labeling, for example), this information would still not be sufficient to provide the activity patterns underlying behavior. The reason is that behavior is often driven by the particular trains of action potentials set up by sensory inputs. This sensory experience is extrinsic to the nervous system and hence inaccessible from just looking at a connectional map.
Response
Brains can encode experiences and learned skills in a form that persists for decades or longer. The physical instantiation of such stable traces of activity is not known, but it seems likely to us that they are embodied in the same way intrinsic behaviors (such as reflexes) are: that is, in the specific pattern of connections between nerve cells. In this view, experience alters connections between nerve cells to record a memory for later recall. Both the sensory experience that lays down a memory and its later recall are indeed trains of action potentials, but in-between, and persisting for long periods, is a stable physical structural entity that holds that memory. In this sense, a map of all the things the brain has put to memory is found in the structure—the connectional map. An ‘activity map’ of the brain that only shows trains of action potentials would certainly be an incomplete map, as most behaviors and memories will not be visible in any finite recording session. Decoding the way experience via electrical activity becomes stably embedded in physical neuronal networks is the unmet challenge that connectomics attempts to solve.
In our view, more challenging perhaps will be ferreting out the relationships between neural circuits and behaviors that are intrinsic and unlearned. In most animals, the behavioral repertoire is mostly genetically encoded. Such inherited circuits may have arbitrary features that came about accidentally at some instant in an animal’s evolution. We therefore think that genetically encoded behaviors could be instantiated in quite diverse ways compared to a more limited number of rules governing experience-dependent formation of circuits. From this perspective, the structural underpinnings of a behavior of Caenorhabditis elegans may actually be more difficult to decode than the circuit underlying a learned behavior in a mammal.
Number nine: signals without synapses and synapses without signals
There is abundant evidence that neurotransmitter released at a synapse is not always restricted to the ‘intended’ postsynaptic target. Spillover of glutamate from excitatory synapses has been shown to affect nearby postsynaptic sites7. It is also well known that neuronal activity can affect the behavior of nearby glial cells, which can then convey this activity to other glial cells through junctions between glia. In all brains, there are also many signals that pass between brain cells and other organ systems via hormones. Steroids originating in the adrenal cortex have effects on brain function as do those from sex organs. Growth hormone, thyroid hormone, insulin, leptin and many others also affect brain function. Furthermore, some forms of neurotransmitter release do not rely on classical synapses8. Lastly, neurons often release peptides that act over large areas, using volume transmission as opposed to restricted transmission at adjacent synapses9. All of the aforementioned examples show that brain function is profoundly affected by chemical cues in ways that would be difficult or impossible to infer from anatomical wiring diagrams.
Conversely, it is generally accepted that a substantial fraction of excitatory synapses can be structurally present but functionally silent10. These synapses can be switched on via a Hebbian learning step. Obviously in the absence of knowing which synapses are silent, a connectome provides a distorted picture of the functionally useful connections in the brain.
Response
It is true that that a map of synaptic connectivity is not identical to a map of the signaling pathways of the brain. In terms of signals without synapses, there is no denying the importance of the brain’s chemical milieu on behavior. The ability of pharmacological agents to rapidly induce sleep, tranquility, excitement, hallucinations and so on means that the behavioral state can be dramatically altered probably without any modification to the connectome.
Therefore, we should be careful not to confuse the goals of connectomics with the aims of maps of the activity patterns in a nervous system. The connectome certainly would not be a map of the behavioral state at the moment the brain is preserved. We think the connectome would be much more than that, as it could provide the entire behavioral capability of the brain. But extracting such information may require detailing an unprecedented amount of anatomical data. This anatomically stringent view of connectomics contrasts with interesting recent ideas about determining the connections between brain cells without information about anatomy or physiology11. A connectional matrix minus the anatomy information, of course, can reveal neither the sites of spillover nor the proximities necessary for the spread of peptides, whereas an anatomical connectome could reveal less direct paths for neurotransmitter and neuropeptide signaling. However, the hormonal milieu via blood flow or cerebrospinal fluid would still remain invisible unless the effects of diffusible factors on behavior had a structural correlate12.
There are also anatomical approaches to deal with the problem of synapses without signals. Some of the earliest evidence for a class of silent synapses came from serial electron microscopy reconstructions13, and we imagine that such information might become available for glutaminergic synapses as well. If silent synapses are structurally different from transmitting ones by virtue of the neurotransmitter receptor, then in principle that difference can be revealed. Immunolabeling for AMPA-type glutamate receptors should solve this problem.
Number eight: ‘junk’ synapses
Given the trillions of synapses in a human brain, most individual synapses are functionally negligible. The brain is probably organized such that the precise number and identity of synapses are unimportant, and what really matters is the general likelihood of connectivity. Thus, some fraction of the synaptic connections could rightly be called ‘junk’: they have no functional role but have so little cost that such scraps persist. If junk synapses are common, then going to the trouble of itemizing all the synaptic connections is both a waste of time and a distraction from more important functional questions.
Response
Our view is that it is premature to assign an influence to individual synapses in wiring diagrams when so few complete wiring diagrams have been described. The lesson from genomics is that many noncoding sequences that initially were thought to be unimportant and sometimes called junk turned out to have functional importance. Moreover, we are not convinced that the nervous system tolerates large numbers of synapses that are not serving a useful function. In fact, the synapses observed in the adult may reflect the small percentage of synapses that survive an extended period of synapse elimination during development14.
Number seven: same structure, many functions
Connectomics might be tractable if each behavior was partitioned to a different circuit element (that is, one circuit, one behavior). However, good evidence shows that multiple sensations or behaviors use the same neurons in different ways15. Neurons can rapidly switch their functional roles in response to chemical signals such as peptides, other neuromodulators or activity levels. Because of this flexibility, it is really not possible to assign a neuron to a function without knowing the behavioral state—something that is invisible in connectomics.
Response
Without an understanding of the physiological responses of neurons to various chemical messengers such as peptides, we agree it will be difficult to define the roles of neurons that switch their function depending on the chemical milieu. The extent to which this kind of switching is a general feature of nervous systems is not yet known.
But whatever the case, it is important to emphasize one limitation of connectomics: it is not a replacement for insights gained by physiological or pharmacological studies. Rather, connectomics may associate specific physiological phenomena with specific neural-circuit motifs so that the next time that motif is observed in the same tissue, it will signify a physiological process without the requirement of repeating the physiological analysis each time. Because in many nervous systems the same neuron types are duplicated many thousands or millions of times, it seems likely that the same motifs will be repeated multiple times. Thus, a little physiology may go a long way.
Number six: same function, many structures
What if there is great variability in the circuits that give rise to a single behavioral output? Without a stereotyped pattern of neural circuits that underlie a particular function, it will be impossible to relate structure to function.
Response
In mammals, at least, variability seems to be the rule. Even the pattern of nerve-muscle connections in the same muscle is quite different from one instantiation to the next16. However, variability in connections does not necessarily mean that the result is incomprehensible. In muscle, for example, there is a skewed distribution of the size of motor units that is the same in each muscle even though the exact location and branching pattern of each axon is unique. Thus, each instantiation appears to be a variation on the same common theme, just as every chess match is different, but they all obey the same rules. One of the reasons we believe connectomics is necessary, is that it is the only way to derive network principles despite variability in the connectivity of individual neurons.
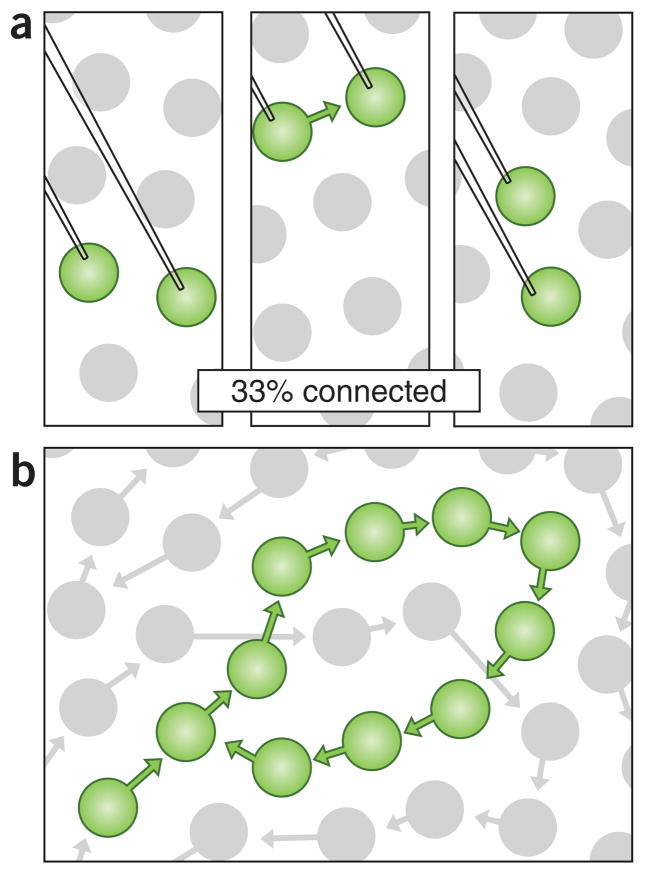
Indeed, deriving network principles without connectomics may be nearly impossible in some cases. Without connectomics, neural circuit diagrams can only be assembled by identifying connected pairs of cells in many different subjects. This approach, although accurate as far as it goes, cannot reveal wiring rules as simple as ‘cell A only connects to cell C when cell B connects to cell C’. Such conditional rules have been revealed from fortuitous circuits that lend themselves to easy analysis. For example, triad synapses in the lateral geniculate nucleus allow the connectivity of cell A, B and C to all be seen in a single electron microscopy section as all the synapses are adjacent17. If more complicated patterns of conditional connectivity exist (as well they might18,19), then there is no way to find these without reconstructing the entire circuit and including all the cellular elements (Fig. 1).
Figure 1.
Potential results from two approaches to studying circuit connectivity. (a) Probing pairs of neurons in multiple subjects determines the probability that neighboring neurons are connected. (b) Connectome of the same tissue reveals network motifs.
Number five: statistical synapses should suffice
Statistical mechanics was a great advance in physics: when many similar events occur at the same time (such as the collisions of molecules in a gas), the behavior of the system can be predicted with accuracy without ascertaining the behavior of each and every particle’s trajectory. Given the trillions of synapses in a brain, is there really any alternative except to take a statistical approach to synaptic connectivity?
Members of the European Union’s initiative on Future and Emerging Technologies have decided, for example, to fund the large ‘Human Brain Project’. In this case, the connectional associations will be determined by characterizing “…the morphologies of different types of neuron present in different regions of the human brain. Combined with modeling, the results would enable the project to predict a large proportion of the short-range connectivity between neurons, without measuring the connectivity experimentally”1. That is, they will create the wiring diagram without having to go to the trouble of obtaining an anatomic connectome.
Response
Because of the heterogeneous nature of brain tissue, statistical approaches to studying neural-circuit wiring are, of course, much more difficult than those for a homogeneous gas. For a statistical model to capture how brains actually generate behavior, ideally it would need to include all the neuronal connectivity motifs. Unfortunately, the number of ways the dozens of different cell types are interconnected, and especially how these connections are contingent on the full cohort of synaptic partners of each cell, is presently unknown. But the advocates of statistical approaches to studying connectivity are not unduly concerned because they believe that models that combine a limited amount of connectivity data with the right set of learning rules will produce fully functional neural circuits.
We, however, think the only way to know whether the results of this strategy are actually consistent with a biological brain, is to compare the predicted wiring to an actual connectome. This is why we submit that ‘measuring the connectivity experimentally’ is a good idea: it provides all the circuit motifs; with analysis, it may also provide the physical instantiation of the learning rules (see below) and is a direct path to building a working model of the brain.
Number four: the mind is no match for the complexity of the brain
Might it be the case that ‘connectomisists’ have bit off more than they can chew? Might it be that the brain’s structural complexity far outstrips the complexity of the thoughts that emanate from even the best and the brightest brains? Although some may hope that there are organizing principles and regularities that will permit substantial compression of the connectional information into a simpler framework, is there really any biological reason such regularities are inevitable? Could the most succinct description that embodies all that a brain does be the brain itself? If so, then there is little point in mapping the connections in great detail because in the best case, one would end up with a description that would be as complicated and intractable as the brain itself.
Response
The proximate goal of connectomics is to generate detailed renderings of the connections between neurons in large volumes of the brain. The purposes that such connectional maps could be put to are numerous. Comparisons between healthy and diseased brains might point to the physical underpinnings of psychiatric diseases. Comparisons between young and old brains might provide an understanding of what kinds of network changes are associated with development and aging. Comparisons between human and other primate brains may provide insight into what underlies intelligence. So even if ‘understanding’ the brain is not likely to be an early triumph of connectomics, there are potentially many fundamental things that this kind of data will reveal. To put it perhaps a bit too bluntly, we think that understanding big data, may be overrated as a goal. Connectomes can immediately provide insights even if understanding lags.
Number three: merely descriptive neuroanatomy, just more expensive
The only hope for curing diseases and uncovering the ways brains work is to get insights into underlying mechanisms. Mechanistic insights come from experiments that test hypotheses by manipulating variables. There is of course a time and a place for descriptive studies in a field, and neuroscience owes Cajal a great debt for his landmark description of the cellular underpinnings of neural tissue, but the time for mere description is long past. Indeed, connectomics sounds modern but is really just a throwback: it is neuroanatomy gussied up with a fancy new word and ultra-expensive machines. Connectomics harkens to a time when description was all we could do. We can do more now.
Response
The Hubble telescope, archaeology, the human genome and much of structural biology are also merely descriptive, but few would argue that they have not provided fundamental insights. The hope of descriptive approaches is that they provide specific data that lead us to general hypotheses (inductive reasoning). They can reveal associations and frameworks that were not readily apparent before. In a variety of fields, big-data initiatives are being used to challenge the sacrosanct ‘hypothesis first’ world view of scientific investigation.
Number two: not much was learned from the connectomes we have
The 10-year effort to generate the connectome of C. elegans20 is widely cited, but it has had less utility than had been imagined originally. No one can claim that the relationship between structure and function has been settled in this very small nervous system. In some respects, connectomes distract from more mechanistic analyses because they reveal many more synaptic interconnections between neurons than would seem necessary. Given the struggle to make sense of the mind of a worm with about 300 neurons, it is very unlikely that we will get anywhere trying to fathom a mammalian nervous system that could be roughly a billion times bigger.
Speaking of big brains, there is already a big Human Connectome project21, so why bother with maps of mice and flies?
Response
At the very least the worm connectome has been helpful in constraining circuit analysis by providing for each neuron the list of upstream and downstream neurons. This is no doubt beneficial, having been the starting point for understanding the neural pathways for touch-induced movements22, for example. But the worm connectome may not be as useful as initially imagined because of its surprising complexity. The high level of interconnectivity among the 300 neurons—a revelation in itself—does not lend itself to easy analysis. The interconnectedness of C. elegans neurons potentially provides for a much more adaptable and diverse behavioral repertoire than one with a simpler wiring diagram but at the expense of easy comprehension by humans.
A particular technical limitation of the worm’s connectome is that its inhibitory and excitatory connections cannot be differentiated in electron microscopy images (unlike the situation for mammals), reducing the overall power of the wiring diagram to reveal new concepts. In addition, there are particular challenges in interpreting a map of connectivity in a highly differentiated nervous system where each neuron has many subcellular compartments, where all the signaling is due to local potentials, and evolution has specialized nearly every cell into a unique type. This specification contrasts to the millions of cells of a single class found in many parts of the (perhaps less evolved) vertebrate nervous system. Thus, the worm’s nervous system may use a fundamentally different strategy than the one predominating in much larger nervous systems in which cell types reflect populations of neurons whose connectivity is organized by experience.
As to the Human Connectome project, this is an important and ambitious multi-institution study to gain information about the organization of the brains of many individuals including many twins. One of its goals is to map the pathways that project between various brain regions (‘projectomics’). However, none of its many goals relate to describing the synaptic connectivity of the brain.
Number one
“If you want to Google on connectome and look [it] up, you can see some gorgeous pictures that are being made of the wiring diagram of the human brain, showing you how the wires move from front to back and up and down, side to side. But again, it’s a static picture. It’d be like, you know, taking your laptop and prying the top off and staring at the parts inside, you’d be able to say, yeah, this is connected to that, but you wouldn’t know how it worked” (Francis Collins, NPR Science Friday; 5 April 2013).
Response
Maybe picking an argument in this case is not so wise, but prying the lid off a computer to see how everything is connected does not seem like such a bad idea, at least as a first step. Without knowing the parts list and what is connected to what, how could one ever really know how it works?
In our view, the ideal brain-imaging technology would provide both a complete map of synaptic connectivity as well as a complete map of the activity of all neurons and synapses in real time during normal behaviors. Even better, would be to do this in a human being who can report on their thoughts while behaving. Unfortunately, we are a long way from such technologies; so what do we do in the meantime?
We think the computer analogy gives us a lead. The computer can be turned off and on without losing much data because the instructions that make it work are embedded in its ‘static’ physicality. A deep understanding of how information is stably stored in the structure of hard discs, the input and output wires of each chip, the physical structures that explain the working of those chips and so on would be enormously helpful in making sense of a computer. Might the same be said of nervous systems?
We think that the static connectivity of the brain has embedded within it much of the ‘instruction book’ that guides the billions of impulses through networks of neurons to ultimately generate outward behavior. We see a connection between the relationship between the connectome and the brain’s functional properties to not only computers but also to the way the static human genome encodes much of how an organism works.
Structural defects in the genome that give rise to a range of diseases are perhaps a good analogy for defects in wiring that give rise to diseases of brain function. In each case, tracing the causal linkages between the static structure (be it genotype or connectotype) and the ultimate phenotype (be it cancer or schizophrenia) is difficult. But this difficulty has not deterred cancer biologists from seeking the ultimate causes (the physical genes) that affect the likelihood of cancer, and we think should not deter neuroscientists from seeking the physical wiring abnormalities that underlie brain disorders, even if this requires high resolution connectomics. In both situations, the structural data is overwhelming but nonetheless holds fundamental truths.
To quote Francis Collins referring to the human genome project: “When you have for the first time in front of you this 3.1-billion-letter instruction book that conveys all kinds of information and all kinds of mystery about humankind, you can’t survey that going through page after page without a sense of awe. I can’t help but look at those pages and have a vague sense that this is giving me a glimpse of God’s mind”23. We likewise feel that the static maps of the brain may engender awe and it might not be so unrealistic to hope that in staring into such a map we might get a glimpse of the human mind.
Why we still want to do connectomics
The large (but we are sure still incomplete) list of arguments above might seem discouraging enough to put a damper on all but the most irrationally enthusiastic proponents of connectomics. In addition to the reasons we provided in our rebuttals, we remain committed to pursuing connectomics because of the absence of one fundamental kind of information about nervous systems that we do not think is attainable in any other way.
What Cajal did not do for neuroscience
Thanks to both his genius and the extraordinary power of the sparse labeling of the Golgi stain, Cajal developed a view about neurons that has had a dominating effect on the way in which we imagine neural circuits to work. Schematic textbook diagrams of retinal circuits, cortical circuits, cerebellar circuits, among many others, provide students with a sense of which cell types connect with one another. These diagrams also serve as a model of how information flows through a circuit of cells because many of these diagrams include arrows that harken back to the arrows found in the original Cajal diagrams (Fig. 2a).
Figure 2.
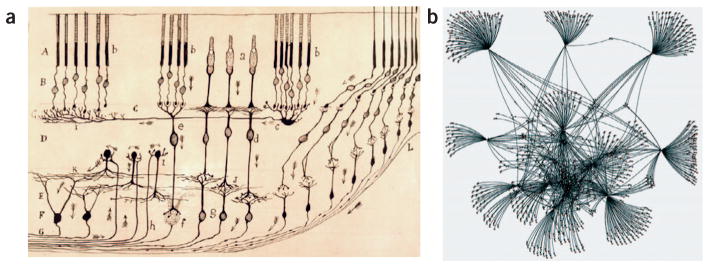
Two types of wiring diagrams. (a) Schematic of mammalian retina drawn by Santiago Ramón y Cajal in 1901. Image courtesy of J. De Carlos. (b) Dense connectivity of the Twitter network of David Rodrigues as rendered by D. Rodrigues (http://www.flickr.com/photos/dr/2048034334/).
The sense one might take from these kinds of diagrams is that this kind of structural class-wise connectivity information explains the way a neural circuit works. One may quibble over the extent to which the details of these connectional maps have been worked out, but the implication is that knowing this kind of neuronal-class connectivity is sufficient to get some understanding about how neurons process information.
In certain cases, this view is probably correct. In an animal in which the majority of neurons are each genetically unique and each has a stereotyped connectivity, connections of individual neurons are the same thing as a class-wise wiring diagram (because to a first approximation each cell is its own class). Thus, one can imagine a detailed map describing exactly which cells are connected, and that map would be exactly the same as what actually exists in the animal.
But in other cases these diagrams are quite different from the actual connectional array. They lack both quantitative information and ignore the different wiring patterns of the cells of the same class. Thus, a retinal schematic diagram shows what classes of cells are interconnected but does not show critical details such as how many different amacrine cells converge on each retinal ganglion cell or which particular amacrine cells are making those connections. A cortical schematic diagram does not show whether two interconnected pyramidal cells have a common third input. A cerebellar schematic diagram does not show whether the cohort of parallel fibers connected to a given Purkinje cell predicts which parallel fibers will connect with adjacent Purkinje cells.
This kind of information is, however, being addressed by new generation of connectomic circuit maps that reveal connectivity between dozens or hundreds of cells in a single piece of tissue17,24–26. These new efforts and several earlier attempts20,27, are beginning to shift the paradigm from class-wise connectivity questions to questions about the particular pattern of connections in a cell class. Variability in the connections of cells of the same class, is the essential feature that is lacking in classical wiring diagrams but that exists in actual neural circuits. Because the vertebrate brain is mostly composed of many copies of each of many cell types (unique neurons such as the Mauthner cell in teleosts are quite exceptional), this aspect of neural circuit organization cannot be ignored. In sum, Cajal’s use of sparse labeling did not provide a means for coming to grips with the way redundant populations are used in circuits. Connectomics, however, does.
Engramics?
In 1904, Richard Semon, a German evolutionary biologist, coined the term ‘engram’ as the physical memory trace that is somehow embedded in an organism after an experience. He was motivated to think about the physicality of memory in an attempt to formulate a means of inheritance of acquired characteristics28. Although this Lamarckian notion is refuted for gene-based inheritance, there is little doubt that humans do acquire information (by learning) during their lives that affects their behaviors and then pass this information on to their children (among others) to alter their behaviors in nongenetic ways. A longstanding goal of neuroscience is to determine the physical basis of an engram.
An interesting feature of the highly redundant populations of neurons in mammalian nervous systems is that they undergo dramatic changes in their connectivity in early postnatal life. Because these alterations are activity-dependent and transform diffusely connected networks into many distinct subnetworks, this process could be the physical underpinnings of memory14.
Regardless of the mechanism, we wonder whether eventually a subfield of neuroscience will exist that is devoted to understanding the encoding and decoding of experience-based changes in neural networks. Engramics would certainly require a hefty amount of connectomics along with sophisticated analysis and ultimately simulations to test the idea that a particular neural network encodes a particular memory.
What is ahead for connectomics?
Assuming that connectomics has a future, several mostly technical obstacles will need attention.
Statistical and analytic tools
In many ways, maps of complete connectomes might look less like the canonical wiring diagrams of Cajal and more like modern renderings of social networks (Fig. 2b). How to compare and analyze these connectional graphs of the brain is a new challenge that will require new mathematical techniques for analyzing graphs, new statistical tests to compare one circuit with another and finally a cadre of neuroscientists who will find mining big data appealing. Ultimately, however, these tools will only be useful if connectomes can be produced easily.
Faster data collection
There are two reasons to collect connectomics data faster. First, the production of connectomes must eventually be easy enough so that multiple experimental conditions can be compared and findings can be replicated. Second, the variety of questions that can be asked increases as volumes enlarge and multiple samples can be processed. If a complete map of every synapse in a human brain is wanted, a serious obstacle is the fact that at current speeds it would take 10 million years to complete.
How fast can we go?
The best technology currently available for producing connectomics data sets is high-throughput electron microscopy. Large-scale electron microscopy data sets are now being produced that are about 300 cubic micrometers. For many circuits, this size is large enough to encompass multiple complete axonal and dendritic arbors. There are a number of ways that this technology could be modified to produce a 10–50 fold increase in data-acquisition rates in the next few years. However, the current bottleneck for obtaining connectomics data is not image acquisition but image segmentation29. Many connectomics data sets rely on a human painting voxels on a computer screen.
One potential speedup is crowdsourcing (http://eyewire.org/). Alternatively learning algorithms can use human segmentations to guide computer efforts30. At the moment the results still require substantial human editing. All of these directions are being pursued, with competitions between groups stimulating new approaches to this daunting problem (http://brainiac.mit.edu/SNEMI3D/). Ironically, much of this effort in image analysis is to make machines do what human brains do easily—something that we might better understand once connectomics analysis of the visual system is complete.
Proteome meets the connectome
The structural mapping of connections will only be useful if the cell type of each of the neurons involved in a circuit is known. Serial reconstruction of a neuron’s shape and location will in some cases be sufficient for the type of neuron to be identified. But it is likely that there is heterogeneity among neurons that look alike. Fortunately the past few decades have seen an explosion in knowledge of the molecular classes of nerve cells31. What remains to be done is finding ways to insert that kind of information into mapping studies. Correlative fluorescence immunostaining with electron microscopy is one approach32. Other techniques are certainly on the horizon.
Conclusion
Despite the many arguments against undertaking a connectomics analysis, we think it must be done for three reasons. First, the conditional patterns of synaptic connectivity generated during development and by experience are inaccessible to techniques that sample from only a few cells at a time. Second, neuroscientists cannot claim to understand brains as long as the network level of brain organization is uncharted; without this detailed information, neuronal physiology is connected to systems physiology by a black box. Third, there is a high likelihood that such exploration will reveal unexpected properties by shining a light (or electrons) on this most mysterious tissue.
Acknowledgments
Our work was supported by a Conte Center grant (US National Institute of Mental Health), the US National Institutes of Health, the Gatsby Charitable Trust and Center for Brain Science Harvard University. We thank D. Rodrigues for use of his rendering of his Twitter network and J. De Carlos for Cajal’s drawing of the retina.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Contributor Information
Joshua L Morgan, Email: joshmorgan@fas.harvard.edu.
Jeff W Lichtman, Email: jeff@mcb.harvard.edu.
References
- 1.Markram H, et al. A Report to the European Commission. 2012 Apr; < http://www.humanbrainproject.eu/files/HBP_flagship.pdf>.
- 2.Collins F, Prabhakar A. The White House Blog. 2013 Apr 2; < http://www.whitehouse.gov/blog/2013/04/02/brain-initiative-challenges-researchers-unlock-mysteries-human-mind>.
- 3.Pautasso M. Sustainability. 2012;4:3234–3247. [Google Scholar]
- 4.Lichtman JW, Denk W. Science. 2011;334:618–623. doi: 10.1126/science.1209168. [DOI] [PubMed] [Google Scholar]
- 5.Masland RH. Curr Opin Neurobiol. 2001;11:431–436. doi: 10.1016/s0959-4388(00)00230-0. [DOI] [PubMed] [Google Scholar]
- 6.Lichtman JW, Sanes JR. Curr Opin Neurobiol. 2008;18:346–353. doi: 10.1016/j.conb.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diamond JS. Nat Neurosci. 2002;5:291–292. doi: 10.1038/nn0402-291. [DOI] [PubMed] [Google Scholar]
- 8.Oláh S, et al. Nature. 2009;461:1278–1281. doi: 10.1038/nature08503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bargmann CI. Bioessays. 2012;34:458–465. doi: 10.1002/bies.201100185. [DOI] [PubMed] [Google Scholar]
- 10.Kerchner G, Nicoll R. Nat Rev Neurosci. 2008;9:813–825. doi: 10.1038/nrn2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zador AM, et al. PLoS Biol. 2012;10:e1001411. doi: 10.1371/journal.pbio.1001411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butcher AJ, et al. J Biol Chem. 2011;286:11506–11518. doi: 10.1074/jbc.M110.154526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jahromi SS, Atwood HL. J Cell Biol. 1974;63:599–613. doi: 10.1083/jcb.63.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lichtman JW, Colman H. Neuron. 2000;25:269–278. doi: 10.1016/s0896-6273(00)80893-4. [DOI] [PubMed] [Google Scholar]
- 15.Weimann JM, Marder E. Curr Biol. 1994;4:896–902. doi: 10.1016/s0960-9822(00)00199-8. [DOI] [PubMed] [Google Scholar]
- 16.Lu J, Tapia JC, White OL, Lichtman JW. PLoS Biol. 2009;7:13. doi: 10.1371/journal.pbio.1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherman SM, Guillery RW. J Neurophysiol. 1996;76:1367–1395. doi: 10.1152/jn.1996.76.3.1367. [DOI] [PubMed] [Google Scholar]
- 18.Seung HS. Neuron. 2009;62:17–29. doi: 10.1016/j.neuron.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 19.Song S, Sjöström PJ, Reigl M, Nelson S, Chklovskii DB. PLoS Biol. 2005;3:e68. doi: 10.1371/journal.pbio.0030068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White JG, Southgate E, Thomson JN, Brenner S. Phil Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 21.Sporns O, Tononi G, Kötter R. PLoS Comput Biol. 2005;1:e42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chalfie M, Sulston JE, Thomson JN, White G. J Neurosci. 1985;5:956–964. doi: 10.1523/JNEUROSCI.05-04-00956.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swinford S. The Sunday Times of London. Jun 11, 2006. [Google Scholar]
- 24.Anderson JR, et al. Mol Vis. 2011;17:355–379. [PMC free article] [PubMed] [Google Scholar]
- 25.Bock DD, et al. Nature. 2011;471:177–182. doi: 10.1038/nature09802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Briggman KL, Helmstaedter M, Denk W. Nature. 2011;471:183–188. doi: 10.1038/nature09818. [DOI] [PubMed] [Google Scholar]
- 27.Freed MA, Sterling P. J Neurosci. 1988;8:2303–2320. doi: 10.1523/JNEUROSCI.08-07-02303.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Semon RW, Simon L. The Mneme. G. Allen & Unwin Ltd; 1921. [Google Scholar]
- 29.Helmstaedter M. Nat Methods. 2013;10:501–507. doi: 10.1038/nmeth.2476. [DOI] [PubMed] [Google Scholar]
- 30.Kaynig V, Vazquez-Reina A. IEEE Trans Med Imaging. 2012;1:1–7. [Google Scholar]
- 31.Lein ES, et al. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 32.Micheva KD, Busse B, Weiler NC, O’Rourke N, Smith SJ. Neuron. 2010;68:639–653. doi: 10.1016/j.neuron.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]