Abstract
The objective of this study was to develop a novel peptide nucleic acid (PNA) probe for Stenotrophomonas maltophilia identification by fluorescence in situ hybridization (FISH). The probe was evaluated using 33 human and veterinary clinical S. maltophilia isolates and 45 reference strains representing common bacterial species in the respiratory tract. The probe displayed 100% sensitivity and 100% specificity on pure cultures and allowed detection in sputum from cystic fibrosis patients. The detection limit was 104 CFU/mL in spiked tracheal aspirate and bronchoalveolar lavage from healthy horses. Altogether the study shows that this species-specific PNA FISH probe facilitates rapid detection of S. maltophilia in biological specimens.
Keywords: Cystic fibrosis, identification, PNA FISH, Stenotrophomonas maltophilia
Stenotrophomonas maltophilia is a Gram-negative bacterium ubiquitous in nature 1. It is capable of causing serious disease in immuno-compromised patients and has been reported in cystic fibrosis (CF) patients with a yearly prevalence of up to 25% 2,3. In veterinary medicine, an increasing number of studies indicate possible associations with respiratory disease 4,5, bacterial cystitis and endocarditis 6, urinary tract infections 7 and lymphadenitis 8. Within the genus, S. maltophilia is the only species that is known to colonize and cause respiratory tract infections in humans and animals.
The objective of this study was to design and evaluate a novel peptide nucleic acid (PNA) probe for S. maltophilia identification by fluorescence in situ hybridization (FISH). A PNA probe targeting 16S rRNA (Flu-OO-CGCCATGGATGTTCC, 5′-3′) specific for S. maltophilia and conjugated with fluorescein isothiocyanate (FITC) was added a PNA blocker probe (Ac-CCACGGATGTTCC, 5′-3′) to prevent cross-reaction with closely related species such as Xanthomonas campestris (Panagene, Daejeon, Korea). Sensitivity and specificity of the probe were evaluated using 35 human and veterinary clinical S. maltophilia isolates, and 43 reference and clinical strains representing common bacterial species in the airways of humans and animals (Table1). All strains were cultured on solid media (5% calf blood agar, chocolate agar or nutrient/yeast/glycerol agar as deemed appropriate) and sub-cultured in tryptic soy broth. Ten microlitres of broth were placed onto a microscopic slide prepared with one drop of fixation solution (phosphate-buffered saline with detergent) and fixed by heating (methanol fixation for sputum smears). After adding one drop of hybridization solution (AdvanDx, Woburn, MA, USA) containing the S. maltophilia PNA probe, a coverslip was applied and hybridization performed by incubating the slides at 55°C for 30 min (90 min for sputum smears). Limited to Gram-negative strains, slides were immersed in preheated deionized water (55°C) for 1 min. All slides were then placed in a wash jar with a preheated (55°C) wash solution (diluted Tris-buffered saline with detergent) in a water bath for 30 min, coverslips removed. After air drying, a drop of mounting medium (photobleaching inhibitor in glycerol) and a coverslip were applied. A positive control slide with S. maltophilia and a negative control slide with the relevant strain hybridized with a universal PNA FISH probe (BacUni PNA, AdvanDx) according to manufacturer's instructions were included in all runs. All slides were evaluated within 2 h under a fluorescence microscope (×60 objective, Olympus BX51, Ballerup, Denmark; Mercury U-LH100HG 100 W lamp) equipped with an FITC/Texas Red Dual Band Filter. Fluorescence images were obtained using an Olympus DP72 camera (1360 × 1024 pixels, 1 s exposure). Samples were considered positive when single cells had a strong fluorescence and clear morphology (Fig.1). The probe yielded positive results with all target organisms and negative results with all non-target organisms (100% sensitivity and 100% specificity).
Table 1.
Reference and clinical strains used for probe evaluation.
| Proteus mirabilis | ATCC 12453 | Streptococcus dysgal. subsp. dysgalactiaeT | CCUG 27301 |
| Bacillus cereusa | S31A8a | Streptococcus dysgal. subsp. equisimilis | ATCC 12388 |
| Escherichia coli | ATCC 35218 | Streptococcus equi subsp. zooepidemicus | CCUG 23256 |
| Enterococcus faecalisT | ATCC 19433 | Streptococcus equi subsp. equi | ATCC 9528 |
| Enterobacter aerogenesT | ATCC 13048 | Streptococcus suisT | CCUG 7984 |
| Serratia marcescens | ATCC 14756 | Streptococcus bovisT | CCUG 17828 |
| Citrobacter freundii | ATCC 43864 | Streptococcus pneumoniae | ATCC 10015 |
| Staphylococcus epidermidis | ATCC 12228 | Actinobacillus equuli subsp. equulia | S55G3a |
| Staphylococcus aureusT | ATCC 12600 | Alcaligenes spp.a | S61E5a |
| Hafnia alveia | S24B4a | Corynebacterium diphtheriae | ATCC 27010 |
| Pasteurella multocida | ATCC 12945 | Bordetella bronchisepticaa | S31B7a |
| Neisseria meningitidis | ATCC 13102 | Klebsiella pneumoniae subsp. pneumoniaeT | ATCC 13883 |
| Acinetobacter baumanniiT | ATCC 19606 | Klebsiella pneumoniae subsp. pneumoniae | ATCC 10031 |
| Moraxella bovisa | S31B1a | Xanthomonas campestris pv. campestris | Strain 8004 |
| Moraxella catarrhalis | ATCC 25240 | Pseudomonas fluorescensT | ATCC 13525 |
| Moraxella nonliquefaciensT | ATCC 19975 | Pseudomonas alcaligenesT | ATCC 14909 |
| Haemophilus influenzae | ATCC 19418 | Pseudomonas putida | ATCC 49128 |
| Burkholderia cepacia | ATCC 25416 | Pseudomonas stutzeriT | ATCC 17588 |
| Burkholderia multivorans | CF 54b | Pseudomonas aeruginosaT | ATCC 10145 |
| Stenotrophomonas acidaminiphilaT | ATCC 700916 | Pseudomonas aeruginosa | PAO1b, CF15b, CF16b |
| Stenotrophomonas maltophiliaT | ATCC 13637 | ||
| Stenotrophomonas maltophilia | Veterinary clinical strains n = 22a and human clinical strains n = 13b | ||
From the Department of Veterinary Disease Biology, University of Copenhagen, Denmark.
From the Department for Clinical Microbiology, University Hospital of Copenhagen, Denmark.
Figure 1.
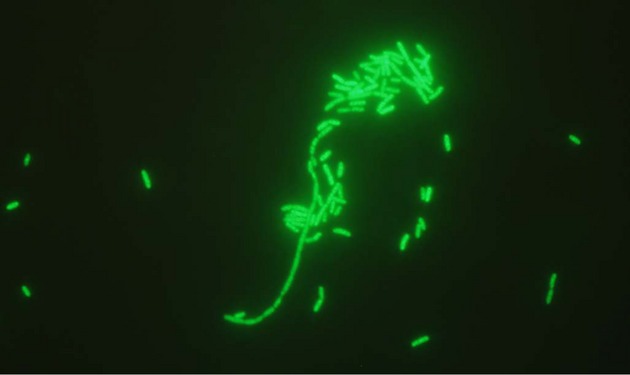
PNA FISH (peptide nucleic acid fluorescence in situ hybridization) microscopy of Stenotrophomonas maltophilia.
The probe was tested on spiked respiratory samples of equine origin to determine the detection limit. Tracheal aspirate (TA) and bronchoalveolar lavage (BAL) samples from three healthy horses were spiked with S. maltophilia ATCC 13637. Ten-fold dilution series were prepared from the spiked TA/BAL samples and PNA FISH smears were prepared for each dilution as described above but without fixation solution. The detection limit was determined by parallel plating of the dilutions on blood agar for colony counts. Fluorescence microscopy of the smears showed at least one to ten S. maltophilia cells in most fields when the concentration in the sample was 104 CFU/mL. The probe specificity was further tested on sputum samples from seven patients with CF. The samples underwent routine culture at the University Hospital of Copenhagen, Denmark. The probe produced positive results in all four samples from S. maltophilia-infected patients and negative results in all control samples (two patients infected with Pseudomonas aeruginosa, one patient infected with Burkholderia multivorans). No background fluorescence was observed.
Isolation and identification of S. maltophilia can be problematic 9–13. Selective differential media have been recommended for improved detection of S. maltophilia from non-sterile sites such as respiratory secretions 14–16. Problems related to misidentification of S. maltophilia by phenotypic methods can be overcome by the use of molecular methods. Pinot et al. 17 used vancomycin, imipenem, amphotericin B medium agar for isolation and multiplex PCR for identification of S. maltophilia. Hogardt et al. 18 designed a species-specific DNA probe for S. maltophilia identification and demonstrated that the probe could be used successfully on sputum and throat samples from CF patients. However, the limit for microscopic detection of bacteria within sputum was 4 × 105 CFU/mL and the sensitivity of the DNA FISH method (90%) was lower than that of the PNA FISH approach described in this study. PNA probes are small in size with a non-charged polyamide backbone that renders them easy to hybridize and increases the binding strength compared to DNA probes 19. PNA probes have better sensitivity and specificity, show improved penetration into cells and through biofilm matrices and are not susceptible to bacterial endonucleases which may be present in clinical samples 20. PNA FISH is a very fast (less than 90 min) and reliable molecular identification method which is easy to perform with very little hands-on time. Furthermore, FISH is useful for in situ detection of this microorganism directly in clinical samples and mixed bacterial populations without prior cultivation. Thus, the S. maltophilia PNA FISH probe described in this study has important applications for studies of biofilm infections and S. maltophilia colonization in patients with CF, where colonization and chronic infection with S. maltophilia is commonly reported.
In conclusion, the S. maltophilia PNA FISH probe demonstrated excellent sensitivity and specificity when tested against clinically relevant bacteria occurring in the respiratory tract of humans and animals. The PNA FISH assay can be implemented in diagnostic laboratories for rapid, simple and reliable in situ identification of S. maltophilia in clinical specimens. It can only be a valuable tool for research aimed at understanding the role played by this organism in CF and in equine respiratory tract infections. For this purpose, further studies are warranted to evaluate the use of the probe for studying spatial distribution of S. maltophilia in polymicrobial biofilms.
Acknowledgments
Xanthomonas campestris pv. campestris strain 8004 was kindly provided by Mari-Anne Newmann, Section for Transport Biology, University of Copenhagen, Denmark.
Conflict of Interest
None declared.
References
- Brooke JS. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin Microbiol Rev. 2012;25:2–41. doi: 10.1128/CMR.00019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalboge CS, Hansen CR, Pressler T, Hoiby N, Johansen HK. Chronic pulmonary infection with Stenotrophomonas maltophilia and lung function in patients with cystic fibrosis. J Cyst Fibros. 2011;10:318–325. doi: 10.1016/j.jcf.2011.03.006. [DOI] [PubMed] [Google Scholar]
- Ryan RP, Monchy S, Cardinale M, et al. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat Rev Microbiol. 2009;7:514–525. doi: 10.1038/nrmicro2163. [DOI] [PubMed] [Google Scholar]
- Albini S, Abril C, Franchini M, Huessy D, Filioussis G. Stenotrophomonas maltophilia isolated from the airways of animals with chronic respiratory disease. Schweiz Arch Tierheilkd. 2009;151:323–328. doi: 10.1024/0036-7281.151.7.323. [DOI] [PubMed] [Google Scholar]
- Winther L, Andersen RM, Baptiste KE, Aalbaek B, Guardabassi L. Association of Stenotrophomonas maltophilia infection with lower airway disease in the horse: a retrospective case series. Vet J. 2010;186:358–363. doi: 10.1016/j.tvjl.2009.08.026. [DOI] [PubMed] [Google Scholar]
- Berger S, Froehlich W. Stenotrophomonas maltophilia as a causative agent of bacterial infections in the horse: two clinical cases. Pferdeheilkunde. 2011;27:381–385. [Google Scholar]
- Kralova-Kovarikova S, Husnik R, Honzak D, Kohout P, Fictum P. Stenotrophomonas maltophilia urinary tract infections in three dogs: a case report. Vet Med. 2012;57:380–383. [Google Scholar]
- Johnson E, Al-Busaidy R, Hameed M. An outbreak of lymphadenitis associated with StenotrophomonasXanthomonasmaltophilia in Omani goats. J Vet Med Ser B Infect Dis Vet Public Health. 2003;50:102–104. doi: 10.1046/j.1439-0450.2003.00643.x. [DOI] [PubMed] [Google Scholar]
- Anderson SW, Stapp JR, Burns JL, Qin X. Characterization of small-colony-variant Stenotrophomonas maltophilia isolated from the sputum specimens of five patients with cystic fibrosis. J Clin Microbiol. 2007;45:529–535. doi: 10.1128/JCM.01444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittar F, Rolain JM. Detection and accurate identification of new or emerging bacteria in cystic fibrosis patients. Clin Microbiol Infect. 2010;16:809–820. doi: 10.1111/j.1469-0691.2010.03236.x. [DOI] [PubMed] [Google Scholar]
- Burdge D, Noble M, Campbell M, Krell V, Speert D. Xanthomonas maltophilia misidentified as Pseudomonas cepacia in cultures of sputum from patients with cystic fibrosis: a diagnostic pitfall with major clinical implications. Clin Infect Dis. 1995;20:445–448. doi: 10.1093/clinids/20.2.445. [DOI] [PubMed] [Google Scholar]
- Carmody LA, Spilker T, LiPuma JJ. Reassessment of Stenotrophomonas maltophilia phenotype. J Clin Microbiol. 2011;49:1101–1103. doi: 10.1128/JCM.02204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson-Stadler LA, Mihaylova SA, Moore ERB. Stenotrophomonas interspecies differentiation and identification by gyrB sequence analysis. FEMS Microbiol Lett. 2012;327:15–24. doi: 10.1111/j.1574-6968.2011.02452.x. [DOI] [PubMed] [Google Scholar]
- Foster NF, Chang BJ, Riley TV. Evaluation of a modified selective differential medium for the isolation of Stenotrophomonas maltophilia. J Microbiol Methods. 2008;75:153–155. doi: 10.1016/j.mimet.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Goncalves-Vidigal P, Grosse-Onnebrink J, Mellies U, Buer J, Rath PM, Steinmann J. Stenotrophomonas maltophilia in cystic fibrosis: improved detection by the use of selective agar and evaluation of antimicrobial resistance. J Cyst Fibros. 2011;10:422–427. doi: 10.1016/j.jcf.2011.06.010. [DOI] [PubMed] [Google Scholar]
- Kerr K, Denton M, Todd N, Corps C, Kumari P, Hawkey P. A new selective differential medium for isolation of Stenotrophomonas maltophilia. Eur J Clin Microbiol Infect Dis. 1996;15:607–610. doi: 10.1007/BF01709373. [DOI] [PubMed] [Google Scholar]
- Pinot C, Deredjian A, Nazaret S, et al. Identification of Stenotrophomonas maltophilia strains isolated from environmental and clinical samples: a rapid and efficient procedure. J Appl Microbiol. 2011;111:1185–1193. doi: 10.1111/j.1365-2672.2011.05120.x. [DOI] [PubMed] [Google Scholar]
- Hogardt M, Trebesius K, Geiger AM, Hornef M, Rosenecker J, Heesemann J. Specific and rapid detection by fluorescent in situ hybridization of bacteria in clinical samples obtained from cystic fibrosis patients. J Clin Microbiol. 2000;38:818–825. doi: 10.1128/jcm.38.2.818-825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellestor F, Paulasova P, Hamamah S. Peptide nucleic acids (PNAs) as diagnostic devices for genetic and cytogenetic analysis. Curr Pharm Des. 2008;14:2439–2444. doi: 10.2174/138161208785777405. [DOI] [PubMed] [Google Scholar]
- Bjarnsholt T, Jensen PO, Fiandaca MJ, et al. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr Pulmonol. 2009;44:547–558. doi: 10.1002/ppul.21011. [DOI] [PubMed] [Google Scholar]


