Abstract
Summary: Here we introduce ccSOL omics, a webserver for large-scale calculations of protein solubility. Our method allows (i) proteome-wide predictions; (ii) identification of soluble fragments within each sequences; (iii) exhaustive single-point mutation analysis.
Results: Using coil/disorder, hydrophobicity, hydrophilicity, β-sheet and α-helix propensities, we built a predictor of protein solubility. Our approach shows an accuracy of 79% on the training set (36 990 Target Track entries). Validation on three independent sets indicates that ccSOL omics discriminates soluble and insoluble proteins with an accuracy of 74% on 31 760 proteins sharing <30% sequence similarity.
Availability and implementation: ccSOL omics can be freely accessed on the web at http://s.tartaglialab.com/page/ccsol_group. Documentation and tutorial are available at http://s.tartaglialab.com/static_files/shared/tutorial_ccsol_omics.html.
Contact: gian.tartaglia@crg.es
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Algorithms for prediction of protein solubility (Wilkinson and Harrison, 1991) and aggregation (Fernandez–Escamilla et al., 2004) provide a solid basis to investigate physico-chemical determinants of amyloid fibril formation and associated diseases (Conchillo–Solé et al., 2007; Tartaglia et al., 2004). In the past years, an in vitro reconstituted translation system allowed the large-scale investigation of Escherichia coli proteins solubility (Niwa et al., 2009), thus providing the opportunity for the development of predictive methods such as ccSOL (Agostini et al., 2012). In ccSOL, coil/disorder, hydrophobicity, hydrophilicity, β-sheet and α-helical propensities are combined together into a solubility propensity score that is useful to investigate protein expression (Baig et al., 2014) as well as bacterial evolution (Warnecke, 2012). Other methods have been developed to predict protein solubility based on amino acid characteristics. For instance, PROSO II (Smialowski et al., 2012) exploits occurrence of monopeptides and dipeptides to estimate heterologous expression in E.coli. PROSO II was trained on the pepcDB database [now Target Track (Berman et al., 2009)] that stores target and protocol information provided by Protein Structure Initiative centers. Both ccSOL and PROSO II perform accurate predictions when used to respectively predict endogenous or heterologous soluble expressions [ccSOL: 76% accuracy; PROSO II: 75% accuracy (Smialowski et al., 2012)]. We found that the experimental status of several Target Track entries (http://sbkb.org/tt/) has been recently updated and new data are available to train predictive methods (see Supplementary Material). Here, we introduce a novel implementation of the ccSOL method, called ccSOL omics, to perform large-scale predictions of endogenous and heterologous expression in E.coli. Our algorithm has been trained on non-redundant Target Track entries to identify soluble and insoluble regions within protein sequences. We envisage that ccSOL omics will be useful for protein engineering studies, as it allows the investigation of sequence variants in large datasets.
2 WORKFLOW AND IMPLEMENTATION
The ccSOL omics server allows the investigation of large protein datasets (see Supplementary Material). Once the user provides sequences in FASTA format, the algorithm calculates:
Solubility profiles. To identify soluble fragments within each polypeptide chain, protein sequences are divided into elements and individual solubility propensities are calculated. Starting from the N-terminus of a protein, we use a sliding window of 21 amino acids that is moved one residue at a time until the C-terminus is reached. The solubility propensity profile of each fragment is calculated as defined in our previous publication (Agostini et al., 2012).
Sequence susceptibility. For each sequence analyzed, the algorithm computes the effect of single amino acid mutations at different positions. This approach is particularly useful to identify regions susceptible to solubility change upon mutation. All variants are reported along with their scores, which provides a basis to engineer protein sequences and test hypotheses such as the occurrence of specific mutations in pathology.
Solubility score. The solubility profile represents a unique signature containing information on all fragments arranged in sequential order. In our approach, the profile is used to estimate solubility upon expression in the E.coli system. As sequences have different lengths, we exploit a method based on Fourier’s transform (Bellucci et al., 2011; Tartaglia et al., 2007) that allows comparison of polypeptide chains with different sizes. Using 100 Fourier’s coefficients, we trained an algorithm that has the same architecture developed for the analysis of protein expression levels in E.coli [i.e. neural network approach (Tartaglia et al., 2009)].
Reliability score. The webserver provides a confidence score based on statistical analysis of both training and testing sets (i.e. sequence range used to validate the method; see Supplementary Material).
All the aforementioned analyses are performed for each submitted protein set if the number of entries is <500. Because of the intense CPU usage, sequence susceptibility scores are not computed for datasets >500 entries.
3 PERFORMANCES
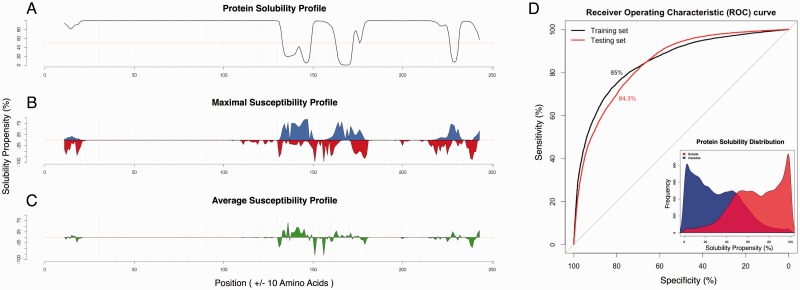
Expression of human prion (PrP) in E.coli is particularly difficult, as the protein accumulates in inactive aggregates (Baneyx and Mujacic, 2004). ccSOL omics correctly predicts that PrP is insoluble and identifies the fragment 130–170 as the least soluble (Fig. 1A–C) together with region 231–253 (not present in the mature form). This finding is very well in agreement with what has been previously reported in literature (Tartaglia et al., 2005, 2008). Moreover, the analysis of susceptible fragments identifies a number of experimentally validated mutations (e.g. G131V, S132I, R148H, V176I and D178N) associated with lower solubility and located in the region promoting PrP aggregation (Corsaro et al., 2012) [see Supplementary Material]. As for the large-scale performances of ccSOL omics, we used a 10-fold cross-validation on Target Track [total of 36 990 entries with 30% redundancy (Fu et al., 2012)] and observed 79% accuracy in discriminating between soluble and insoluble proteins. Furthermore, we tested the algorithm on three independent datasets containing protein expression data [total of 31 760 entries taken from E.coli (Niwa et al., 2009), SOLpro (Magnan et al., 2009) and PROSO II (Smialowski et al., 2012)] and found 74% accuracy (Fig. 1D; see also Supplementary Material).
Fig. 1.
Human Prion Solubility and ccSOL Performances. (A) Starting from the N-terminus, ccSOL computes the solubility profile using a sliding window moved toward the C-terminus. ccSOL identifies the fragment 130–170 as the most insoluble within the C-terminus of human PrP (region 231–253 is not present in the mature form of the protein). (B, C) Maximal and average susceptibility upon single-point mutation. (D) We trained on the Target Track set (AUROC = 85.5%) and tested on E.coli [AUROC = 93.3%; (Niwa et al., 2009)], SOLpro [AUROC = 85.7%; (Magnan et al., 2009)] and PROSO II [AUROC = 82.9%; (Smialowski et al., 2012)] proteins. Inset: overall score distribution for soluble (red) and insoluble (blue) proteins
4 CONCLUSIONS
The ccSOL omics algorithm shows excellent performances in predicting solubility of endogenous and heterologous genes in E.coli. We hope that the webserver will be useful for biotechnological purposes, as it could be for instance used to design fusion tags for soluble expression. Although accurate, our calculations are based on sequence features, and integration with structural characteristics will dramatically increase the predictive power. We plan to combine ccSOL omics with information on chaperone (Tartaglia et al., 2010) and RNA (Bellucci et al., 2011; Choi et al., 2009) interactions, as these molecules greatly contribute to the solubility of protein products (Cirillo et al., 2014; Zanzoni et al., 2013).
ACKNOWLEDGEMENTS
We thank A. Zanzoni and G. Bussotti for stimulating discussions.
Funding: The research leading to these results has received funding from the European Union Seventh Framework Programme (FP7/2007–2013), through the European Research Council, under grant agreement RIBOMYLOME 309545, and from the Spanish Ministry of Economy and Competitiveness (SAF2011-26211). We also acknowledge support from the Spanish Ministry of Economy and Competitiveness, ‘Centro de Excelencia Severo Ochoa 2013–2017’ (SEV-2012-0208).
Conflicts of interest: none declared.
REFERENCES
- Agostini F, et al. Sequence-based prediction of protein solubility. J. Mol. Biol. 2012;421:237–241. doi: 10.1016/j.jmb.2011.12.005. [DOI] [PubMed] [Google Scholar]
- Baig F, et al. Dynamic transcriptional response of Escherichia coli to inclusion body formation. Biotechnol. Bioeng. 2014;111:980–999. doi: 10.1002/bit.25169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baneyx F, Mujacic M. Recombinant protein folding and misfolding in Escherichia coli. Nat. Biotechnol. 2004;22:1399–1408. doi: 10.1038/nbt1029. [DOI] [PubMed] [Google Scholar]
- Bellucci M, et al. Predicting protein associations with long noncoding RNAs. Nat. Methods. 2011;8:444–445. doi: 10.1038/nmeth.1611. [DOI] [PubMed] [Google Scholar]
- Berman HM, et al. The protein structure initiative structural genomics knowledgebase. Nucleic Acids Res. 2009;37:D365–D368. doi: 10.1093/nar/gkn790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SI, et al. RNA-mediated chaperone type for de novo protein folding. RNA Biol. 2009;6:21–24. doi: 10.4161/rna.6.1.7441. [DOI] [PubMed] [Google Scholar]
- Cirillo D, et al. Discovery of protein-RNA networks. Mol Biosyst. 2014;10:1632–1642. doi: 10.1039/c4mb00099d. [DOI] [PubMed] [Google Scholar]
- Conchillo-Solé O, et al. AGGRESCAN: a server for the prediction and evaluation of ‘hot spots’ of aggregation in polypeptides. BMC Bioinformatics. 2007;8:65. doi: 10.1186/1471-2105-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsaro A, et al. Role of prion protein aggregation in neurotoxicity. Int. J. Mol. Sci. 2012;13:8648–8669. doi: 10.3390/ijms13078648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Escamilla AM, et al. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- Fu L, et al. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnan CN, et al. SOLpro: accurate sequence-based prediction of protein solubility. Bioinformatics. 2009;25:2200–2207. doi: 10.1093/bioinformatics/btp386. [DOI] [PubMed] [Google Scholar]
- Niwa T, et al. Bimodal protein solubility distribution revealed by an aggregation analysis of the entire ensemble of Escherichia coli proteins. Proc. Natl Acad. Sci. USA. 2009;106:4201–4206. doi: 10.1073/pnas.0811922106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smialowski P, et al. PROSO II – a new method for protein solubility prediction. FEBS J. 2012;279:2192–2200. doi: 10.1111/j.1742-4658.2012.08603.x. [DOI] [PubMed] [Google Scholar]
- Tartaglia GG, et al. The role of aromaticity, exposed surface, and dipole moment in determining protein aggregation rates. Protein Sci. 2004;13:1939–1941. doi: 10.1110/ps.04663504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia GG, et al. Prediction of aggregation rate and aggregation-prone segments in polypeptide sequences. Protein Sci. 2005;14:2723–2734. doi: 10.1110/ps.051471205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia GG, et al. Prediction of local structural stabilities of proteins from their amino acid sequences. Structure. 2007;15:139–143. doi: 10.1016/j.str.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Tartaglia GG, et al. Prediction of aggregation-prone regions in structured proteins. J. Mol. Biol. 2008;380:425–436. doi: 10.1016/j.jmb.2008.05.013. [DOI] [PubMed] [Google Scholar]
- Tartaglia GG, et al. A relationship between mRNA expression levels and protein solubility in E. coli. J. Mol. Biol. 2009;388:381–389. doi: 10.1016/j.jmb.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Tartaglia GG, et al. Physicochemical determinants of chaperone requirements. J. Mol. Biol. 2010;400:579–588. doi: 10.1016/j.jmb.2010.03.066. [DOI] [PubMed] [Google Scholar]
- Warnecke T. Loss of the DnaK-DnaJ-GrpE chaperone system among the aquificales. Mol. Biol. Evol. 2012;29:3485–3495. doi: 10.1093/molbev/mss152. [DOI] [PubMed] [Google Scholar]
- Wilkinson DL, Harrison RG. Predicting the solubility of recombinant proteins in Escherichia coli. Nat. Biotechnol. 1991;9:443–448. doi: 10.1038/nbt0591-443. [DOI] [PubMed] [Google Scholar]
- Zanzoni A. Principles of self-organization in biological pathways: a hypothesis on the autogenous association of alpha-synuclein. Nucleic Acids Res. 2013;41:9987–9998. doi: 10.1093/nar/gkt794. [DOI] [PMC free article] [PubMed] [Google Scholar]