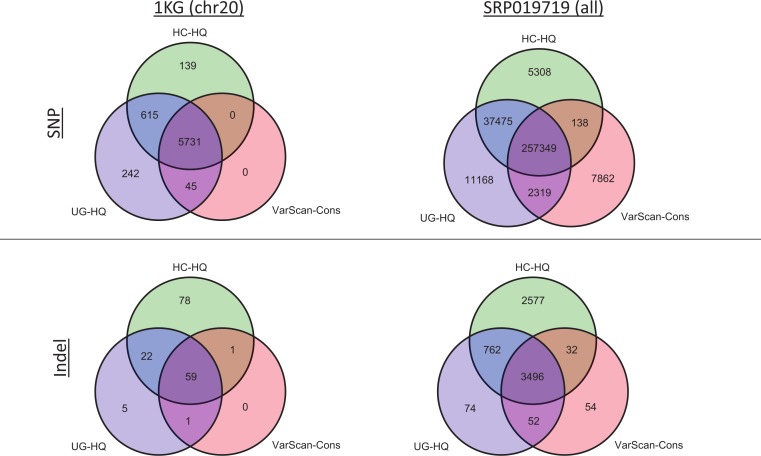
Figure 8. Exome variant caller overlap (coding variants).
All coding variants were tabulated for all exome samples (1KG n = 12; SRP019719 n = 15). Only coding variants on chromosome 20 were considered for the 1000 Genomes (1KG) samples, but all coding variants were considered for the SRP019719 samples. In order to simplify presentation of these results, we focused on the highest quality variant calls for each variant calling strategy: GATK UnifiedGenotyper with low-quality variants removed (UG-HQ, blue), GATK HaplotypeCaller with low-quality variants removed (HC-HQ, green), and VarScan using a custom set of conservative parameters (VarScan-Cons, red). Similarly, only variants subject to GATK indel realignment and quality score recalibration (“Full Pipeline”) are considered for this comparison. To show the different concordance rates, SNPs are presented at the figure and indels are presented at the bottom of the figure. Almost all VarScan-Cons variants were also called by GATK (either HaplotypeCaller or UnifiedGenotyper). All three variant callers called a similar number of SNPs, but GATK HaplotypeCaller called more indels than either GATK UnifiedGenotyper or VarScan-Cons.