Abstract
Protein kinases phosphorylate substrates in the context of specific phosphorylation site sequence motifs. The knowledge of the specific sequences that are recognized by kinases is useful for mapping sites of phosphorylation in protein substrates and facilitates the generation of model substrates to monitor kinase activity. Here, we have adapted a positional scanning peptide library method to a microarray format that is suitable for the rapid determination of phosphorylation site motifs for tyrosine kinases. Peptide mixtures were immobilized on glass slides through a layer of a tyrosine-free Y33F mutant avidin to facilitate the analysis of phosphorylation by radiolabel assay. A microarray analysis provided qualitatively similar results in comparison with the solution phase peptide library “macroarray” method. However, much smaller quantities of kinases were required to phosphorylate peptides on the microarrays, which thus enabled a proteome scale analysis of kinase specificity. We illustrated this capability by microarray profiling more than 80% of the human nonreceptor tyrosine kinases (NRTKs). Microarray results were used to generate a universal NRTK substrate set of 11 consensus peptides for in vitro kinase assays. Several substrates were highly specific for their cognate kinases, which should facilitate their incorporation into kinase-selective biosensors.
Keywords: peptide microarrays, protein kinases, enzyme specificity, peptide libraries, nonreceptor tyrosine kinases, kinase inhibitors, drug discovery
Introduction
Protein kinases comprise a large group of molecules with diverse roles in cellular signaling networks.1 Because protein kinases are often mutated, amplified, or otherwise deregulated in cancer, there has been significant effort geared toward the identification of kinase inhibitors for therapeutic use.2 The identification and optimization of kinase inhibitor lead compounds requires an efficient enzymatic assay to measure the activity of the kinase of interest.3 For a high-throughput screening campaign, the availability of an efficient substrate can drastically reduce the amount of enzyme that is required for the screening. Given that the success rate for producing active kinases in bacteria is rather low (∼10%),4 insect and mammalian cell expression systems are generally required, which means that kinase production on a large scale is often costly. The availability of an efficient substrate enables the screening of a larger compound library. In addition to their uses in drug discovery, optimized peptide substrates have been used as probes to monitor the dynamics of protein kinase activity in living cells. Highly specific optimized peptides have been used to assay kinases in crude systems such as cell lysates.5,6 In addition, the incorporation into fluorescent biosensors enables researchers to report on the spatiotemporal dynamics of kinase activity in individual living cells.7 High affinity consensus peptides can also be used as structural probes to examine kinase–substrate interactions8,9 and to monitor activity-dependent conformational changes.10
Assays for protein kinases generally involve the use of non-native model substrates, including proteins (myelin basic protein, casein, etc.) or peptides.11 Peptide substrates can be generated from the sequence that flanks the phosphorylation site of a known protein substrate or discovered by trial and error. Peptide library approaches based on the analysis of peptide mixtures or arrays can facilitate the screening of large numbers of peptide sequences to produce optimized consensus kinase substrates.12−14 We have reported a positional scanning peptide library that comprehensively substitutes each of the 20 amino acid residues at each of nine positions that surround a central phosphorylation site.15 The miniaturization of the method to a 1536 well plate format allowed us to determine phosphorylation consensus sequences for the majority of kinases from the budding yeast Saccharomyces cerevisiae;16 however, the necessity to produce nontrivial quantities of kinase (generally tens of μg per analysis) and the limited capacity for multiplexing has hampered our ability to rapidly screen large numbers of kinases.
An alternative approach to the analysis of kinase specificity is to use peptide microarrays.17,18 In this approach, substrate peptides that are immobilized on glass slides in a high-density format are treated with kinase and ATP, and the relative extent of phosphorylation of each peptide is determined either using autoradiography or a fluorescence detection strategy. Peptide microarrays have a potential advantage in that they require very small amounts of kinase, which theoretically could enable large scale analysis. Current microarray strategies typically use a set of (often thousands of) defined peptide sequences that are typically derived from authentic proteomes. Such peptide microarrays have been used both to profile individual kinases and to dissect the activation of specific kinases within crude cell lysates.18−20 One disadvantage of the current strategies is that many proteome-derived peptides have multiple potential sites of phosphorylation, which causes ambiguity in the assignment of specific sites to a given kinase. In addition, peptides that are derived from authentic protein substrates of kinases can lack key determinants of specificity and thus make inefficient substrates, as in the case of the phosphorylation of the C-terminal tail of Src family kinases by their upstream regulators CSK and CHK.21
Here, we describe the miniaturization of our positional scanning peptide library to a microarray format. As a proof-of-concept experiment, we have profiled the entire set of human nonreceptor tyrosine kinases (NRTKs), which establishes consensus motifs for 26 enzymes. On the basis of the peptide microarray data, we have generated consensus peptides for each NRTK family and determined a selectivity profile for each substrate. These peptides provide a universal set of efficient peptide substrates to enable the screening for NRTK inhibitors as well as a general set of tools for the analysis of NRTK signaling pathways.
Experimental Section
Peptide Microarray Fabrication
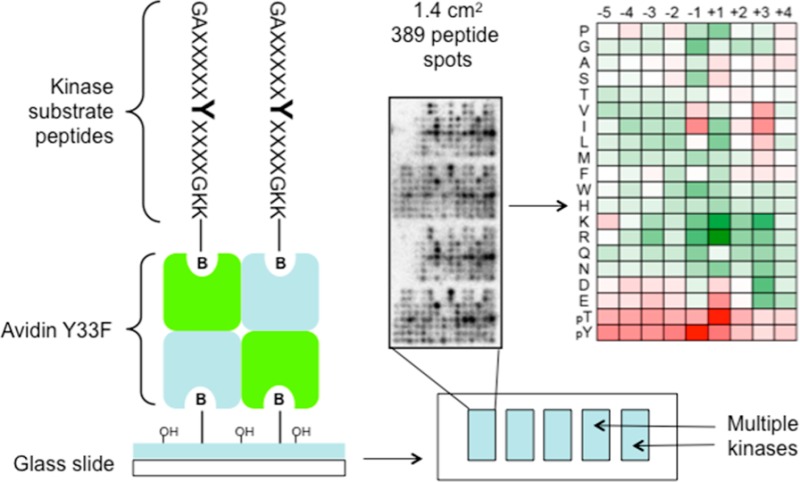
Nonglycosylated chicken avidin Y33F mutant22 was produced in E. coli BL-21(AI) cells using a pET101/D-TOPO-based expression plasmid where the original avidin signal peptide was replaced by a bacterial signal peptide as described.23 The protein was produced in a Labfors Infors 3 fermenter by using a fed-batch method,24 purified by 2-iminobiotin affinity chromatography, and verified by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Avidin-Y33F was diluted in a binding buffer (10 mM Tris-HCl, pH 7.5, 140 mM NaCl, 10% glycerol, 0.1% Triton X-100) to 20–50 μg/mL. The avidin solution (200 μL) was spread onto high-density biotin-coated glass slides (Microsurfaces, Inc.), and the slides were incubated in a humidified chamber for 30 min. The excess avidin was removed by washing extensively with Tris-buffered saline with Tween 20 (TBST) (10 mM Tris-HCl, pH 7.5, 140 mM NaCl, 0.1% Tween 20) followed by a brief rinse with ddH2O. Slides were then blown dry with nitrogen gas and stored at 4 °C until printing. Microarrays were prepared by spotting onto the slides 189 peptide mixtures (synthesized by Anaspec, Inc.) with the general sequence G-A-X-X-X-X-X-Y-X-X-X-X-G-A–K-K (biotin), in which X is an equimolar mixture of the 18 amino acids, excluding Cys and Tyr. For each X position, 21 distinct mixtures were used in which one of the 19 unmodified amino acids (excluding Cys), phosphothreonine (pThr), or phosphotyrosine (pTyr) was incorporated as a single fixed residue at that position. The peptides were diluted from 10 mM dimethyl sulfoxide (DMSO) stock solutions to a final concentration of 1 mM in the spotting buffer (10 mM Tris-HCl, pH 7.5, 140 mM NaCl, 15% glycerol, 0.01% Triton X-100). The peptides were spotted in a humidified chamber using a GeneMachines Omnigrid robotic printer. Four duplicate sets of peptides were spotted on each slide, and each set of 396 peptides occupied an 18 mm × 8 mm area with an interspot distance of 0.5 mm. Following printing, the slides were incubated for at least 1 h in a sealed 50 mL conical tube. The excess peptide was removed by washing with TBST that contained 5 mM biotin once followed by TBST three times, for 5 min per wash. After being rinsed with ddH2O, the slides were blown dry under a stream of nitrogen and stored at 4 °C. The printed slides were used within a few weeks.
Production of Protein Kinases
Gateway entry vectors (pDONR221) that harbored the full length coding sequences for 26 human NRTKs were purchased from Life Technologies. A plasmid that harbored the human FER coding sequence was obtained from Nora Heisterkamp (Children’s Hospital, Los Angeles), and TEC cDNA was obtained from Open Biosystems. The entry vectors for these two NRTKs were generated by a polymerase chain reaction (PCR) amplification of the corresponding cDNA with the primers that added the attB adapters, followed by recombination into pDONR221 using BP clonase as per the manufacturer’s instructions. NRTK coding sequences were recombined using LR clonase (Life Technologies) as directed by the manufacturer into pDEST27, a mammalian expression vector that produces N-terminal GST fusion proteins. To produce kinases, HEK293T cells were transiently transfected in 6 cm plates with 8 μg of the appropriate expression vector using 20 μL of Lipofectamine 2000 (Life Technologies) as directed by the manufacturer. After 48 h, the plates were lysed in 500 μL of lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 1 mM ethylenediaminetetraacetic acid (EDTA), 1 mM ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid (EGTA), 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 μg/mL leupeptin, 2 μg/mL pepstatin A, and 10 μg/mL aprotinin). The lysates were cleared by centrifugation at 4 °C for 10 min at 13 000 × g and incubated with glutathione Sepharose 4B resin (GE Healthcare, 15 μL per plate) while being mixed for 2 h at 4 °C. The beads were pelleted (2 min at 4000 × g) and washed twice with lysis buffer and then twice with a wash buffer (50 mM N-(2-hydroxyethyl)piperazine-N′-ethanesulfonic acid (HEPES), pH 7.4, 100 mM NaCl, 5 mM β-glycerophosphate, 0.1 mM Na3VO4, 0.01% Igepal CA630, 10% glycerol). The kinases were eluted from the beads in batch by incubating with two rounds of 0.5 mg/mL reduced glutathione (GSH) in a wash buffer (20 μL per plate). The purified kinases were quantified when a Coomassie-stained polyacrylamide gel was scanned with a LI-COR Odyssey imager using bovine serum albumin (BSA) as a standard. For three of the kinases (BMX, PTK6, and TEC), the transfected cells were treated with 50 μM of sodium pervanadate 30 min prior to lysis to activate the kinases. For these kinases, we generated inactive mutants using the QuikChange method (Stratagene) by converting the catalytic loop Asp residue (equivalent to the residue Asp166 of cAMP-dependent protein kinase) to Asn.25 The mutant expression constructs were verified by Sanger sequencing prior to use. Active JAK1 and JAK3 catalytic domains and full length CSK used for microarray screening were produced from a baculovirus expression system in insect cells as described.26,27 The bacterially-expressed FES (generated in Stefan Knapp’s group) and insect cell-expressed full length ABL (generated by Anthony Koleske’s group) used for the microarray screens were previously described.28,29 LCK, TXK, and JAK2 that were used for microarray screening were purchased from Signalchem.
Peptide Microarray Screening
Peptide sets were partitioned by the slide being marked with a PAP pen (Abcam). The purified kinase (1–10 ng) in 20 μL of kinase reaction buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 1 mM DTT, 0.5% Triton X-100, and 0.5–1 μCi [γ-33P]ATP) was added onto one duplicate peptide set, and the slide was incubated in a humidified chamber at 30 °C for 30 min. Following incubation, the slide was washed three times with a quenching buffer (10 mM Tris-HCl, pH 7.5, 140 mM NaCl, 0.1% SDS) and two times with 2 M NaCl, for 10 min per wash. The slides were then rinsed with ddH2O, dried under a stream of nitrogen, covered in plastic wrap, and exposed to a phosphor storage screen. Following exposure, the phosphor screen was scanned on a phosphor imager (Bio Rad), and the radiolabel incorporation into the peptide spots was quantified with QuantityOne software. The spot intensities were normalized so that the average value within a position in the peptide was assigned to unity. Each kinase was screened two times, and normalized values were averaged across the two runs.
Radiolabel Kinase Assays with Consensus Peptide Substrates
The peptides were produced on a Creosalus Tetras automated peptide synthesizer using standard Fmoc protocols.30 All of the protected amino acids were from Advanced Chemtech, and the synthesis resin (Fmoc-Lys(biotin-LC)-Wang resin) was from Anaspec. The peptides were purified by preparative reversed-phase high-performance liquid chromatography (HPLC) (acetonitrile–water gradient) and verified by matrix-assisted laser desorption ionization mass spectometry (MALDI-MS) prior to use. The peptides (5 μM, diluted from 1 mM aqueous stock solutions) in the reaction buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 1 mM DTT, 50 μM ATP with 0.5 μCi/μL [γ-33P]ATP) were incubated in sealed 384 well plates (20 μL per well) for 30 min at 30 °C. The plate was then chilled on ice, and 2 μL aliquots from each well were transferred to a streptavidin-coated membrane (SAM2 Biotin Capture Membrane, Promega) simultaneously using a slot pin replicator (V&P Scientific). The membrane was washed three times with 10 mM Tris-HCl, pH 7.5, 140 mM NaCl, 0.1% SDS, twice with 2 M NaCl, twice with 2 M NaCl with 1% H3PO4, and twice with water. The membrane was then air-dried and analyzed by phosphor imaging to determine the relative levels of radiolabel incorporation into each peptide. To determine the kinetic constants for the individual substrates, 30 μL reactions were performed in the reaction buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10 mM MgCl2, 50 μM ATP with 0.5 μCi/μL [γ-33P]ATP). Substrate concentration was varied in two-fold increments over the following ranges: ABLtide, 1–32 μM; LYNtide, 1–32 μM (LYN kinase assay) and 1–64 μM (SRC kinase assay); SRCtide, 1–64 μM; FEStide, 1–64 μM; SYKtide, 0.125–8 μM. The kinase concentrations were as follows: ABL, 5.5 nM; LYN, 4.7 nM; SRC, 8.5 nM; FES, 21 nM; SYK, 2.6 nM. Following incubation at 30 °C for 5 min, 20 μL aliquots were transferred to P81 phosphocellulose filters. After being extensively washed with 0.85% H3PO4, the filters were immersed briefly in acetone, air-dried, and analyzed by scintillation counting.
Results and Discussion
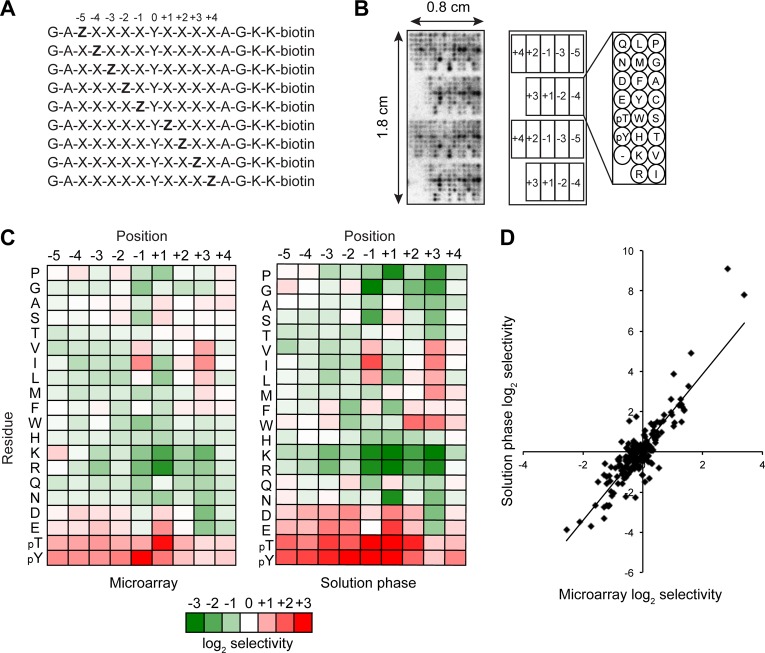
To facilitate the rapid identification of phosphorylation site motifs for large numbers of tyrosine kinases, we adapted a positional scanning peptide library (PSPL) method15 to a high-density microarray format. While collections of thousands of kinase peptide substrates18 and smaller sets of peptide mixtures17 were analyzed previously in a microarray format, the PSPL approach involves a systematic substitution of all possible residues at positions proximal to the phosphorylation site and is thus well-suited to generate optimized substrate sequences. The PSPL consists of 189 C-terminally biotinylated peptide mixtures in which a central Tyr residue is flanked by a degenerate sequence31 (Figure 1A). In each mixture, a single residue is fixed at one of 9 positions surrounding the Tyr residue. Multiple strategies have been developed to immobilize peptides and proteins on glass slides for microarray analysis.32 We chose to use avidin-coated slides to immobilize peptides via their biotin groups. This method facilitated the comparison of the same PSPL used in both microarray and conventional formats. In addition, the avidin provides an intervening hydrophilic layer that is necessary for kinases to phosphorylate bound peptides.33 To avoid potential complications from the phosphorylation of avidin itself, we utilized an avidin mutant (Y33F) in which its sole Tyr residue is changed to Phe, but retains a high affinity for biotin.22 We generated the avidin-coated slides by incubating commercially available biotin-coated slides with avidin-Y33F. Because avidin is a tetramer, attachment to the biotin-coated slide occurs in a manner that leaves additional binding sites unoccupied and available for peptide immobilization. The peptides were spotted in duplicate at high density using a robotic printer (Figure 1B).
Figure 1.
Peptide microarray method for the determination of tyrosine kinase phosphorylation motifs. (A) Sequence of PSPL components used to generate microarrays. Degenerate positions indicated by X have an equimolar mixture of the 18 amino acids, excluding Cys and Tyr, and biotin is appended via an aminohexanoic acid spacer to the ε-amine of the C-terminal Lys residue, which enables binding to an avidin-coated surface. Positions marked Z indicate a single amino acid, one of 19 amino acids (excluding Cys), pThr (pT), or pTyr (pY). (B) Peptide microarray image for the NRTK FES, which shows the dimensions and layout of peptides on the array. The full set of 189 peptides is spotted in duplicate for each analysis. (C) A comparison of microarray data with published solution phase PSPL data for FES. Spot intensities were normalized so that the average value within a position was equal to one. Heat maps (generated with Microsoft Excel) show the log2 transformed data with positive selections shown in red and negative selections shown in green. Both microarray and solution phase data are the averages from two separate runs. (D) Correlation between spot intensities for corresponding peptides from FES microarray and solution phase assays. Data were normalized as in panel C.
We initially tested the microarray format using the NRTK FES, which we have previously profiled using the solution phase PSPL method.28 The peptide microarrays were incubated with FES and radiolabeled ATP, washed extensively to remove any unbound radiolabel, and exposed to a phosphor screen to quantify the peptide phosphorylation. We found that the results from the microarray format were generally similar to those obtained from the solution phase method (Figure 1C). The overall correlation between the normalized spot intensities for all peptide signals across the two methods (R2 = 0.77, Figure 1D) is comparable to the correlation between replicates for the solution phase PSPL that we previously reported (R2 = 0.81).16 However, we found that the microarray format had a lower signal-to-noise ratio, which resulted in overall smaller normalized selectivity values. In addition, the signals on the microarrays for peptides with fixed Met residues were lower than expected, presumably due to oxidation through the exposure of the slides to air during storage. The peptides that contained Cys produced uniformly high signals independent of position (Figure 1B), and these peptides were removed from subsequent microarrays. As previously noted for the PSPL method,21 the peptides that contained Tyr at fixed positions (Figure 1B) provided high signals as well, a likely artifact of having two potential sites of phosphorylation. Overall, however, there was good agreement between the microarray and solution phase methods, in particular with respect to the residues with strong positive selection. These results suggested that microarrays offer a rapid alternative to solution phase PSPLs for peptide substrate optimization.
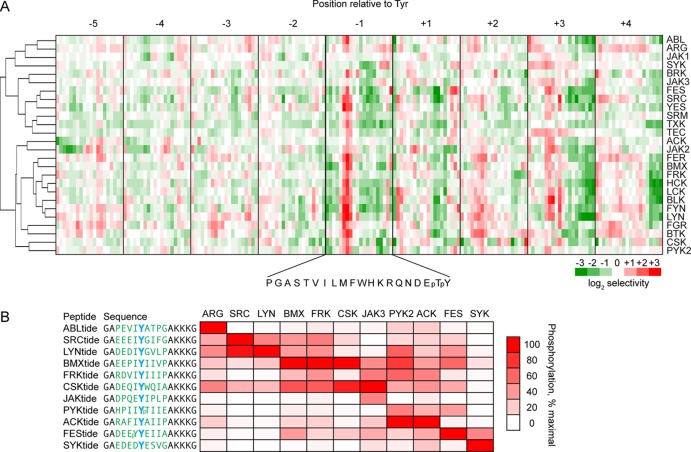
Using Gateway recombination, we generated a set of mammalian expression vectors to produce 29 of the 32 human NRTKs in full-length form as N-terminal GST fusion proteins. Kinases were produced by transient overexpression in HEK293T cells and were affinity purified from cell lysates. We found that 18 of the 29 NRTKs produced in this manner were sufficiently active to provide a robust signal using the peptide microarrays. In three cases (BMX, PTK6, and TEC), it was necessary to treat the cells with the protein tyrosine phosphatase inhibitor pervanadate prior to lysis in order to produce active kinases. Because we were concerned that pervanadate treatment could increase the risk of contamination with endogenous kinase activities that could influence our results, we generated inactive mutant forms of those three kinases. No signal was detectable on the microarrays for any of the inactive kinase mutants (not shown), which suggests that the signals observed with the wild-type (WT) kinases were attributable to the intended kinase. For seven additional NRTKs, we performed microarray analysis using active kinase that was purified from insect cells, three of which came from a commercial source. The expression systems used for each kinase are listed in Table 1 of the Supporting Information. Typically low nanogram quantities of kinase were used for each microarray run, compared to the microgram quantities that are required for solution phase PSPL. Overall, we generated data for 26 NRTKs. Normalized, quantified spot intensities for each kinase are depicted as a heat map in Figure 2, panel A, and the most strongly selected residues are listed in Table 1 (normalized data are provided in table form as Supplementary Data Set 1, Supporting Information).
Figure 2.
Microarray analysis of human NRTKs. (A) Clustered heat map showing peptide microarray data for 26 NRTKs. Shown are the averages of two separate assays performed on duplicate peptide sets for each kinase. Data were normalized as in Figure 1. Cluster analysis by complete linkage algorithm was done using Cluster 3.0. The heat map and dendrogram were prepared using Java TreeView 1.1.6r4.48 (B) Radiolabel kinase assay that used consensus peptide substrates. Solution phase kinase assays were performed with the indicated NRTKs on consensus peptide substrates with the indicated sequences generated based on the peptide microarray data. The phosphoacceptor Tyr is shown in blue, the variable sequence in green, and the fixed sequence common to all peptides in black. All peptides had C-terminal biotinyllysine residues (not shown). Peptide phosphorylation was assessed by the phosphor imager analysis of radiolabel incorporation from [γ-33P]ATP. Heat map (generated in Microsoft Excel) shows the percentage level of phosphorylation relative to the peptide that was phosphorylated to the greatest extent by a given kinase. Each assay was performed in duplicate, and the data show the average of three separate runs, except for the kinase FRK (average of two separate runs).
Table 1. Comparison of Phosphorylation Motifs Identified by Microarray Analysis with Prior Studiesa.
| preferred residues by position |
||||||||
|---|---|---|---|---|---|---|---|---|
| kinase | method | –3 | –2 | –1 | 0 | +1 | +2 | +3 |
| ABL | microarray | I | Y | P | ||||
| OPL38 | I | Y | A | P | ||||
| ACK | microarray | A | Y | A | I/L | |||
| ARG | microarray | E | I | Y | A | T | P | |
| BLK | microarray | I/L/V | Y | A/G/D | F/I/L | |||
| phage35 | I | Y | D/E | L | ||||
| BMX | microarray | I/L/V/pY | Y | I/W/pT | I | |||
| PSPL37 | N/D/E | I/L/pY | Y | W | W | |||
| BRK | microarray | I | I/F | Y | W | |||
| BTK | microarray | L | I/L/V | Y | W | I | P | |
| CSK | microarray | S | I/V | Y | pY | H/I/Q/pY | F/M | |
| OPL47 | I | Y | F | |||||
| FER | microarray | I/L/M | Y | E | I/L | I/V | ||
| FES | microarray | E | pY | I/V/pY | Y | E/pT/pY | I/V | |
| OPL38 | E | E | I | Y | E | E | I | |
| FGR | microarray | I/V | Y | G | F | |||
| OPL47 | I/V | Y | ||||||
| FRK | microarray | I/L/V | Y | pY | I | I | ||
| FYN | microarray | E | D | I/L/V | Y | G/W/pY | I | F/I/L |
| HCK | microarray | I | I/V | Y | G | F/P | ||
| JAK1 | microarray | S/G | Y | F/M | V | |||
| JAK2 | microarray | K/P | P | I | Y | F/I/V | I/L/P | |
| JAK3 | microarray | P | D/E | Y | F/I | L | ||
| LCK | microarray | I/V | Y | G/A | F/I/L | |||
| OPL38 | I/L/V | Y | G | F/I/L/V | ||||
| LYN | microarray | E | I/L/V | Y | G/E | I | F/I | |
| OPL47 | E | I | Y | E | ||||
| phage35 | D/E | I/L | Y | E/D | L | |||
| PYK2 | microarray | I/L/V | Y | A/pT | I/pY | I/V | ||
| SRC | microarray | E | I/L/V | Y | G/E | I | F/I | |
| OPL38 | E | E | I/V | Y | G/E | F | ||
| SRM | microarray | I | Y | V | ||||
| SYK | microarray | pT | D/E/pT | Y | E | V | ||
| phage35 | D/E | S | D | Y | E | |||
| TEC | microarray | I/L | Y | E/A | ||||
| TXK | microarray | P | I/L/V | Y | V/pT | |||
| YES | microarray | E | I/L/V | Y | E/G | I | F/L | |
For the microarray data, residues at the indicated position, with normalized selectivity values >1.7, calculated as described in the Experimental section, are shown. JAK2 and SYK had no residues that scored at that threshold, so a lower stringency cutoff (1.5) was used. For results from the literature, residues highlighted in the relevant citation are shown. OPL, oriented peptide library.
For kinases that were previously characterized by other peptide screening methods, the microarray analysis showed generally good agreement with the published consensus sequences (Table 1). Almost all of the NRTKs examined were strongly selective for aliphatic residues, in particular Ile, at the −1 position relative to the phosphoacceptor Tyr. Exceptions to this rule included SYK, which as previously reported, strongly prefers acidic residues at the −1 position,34−36 and JAK kinases, which appeared to select both aliphatic and acidic residues. Also, in agreement with previous reports using PSPL analysis, both FES and BMX selected pTyr at the −1 position, which is consistent with potential substrate priming by other tyrosine kinases.28,37 In addition, the various kinases favored or disfavored distinct sets of residues at the −3, +1, and +3 positions. A clustering of the kinases based on their specificity profiles (Figure 2A) largely recapitulated a human NRTK dendrogram based on sequence similarity;1 however, there were some notable exceptions where closely related kinases differed in their phosphorylation site preferences. For example, JAK2 had a phosphorylation motif that differed from those of JAK1 and JAK3 (Figure 2A and Table 1).
The distinct phosphorylation site preferences of the various kinases suggests the feasibility of the generation of selective peptide substrates for each group of NRTK. To test our capacity to generate selective substrates from the microarray data, we generated an initial set of 11 consensus peptide substrates that incorporated residues that were most favorably selected by their corresponding kinases at the −4 through +4 positions, flanked by a linker sequence that was identical for each peptide. We then performed solution phase radiolabel kinase assays in which we determined the relative phosphorylation rates for the full set of 11 peptides by 11 representative kinases (Figure 2B). A comparison of the rates across all peptides tested for a given kinase showed that in most cases (7 out of 11) the consensus peptide was the best substrate of its intended kinase. In the other four cases, we noted that the best substrate for the kinase was phosphorylated no more than 2.5-fold faster than was the peptide designed for that kinase. For example, FRK phosphorylated FRKtide at 42% of the rate of its best substrate, BMXtide. While in four cases (ARG, JAK3, SYK, and FES), the designed peptide appeared to be highly specific for its cognate kinase, in most cases, a given peptide was phosphorylated efficiently by multiple kinases. For example, BMXtide, though designed as a BMX substrate, was efficiently phosphorylated by 6 of the 11 kinases. These results confirm the utility of peptide library screening in the rapid optimization of peptide substrates.
To further characterize the consensus peptide substrates, we determined the steady state kinetic constants (kcat and Km) for several kinase–peptide combinations by measuring the initial rates of phosphorylation over a range of substrate concentrations (Table 2). We found the Km values to be in a range (1.3–18 μM) similar to or lower than those of the previously reported consensus peptide substrates for tyrosine kinases.34,35,38−41 Notably, we confirmed that peptide substrates could discriminate at least to some extent between the highly related kinases SRC and LYN, which is consistent with a recent report of differential protein substrate targeting by different SRC-family kinases.42
Table 2. Kinetic Analysis of Consensus Peptide Substrate Phosphorylation by NRTKs.
| kinase | peptide | kcat, s–1 | Km, μM | kcat/Km, μM–1 s–1 |
|---|---|---|---|---|
| ABL | ABLtide | 0.33 | 10 | 0.032 |
| SRC | SRCtide | 2.9 | 14 | 0.21 |
| LYNtide | 2.0 | 18 | 0.11 | |
| LYN | SRCtide | 5.4 | 27 | 0.20 |
| LYNtide | 3.0 | 7.1 | 0.42 | |
| FES | FEStide | 0.76 | 18 | 0.042 |
| SYK | SYKtide | 0.45 | 1.3 | 0.35 |
Conclusions
Here, we have used peptide microarrays to characterize the phosphorylation site specificity of almost all human NRTKs. A key advantage of microarray methods over “macroarray” peptide libraries is that only small amounts (ng quantities) of kinase are required. The requirement for such quantities facilitates the use of mammalian expression systems, which are straightforward to establish and have a high success rate for producing active kinase, yet have low yields compared to those of other expression systems. We found that small scale transfections in HEK293T cells (one 6 cm diameter plate of cells) typically produced μg quantities of kinase, a large excess over what was needed to perform microarray analysis. In our study, 62% of the NRTKs (18 out of 29) produced in HEK293T cells as GST fusion proteins were sufficiently active for microarray experiments.
While peptide microarray analysis produced qualitatively similar results to the solution phase PSPL analysis, we did observe a lower signal-to-noise ratio with the microarray format. Multiple factors may contribute to the decreased relative signal observed on microarrays. For example, steric hindrance from the microarray surface may reduce the phosphorylation efficiencies of immobilized peptides. In addition, the smaller spot size on the microarray compared to that on a macroarray may produce intrinsically higher noise because background fluctuations are averaged over a much smaller area. Alternative detection strategies that use pTyr specific antibodies or phosphate-binding fluorescent dyes have also been used with peptide microarrays and may provide enhanced sensitivity.17,43 However, antibodies in particular warrant caution because their binding affinities may be influenced by the sequence surrounding the pTyr residue, which could cause erroneous results.
We produced a set of 11 consensus peptide substrates to assay NRTKs. Most (7) of these consensus peptides were indeed optimally phosphorylated by their intended kinases. A number of factors could have contributed to our failure to generate optimal substrates for some of the kinases. Peptide immobilization on glass slides, for example, could produce different results compared to those of a solution phase kinase assay for some kinases. In addition, residues favored at particular positions by some NRTKs may be dependent on the surrounding sequence context, which would not be uncovered by using PSPLs alone. Despite these limitations, we did generate efficient substrates for each NRTK, which should serve as a useful tool for future inhibitor screening. While the consensus peptides were designed for maximal efficiency as opposed to selectivity, several peptides did appear to be highly specific substrates for their corresponding NRTKs. Theoretically, our data could be used to generate more specific substrates by intentionally incorporating residues that are deselected by noncognate kinases. Enhanced specificity may also be achieved by the incorporation of docking motifs that interact outside of the kinase active site.6,44−46 Such highly specific substrate sequences have potential applications in the development of biosensors to monitor kinase activity in complex systems.
Acknowledgments
We thank Valerie Reinke (Yale) for the use of the microarray printer. We thank Laurie Parker (Purdue University) for the valuable discussions. We thank Reuben Shaw (Salk Institute), Nora Heisterkamp (Childrens Hospital, Los Angeles), and Stefan Knapp (Oxford) for sharing the clones and reagents. We thank Daisy Sio Seng Lio (University of Melbourne) for the purification and characterization of recombinant Csk expressed in baculovirus-infected Sf9 cells. We also thank Dr. David Morgan (University of California, San Francisco) for the gift of the recombinant Csk baculovirus. This work was supported by National Institutes of Health Grants No. R21 RR022859, R21 CA147993, and R01 GM104047 to B.E.T.; Grant No. R01 AI075133 to T.J.B.; and Grant No. R01 GM102262 to T.J.B. and B.E.T. We also acknowledge the support from Grant No. 1050486 from the National Heath Medical Research Council of Australia to H.-C.C. Y.D. was supported by a James Hudson Brown–Alexander Brown Coxe postdoctoral fellowship from Yale University School of Medicine.
Supporting Information Available
Supplementary table shows all NRTKs that were analyzed and the systems used for their expression. Supplementary data set includes quantified peptide microarray data for all NRTKs. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Manning G.; Whyte D. B.; Martinez R.; Hunter T.; Sudarsanam S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [DOI] [PubMed] [Google Scholar]
- Fedorov O.; Muller S.; Knapp S. The (un)targeted cancer kinome. Nat. Chem. Biol. 2010, 6, 166–169. [DOI] [PubMed] [Google Scholar]
- Zaman G. J.; Garritsen A.; de Boer T.; van Boeckel C. A. Fluorescence assays for high-throughput screening of protein kinases. Comb. Chem. High Throughput Screening 2003, 6, 313–320. [DOI] [PubMed] [Google Scholar]
- Park J.; Hu Y.; Murthy T. V.; Vannberg F.; Shen B.; Rolfs A.; Hutti J. E.; Cantley L. C.; Labaer J.; Harlow E.; Brizuela L. Building a human kinase gene repository: Bioinformatics, molecular cloning, and functional validation. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 8114–8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shults M. D.; Janes K. A.; Lauffenburger D. A.; Imperiali B. A multiplexed homogeneous fluorescence-based assay for protein kinase activity in cell lysates. Nat. Methods 2005, 2, 277–283. [DOI] [PubMed] [Google Scholar]
- Placzek E. A.; Plebanek M. P.; Lipchik A. M.; Kidd S. R.; Parker L. L. A peptide biosensor for detecting intracellular Abl kinase activity using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Biochem. 2010, 397, 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X.; Herbst-Robinson K. J.; Zhang J. Visualizing dynamic activities of signaling enzymes using genetically encodable FRET-based biosensors from designs to applications. Methods Enzymol. 2012, 504, 317–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock A. N.; Debreczeni J.; Amos A. L.; Knapp S.; Turk B. E. Structure and substrate specificity of the Pim-1 kinase. J. Biol. Chem. 2005, 280, 41675–41682. [DOI] [PubMed] [Google Scholar]
- Chen C.; Ha B. H.; Thevenin A. F.; Lou H. J.; Zhang R.; Yip K. Y.; Peterson J. R.; Gerstein M.; Kim P. M.; Filippakopoulos P.; Knapp S.; Boggon T. J.; Turk B. E. Identification of a major determinant for serine–threonine kinase phosphoacceptor specificity. Mol. Cell 2014, 53, 140–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y.; Couch B. A.; Koleske A. J.; Turk B. E. A peptide photoaffinity probe specific for the active conformation of the Abl tyrosine kinase. ChemBioChem 2012, 13, 2510–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter A. N. Assays of protein kinases using exogenous substrates. Curr. Protoc. Mol. Biol. 2001, Chapter 18, Unit 18.17. [DOI] [PubMed] [Google Scholar]
- Songyang Z.; Blechner S.; Hoagland N.; Hoekstra M. F.; Piwnica-Worms H.; Cantley L. C. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr. Biol. 1994, 4, 973–982. [DOI] [PubMed] [Google Scholar]
- Tegge W. J.; Frank R. Analysis of protein kinase substrate specificity by the use of peptide libraries on cellulose paper (SPOT-method). Methods Mol. Biol. 1998, 87, 99–106. [DOI] [PubMed] [Google Scholar]
- Rodriguez M.; Li S. S.; Harper J. W.; Songyang Z. An oriented peptide array library (OPAL) strategy to study protein–protein interactions. J. Biol. Chem. 2004, 279, 8802–8807. [DOI] [PubMed] [Google Scholar]
- Hutti J. E.; Jarrell E. T.; Chang J. D.; Abbott D. W.; Storz P.; Toker A.; Cantley L. C.; Turk B. E. A rapid method for determining protein kinase phosphorylation specificity. Nat. Methods 2004, 1, 27–29. [DOI] [PubMed] [Google Scholar]
- Mok J.; Kim P. M.; Lam H. Y.; Piccirillo S.; Zhou X.; Jeschke G. R.; Sheridan D. L.; Parker S. A.; Desai V.; Jwa M.; Cameroni E.; Niu H.; Good M.; Remenyi A.; Ma J. L.; Sheu Y. J.; Sassi H. E.; Sopko R.; Chan C. S.; De Virgilio C.; Hollingsworth N. M.; Lim W. A.; Stern D. F.; Stillman B.; Andrews B. J.; Gerstein M. B.; Snyder M.; Turk B. E. Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci. Signaling 2010, 3, ra12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uttamchandani M.; Chan E. W.; Chen G. Y.; Yao S. Q. Combinatorial peptide microarrays for the rapid determination of kinase specificity. Bioorg. Med. Chem. Lett. 2003, 13, 2997–3000. [DOI] [PubMed] [Google Scholar]
- Schutkowski M.; Reimer U.; Panse S.; Dong L.; Lizcano J. M.; Alessi D. R.; Schneider-Mergener J. High content peptide microarrays for deciphering kinase specificity and biology. Angew. Chem., Int. Ed. 2004, 43, 2761–2674. [DOI] [PubMed] [Google Scholar]
- Rychlewski L.; Kschischo M.; Dong L.; Schutkowski M.; Reimer U. Target specificity analysis of the Abl kinase using peptide microarray data. J. Mol. Biol. 2004, 336, 307–311. [DOI] [PubMed] [Google Scholar]
- Diks S. H.; Kok K.; O’Toole T.; Hommes D. W.; van Dijken P.; Joore J.; Peppelenbosch M. P. Kinome profiling for studying lipopolysaccharide signal transduction in human peripheral blood mononuclear cells. J. Biol. Chem. 2004, 279, 49206–49213. [DOI] [PubMed] [Google Scholar]
- Ia K. K.; Jeschke G. R.; Deng Y.; Kamaruddin M. A.; Williamson N. A.; Scanlon D. B.; Culvenor J. G.; Hossain M. I.; Purcell A. W.; Liu S.; Zhu H. J.; Catimel B.; Turk B. E.; Cheng H. C. Defining the substrate specificity determinants recognized by the active site of C-terminal Src kinase-homologous kinase (CHK) and identification of beta-synuclein as a potential CHK physiological substrate. Biochemistry 2011, 50, 6667–6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marttila A. T.; Hytonen V. P.; Laitinen O. H.; Bayer E. A.; Wilchek M.; Kulomaa M. S. Mutation of the important Tyr-33 residue of chicken avidin: Functional and structural consequences. Biochem. J. 2003, 369, 249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hytonen V. P.; Laitinen O. H.; Airenne T. T.; Kidron H.; Meltola N. J.; Porkka E. J.; Horha J.; Paldanius T.; Maatta J. A.; Nordlund H. R.; Johnson M. S.; Salminen T. A.; Airenne K. J.; Yla-Herttuala S.; Kulomaa M. S. Efficient production of active chicken avidin using a bacterial signal peptide in Escherichia coli. Biochem. J. 2004, 384, 385–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maatta J. A.; Eisenberg-Domovich Y.; Nordlund H. R.; Hayouka R.; Kulomaa M. S.; Livnah O.; Hytonen V. P. Chimeric avidin shows stability against harsh chemical conditions—biochemical analysis and 3D structure. Biotechnol. Bioeng. 2011, 108, 481–490. [DOI] [PubMed] [Google Scholar]
- Gibbs C. S.; Zoller M. J. Rational scanning mutagenesis of a protein kinase identifies functional regions involved in catalysis and substrate interactions. J. Biol. Chem. 1991, 266, 8923–8931. [PubMed] [Google Scholar]
- Boggon T. J.; Li Y.; Manley P. W.; Eck M. J. Crystal structure of the Jak3 kinase domain in complex with a staurosporine analog. Blood 2005, 106, 996–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K. C.; Lio D. S.; Dobson R. C.; Jarasrassamee B.; Hossain M. I.; Roslee A. K.; Ia K. K.; Perugini M. A.; Cheng H. C. Development of the procedures for high-yield expression and rapid purification of active recombinant Csk-homologous kinase (CHK): Comparison of the catalytic activities of CHK and CSK. Protein Expression Purif. 2010, 74, 139–147. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P.; Kofler M.; Hantschel O.; Gish G. D.; Grebien F.; Salah E.; Neudecker P.; Kay L. E.; Turk B. E.; Superti-Furga G.; Pawson T.; Knapp S. Structural coupling of SH2-kinase domains links Fes and Abl substrate recognition and kinase activation. Cell 2008, 134, 793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanis K. Q.; Veach D.; Duewel H. S.; Bornmann W. G.; Koleske A. J. Two distinct phosphorylation pathways have additive effects on Abl family kinase activation. Mol. Cell. Biol. 2003, 23, 3884–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellings D. A.; Atherton E. Standard Fmoc protocols. Methods Enzymol. 1997, 289, 44–67. [DOI] [PubMed] [Google Scholar]
- Huang X.; Finerty P. Jr.; Walker J. R.; Butler-Cole C.; Vedadi M.; Schapira M.; Parker S. A.; Turk B. E.; Thompson D. A.; Dhe-Paganon S. Structural insights into the inhibited states of the Mer receptor tyrosine kinase. J. Struct. Biol. 2009, 165, 88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H.; Chen G. Y.; Yao S. Q. Recent advances in microarray technologies for proteomics. Chem. Biol. 2013, 20, 685–699. [DOI] [PubMed] [Google Scholar]
- MacBeath G.; Schreiber S. L. Printing proteins as microarrays for high-throughput function determination. Science 2000, 289, 1760–1763. [DOI] [PubMed] [Google Scholar]
- Lipchik A. M.; Killins R. L.; Geahlen R. L.; Parker L. L. A peptide-based biosensor assay to detect intracellular Syk kinase activation and inhibition. Biochemistry 2012, 51, 7515–7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R.; Baumann G.; Gram H. Catalytic specificity of phosphotyrosine kinases Blk, Lyn, c-Src, and Syk as assessed by phage display. J. Mol. Biol. 1996, 260, 664–677. [DOI] [PubMed] [Google Scholar]
- Brunati A. M.; Donella-Deana A.; Ruzzene M.; Marin O.; Pinna L. A. Site specificity of p72syk protein tyrosine kinase: Efficient phosphorylation of motifs recognized by Src homology 2 domains of the Src family. FEBS Lett. 1995, 367, 149–152. [DOI] [PubMed] [Google Scholar]
- Chen S.; Jiang X.; Gewinner C. A.; Asara J. M.; Simon N. I.; Cai C.; Cantley L. C.; Balk S. P. Tyrosine kinase BMX phosphorylates phosphotyrosine-primed motif mediating the activation of multiple receptor tyrosine kinases. Sci. Signaling 2013, 6, ra40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songyang Z.; Carraway K. L. 3rd; Eck M. J.; Harrison S. C.; Feldman R. A.; Mohammadi M.; Schlessinger J.; Hubbard S. R.; Smith D. P.; Eng C.; et al. Catalytic specificity of protein–tyrosine kinases is critical for selective signalling. Nature 1995, 373, 536–539. [DOI] [PubMed] [Google Scholar]
- Li M.; Luraghi P.; Amour A.; Qian X. D.; Carter P. S.; Clark C. J.; Deakin A.; Denyer J.; Hobbs C. I.; Surby M.; Patel V. K.; Schaefer E. M. Kinetic assay for characterization of spleen tyrosine kinase activity and inhibition with recombinant kinase and crude cell lysates. Anal. Biochem. 2009, 384, 56–67. [DOI] [PubMed] [Google Scholar]
- Nair S. A.; Kim M. H.; Warren S. D.; Choi S.; Songyang Z.; Cantley L. C.; Hangauer D. G. Identification of efficient pentapeptide substrates for the tyrosine kinase pp60c-Src. J. Med. Chem. 1995, 38, 4276–4283. [DOI] [PubMed] [Google Scholar]
- Wang C.; Lee T. R.; Lawrence D. S.; Adams J. A. Rate-determining steps for tyrosine phosphorylation by the kinase domain of v-fps. Biochemistry 1996, 35, 1533–1539. [DOI] [PubMed] [Google Scholar]
- Takeda H.; Kawamura Y.; Miura A.; Mori M.; Wakamatsu A.; Yamamoto J.; Isogai T.; Matsumoto M.; Nakayama K. I.; Natsume T.; Nomura N.; Goshima N. Comparative analysis of human SRC-family kinase substrate specificity in vitro. J. Proteome Res. 2010, 9, 5982–5993. [DOI] [PubMed] [Google Scholar]
- Martin K.; Steinberg T. H.; Cooley L. A.; Gee K. R.; Beechem J. M.; Patton W. F. Quantitative analysis of protein phosphorylation status and protein kinase activity on microarrays using a novel fluorescent phosphorylation sensor dye. Proteomics 2003, 3, 1244–1255. [DOI] [PubMed] [Google Scholar]
- Harvey C. D.; Ehrhardt A. G.; Cellurale C.; Zhong H.; Yasuda R.; Davis R. J.; Svoboda K. A genetically encoded fluorescent sensor of ERK activity. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 19264–19269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicena P.; Miller W. T. Processive phosphorylation of p130Cas by Src depends on SH3–polyproline interactions. J. Biol. Chem. 2001, 276, 28190–28196. [DOI] [PubMed] [Google Scholar]
- Fernandes N.; Bailey D. E.; Vanvranken D. L.; Allbritton N. L. Use of docking peptides to design modular substrates with high efficiency for mitogen-activated protein kinase extracellular signal-regulated kinase. ACS Chem. Biol. 2007, 2, 665–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzzene M.; Songyang Z.; Marin O.; Donella-Deana A.; Brunati A. M.; Guerra B.; Agostinis P.; Cantley L. C.; Pinna L. A. Sequence specificity of C-terminal Src kinase (CSK)—a comparison with Src-related kinases c-Fgr and Lyn. Eur. J. Biochem. 1997, 246, 433–439. [DOI] [PubMed] [Google Scholar]
- Saldanha A. J. Java Treeview—extensible visualization of microarray data. Bioinformatics 2004, 20, 3246–3248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.