Abstract
PSII undergoes photodamage, which results in photoinhibition—the light-induced loss of photosynthetic activity. The main target of damage in PSII is the reaction center protein D1, which is buried in the massive 1.4 MDa PSII holocomplex. Plants have evolved a PSII repair cycle that degrades the damaged D1 subunit and replaces it with a newly synthesized copy. PSII core proteins, including D1, are phosphorylated in high light. This phosphorylation is important for the mobilization of photoinhibited PSII from stacked grana thylakoids to the repair machinery in distant unstacked stroma lamellae. It has been recognized that the degradation of the damaged D1 is more efficient after its dephosphorylation by a protein phosphatase. Recently a protein phosphatase 2C (PP2C)-type PSII core phosphatase (PBCP) has been discovered, which is involved in the dephosphorylation of PSII core proteins. Its role in PSII repair, however, is unknown. Using a range of spectroscopic and biochemical techniques, we report that the inactivation of the PBCP gene affects the growth characteristic of plants, with a decreased biomass and altered PSII functionality. PBCP mutants show increased phosphorylation of core subunits in dark and photoinhibitory conditions and a diminished degradation of the D1 subunit. Our results on D1 turnover in PBCP mutants suggest that dephosphorylation of PSII subunits is required for efficient D1 degradation.
Keywords: PBCP, Photoinhibition, PSII repair cycle, STN8
Introduction
In plants and algae, reversible protein phosphorylation optimizes many aspects of photosynthetic light harvesting and conversion in changing environmental conditions (Allen 1992, Pesaresi et al. 2011, Puthiyaveetil et al. 2012, Rochaix 2013). Central to this regulation is the phosphorylation of PSII, the water-oxidizing photosystem of oxygenic photosynthesis and its bound light-harvesting antenna system. PSII becomes phosphorylated on its peripheral antenna or on its core reaction center subunits in response to changes in light quality and quantity. Phosphorylation of PSII core proteins is primarily controlled by light intensity (Elich et al. 1992, Tikkanen and Aro 2012, Tikkanen and Aro 2014) and is associated with the repair of damaged PSII.
The PSII holocomplex is 1.4 MDa in size. It comprises a peripheral light-harvesting antenna and an internal reaction center core. The peripheral antenna is made up of light-harvesting complex II (LHCII), which is trimeric. LHCII is tethered to the core through monomeric minor light-harvesting complexes known as CP29, CP26 and CP24. The core is made up of reaction center proteins D1 and D2 and an internal antenna comprising CP43 and CP47 proteins. Other PSII subunits are involved in dimerization and water-splitting functions that complement the PSII holocomplex. The D1 subunit, buried in the dimeric core, is the main target of photodamage in PSII (Kyle et al. 1984). In plants and algae, the damaged D1 subunit is degraded and replaced with a newly synthesized copy through a robust repair mechanism known as the PSII repair cycle (Melis 1999, Nixon et al. 2010). The PSII repair cycle maintains photosynthetic efficiency in high light, without which the photosynthetic function and efficiency collapse (Melis 1999). In plants and algae, functional PSII is tucked away in the tightly stacked granal regions of the thylakoid membrane (Andersson and Anderson 1980, Albertsson 2001). The PSII repair machinery, however, is located in the unstacked regions of the membrane. This spatial organization poses special problems for the efficient operation of the repair process since PSII units in stacked grana are rather immobile (Kirchhoff et al. 2008, Mullineaux 2008). How the damaged PSII is mobilized from the stacked regions to the unstacked regions of the thylakoid membrane and how the repair machinery gains access to the centrally located, damaged D1 is unclear (Kirchhoff 2013). Phosphorylation of core PSII subunits has been suggested to provide answers to these questions. Phosphorylation imparts increased mobility to damaged photosystems and causes disassembly of the PSII holocomplex (Tikkanen et al. 2008, Goral et al. 2010, Fristedt and Vener 2011, Herbstova et al. 2012, Puthiyaveetil and Kirchhoff 2013). A phosphorylation map of the PSII indeed reveals the location of phosphosites at the antenna attachment sites and at the monomer–monomer interface (Puthiyaveetil and Kirchhoff 2013). This strategic location of phosphosites is likely to enable the disassembly of PSII by antenna dissociation and monomerization of the dimeric cores. A smaller particle size of the disassembled PSII may eventually account for its increased mobility in the crowded granal regions (Goral et al. 2010, Herbstova et al. 2012). It has been suggested that phosphorylated PSII core proteins are dephosphorylated before they are recycled in the PSII repair cycle (Koivuniemi et al. 1995). In particular for the degradation of the D1 to proceed, they need to be dephosphorylated by a protein phosphatase (Rintamaki et al. 1996).
Key kinases and phosphatases in the reversible phosphorylation of PSII have now been identified. Chlamydomonas STT7 (state transition thylakoid 7) kinase, with the STN7 (state transition 7) homolog in plants, is the serine/threonine kinase that phosphorylates LHCII in state transitions (Depege et al. 2003, Bellafiore et al. 2005). STT7/STN7 also phosphorylates the monomeric antenna CP29, and phospho-CP29 is part of the mobile LHCII complex in algal state transitions (Kargul et al. 2005, Takahashi et al. 2006). Phosphorylation of plant CP29, however, is implicated in the disassembly of PSII in high light (Fristedt and Vener 2011). A second, paralogous kinase known as STL1 in Chlamydomonas, or STN8 in plants, is largely responsible for the phosphorylation of the PSII core subunits D1, D2 and CP43, and PsbH subunits (Bonardi et al. 2005). CP29 also harbors STN8-dependent phosphorylation sites, which may aid in the antenna dissociation during PSII repair (Fristedt and Vener 2011). Since phosphorylation on serine or threonine residues is stable and long lived under physiological conditions, dedicated serine/threonine phosphatases are usually found along with the kinases to ensure efficient reversibility of the protein phosphorylation reactions (Cohen 2002). PSII core proteins are dephosphorylated by specific intrinsic and extrinsic phosphatases, which are independent of the LHCII phosphatase (Sun et al. 1989, Rokka et al. 2000, Vener et al. 2000). It has been shown that a putative membrane-associated, extrinsic PSII core phosphatase would be easily washed off with NaCl-containing buffers, be inhibited by chelators of divalent cations and be slightly stimulated by the reducing agent dithiothreitol (DTT) (Sun et al. 1989). A PSII core phosphatase, exactly fitting this description, has now been identified (Samol et al. 2012). PSII core phosphatase (PBCP) is a protein phosphatase 2C (PP2C)-type membrane-extrinsic, Mn2+-dependent and DTT-stimulated protein phosphatase that is involved in dephosphorylation of core proteins. Interestingly, with the absence of a membrane-intrinsic domain, PBCP is related to the LHCII phosphatase TAP38/PPH1. In PBCP knockout mutants of Arabidopsis, PSII core proteins remain largely phosphorylated (Samol et al. 2012).
The significance of PBCP for PSII repair is currently unknown. In order to understand the role of PBCP and, in general, test the role of PSII core dephosphorylation, we characterized two independent T-DNA insertion lines of the PBCP gene of Arabidopsis thaliana, which have been genotyped to homozygosity (Samol et al. 2012). A T-DNA insertion is harbored in the fourth exon of pbcp-1 and in the 5′-untranslated region of pbcp-2. Using a range of spectroscopic and biochemical techniques, we report on the characteristics of PBCP knockout plants. PBCP mutants show increased PSII core phosphorylation in dark and in photoinhibitory conditions, diminished degradation of the D1 protein, decreased biomass and altered PSII functionality.
Results
PSII core phosphorylation in PBCP knockout mutants
Since the PBCP enzyme catalyzes the dephosphorylation of PSII core subunits, it is expected that PBCP mutants would show higher phosphorylation levels. This was already demonstrated for plants treated with low white, blue and far-red lights (Samol et al. 2012). Here we extend this analysis to dark- and high light- (HL) incubated samples. PSII core phosphorylation was detected by Western immunoblot analysis with anti-phosphothreonine antibodies on thylakoids isolated from protoplasts (Fig. 1A). As a control, we included the stn7/stn8 double mutant showing that the designated subunits are targets of the thylakoid STN kinases. Relative changes in the phosphorylation level of the D1, D2 and CP43 subunits were quantified by normalizing the band intensities to that of the wild-type (WT) dark value (Fig. 1B). For WT samples, HL treatment leads to an approximately 35% increased phosphorylation level for D1, an approximately 70% increase for D2 and an approximately 260% increase for CP43. These HL-induced increases in PSII core phosphorylation are in agreement with the literature (Bonardi et al. 2005, Tikkanen et al. 2008, Fristedt and Vener 2011). In the pbcp-1 mutant, the phosphorylation level in dark-adapted samples is significantly higher compared with their WT counterparts (∼40% higher for D1, ∼90% for D2 and ∼150% for CP43). Overall the PSII phosphorylation level in dark-adapted thylakoid membranes of the pbcp-1 mutant is similar to the level found for HL-treated WT samples (Fig. 1B). For the pbcp-1 mutant, HL treatment leads to a further increase in PSII core phosphorylation values (Fig. 1).
Fig. 1.
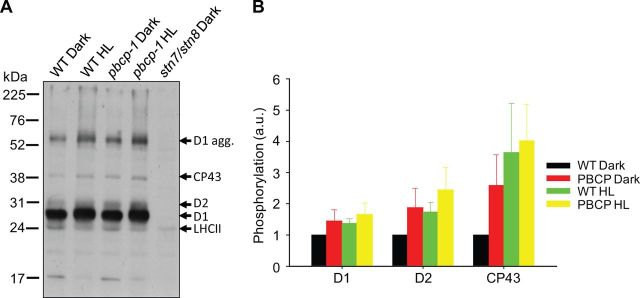
PBCP mutants show elevated phosphorylation in the dark and in HL. (A) A representative immunoblot of the thylakoid phosphoproteins is shown. (B) The phosphorylation levels of D1, D2 and CP43, calculated from the anti-phosphothreonine immunoblot, is graphically shown. The levels of phosphorylation in pbcp-1 dark, WT HL and pbcp-1 HL are shown as ratios to the WT dark. The level in WT dark was taken as ‘1’.
Thylakoid membrane composition
Next we analyzed the concentration of major protein complexes in isolated thylakoid membranes of PBCP mutants. The amount of protein complexes quantified on a Chl basis is summarized in Table 1. The amounts of PSI and Cyt b6f complexes are very similar in the WT and pbcp-1 mutant. From quantitative SDS–PAGE it follows that the amount of the 25 kDa band (mainly LHCII) is also very similar. The only significant difference between the PBCP mutant and the WT is apparent for PSII. The phosphatase mutant contains 20% more PSII (lower Chl/PSII ratio). From the comparison of the LHCII and PSII data it follows that pbcp-1 has a lower LHCII/PSII ratio (about a 15% decrease compared with the WT), which is in line with the higher Chl a/b ratio (Table 1).
Table 1.
Composition of thylakoid membranes in the WT and pbcp-1 mutant
| Chl/PSIIa | Chl/LHCIIb | LHCII/PSIIc | Chl/Cyt b6fd | Chl/PSIe | Chl a/Chlb | |
|---|---|---|---|---|---|---|
| WT | 308 ± 43 | 27.9 ± 3.5 | 11.1 ± 2.1 | 893 ± 145 | 522 ± 20 | 3.26 ± 0.11 |
| pbcp-1 | 258 ± 26 | 26.8 ± 2.4 | 9.6 ± 1.6 | 871 ± 155 | 516 ± 7 | 3.42 ± 0.14 |
Data represent the mean ± SD of 3–6 biological replicates per genotype.
a Expressed as mol Chl per mol Cyt b559.
b Expressed as mol Chl to mol 25 kDa band deduced from quantitative SDS–PAGE. This gives the amount of monomeric LHCII.
c Calculated as the Chl/PSII/Chl/LHCII ratio.
d Average of Cyt f and Cyt b6 content (Cyt b6 was divided by two because each Cyt b6f molecule contains two haems.
e Expressed as mol Chl per mol P700.
PSII antenna organization
The protein composition data presented above show a slight decrease in the PSII antenna proteins in PBCP mutants. Therefore we analyzed the functional PSII antenna size by Chl fluorescence spectroscopy. In a first set of experiments the functional PSII antenna size was studied by Chl fluorescence induction analysis on isolated thylakoid membranes in the presence of the PSII inhibitor DCMU (Lavergne and Trissl 1995, Lazar 1999). Fig. 2A shows normalized induction curves of the variable Chl a fluorescence (Fv) for dark-adapted WT and mutant samples, respectively. Fv indicates the reduction level of the primary quinone acceptor of PSII, QA. All measurements start with dark incubation that leads to complete oxidation of QA. Thus Fv is zero. The light-induced transition from Fv = 0 to Fv = 1 (Fig. 2A) reflects the reduction of QA to QA−. The PSII antenna size can be estimated as the inverse time required for a 60% increase in Fv (Rappaport et al. 2007). These numbers (rates) are given in Fig. 2A. While the WT curves (black) cluster in one group, the curves for pbcb-1 cluster in two sets (highlighted by purple and red colors). This non-homogenous behavior for the pbcp-1 mutant was also observed for growth phenotype data shown below, and will be discussed later. Regardless, the rate constant for data set 1 reveals an approximately 20% reduction in PSII antenna size for the phosphatase mutant in accordance with the compositional analysis presented above.
Fig. 2.
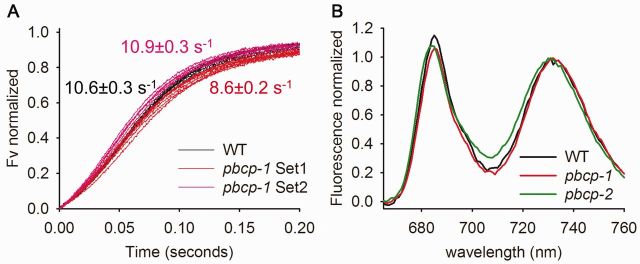
PBCP mutants show smaller PSII antenna. Normalized fluorescence induction curves of the variable Chl a fluorescence in the WT and PBCP mutants are given. (A) Normalized variable fluorescence (Fv) is plotted against the time. (B) Normalized low temperature 77K fluorescence emission in the WT and PBCP mutants. Data represent the average of 3–4 independent measurements.
The Chl fluorescence studies are complemented by low temperature (77K) fluorescence spectroscopy recorded on shock-frozen leaves (Fig. 2B). The spectra were normalized to the peak at around 735 nm. The 735 nm band is emitted from PSI/LHCI (Andreeva et al. 2003, Haferkamp et al. 2010). The peak around 685 nm originates from the PSII core (Andreeva et al. 2003, Haferkamp et al. 2010). Changes in the 685 nm emissions relative to the 735 nm fluorescence indicate changes in the PSII antenna size. Thus for both the pbcp-1 and pbcp-2 mutants, the lower 685 nm/735 nm ratio relative to the WT (Fig. 2B) provides evidence for a slight decrease in PSII antenna size. Overall the compositional as well as the functional analysis of PSII antenna organization consistently shows a small (15–20%) decrease in PSII antenna size in PBCP mutants.
Phenotypic analysis of PBCP knockout mutants
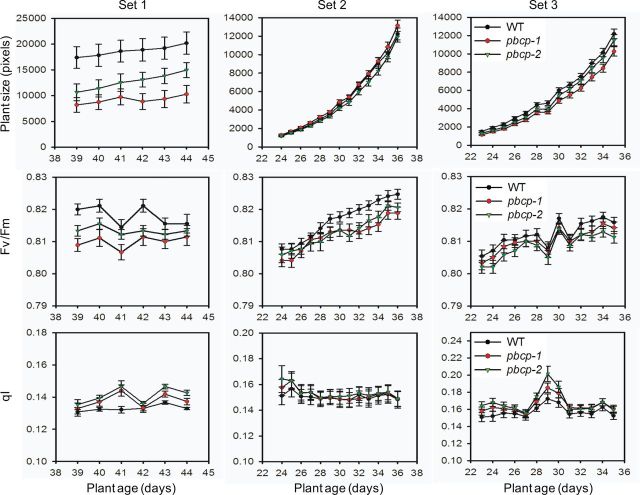
We examined whether impaired PSII core dephosphorylation in PBCP mutants influences their growth phenotype. In a first set of experiments we measured the above-ground biomass of mature, 6-week-old plants by weighing. The total above-ground biomass for the WT is 1.92 ± 0.29 g (mean ± SE) per plant, 0.99 ± 0.17 g for the pbcp-1 mutant and 1.28 ± 0.17 g for the pbcp-2 mutant. It follows that the biomass is only about 52% of that of the WT in the pbcp-1 mutant and about 67% in the pbcp-2 mutant. The difference between the WT and pbcp-1 is statistically significant (P = 0.03) but that for pbcp-2 is not (P = 0.08).
For further insights into the growth phenotype of PBCP knockout mutant plants, we measured detailed growth curves in a phenomics facility. The facility allows fully automated acquisition of Chl fluorescence images. In addition to total (above-ground) plant size, the maximal photosynthetic efficiency of PSII (Fv/Fm parameter) and the Chl fluorescence parameter qI (photoinhibition-dependent quenching) (Krause and Jahns 2003) were determined. The qI parameter measures photoinhibition of PSII and was probed after a 5 min illumination period (see the Materials and Methods). The phenomics experiment was designed to study the plants under standard non-stress growth conditions, i.e. under normal growth light illumination. The measurements were performed at the end of the night period. Fig. 3 shows examples of plant growth phenotypes. In this set of experiments, the two phosphatase mutants show retarded growth. A quantitative analysis of such images for the three sets of plants (Set 1–3) is given in Fig. 4. The top leftmost panel shows that both mutants have a slight growth phenotype. Examination of the Fv/Fm and qI parameters (Fig. 4, middle and bottom leftmost panels) reveals that in both mutants PSII is slightly more stressed by light, compared with the WT. However, we repeated the experiment and found that the retarded growth is nearly absent in the second round of the experiment (Set 2, top panel). The third round (Set 3), however, was similar to the first round (Set 1), showing differences between the WT and mutants. This inconsistency in the behavior of PBCP mutants will be discussed later.
Fig. 3.
PBCP mutants show retarded growth under normal light conditions. Images of representative plants taken at different ages (indicated at the top) after germination are shown. Plants are grown under standard growth conditions (illumination for 9 h with 220 µmol quanta m−2 s−1 at 23°C, dark temperature 21°C). The statistical analysis of plant growth is given in Fig. 4.
Fig. 4.
PBCP mutants show altered development and PSII functionality. Quantitative analysis of growth, PSII quantum efficiency and photoinhibitory parameters are shown for the WT and PBCP mutants. Each column of panels represents measurements from a single round of experiments (Set 1–3). The top row of panels shows the growth curve; the middle row, the PSII quantum efficiency (Fv/Fm); and the bottom row, the photoinhibitory parameter (qI).
Impaired D1 degradation in PBCP mutants
A possible explanation for the impaired growth and the slightly increased level of photoinhibition in the phosphatase mutants is that the efficiency of the PSII repair cycle is affected. To test this, we measured the photochemical efficiency of PSII and the amount of D1 in detached leaves after 60 min of HL treatment. We then compared these parameters with those of the dark-adapted leaves. Experiments were performed in the presence or absence of the plastidial protein synthesis inhibitor lincomycin. Lincomycin inhibits the biosynthesis of the D1 subunit and allows study of the net degradation of D1 (Tyystjarvi and Aro 1996). A 60 min HL treatment (∼1,000 µmol quanta m−2 s−1) without lincomycin causes photoinhibiton to PSII monitored as a decline in the maximal photochemical efficiency parameter Fv/Fm from approximately 0.75 to approximately 0.57 (Table 2). No significant differences were noted between the WT and phosphatase mutants. Addition of lincomycin leads to stronger photoinhibition, i.e. Fv/Fm further decreases to about 0.47. Again no statistically significant changes are apparent between the three genotypes. Essentially similar results were obtained for HL treatment with 1,500 µE m−2 s−1 (results not shown).
Table 2.
Maximal photochemical quantum efficiency of PSII (Fv/Fm) in dark- and HL-treated leaves of WT and PBCP mutants
| Dark | HL | Dark + lincomycin | HL + lincomycin | |
|---|---|---|---|---|
| WT | 0.75 ± 0.03 | 0.58 ± 0.13 | 0.75 ± 0.02 | 0.47 ± 0.125 |
| pbcp-1 | 0.74 ± 0.03 | 0.55 ± 0.11 | 0.74 ± 0.01 | 0.48 ± 0.06 |
| pbcp-2 | 0.75 ± 0.01 | 0.53 ± 0.05 | 0.76 ± 0.01 | 0.47 ± 0.07 |
Data represent the mean ± SD of 8–10 measurements performed on 4–5 plants per genotype.
There were no significant changes in all conditions for both genotypes relative to the WT (P > 0.05) as determined by t-test.
D1 degradation was analyzed by Western blot analysis of HL-treated leaves in the presence of lincomycin (Fig. 5). For WT leaves, 60 min of HL (∼1,500 µmol quanta m−2 s−1) leads to a decrease in the D1 level by about 25%. This is in accordance with the literature (e.g. Kirchhoff et al. 2011). In contrast, no such decline is seen for the PBCP mutant (Fig. 5). It thus follows that D1 degradation is impaired in plants that cannot dephosphorylate PSII core subunits.
Fig. 5.
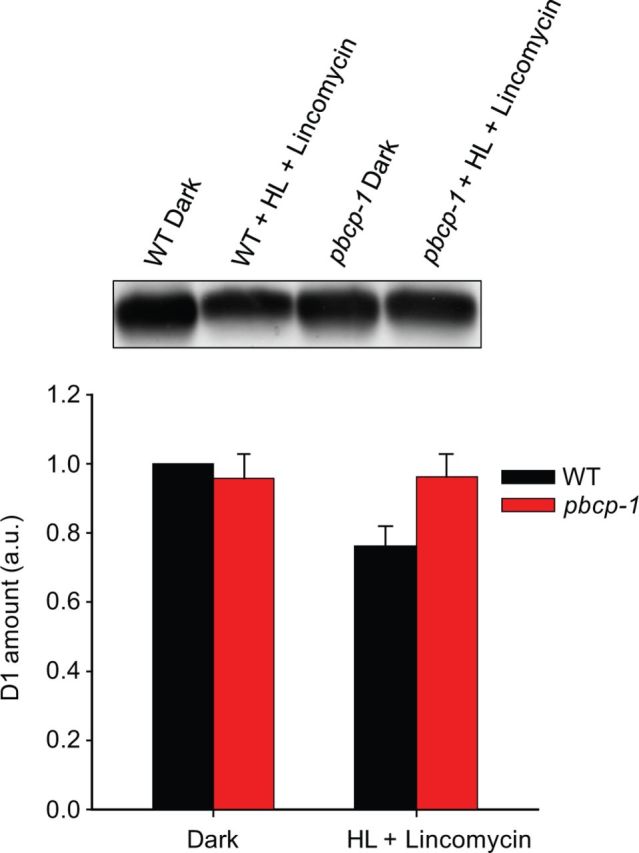
PBCP mutants show diminished D1 degradation in the presence of lincomycin under HL. The amount of D1 in the dark and in HL in the presence of lincomycin, as quantified from two anti-D1 immunoblots, is shown graphically. The D1 amounts in pbcp-1 dark, WT HL + lincomycin, and pbcp-1 HL + lincomycin are shown as ratios to the WT dark amount, while the WT dark amount was kept as ‘1’. A representative D1 immunoblot is shown at the top.
Discussion
The structural and functional parameters examined in this study indicate that deletion of the PSII core phosphatase has only a modest impact on the photosynthetic machinery. In detail, the compositional analysis reveals no significant changes in the abundance of the major thylakoid membrane protein complexes (LHCII, PSI and the Cyt b6f complex). Only the amount of PSII (on a Chl basis) is slightly increased by about 20% in the PBCP mutant. It therefore results in a small (15%) decrease in the LHCII/PSII ratio in the mutant. In accordance with this altered LHCII/PSII ratio, the functional PSII antenna size, probed by Chl fluorescence induction analysis and 77K spectra, is slightly reduced in PBCP mutants. As shown in Fig. 2A, plotting the individual fluorescence induction curves in the presence of DCMU reveals clustering of the data into two sets; one set with a slightly higher and one set with a slightly lower apparent PSII antenna size compared with the WT (expressed by the rate constants given in Fig. 2A). This inconsistency in the behavior of PBCP mutants has already been noted in an earlier study (Samol et al. 2012), and will be discussed further below.
Fig. 1 shows that PSII core subunits are already significantly phosphorylated in dark-adapted samples. This is a well-documented observation and can be attributed mainly to the activity of STN kinases (Bonardi et al. 2005, Fristedt et al. 2009, Fristedt et al. 2010, Pesaresi et al. 2011, Tikkanen and Aro 2012). Under HL, the PSII core phosphorylation increases, most probably because the STN8 kinase is more active under this condition. In the pbcp-1 knockout mutant, the overall PSII core phosphorylation is increased (Fig. 1) because the equilibrium between STN8-dependent phosphorylation and PBCP-dependent dephosphorylation is drastically shifted to the phosphorylation reaction. An interesting detail is that the phosphorylation level can still be increased in the phosphatase mutant by HL. If PBCP is the only PSII core phosphatase, then PSII core proteins should already be maximally phosphorylated in the PBCP knockout mutants in the dark due to a constant and unopposed residual activity of STN8 (see above). No difference in phosphorylation levels should therefore be expected between dark-adapted and HL-incubated mutant samples. The fact that this is not the case, as HL causes a further increase in phosphorylation, points to the existence of additional PSII core phosphatases. Since phosphoserines and phosphothreonines are exceptionally stable at room temperature, it is unlikely that any non-enzymatically catalyzed dephosphorylation of PSII can explain the difference between HL-treated and dark-adapted samples in the pbcp-1 mutant. The possibility of a second, membrane-intrinsic PSII core phosphatase has been raised by some studies (Sun et al. 1989, Allen 1992, Vener et al. 2000). This putative second core phosphatase has been suggested to be regulated by a lumen-located immunophilin known as TLP40 (Vener et al. 2000). The modest effects of PBCP mutants manifested in our study, the pattern of phosphorylation presented in Fig. 1 and some of the odd features of the PBCP revealed in an earlier study (Samol et al. 2012) support this scenario of an undiscovered PSII core phosphatase. Only the identification and characterization of this second (or more) phosphatase(s) will help to address the complete ramifications of PSII core dephosphorylation in the PSII repair cycle and in the functionality of PSII.
An important observation from our study is that the increased PSII core phosphorylation in PBCP knockouts correlates with a diminished D1 degradation in HL-treated leaves in the presence of lincomycin (Fig. 5). This observation supports the notion that dephosphorylation of PSII core subunits is required for efficient D1 degradation by FtsH and Deg proteases (Rintamaki et al. 1996). The missing capability to dephosphorylate the PSII core proteins thus retards D1 degradation in the phosphatase mutants. It is interesting to note that the amounts of D1 in dark-treated WT and PBCP mutants are similar (Fig. 5). This suggests that the damaged D1 does not accumulate in the long term in phosphatase mutants grown under non-stressed conditions and points to the possibility that D1 is in fact degraded albeit less efficiently and with a slow pace by the same proteases or by a different degradation pathway (Bonardi et al. 2005, Pesaresi et al. 2011).
The retarded D1 degradation in the phosphatase mutant seems to have no significant impact on the PSII functionality in the short term (60 min HL treatment) since the HL-induced decrease in the maximal photochemical PSII yield is not different in the WT and mutants (Table 2). This was unexpected because retarded D1 degradation must lead to slower repair of damaged PSII and thus a net decline in functional PSII centers. A possible explanation for the apparent tolerance of the mutant to PSII photoinhibition is its smaller PSII antenna size (see earlier discussion). Due to a smaller antenna, the mutant experiences lower excitation pressure and thus is less vulnerable to photodamage. However, this presumed photoprotective effect of a smaller antenna size on the rate of PSII photoinhibition is controversial, with some reports showing a photoprotective effect while others show no impact (reviewed in Tyystjarvi 2013). Thus whether the smaller antennae of PBCP mutants are photoprotective or not remains to be settled. In contrast to the lack of a photoinhibition phenotype in the short experiment, differences between the WT and PBCP mutants are apparent for the long-term experiments performed in the phenomics facility (Figs 3, 4; see also the biomass data). In two of three experiments both PBCP mutants show retarded growth, compared with the WT. Interestingly our results contrast with the original description on PBCP (Samol et al. 2012), which does not report any growth defects. In all three experiments PBCP mutants also have slightly lower Fv/Fm (dark-adapted state) and higher qI parameters. Both indicate higher photoinhibition in the PBCP mutants. An inefficient operation of the PSII repair cycle, due to the diminished degradation of the D1 protein, might explain the growth phenotype of the PBCP mutant. At this point it cannot be ruled out that the inactivation of the PBCP gene causes pleiotropic effects on growth. Pleiotropic effects have been noted in mutants of other chloroplast phosphatases and kinases. The inactivation of the state transition phosphatase TAP38 unexpectedly improves photosynthetic performance and plant growth in constant light conditions (Pribil et al. 2010). Lack of the state transition kinase STN7 produces plants with an over-reduced plastoquinone pool, which causes pleotropic effects on chloroplast gene regulation (Pesaresi et al. 2009). The stn7 mutant also has a severe growth phenotype under fluctuating light intensity (Tikkanen et al. 2010). Since state transition is a light quality phenomenon, it is not clear why lack of STN7 produces a light quantity phenotype. Similarly, the inability to adjust the stoichiometry of photosystems has indirect consequences for state transitions (Allen et al. 2011). The rice stn8 mutant shows slower growth (Nath et al. 2013), which was not observed with the Arabidopsis STN8 mutant. It remains to be determined whether the rice phenotype is a pleiotropic effect.
An unresolved observation in our study is that the PBCP mutants sometimes show contradictory phenotypes. In particular, retarded growth was not observed in measurements with Set 2 (Fig. 4). An inconsistent response in the PSII repair cycle has also been noted in the previous characterization of PBCP mutants (Samol et al. 2012). Currently there is no explanation for this variability that was also apparent for PSII antenna size (Fig. 2). One possibility is that another phosphatase is involved, which is activated to a greater or lesser extent in the absence of PBCP. Future studies should examine this intricacy of PBCP in order to understand fully the plasticity of reversible protein phosphorylation in regulating photosynthetic processes.
Materials and Methods
Plant material and membrane preparations
Arabidopsis thaliana WT and PBCP knockout mutant seedlings were grown from seeds on soil at 24°C and a photon flux density of 150 µE m−2 s−1 with an 8 h day and 16 h dark photoperiod unless otherwise specified. Protoplasts were isolated from detached leaves using a cellulose and macerozyme enzyme solution, essentially as described in Herbstova et al. (2012). Thylakoid membranes were obtained by pottering intact protoplasts in a hypotonic buffer containing 40 mM HEPES (pH 7.6), 80 mM KCl, 7 mM MgCl2, and the protease inhibitors Pefablock SC (Roche), leupeptin and antipain (Sigma-Aldrich). Thylakoids were pelleted (at 3,200 × g for 10 min at 4°C) and resuspended in a storage buffer containing 50 mM HEPES (pH 7.5), 0.1 M sorbitol and 2 mM MgCl2.
Phosphorylation analysis
Protoplasts were incubated either in the dark or in HL (1,500 µE m−2 s−1) for 1 h, while being slowly agitated. Thylakoids were isolated from protoplasts as described earlier. Thylakoid storage buffers contained 10 mM NaF and phosSTOP (Roche) to prevent dephosphorylation of phosphorylated thylakoid proteins. Laemmli sample buffer-treated thylakoids were separated on an 11.5% SDS–urea–polyacrylamide gel. Samples were loaded on the gel on an equal Cyt b559 (PSII) basis. Separated proteins were then electroblotted onto a PVDF (polyvinylidene fluoride) membrane using standard Western blotting protocols. The membrane was then blocked in 5% bovine serum albumin (BSA) overnight at 4°C, washed and then probed with anti-phosphothreonine antibody (Zymed) for 90 min at room temperature. Horseradish peroxidase-conjugated secondary antibody was used in the immunodetection. Immunoreactive bands were detected by fluorography using the ECL detection kit (GE Healthcare). Phosphorylation levels were determined from densitometric quantification of the bands by using ImagePro Plus software.
D1 quantification
For D1 quantification, detached leaves of WT and PBCP mutants were incubated in 1 mM lincomycin solution overnight in the dark. Leaves were incubated in such a way that only the petioles made contact with the lincomycin solution. The maximal photochemical efficiency of PSII was measured as Fv/Fmax with a pulse-modulated fluorimeter (Hansatech OxyLab). Leaves were then ground in liquid nitrogen and the total protein was extracted with 1× Laemmli sample buffer (without the bromophenol blue). The extract was centrifuged at 13,000 r.p.m. in a microcentrifuge for 10 min to remove unsolubilized plant material. The supernatant was then transferred to new tubes and used for the D1 quantification. The Chl concentration of the samples was determined by the Porra method. Samples equivalent to 1.5 µg of Chls were then subjected to SDS–urea–PAGE. Separated proteins were electroblotted onto a PVDF membrane and the membrane was blocked in 5% non-fat dry milk solution overnight at 4°C. The membrane was probed with antibody raised against the C-terminus of the D1 protein (Agrisera). Immunoreactive bands were detected by fluorography as described earlier.
Chl fluorescence induction kinetics
Chloroplasts were isolated from Arabidopsis leaves as described in Daum et al. (2010), with a slight modification of the second spin from 2 min to 5 min. Chloroplasts were then lysed with the shock buffer, containing 25 mM HEPES (pH 7.5), 40 mM KCl and 7 mM MgCl2, to obtain thylakoids. Thylakoids were resuspended in double concentration buffer (shock buffer + 600 mM sorbitol). To perform the measurements, thylakoids at a concentration of 10 µg ml−1 were taken in a 1,500 µl final volume (1:1 mixture of shock buffer and double concentration buffer), and DCMU was added to a final concentration of 100 µM. These were then mixed and incubated for 2 min at room temperature to allow oxidation of QA. Fluorescence induction kinetics were recorded with a home-built instrument as described in Haferkamp et al. (2010). Samples were excited with 530 nm light that allows homogenous light distribution (Rappaport et al. 2007) in the cuvette. Sample preparation and measurements were conducted in a dark room to avoid reduction of QA before the measurements. Fluorescence induction kinetics were mathematically analyzed as described in Kirchhoff et al. (2004). Each measurement was performed in duplicate, using separate samples from the same preparation. In total, 3–6 biological replicates were measured.
Low-temperature fluorescence spectroscopy
Low temperature fluorescence emission spectra were obtained with a Horiba Jobin Yvon FluoroMax 4 spectrofluorometer (λex = 475 nm, λem = 650–800 nm; bandwidth = 5 nm). Leaf material from dark-adapted and HL-treated plants was ground in a buffer medium (450 mM sorbitol, 10 mM EDTA, 10 mM NaHCO3, 20 mM Tricine and 0.1% BSA; pH 8.2).
Phenomics analysis
The photosynthetic parameters were determined for intact Arabidopsis plants in the WSU phenomics facility consisting of a greenhouse (artificial illumination) and an optical screening robot. Nine pbcp-1, pbcp-2 and WT plants were grown in the greenhouse with a 9 h light period of 220 µE m−2 s−1 illumination at daily temperatures of 21°C in the dark and 23°C in the light. The Arabidopsis plants were acclimated to measurement chamber conditions for 5 d before the measurement. Measurements were performed daily in the latter half of the dark period. The Fluorcam XYZ system (PSI company) is a mobile and programmable fluorescence imaging robot capable of moving throughout the growth chamber on a gantry, performing automated measurements utilizing blue (455 nm) and red (618 nm) light-emitting diodes (LEDs) to excite Chls. A Fluorcam 2701 LU camera, equipped with a fluorescence filter, was used to capture the emitted fluorescence. The qI parameter was determined as non-photosynthetic quenching (NPQ) that does not recover within 3 min after a 5 min illumination (220 µE m−2 s−1), i.e. when the fast qE (energy-dependent quenching) component relaxes. It was checked that after the 3 min of darkness the NPQ level is stable, which indicates qE relaxation.
Determination of the biomass for WT and PCBP mutant plants of Arabidopsis thaliana
The respective WT, pbcp-1 and pbcp-2 mutant plants were harvested after a 6 week growth period and weighed. Nine plants were used for each genotype. The mean and SE for the fresh weight was calculated for each genotype. A t-test was used for statistical analysis of the data.
Funding
This work was funded by the National Science Foundation [NSF-MCB115871]; the United States–Israel Binational Agricultural Research and Development Fund [BARD US-4334-10]; the US National Institute of Food and Agriculture [NIFA; #11D-3037–3784].
Acknowledgments
Robert Yarbrough is acknowledged for proofreading the manuscript.
Glossary
Abbreviations
- DTT
dithiothreitol
- HL
high light
- LHCII
light-harvesting complex II
- NPQ
non-photochemical quenching
- PBCP
PSII core phosphatase
- PPH1
protein phosphatase 1
- QA
quinone A
- QB
quinone B
- qE
energy-dependent quenching
- qI
photoinhibition-dependent quenching
- STT7
state transition thylakoid 7
- STL1
state transition less 1
- STN7
state transition 7
- STN8
state transition 8
- TAP38
thylakoid-associated phosphatase of 38 kDa
- WT
wild type
Disclosures
The authors have no conflicts of interest to declare.
References
- Albertsson P. A quantitative model of the domain structure of the photosynthetic membrane. Trends Plant Sci. 2001;6:349–358. doi: 10.1016/s1360-1385(01)02021-0. [DOI] [PubMed] [Google Scholar]
- Allen JF. Protein phosphorylation in regulation of photosynthesis. Biochim. Biophys. Acta. 1992;1098:275–335. doi: 10.1016/s0005-2728(09)91014-3. [DOI] [PubMed] [Google Scholar]
- Allen JF, Santabarbara S, Allen CA, Puthiyaveetil S. Discrete redox signaling pathways regulate photosynthetic light-harvesting and chloroplast gene transcription. PLoS One. 2011;6:e26372. doi: 10.1371/journal.pone.0026372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson B, Anderson JM. Lateral heterogeneity in the distribution of chlorophyll–protein complexes of the thylakoid membranes of spinach chloroplasts. Biochim. Biophys. Acta. 1980;593:427–440. doi: 10.1016/0005-2728(80)90078-x. [DOI] [PubMed] [Google Scholar]
- Andreeva A, Stoitchkova K, Busheva M, Apostolova E. Changes in the energy distribution between chlorophyll–protein complexes of thylakoid membranes from pea mutants with modified pigment content. I. Changes due to the modified pigment content. J. Photochem. Photobiol. B. 2003;70:153–162. doi: 10.1016/s1011-1344(03)00075-7. [DOI] [PubMed] [Google Scholar]
- Bellafiore S, Barneche F, Peltier G, Rochaix JD. State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature. 2005;433:892–895. doi: 10.1038/nature03286. [DOI] [PubMed] [Google Scholar]
- Bonardi V, Pesaresi P, Becker T, Schleiff E, Wagner R, Pfannschmidt T, et al. Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature. 2005;437:1179–1182. doi: 10.1038/nature04016. [DOI] [PubMed] [Google Scholar]
- Cohen P. The origins of protein phosphorylation. Nat. Cell Biol. 2002;4:E127–E130. doi: 10.1038/ncb0502-e127. [DOI] [PubMed] [Google Scholar]
- Daum B, Nicastro D, Austin J, 2nd, McIntosh JR, Kuhlbrandt W. Arrangement of photosystem II and ATP synthase in chloroplast membranes of spinach and pea. Plant Cell. 2010;22:1299–1312. doi: 10.1105/tpc.109.071431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depege N, Bellafiore S, Rochaix JD. Rote of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science. 2003;299:1572–1575. doi: 10.1126/science.1081397. [DOI] [PubMed] [Google Scholar]
- Elich TD, Edelman M, Mattoo AK. Identification, characterization, and resolution of the in vivo phosphorylated form of the D1 photosystem II reaction center protein. J. Biol. Chem. 1992;267:3523–3529. [PubMed] [Google Scholar]
- Fristedt R, Granath P, Vener AV. A protein phosphorylation threshold for functional stacking of plant photosynthetic membranes. PLoS One. 2010;5:e10963. doi: 10.1371/journal.pone.0010963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristedt R, Vener AV. High light induced disassembly of photosystem II supercomplexes in Arabidopsis requires STN7-dependent phosphorylation of CP29. PLoS One. 2011;6:e24565. doi: 10.1371/journal.pone.0024565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristedt R, Willig A, Granath P, Crevecoeur M, Rochaix JD, Vener AV. Phosphorylation of photosystem II controls functional macroscopic folding of photosynthetic membranes in Arabidopsis. Plant Cell. 2009;21:3950–3964. doi: 10.1105/tpc.109.069435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goral TK, Johnson MP, Brain AP, Kirchhoff H, Ruban AV, Mullineaux CW. Visualizing the mobility and distribution of chlorophyll proteins in higher plant thylakoid membranes: effects of photoinhibition and protein phosphorylation. Plant J. 2010;62:948–959. doi: 10.1111/j.0960-7412.2010.04207.x. [DOI] [PubMed] [Google Scholar]
- Haferkamp S, Haase W, Pascal AA, van Amerongen H, Kirchhoff H. Efficient light harvesting by photosystem II requires an optimized protein packing density in grana thylakoids. J. Biol. Chem. 2010;285:17020–17028. doi: 10.1074/jbc.M109.077750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbstova M, Tietz S, Kinzel C, Turkina MV, Kirchhoff H. Architectural switch in plant photosynthetic membranes induced by light stress. Proc. Natl Acad. Sci. USA. 2012;109:20130–20135. doi: 10.1073/pnas.1214265109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kargul J, Turkina MV, Nield J, Benson S, Vener AV, Barber J. Light-harvesting complex II protein CP29 binds to photosystem I of Chlamydomonas reinhardtii under State 2 conditions. FEBS J. 2005;272:4797–4806. doi: 10.1111/j.1742-4658.2005.04894.x. [DOI] [PubMed] [Google Scholar]
- Kirchhoff H. Structural constraints for protein repair in plant photosynthetic membranes. Plant Signal. Behav. 2013;8:e23634. doi: 10.4161/psb.23634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff H, Borinski M, Lenhert S, Chi L, Buchel C. Transversal and lateral exciton energy transfer in grana thylakoids of spinach. Biochemistry. 2004;43:14508–14516. doi: 10.1021/bi048473w. [DOI] [PubMed] [Google Scholar]
- Kirchhoff H, Haferkamp S, Allen JF, Epstein DB, Mullineaux CW. Protein diffusion and macromolecular crowding in thylakoid membranes. Plant Physiol. 2008;146:1571–1578. doi: 10.1104/pp.107.115170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff H, Hall C, Wood M, Herbstova M, Tsabari O, Nevo R, et al. Dynamic control of protein diffusion within the granal thylakoid lumen. Proc. Natl Acad. Sci. USA. 2011;108:20248–20253. doi: 10.1073/pnas.1104141109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivuniemi A, Aro EM, Andersson B. Degradation of the D1- and D2-proteins of photosystem II in higher plants is regulated by reversible phosphorylation. Biochemistry. 1995;34:16022–16029. doi: 10.1021/bi00049a016. [DOI] [PubMed] [Google Scholar]
- Krause GH, Jahns P, editors. Dordrecht: Kluwer Academic Publishers; 2003. Pulse Amplitude Modulated Fluorometry and its Application in Plant Science. [Google Scholar]
- Kyle DJ, Ohad I, Arntzen CJ. Membrane protein damage and repair: selective loss of a quinone-protein function in chloroplast membranes. Proc. Natl Acad. Sci. USA. 1984;81:4070–4074. doi: 10.1073/pnas.81.13.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavergne J, Trissl HW. Theory of fluorescence induction in photosystem II: derivation of analytical expressions in a model including exciton-radical-pair equilibrium and restricted energy transfer between photosynthetic units. Biophys. J. 1995;68:2474–2492. doi: 10.1016/S0006-3495(95)80429-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar D. Chlorophyll a fluorescence induction. Biochim. Biophys. Acta. 1999;1412:1–28. doi: 10.1016/s0005-2728(99)00047-x. [DOI] [PubMed] [Google Scholar]
- Melis A. Photosystem-II damage and repair cycle in chloroplasts: what modulates the rate of photodamage? Trends Plant Sci. 1999;4:130–135. doi: 10.1016/s1360-1385(99)01387-4. [DOI] [PubMed] [Google Scholar]
- Mullineaux CW. Factors controlling the mobility of photosynthetic proteins. Photochem. Photobiol. 2008;84:1310–1316. doi: 10.1111/j.1751-1097.2008.00420.x. [DOI] [PubMed] [Google Scholar]
- Nath K, Poudyal RS, Eom JS, Park YS, Zulfugarov IS, Mishra SR, et al. Loss-of-function of OsSTN8 suppresses the photosystem (PS) II core protein phosphorylation and interferes with PSII repair mechanism in rice (Oryza sativa) Plant J. 2013;76:675–686. doi: 10.1111/tpj.12331. [DOI] [PubMed] [Google Scholar]
- Nixon PJ, Michoux F, Yu J, Boehm M, Komenda J. Recent advances in understanding the assembly and repair of photosystem II. Ann. Bot. 2010;106:1–16. doi: 10.1093/aob/mcq059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi P, Hertle A, Pribil M, Kleine T, Wagner R, Strissel H, et al. Arabidopsis STN7 kinase provides a link between short- and long-term photosynthetic acclimation. Plant Cell. 2009;21:2402–2423. doi: 10.1105/tpc.108.064964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi P, Pribil M, Wunder T, Leister D. Dynamics of reversible protein phosphorylation in thylakoids of flowering plants: the roles of STN7, STN8 and TAP38. Biochim. Biophys. Acta. 2011;1807:887–896. doi: 10.1016/j.bbabio.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Pribil M, Pesaresi P, Hertle A, Barbato R, Leister D. Role of plastid protein phosphatase TAP38 in LHCII dephosphorylation and thylakoid electron flow. PLoS Biol. 2010;8:e1000288. doi: 10.1371/journal.pbio.1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthiyaveetil S, Ibrahim IM, Allen JF. Oxidation–reduction signalling components in regulatory pathways of state transitions and photosystem stoichiometry adjustment in chloroplasts. Plant Cell Environ. 2012;35:347–359. doi: 10.1111/j.1365-3040.2011.02349.x. [DOI] [PubMed] [Google Scholar]
- Puthiyaveetil S, Kirchhoff H. A phosphorylation map of the photosystem II supercomplex C2S2M2. Front. Plant Sci. 2013;4:459. doi: 10.3389/fpls.2013.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport F, Beal D, Joliot A, Joliot P. On the advantages of using green light to study fluorescence yield changes in leaves. Biochim. Biophys. Acta. 2007;1767:56–65. doi: 10.1016/j.bbabio.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Rintamaki E, Kettunen R, Aro EM. Differential D1 dephosphorylation in functional and photodamaged photosystem II centers. Dephosphorylation is a prerequisite for degradation of damaged D1. J. Biol. Chem. 1996;271:14870–14875. doi: 10.1074/jbc.271.25.14870. [DOI] [PubMed] [Google Scholar]
- Rochaix JD. Redox regulation of thylakoid protein kinases and photosynthetic gene expression. Antioxid. Redox Signal. 2013;18:2184–2201. doi: 10.1089/ars.2012.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokka A, Aro EM, Herrmann RG, Andersson B, Vener AV. Dephosphorylation of photosystem II reaction center proteins in plant photosynthetic membranes as an immediate response to abrupt elevation of temperature. Plant Physiol. 2000;123:1525–1536. doi: 10.1104/pp.123.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samol I, Shapiguzov A, Ingelsson B, Fucile G, Crevecoeur M, Vener AV, et al. Identification of a photosystem II phosphatase involved in light acclimation in Arabidopsis. Plant Cell. 2012;24:2596–2609. doi: 10.1105/tpc.112.095703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Bailey D, Jones MW, Markwel J. Chloroplast thylakoid protein phosphatase is a membrane surface-associated activity. Plant Physiol. 1989;89:238–243. doi: 10.1104/pp.89.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Iwai M, Takahashi Y, Minagawa J. Identification of the mobile light-harvesting complex II polypeptides for state transitions in Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA. 2006;103:477–482. doi: 10.1073/pnas.0509952103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikkanen M, Aro EM. Thylakoid protein phosphorylation in dynamic regulation of photosystem II in higher plants. Biochim. Biophys. Acta. 2012;1817:232–238. doi: 10.1016/j.bbabio.2011.05.005. [DOI] [PubMed] [Google Scholar]
- Tikkanen M, Aro EM. Integrative regulatory network of plant thylakoid energy transduction. Trends Plant Sci. 2014;19:10–17. doi: 10.1016/j.tplants.2013.09.003. [DOI] [PubMed] [Google Scholar]
- Tikkanen M, Grieco M, Kangasjarvi S, Aro EM. Thylakoid protein phosphorylation in higher plant chloroplasts optimizes electron transfer under fluctuating light. Plant Physiol. 2010;152:723–735. doi: 10.1104/pp.109.150250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikkanen M, Nurmi M, Kangasjarvi S, Aro EM. Core protein phosphorylation facilitates the repair of photodamaged photosystem II at high light. Biochim. Biophys. Acta. 2008;1777:1432–1437. doi: 10.1016/j.bbabio.2008.08.004. [DOI] [PubMed] [Google Scholar]
- Tyystjarvi E. Photoinhibition of photosystem II. Int. Rev. Cell Mol. Biol. 2013;300:243–303. doi: 10.1016/B978-0-12-405210-9.00007-2. [DOI] [PubMed] [Google Scholar]
- Tyystjarvi E, Aro EM. The rate constant of photoinhibition, measured in lincomycin-treated leaves, is directly proportional to light intensity. Proc. Natl Acad. Sci. USA. 1996;93:2213–2218. doi: 10.1073/pnas.93.5.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vener AV, Rokka A, Fulgosi H, Andersson B, Herrmann RG. A cyclophilin-regulated PP2A-like protein phosphatase in thylakoid membranes of plant chloroplasts. Biochemistry. 2000;39:2130. doi: 10.1021/bi005102g. [DOI] [PubMed] [Google Scholar]