Abstract
Actinobaculum schaalii is an emerging uropathogen. So far, its identification has been performed with 16S rRNA gene sequencing or PCR. The diagnosis has often been delayed due to fastidious growth and identification problems. Eleven clinical isolates of A. schaalii from bloodstream infections that were initially identified with 16S rRNA sequencing analysis were recovered and later identified with matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF). We present a review of bacteriological data of these patients, an algorithm for fast laboratory work-up and advocate the use of sensitized culture of urine to allow better recovery of A. schaalii in susceptible patients.
Keywords: Actinobaculum schaalii infection, bacteriological diagnostics, emerging infections, Gram-positive rod, urinary tract infection
Introduction
Actinobaculum schaalii is not yet a well-known bacterium but has been increasingly reported as an emerging pathogen causing urinary tract infections (UTIs) in patients with underlying uropathologies 1–8 or infections of other organs 9–11. This pathogen has also been reported to cause urinary infections in children with renal or neurological disorders 12,13. Due to its fastidious growth in microaerophilic environments A. schaalii may be overlooked when traditional culture methods are applied. In addition, A. schaalii may be neglected and misinterpreted as a representative of normal microbiota of mucosa or skin 3,6,7, especially when present as a co-isolate with ‘classical’ pathogens of UTIs, such as Escherichia coli. In many reported cases diagnosis was delayed, and the final identification to the species level was achieved after 16S rRNA gene sequencing 1–13. What makes this pathogen important is its peculiar antimicrobial resistance to ciprofloxacin and co-trimoxazole, the drugs often prescribed empirically for the treatment of UTIs.
Urine from urological or immunocompromised patients can be cultured using a sensitized procedure. This procedure includes, in addition to the conventional agar for Gram-negative bacteria, the cultivation on a chocolate agar with a 10 μL loop to increase sensitivity.
Below we present results that advocate the use of a sensitized culture in risk-group patients. We also demonstrate the key metabolic reactions, an algorithm to increase the rate of laboratory work-up, and the suitability of matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) for identification of A. schaalii.
Materials and Methods
Eleven clinical isolates collected from 2008 through 2012 were from five district laboratories. All were isolated from the routine blood culture (bioMerieux, Marcy L’Etoile, France) and initially identified using 16S rRNA gene sequencing (Huslab, Helsinki, Finland) according to the previously published protocol 14. Ethics approval was not requested because the data presented were collected only from laboratory records.
All isolates were stored at −70°C before being retrieved and cultured on anaerobe basal agar with 10% horse blood (ABA; Oxoid Ltd, Basingstoke, UK), chocolate, and sheep blood agar at +35°C in a CO2 atmosphere. Retrospectively, phenotypical identification of all the strains was performed with Rapid ID32A; API Coryne and VITEK 2 (bioMerieux). The susceptibility testing was performed from a suspension of McFarland 6 with E-tests (bioMerieux) or with disc diffusion on ABA, 48 h.
Before MALDI-TOF analysis the isolates were sub-cultured on fastidious anaerobe agar (FAA) with 5% sheep blood and incubated for 48 h. Each isolate was initially inoculated directly from a colony onto a steel target plate on two consecutive spots. One microlitre of matrix solution (α-cyano-4-hydroxycinnamic acid in 50% acetonitrile, 2.5% trifluoroacetic acid; Bruker Daltonics, Bremen, Germany) was added. The identification was performed by MALDI-TOF MS using Microflex LT instrument with Flex Control 3.0 software and Maldi Biotyper DB Update_V3.2.1.1 (Bruker Daltonics) at the Laboratory of Clinical Microbiology, Turku University Hospital. If both scores from the initial run gave a value of <2.0, the bacterial material was extracted as specified by the manufacturer and re-analysed.
To test the sensitized culture midstream urine from a young healthy male was collected and artificial urine samples were prepared from three isolates with serial dilutions of 103–106 CFU/mL. These samples and unspiked urine were cultured on ABA, chocolate agar and ChromID™CPS3 (bioMerieux) with a 10 μL loop. The results were read 24 and 48 h later.
Results and Discussion
The patient-related data and laboratory identification of recovered isolates are presented in Table1. All 11 strains were successfully identified to the species level (correct identification with a score value ≥2.0) but 10/11 strains required an extraction step with 70% formic acid for reliable identification. Only one strain (S-21528) was reliably identified to species level directly from the colony.
Table 1.
Patient-related data, biochemical and MALDI-TOF identification of 11 Actinobaculum schaalii isolates
| Isolate | Reported results from urine within 2 week proximity | Growth from aerobic or anaerobic bottle | Concomitant isolates in blood | Maldi Biotyper A. schaalii confirmed, score | Vitek | APICoryne | Rapid ID 32A |
|---|---|---|---|---|---|---|---|
| S-21495 | Corynebacterium spp. | 1 anaerobic bottle | Peptoniphilus asacchari in 1 anaerobic bottle | 2.219 | 6764100010641 | 2050721 | 0410477717 |
| S-21528 | Oligella urethralis and Aerococcus spp. | 1 anaerobic bottle | Aerococcus species in 1 aerobic bottle | 2.263 | 2601000000401 | 0010721 | 0426477717 |
| M-21846 | 1. No growth Lactobacillus spp. |
1 aerobic and 1 anaerobic bottle each | No | 2.124 |
6727102010661 6764102010621 |
2050721 | 0436477717 |
| J-20772 | Aerobic mixed culture | 1 anaerobic bottle | No | 2.070 | 6765100010441 | 2050721 | 0416477717 |
| P-132 | Klebsiella pneumoniae | 2 aerobic bottles | No | 2.117 | 6625100010401 | 2050721 | 0436077717 |
| P-77 | Klebsiella oxytoca | 1 aerobic bottle | No | 2.115 | 6625000010401 | 0050220 | 0436477717 |
| K-26636 | Aerobic mixed culture | 2 anaerobic bottles | No | 2.202 | 6724100010641 | 0050721 | 0414477717 |
| K-27888 | Klebsiella pneumoniae | 2 aerobic and 1 anaerobic bottles |
Enterococcus faecalison in 1 anaerobic bottle Atopodium parvulum in the same anaerobic bottle as A. schaalii |
2.004 | 2721000000401 | 4050320 | 0400271717 |
| K-27197 | 1. No growth 2. Mixed aerobic culture |
1 aerobic and 1 anaerobic bottle | Peptosteptococcus tetradius in 1 anaerobic bottle | 2.151 | 6725102010661 | 4010621 | 0434477717 |
| T-41163 | 1. Mixed aerobic culture Lactobacillus spp. |
1 anaerobic bottle | No | 2.056 |
6764100010401 1 |
4150621 | 0434477717 |
| T-42442 | Enterococcus faecalis | 1 anaerobic bottle | No | 2.183 | 2721000000001 | 4010000 | 0416073714 variability in reactions |
The simulated urine samples produced visible growth on ABA already after 24 h but on the sheep blood and chocolate agars the cultivation required 48 h. No growth was detectable on the CPS3. The number of colonies was proportional to the strength of the spiked urine. The algorithm for the identification of A. schaalii is presented in Fig.1.
Figure 1.
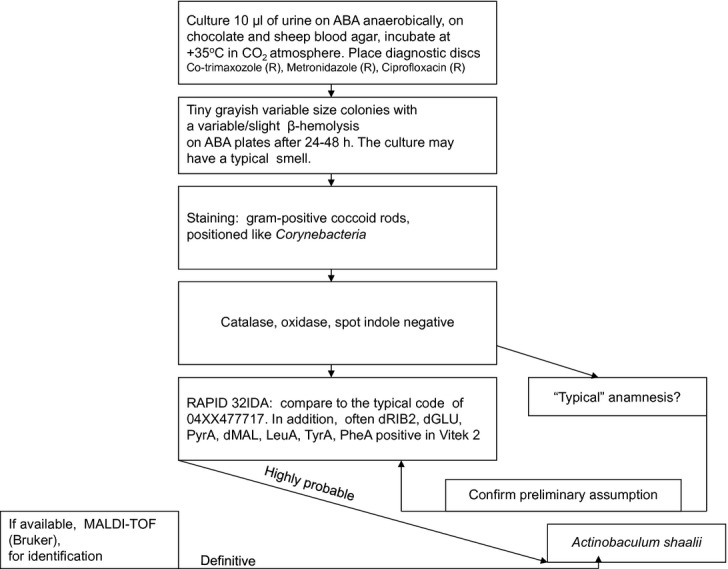
A proposed algorithm for identification of Actinobaculum schaalii from clinical specimens. ABA, anaerobe basal agar.
The retrospective analysis of the patient data indicated that urinalysis was requested from all the patients in whom A. schaalii was isolated from blood; however, aerobic mixed culture and non-significant growth of Corynebacterium species or Lactobacillus was reported in 6/11 patients. In the remainder (5/11) the urinalysis resulted in identification of bacteria associated with UTIs but as expected, A. schaalii was not recovered. In 4/11 patients co-infection with other pathogens was detected in the bloodstream (Table1). RAPID ID 32A provided the most sustained identification profile of 04XXX77717, where the X stands for any reaction (Table1). This profile suggests Capnocytophaga spp. or Micromonas micros, but these species can easily be ruled out. This commercial test strip can also misinterpret A. schaalii for Actinomyces meyeri; however, the latter can be ruled out on the basis that (1) this pathogen is seldom recovered from blood culture, (2) the origin of infection is often related to oral cavity, and (3) the bacteria are not aero-tolerant in comparison with A. schaalii.
All tested isolates were susceptible to penicillin (MIC below 0.008 g/L), cephalexin (disc diffusion, mean 32, range 26–38 mm) and cefuroxime (MIC below 0.016 μ/L) being in agreement with Cattoir et al. 15. In one study this pathogen was reported susceptible 16 but in another study 5 28% of strains were resistant or intermediately susceptible to pivmecillinam. Our strains had an inhibition zone to pivmecillinam in the range 28–38 mm but were unambiguously resistant to co-trimaxozole with no zone of growth inhibition whereas for ciprofloxacin a slight inhibition zone was detectable for some strains (data not shown).
In our opinion culture, phenotypical examination and MALDI-TOF identification are superior to PCR because the genomic material of A. schaalii can be detected with PCR in up to 13% of asymptomatic patients 17. We do not perform microscopy of urine samples of all patients aged over 60 years, as recommended by Nielsen et al. 5. We believe that screening the urine using flow cytometry sorting (FACS) 18 will help identify those samples that require special culture.
In conclusion, we advocate the sensitized culture to increase the yield of A. schaalii recovery from urine in risk-group patients and MALDI-TOF for fast identification.
Acknowledgments
We thank our colleagues for sending us their A. schaalii isolates.
Conflict of Interest
None declared.
References
- Fendukly F, Osterman B. Isolation of Actinobaculum schaalii and Actinobaculum urinale from a patient with chronic renal failure. J Clin Microbiol. 2005;43:3567–3569. doi: 10.1128/JCM.43.7.3567-3569.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm PD, Van Eijk J, Veltman S, Muleman E, Schüllin T. Urosepsis with Actinobaculum schaalii and Aerococcus urinae. J Clin Microbiol. 2006;44:625–654. doi: 10.1128/JCM.44.2.652-654.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M, Prag J, Kemp M, et al. Ten cases of Actinobaculum schaalii infection: clinical relevance, bacterial identification, and antibiotic susceptibility. J Clin Microbiol. 2005;43:5305–5308. doi: 10.1128/JCM.43.10.5305-5308.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinaud C, Gaillard T, Maslin J, et al. Actinobaculum schaalii bacteria in an aged male patient. Med Mal Infect. 2008;38:617–619. doi: 10.1016/j.medmal.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Søby KM, Christensen JJ, Prag J. Actinobaculum schaalii: a common cause of urinary tract infection in the elderly population. Bacteriological and clinical characteristics. Scan J Infect Dis. 2010;42:43–47. doi: 10.3109/00365540903289662. [DOI] [PubMed] [Google Scholar]
- Gomez E, Gustafson DR, Rosenblatt JE, Patel R. Actinobaculum bacteremia: a report of 12 cases. J Clin Microbiol. 2011;49:4311–4313. doi: 10.1128/JCM.00798-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beguelin C, Genne D, Vara A, et al. Actinobaculum schaalii: clinical observation of 20 cases. Clin Microbiol Infect. 2011;17:1027–1031. doi: 10.1111/j.1469-0691.2010.03370.x. [DOI] [PubMed] [Google Scholar]
- García-Bravo M, González-Fernández MB, García-Castro MA, Jaime-Muniesa ML. Urinary tract infection caused by Actinobaculum schaalii in an elderly patient. Rev Esp Quimioter. 2011;24:52–53. [PubMed] [Google Scholar]
- Agudo S, Domingo D, Navarro JL, Arenal N, López-Brea M. Actinobaculum schaalii: a new cause of mastitis. Clin Microbiol Infect. 2009;15:S4. [Google Scholar]
- Hoenigl M, Leitner E, Valentin T, et al. Endocarditis caused by Actinobaculum schaalii. Emerg Infect Dis. 2010;16:1171–1173. doi: 10.3201/eid1607.100349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Bempt I, Van Trappen S, Cleenwerck I, et al. Actinobaculum schaalii causing Fournier’s Gangrene. J Clin Microbiol. 2011;49:2369–2371. doi: 10.1128/JCM.00272-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajkrt D, Simmons-Smit AM, Savelkoul PH, van den Hoek J, Hack WW, van Furth AM. Pyelonephritis caused by Actinobaculum schaalii in a child with pyeloureteral junction obstruction. Eur J Clin Microbiol Infect Dis. 2003;53:679–682. doi: 10.1007/s10096-003-0933-3. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Berlinger L, Liniger B, Grunt S, Agyeman P, Ritz N. Actinobaculum schaalii an emerging pediatric pathogen? BMC Infect Dis. 2012;12:201. doi: 10.1186/1471-2334-12-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalava JM, Mäntymaa L, Ekblad U, et al. Bacterial 16S rDNA polymerase chain reaction in the detection of intra-amnionitic infection. Br J Obstet Gynaecol. 1996;103:664–669. doi: 10.1111/j.1471-0528.1996.tb09835.x. [DOI] [PubMed] [Google Scholar]
- Cattoir V, Varca A, Greub G, Prod’hom G, Legrand P, Lienhard R. In vitro susceptibility of Actinobaculum schaalii to 12 antimicrobial agents and molecular analysis of fluoroquinolone resistance. J Antimicrob Chemother. 2010;65:2514–2517. doi: 10.1093/jac/dkq383. [DOI] [PubMed] [Google Scholar]
- Andersen PK, Søby KM, Bank S, Prag J. In vitro susceptibility of Actinobaculum schaalii to mecillinam. J Antimicrob Chemother. 2011;66:2181–2182. doi: 10.1093/jac/dkr270. [DOI] [PubMed] [Google Scholar]
- Bank S, Jensen A, Hansen TM, Søby KM, Prag J. Actinobaculum schaalii, a common uropathogen in elderly patients, Denmark. Emerg Infect Dis. 2010;16:76–80. doi: 10.3201/eid1601.090761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolkkonen S, Paattiniemi EL, Kärpänoja P, Sarkkinen H. Screening of urine samples by flow cytometry reduces the need for culture. J Clin Microbiol. 2010;48:3117–3121. doi: 10.1128/JCM.00617-10. [DOI] [PMC free article] [PubMed] [Google Scholar]


