Abstract
The emergence of Klebsiella pneumoniae producing carbapenemase (KPC) has now become a global concern. As a part of a nationwide multicentre surveillance study in Cuba, three K. pneumoniae clinical isolates resistant to carbapenems were detected for a 1-month period (September to October 2011). PCR and sequence analysis revealed that the three strains harboured blaKPC-2. They showed resistance or intermediate susceptibility to expanded-spectrum cephalosporins, other β-lactams, a β-lactam/β-lactamase inhibitor combination, and gentamicin. Two strains were susceptible only to colistin, whereas the other strain showing colistin resistance was susceptible to fluoroquinolones. These blaKPC-2-positive K. pneumoniae strains were classified into ST1271 (CC29), a novel clone harbouring blaKPC-2, and were revealed to be genetically identical by PCR-based DNA fingerprinting. The three patients infected with the KPC-producing K. pneumoniae had common risk factors, and had no overseas travel experience outside Cuba, suggesting local acquisition of the resistant pathogen. This is the first report of a KPC-producing K. pneumoniae in Cuba. Although detection of KPC in Enterobacteriaceae is still rare in Cuba, our finding indicated that KPC-producing bacteria are a global concern and highlighted the need to identify these microorganisms in clinical laboratories.
Keywords: Clinical isolates, Cuba, Klebsiella pneumoniae producing carbapenemase, multilocus sequence typing
Introduction
The Klebsiella pneumoniae carbapenemase (KPC) was first described in 1996 in North Carolina, USA. Thereafter, expansion of KPC in clinical isolates has been reported from different continents associated with the global spread of the clonal lineages of K. pneumoniae, such as ST248/ST258 1. The ST258 K. pneumoniae has been described as an international KPC-producing clone 2, and its global spread including Latin American countries has also been reported 3. Because the KPC gene is carried by plasmids, potentially rapid transmission of carbapenem resistance has been recognized as a major threat to the antimicrobial treatment of infections with gram-negative microorganisms 2. The Pan American Health Organization issued an epidemiological alert with the increase of carbapenemase in Enterobacteriaceae from many Latin American countries in 2010 4. Since then, Cuba has initiated a surveillance network for K. pneumoniae clinical isolates from reference hospitals to analyse their antimicrobial susceptibility and genetic mechanisms of drug resistance with special attention to carbapenem resistance, in the ‘Pedro Kourí’ Institute of Tropical Medicine 5. During this national surveillance, three isolates of KPC-producing K. pneumoniae were detected for the first time in Cuba.
Methods
As a part of a nationwide multicentre surveillance study in Cuba, three K. pneumoniae clinical isolates resistant to carbapenems were detected in the ‘Pedro Kourí’ Institute of Tropical Medicine for a 1-month period (September to October 2011). The antimicrobial susceptibility to a wide range of antibiotics was determined using E-test (BioMérieux, Marcy l'Etoile, France) according to the manufacturer's recommendation. MICs were interpreted into susceptible or resistant according to CLSI guidelines, 2012 6, except for colistin, which was judged by EUCAST criteria (susceptible ≤2 g/mL, resistant ≥4 g/mL), (http://www.eucast.org/clinicalbreakpoints/). A double-disc synergy test was performed to detect extended spectrum β-lactamases (ESBLs) 6 and 3-aminophenylboronic acid test was used to screen for production of carbapenemases 7. The presence of genes encoding carbapenemase was determined by PCR using protocols and conditions as described previously 8. Nucleotide sequence of blaKPC was determined by direct sequencing with PCR products by using the BigDye Terminator version 3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA, USA). Sequence type (ST) of K. pneumoniae based on the scheme of multilocus sequence typing (MLST) was determined according to the methods available at the website (www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html). KPC-2 gene sequence of K. pneumoniae strain 354 was deposited to GenBank database under accession no. KJ151293. To analyse genetic diversity of K. pneumoniae isolates, genomic DNA fingerprinting was performed by randomly amplified polymorphic DNA analysis with the use of a single primer (M13, ERIC-2, AP-1, AP-4 and AP-7), and repetitive extragenic palindromic sequence-based PCR (REP-PCR) with a pair of primers (REP1R-I, REP2-I), as described previously 9–13.
Results and Discussion
Three clinical isolates of K. pneumoniae (strains 328, 354, 355) recovered from two provinces of Cuba (Holguin and Havana city) were confirmed to be resistant to imipenem and meropenem. These strains showed resistance or intermediate susceptibility to expanded-spectrum cephalosporins, β-lactam, β-lactam/β-lactamase inhibitor combination (piperacillin/tazobactam), and gentamicin. Two of these strains (354, 355) were susceptible only to colistin, whereas the other strain (328) showing resistance to colistin was susceptible to fluoroquinolones (Table1). The synergy test to detect ESBLs and carbapenemase was positive, suggesting the production of an ESBL and KPC enzyme. The KPC gene was detected in the three strains by PCR, and their sequences were revealed to be identical to blaKPC-2. The three blaKPC-2-positive K. pneumoniae strains were classified into ST1271 (CC29), a single locus variant of ST29. The ST1271 was identified as a novel clone harbouring blaKPC-2 in the present study, because the presence of blaKPC-2 had not been reported in ST1271 as well as ST29 clones. By randomly amplified polymorphic DNA analysis with five different primers and REP PCR, three strains showed the same banding patterns (Fig.1), indicating that these strains are genetically identical and have the same origin. The two colistin-susceptible strains were derived from the same hospital in Havana, exhibited similar resistance patterns, and are suggested to be the same strain that had transmitted via nosocomial infection.
Table 1.
Antimicrobial susceptibility of Klebsiella pneumoniae clinical isolates producing KPC-2 carbapenemase
| Antibiotics | MIC Interpretationa | ||
|---|---|---|---|
| Strain 328 | Strain 354 | Strain 355 | |
| Piperacillin/tazobactam | R | R | R |
| Cefotaxime | R | R | R |
| Ceftazidime | R | R | R |
| Cefoxitin | S | R | R |
| Cefepime | R | R | R |
| Aztreonam | R | R | R |
| Meropenem | R | R | R |
| Imipenem | R | R | I |
| Amikacin | R | R | I |
| Gentamicin | R | R | R |
| Colistin | R | S | S |
| Ciprofloxacin | S | R | R |
| Levofloxacin | S | R | R |
| Nalidixic acid | S | R | R |
| Trimethoprim-sulfamethoxazole | R | R | R |
R, Resistant; S, susceptible; I, Intermediate.
According to CLSI standards (2012).
Figure 1.
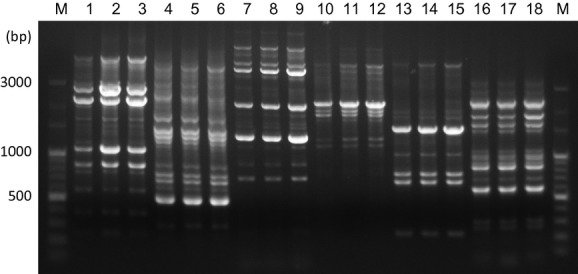
DNA fingerprints obtained by randomly amplified polymorphic DNA analysis with primer M13 (lanes 1–3), ERIC2 (lanes 4–6), AP-1 (lanes 7–9), AP-4 (lanes 10–12), and AP-7 (lanes 13–15), and repetitive extragenic palindromic sequence-based PCR (lanes 16–18). Lanes 1, 4, 7, 10, 13, 16, strain 328; lanes 2, 5, 8, 11, 14, 17, strain 354; lanes 3, 6, 9, 12, 15, 18, strain 355. Lane ‘M’ represents DNA size marker and molecular size (bp) is shown on the left.
The three strains with KPC genes were derived from two separate provinces, i.e. Holguin (eastern Cuba) and Havana city (western Cuba). Patients infected with the KPC-producing K. pneumoniae had no experience of travel outside Cuba, suggesting that local acquisition and a silent dissemination of the KPC-positive K. pneumoniae ST1271 clone in this country. Because intercontinental travel has been directly linked with the spread of KPCs through patients colonized or infected with KPC-producing K. pneumoniae 14,15, international tourism might have played an important role in its emergence in Cuba. ST29 K. pneumoniae strains have been isolated from Europe, e.g. Spain and Greece, and also from Brazil 3. In our previous report on the antimicrobial resistance of K. pneumoniae in Cuba through the national surveillance programme (2009–10), 54 isolates (23.6%) were positive for ESBL, and were classified into 27 STs, showing high clonal diversity 5. However, none of the ST identified in the ESBL-positive isolates belonged to CC29. Therefore, in Cuba, the ST1271 K. pneumoniae strains are genetically distinct from ESBL-positive strains. Although the origin of the ST1271 strains has yet to be determined, it is possible that ST1271 might have been brought from another country recently, associated with acquisition of the KPC gene. Further molecular epidemiological studies on K. pneumoniae isolates, including those from healthy individuals, may provide more information on distribution and transmission of the ST1271 clone.
It was noted in the present study that an ST1271 K. pneumoniae strain showed resistance to colistin. Generally, the colistin resistance rate of K. pneumoniae has been reported to be low 16. During the period of antimicrobial resistance surveillance (2010–12) of Klebsiella spp. in Cuba, we notified 15% colistin resistance, which would be of great concern in clinical settings 17. Our present finding indicates that the colistin resistance has emerged in multidrug-resistant K. pneumoniae. This antibiotic is recognized as a key therapeutic option for carbapenem-resistant bacteria, and is particularly important in countries with limited resources, such as Cuba where tigecycline is not available.
Table2 shows the clinical information of three patients infected with K. pneumoniae producing KPC-2 carbapenemase. These patients had common risk factors, such as prolonged hospitalization, intravenous catheter, previous antimicrobial therapy, and underlying disease. These findings are consistent with previous studies that notified risk factors for acquiring infections with KPC-producing K. pneumoniae 18. In the present study, all three K. pneumoniae producing KPC were isolated from blood specimens and two of the patients died with septic shock. The susceptibility profile of these isolates indicated considerably limited therapeutic options, i.e. colistin in two patients or fluoroquinolone in one patient. The optimal treatment for infections caused by KPC-producing K. pneumoniae has been difficult to describe, resulting in mortality rates of at least 50% 19. Therefore, evaluation of effective antibiotic options and rigorous infection control measures may be necessary to reduce carbapenemase-producing microorganisms.
Table 2.
Clinical information of patients infected with Klebsiella pneumoniae producing KPC-2 carbapenemase
| Cases | Age | Sex | Province | Hospital | Department | Isolation date | Risk factors | Disease | Specimen | Final therapy | Died |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Case-1 (strain 328) | 42 | Male | Holguín (Eastern Cuba) | 1 | Intensive care unit | September 2011 | Previous antimicrobial therapy, urinary catheter, prolonged hospitalization, intravenous catheter disease (Haemorrhage itraparenkimatosa) |
Septicaemia | Blood | Ciprofloxacin | No |
| Case-2 (strain 354) | 49 | Female | Havana city (Western Cuba) | 2 | Marrow transplant unit | October 2011 | Previous antimicrobial therapy, chemotherapy gynaecological surgery prolonged hospitalization, intravenous catheter Underlying disease (Acute myeloid leukaemia) |
Cellulitis in surgery site Septic shock |
Blood | Cefepime, amikacin, vancomycin and amphotericin B | Yes |
| Case-3 (strain 355) | 57 | Female | Havana city (Western Cuba) | 2 | Intensive care unit | October 2011 | Previous antimicrobial therapy, surgery for valve replacement, mechanical ventilation urinary catheter, prolonged hospitalization, intravenous catheter, Underlying disease (Mitral stenosis) |
Pneumonia Paralytic ileus Cardiogenic shock Septic shock |
Blood | Colistin plus amphotericin B | Yes |
KPC, Klebsiella pneumoniae producing carbapenemase.
Detection of the novel KPC-producing K. pneumoniae clone (ST1271) in the present study demonstrated the importance of monitoring hospitalized patients for the further emergence of carbapenem resistance in K. pneumoniae as well as in other gram-negative pathogens. Although carbapenemase-producing Enterobacteriaceae are still rare in Cuba, our finding confirmed that KPC-producing isolates are a global concern, highlighting the need to identify these microorganisms in clinical laboratories.
Acknowledgments
We thank the microbiologists of the Cuban national network of Microbiology who contribute to the surveillance of Klebsiella spp. Fernando Pasteran provided excellent technical assistance in the interpretation of the 3-aminophenylboronic acid test to screen the production of carbapenemases. This study was supported in part by a Grant-in-Aid for Scientific Research (KAKENHI) (grant no. 26460804), Japan.
Conflict of Interest
None declared.
References
- Hirsch E, Tam V. Detection and treatment options for Klebsiella pneumoniae carbapenemases (KPCs): an emerging cause of multidrug-resistant infection. J Antimicrob Chemother. 2010;65:1119–1125. doi: 10.1093/jac/dkq108. [DOI] [PubMed] [Google Scholar]
- Mataseje LF, Bryce E, Roscoe D, et al. Canadian Nosocomial Infection Surveillance Program. Carbapenem-resistant Gram-negative bacilli in Canada 2009–10: results from the Canadian Nosocomial Infection Surveillance Program (CNISP) J Antimicrob Chemother. 2012;67:1359–1367. doi: 10.1093/jac/dks046. [DOI] [PubMed] [Google Scholar]
- Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis. 2009;9:228–236. doi: 10.1016/S1473-3099(09)70054-4. [DOI] [PubMed] [Google Scholar]
- OPS/PAHO. Alerta epidemiológica: Diseminación de carbapenemasas en Klebsiella pneumoniae en Latinoamérica. Available at http://www2.paho.org/hq/dmdocuments/2010/alertas_epi_2010_02_julio_carbapenemasas.pdf (accessed 2 July 2010)
- Quiñones D, Valverde A, Rodríguez-Baños M, et al. High clonal diversity in a non-outbreak situation of clinical ESBL-producing Klebsiella pneumoniae isolates in the first national surveillance program in Cuba. Microb Drug Resist. 2014;20:45–51. doi: 10.1089/mdr.2013.0021. [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute. Enterobacteraceae M02-M07- Minimal Inhibitory Concentration. Wayne, PA: 2012. Performance standards for antimicrobial susceptibility testing; 17th informational supplement. CLSI document M100-S20. Table1. [Google Scholar]
- Pasteran F, Mendez T, Guerriero L, Rapoport M, Corso A. Sensitive screening tests for suspected class A carbapenemase production in species of Enterobacteriaceae. J Clin Microbiol. 2009;47:1631–1639. doi: 10.1128/JCM.00130-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford PA, Bratu S, Urban C, et al. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 β-lactamases in New York City. Clin Infect Dis. 2004;39:55–60. doi: 10.1086/421495. [DOI] [PubMed] [Google Scholar]
- Gräser Y, Klare I, Halle E, et al. Epidemiological study of an Acinetobacter baumannii outbreak by using polymerase chain reaction fingerprinting. J Clin Microbiol. 1993;31:2417–2420. doi: 10.1128/jcm.31.9.2417-2420.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboli AC, Houston ED, Monteforte JS, Wood CA, Hamill RJ. Discrimination of epidemic and sporadic isolates of Acinetobacter baumannii by repetitive element PCR-mediated DNA fingerprinting. J Clin Microbiol. 1994;32:2635–2640. doi: 10.1128/jcm.32.11.2635-2640.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier N, Saulnier P, Chachaty E, Dumontier S, Andremont A. Random amplified polymorphic DNA typing versus pulsed-field gel electrophoresis for epidemiological typing of vancomycin-resistant enterococci. J Clin Microbiol. 1996;34:1096–1099. doi: 10.1128/jcm.34.5.1096-1099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endtz HP, van den Braak N, van Belkum A, et al. Fecal carriage of vancomycin-resistant enterococci in hospitalized patients and those living in the community in The Netherlands. J Clin Microbiol. 1997;35:3026–3031. doi: 10.1128/jcm.35.12.3026-3031.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartelle M, del Mar Tomas M, Pertega S, et al. Risk factors for colonization and infection in a hospital outbreak caused by a strain of Klebsiella pneumoniae with reduced susceptibility to expanded-spectrum cephalosporins. J Clin Microbiol. 2004;4:4242–4249. doi: 10.1128/JCM.42.9.4242-4249.2004. 2: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Price LS, Quinn JP. The spread of Klebsiella pneumoniae carbapenemases: a tale of strains, plasmids, and transposons. Clin Infect Dis. 2009;49:1739–1741. doi: 10.1086/648078. [DOI] [PubMed] [Google Scholar]
- Lascols C, Peirano G, Hackel M, Laupland K, Pitout J. Surveillance and molecular epidemiology of Klebsiella pneumoniae that produce carbapenemases; the first report of OXA-48-like enzymes in North America. Antimicrob Agents Chemother. 2013;57:130–136. doi: 10.1128/AAC.01686-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoeun Suh J, Seong Son J, Ryeon Chung D, Ran Peck K, Soo Ko K, Hoon Song J. Nonclonal emergence of colistin-resistant Klebsiella pneumoniae isolates from blood samples in South Korea. Antimicrob Agents Chemother. 2010;54:560–562. doi: 10.1128/AAC.00762-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiñones D. Surveillance of Klebsiella and Acinetobacter in Cuba. APUA Newsletter. 2012;30:7–10. [Google Scholar]
- Chang M, Arguelo C, Fernando Aguilar F, Freitas A, Campos A. The first report of infection with Klebsiella pneumoniae carrying the blaKPC gene in State of Mato Grosso does Sul, Brazil. Rev Soc Bras Med Trop. 2013;46:114–115. doi: 10.1590/0037-868215502013. [DOI] [PubMed] [Google Scholar]
- Lee G, Burgess D. Treatment of Klebsiella pneumoniae Carbapenemase (KPC) infections: a review of published case series and case reports. Ann Clin Microbiol Antimicrob. 2012;13:11–32. doi: 10.1186/1476-0711-11-32. [DOI] [PMC free article] [PubMed] [Google Scholar]


