Abstract
Recently the rationale for combining targeted therapy with immunotherapy has come to light, but our understanding of the immune response during MAPK pathway inhibitor treatment is limited. We discovered that the immune-microenvironment can act as source of resistance to MAPK pathway-targeted therapy, and moreover during treatment this source becomes reinforced. In particular, we identified macrophage-derived TNFα as a crucial melanoma-growth factor that provides resistance to MAPK pathway inhibitors through the lineage-transcription factor MITF. Most strikingly, in BRAF mutant melanomas of patients and BRafV600E-melanoma allografts MAPK pathway inhibitors increased the number of tumor-associated macrophages, and TNFα and MITF expression. Inhibiting TNFα-signaling with IκB-kinase inhibitors profoundly enhanced the efficacy of MAPK pathway inhibitors by targeting not only the melanoma cells, but also the microenvironment. In summary, we identify the immune-microenvironment as a novel source of resistance and reveal a new strategy to improve the efficacy of targeted therapy in melanoma.
Keywords: Melanoma/skin cancer, cellular responses to anticancer drugs, kinase and phosphatase inhibitors, protein kinases and phosphatases as target for therapy
Introduction
The MAP-kinase (MAPK) signaling pathway consisting of the kinases RAF/MEK/ERK is hyperactivated in up to 90% of melanomas. The dependence of melanoma cells on this activated pathway has been utilized successfully in the clinic by selectively inhibiting the RAF kinase BRAF, which is mutated in ~50% of melanomas (1). The efficacy of these inhibitors is limited however by the onset of resistance, and in the majority of cases this occurs through re-activation of the pathway (2, 3). This is currently addressed by inhibiting the pathway further downstream using MEK inhibitors in combination with BRAF inhibitors (4).
Other forms of resistance that have been described rely on the activation of additional signaling pathways such as signaling downstream of PI3 kinase (PI3K), which can be targeted by selective inhibition (5). Another intracellular event that can cause innate and acquired resistance is the high expression of survival factors. One such survival factor, which we have previously identified, is the melanocytic specific transcription factor MITF (6). MITF dependent resistance is probably due to its central role regulating multiple survival and anti-apoptotic genes (7). Indeed, the MITF target BCL2A1 has been identified to antagonize BRAF inhibition (8). Furthermore, components of the differentiation program that stimulates up-regulation of MITF are also involved in MAPK pathway inhibitor resistance (9).
In addition to these endogenous mechanisms of resistance, secreted factors that originate from the stroma can induce resistance. For instance stromal fibroblast-derived HGF causes activation of receptor tyrosine kinases that act to re-activate the pathway by signaling through RAS (10). One important microenvironment derived cytokine is TNFα, which has been described to block apoptosis in BRAF-depleted melanoma cells (11). TNFα can execute pro-tumorigenic activities in melanoma such as promoting tumor growth, angiogenesis and invasion (12, 13). Furthermore, vascular progression and a more metastatic melanoma phenotype correlate with an increased activity of NFκB, a transcription factor that bedsides other growth factors, cytokines or chemokines is activated by TNFα (14-16). In light of these findings we wanted to study the role of TNFα in melanoma growth and survival as well as resistance to MAPK pathway targeted therapy.
Results
TNFα is required for growth and survival of melanoma cells
Mice expressing BrafV600E in the melanocyte lineage develop melanomas with a median latency of 12 months (17), but we found that lack of TNFα in BrafV600E/TNFα−/− mice significantly delayed the median latency by ~6 months (Figure 1A). Furthermore, when we injected melanoma cells derived from BRafV600E mice (TNFR expressing 4434 cells, Supplementary Fig. S1A) into syngeneic wild type or TNFα−/− mice, the average tumor size in TNFα deficient mice was severely reduced (Figure 1B). These data strongly suggested that TNFα is required for the growth of melanoma cells in vivo. Indeed, TNFα stimulated proliferation of 4434 melanoma cells in vitro (Figure 1C), induced IKB phosphorylation (pIKB) and protected the cells from cell death when they were unable to adhere to extracellular matrix (Figure 1D). One of the key regulators of melanoma cell survival and proliferation is the lineage survival factor MITF. We found that TNFα up-regulated MITF expression in BRafV600E mouse melanoma cells which correlated with reduced caspase-3 cleavage under anoikis conditions (Figure 1E). TNFα induced pIKB and increased MITF expression also in human BRAF mutant TNFR expressing (Supplementary Fig. S1B) melanoma cells, stimulated their growth (not shown) and protected these cells from anoikis (Figure 1E, F, G). Importantly, overexpression of MITF alone significantly reduced cell death and caspase-3 cleavage under anoikis conditions (Figure 1F, G). On the other hand, counteracting the TNFα mediated MITF up-regulation by RNAi abolished the protective effect of TNFα without affecting pIKB (Figure 1H), suggesting that MITF contributes to TNFα mediated survival.
Figure 1. TNFα is an important survival and growth signal for melanoma.
A. Kaplan-Meier plot showing melanoma-free survival (%) of tamoxifen-treated BrafV600E;Tyr::CreERT2 (BRAFV600E) and BrafV600E;Tnfα−/−;Tyr::CreERT2 (BRAFV600E/TNFα−/−) mice, and control mice (Ethanol-treated BrafV600E;Tnfα−/−;Tyr::CreERT2 mice and tamoxifen-treated Tyr::CreERT2 mice). p< 0.0001; Log-rank (Mantel Cox) Test. B. Growth of BRafV600E-4434 melanoma allografts in WT and TNFα−/− mice. C. in vitro growth assay of BRafV600E-4434 melanoma cells treated with BSA or 50ng TNFα once every 3 days. D. Anoikis assay of BRafV600E-4434 melanoma cells for dead cells detected by trypan blue staining. Cells were cultured under non-adherent conditions for 72hrs and treated with BSA or 50ng TNFα. A Western blot for MITF, pIKBα, cleaved caspase3 and ERK2 is shown. E. Western blot of the indicated cell lines for MITF and pIKBα and ERK2 after 24hrs treatment with 50ng TNFα. F. Anoikis assay for untreated or TNFα treated 4434, A375 and 4434- and A375-MITF overexpressing cells. G. Western blot for MITF, pIKBα, cleaved caspase3 and ERK2 of detached A375 and A375-MITF cells treated for 48hrs with 50ng TNFα. H. Anoikis assay for untreated or TNFα stimulated A375 cells transfected with control or MITF specific siRNAs. A Western blot for MITF, pIKBα, cleaved caspase3 and ERK2 is shown.
TNFα regulates MITF expression through canonical NF-kB signaling
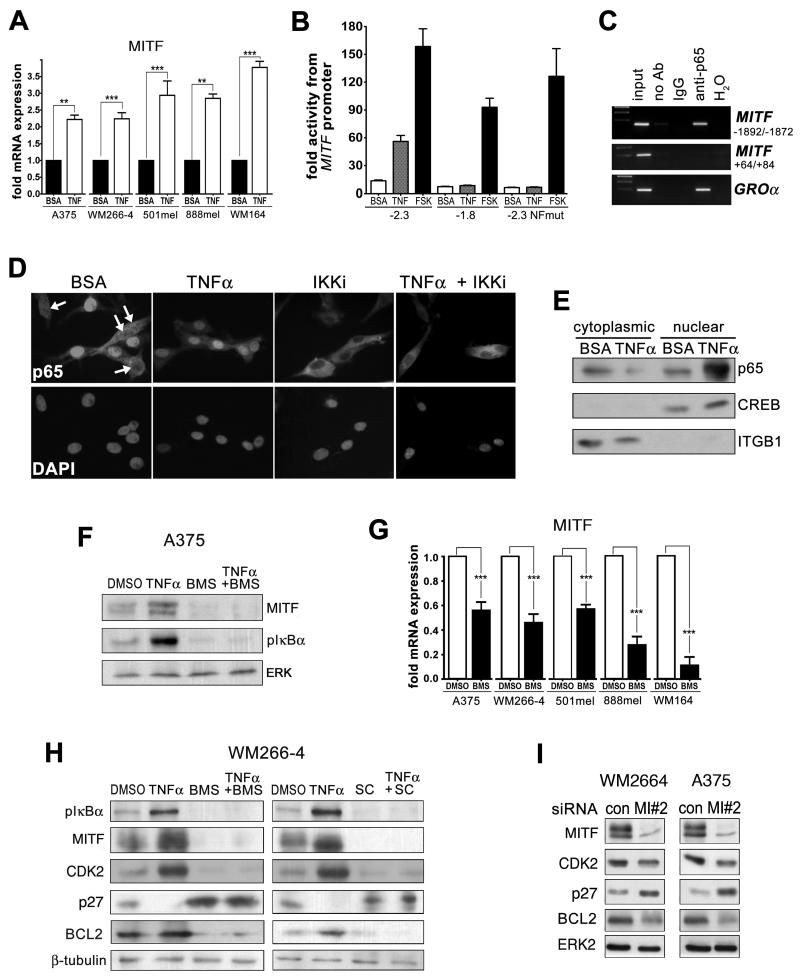
To establish the mechanism of TNFα-mediated MITF regulation, we analyzed MITF mRNA expression in different melanoma cell lines. This revealed that TNFα regulates MITF at transcriptional level (Figure 2A), which was further confirmed by a MITF promoter analysis (Figure 2B). Whereas TNFα efficiently activated a −2.3kb promoter fragment that contains a potential NFκB binding site at −1870/−1879, it failed to elicit a response from a −1.8kb promoter fragment that lacked the site or when the potential site was mutated (Figure 2B, Supplementary Figure S2A, B). A chromatin-IP confirmed that NF-κB/p65 binds to the MITF promoter (Figure 2C). Although TNFα stimulated IκBα phosphorylation and nuclear translocation of NF-κB/p65 in melanoma cells, basal activation of NF-κB signaling was detectable in the absence of exogenous TNFα (Figure 2D, E and F). Inhibition of IκB kinase (IKK) activity using BMS345541 (inhibits IKKα and IKKβ) or SC-514 (IKKβ specific) was able to efficiently block p65 nuclear translocation, led to a reduction in phospho-IkBα, and decreased both protein and mRNA expression of MITF (Figure 2D-G). This indicates that TNFα and IKK/NF-κB signaling contribute to the regulation of MITF expression in BRAF mutant melanoma cells. In line with this, along with diminished MITF expression, IKK inhibition in BRAF mutant melanoma cells resulted in reduced CDK2 and BCL2 expression, while p27 was upregulated (Figure 2H). These are well-characterized MITF target genes (7), and using RNAi we confirmed that MITF regulates the expression of these cell cycle and survival proteins in melanoma cells (Figure 2I, Supplementary Figure S2C).
Figure 2. TNFα regulates MITF expression through IKK.
A. Real time qPCR analysis of a panel of melanoma cell lines treated with 50ng TNFα for 24hrs. B. Different MITF promoter construct activity as detected by luciferase in WM266-4 cells treated with 50ng TNFα for 24hrs. Forskolin (FSK) served as positive control. C. NF-κB/p65 Chromatin-IP from TNFα treated WM266-4 cells. The indicated regions of the M-MITF promoter region or a coding region of the MITF gene were amplified. Amplification of the GROα promoter served as positive control (50). D. Immunofluorescence analysis for NF-kB/p65 in WM266-4 cells treated with 50ng TNFα for 2hrs or 0.5μM BMS-345541 for 2hrs either alone or in combination. E. Western blot of cytoplasmic and nuclear extracts from WM266-4 cells treated with BSAE or 50ng TNFα. F. Western blot of A375 cells treated with TNFα or DMSO and 0.5μM BMS-345541 (IKKi) as indicated for 24hrs. G. Real time qPCR analysis of a panel of melanoma cell lines either untreated or treated with 0.5μM BMS-345541 for 24hrs. H. Western blot of WM266-4 melanoma cells treated with TNFα or DMSO, BMS-345541 (0.5μM) or SC-514 (1μM) as indicated for 24hrs. I. Western blot of WM266-4 and A375 melanoma cells transfected with control or MITF specific siRNAs for 24hrs for MITF, CDK2, p27, BCL2 and ERK2.
Macrophages induce MITF expression through TNFα and significantly impact on melanoma cell growth
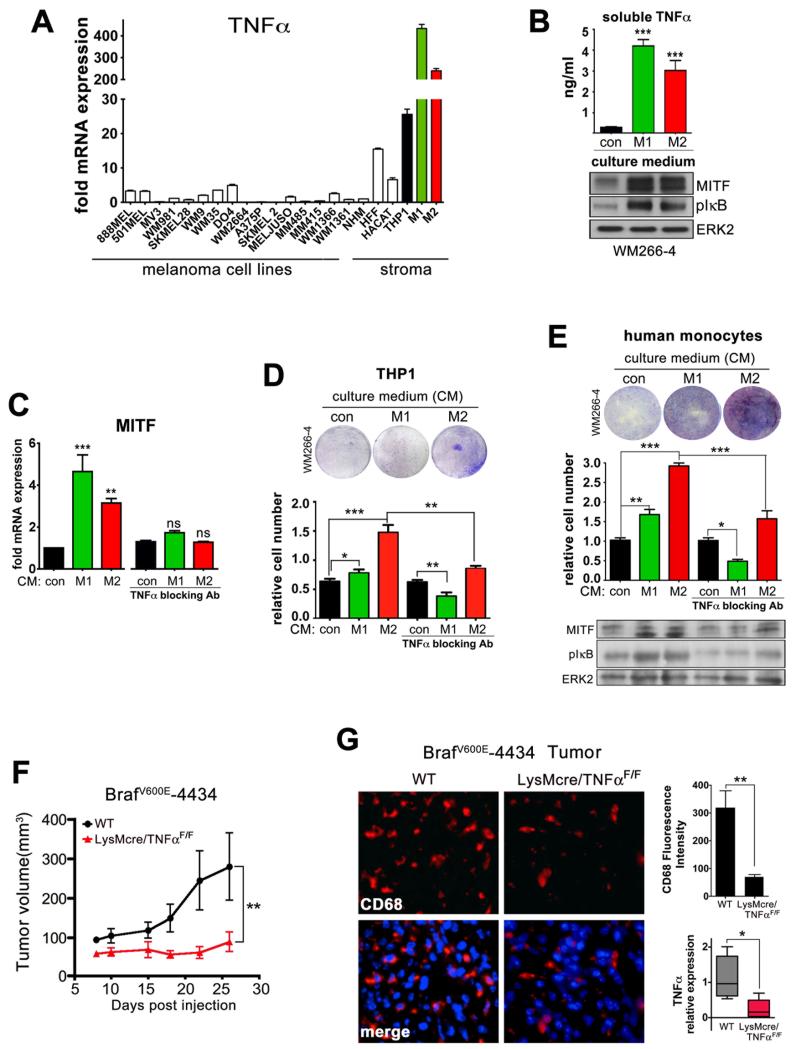
We next wished to identify the source of TNFα expression, and found an average 2-5 fold increase in TNFα mRNA throughout a panel of 16 melanoma cell lines compared to normal melanocytes (NHM) (Figure 3A). However, A375 and WM266-4 cells do not express significant amounts of TNFα, which suggests that the basal IKK/NF-κB activation we observed might be due to other mechanisms such as autocrine signaling through CXCL1, PI3K/AKT signaling or loss of p16CDKN2 (16). Also 4434-BRafV600E cells do not express any TNFα (Supplementary Figure S3A), which is in agreement with the reduced tumor growth in TNFα deficient mice (see Figure 1B). We therefore analyzed stromal cells, including fibroblast, keratinocytes and also macrophages as they are a major source of TNFα (18). Macrophages can polarize into the classically activated M1 and the alternatively activated M2 phenotype (19), and these phenotypes can be generated in vitro by differentiating and polarizing monocytic THP-1 cells through treatment with specific cytokines (Supplementary Figure S3B). We found that both M1 and M2 macrophages were indeed the highest TNFα expressing cells (Figure 3A).
Figure 3. Macrophages induce MITF expression through TNFα, which is required for melanoma growth.
A. Real time qPCR analysis for TNFα in melanoma cell lines and stromal cells: fibroblasts (HFF), keratinocytes (HACAT), THP1 and macrophages (M1, M2) compared to non-transformed melanocytes (NHM). B. TNFα production in conditioned media from undifferentiated and differentiated macrophages detected by ELISA. A Western blot for MITF and pIKBα of WM266-4 lysates following treatment with macrophage conditioned media for 24hrs is shown. C. qPCR gene expression analysis of MITF following treatment with macrophage conditioned media 24hrs with or without the addition of 5ng TNFα blocking antibody for 24hrs. D. Colony formation assay of WM266-4 cells treated with conditioned media from THP1 derived M1 or M2 macrophages or control medium for 3 weeks. E. Colony formation assay of WM266-4 cells treated with conditioned media from human monocyte derived M1 or M2 macrophages or control medium for 3 weeks. F. Growth of BRafV600E-4434 melanoma allografts in WT or LysMcre/TNFαF/F mice. G. CD68 histology of BRafV600E-4434 melanoma samples from WT and LysMcre/TNFαF/F mice. The relative CD68 immunofluorescence intensity (n=3 tumors) and TNFα expression (n=5 tumors) is shown.
In accordance with the TNFα mRNA expression, soluble TNFα was detectable in the medium of M1 and M2 polarized macrophages (Figure 3B), and treatment of WM266-4 cells with conditioned media from either M1 or M2 macrophages led to increased IkBα phosphorylation and increased MITF expression at protein as well as mRNA level (Figure 3B, C, Supplementary Figure S3C). The major driver of the macrophage-induced MITF upregulation was secreted TNFα, as conditioned media after addition of a TNFα blocking antibody no longer induced MITF expression (Figure 3C).
Exposure of melanoma cells to conditioned medium from M1 macrophages for three weeks had a slight growth promoting effect, but growth was suppressed when TNFα-action was inhibited by a blocking antibody (Figure 3D). On the other hand, M2 macrophage-derived conditioned medium stimulated growth (Figure 3D). However, depletion of TNFα using a blocking antibody significantly reduced this growth promoting effect (Figure 3D). Importantly, similar results were obtained when using human peripheral-blood monocyte derived macrophages (Figure 3E, Supplementary Figure S3D). On the other hand, keratinocytes and fibroblasts which express 5-10fold more TNFα than melanoma cells, but 10-80fold less than macrophages (Figure 3A) did not support melanoma cell growth in a TNFα dependent manner (Supplementary Figure S3E).
Macrophage recruitment to melanoma is well documented and has been linked to UV-induced melanomagenesis in mice (20). Using publically available gene expression data sets (21, 22), we found that the expression of macrophage markers was significantly upregulated during melanoma progression (Supplementary Figure S4A-C), indicating the availability of this potential TNFα source in the tumor microenvironment. To assess the importance of macrophage-derived TNFα for melanoma growth in vivo we used LysMcre/TNFαF/F mice, in which Cre-mediated recombination results in loss of TNFα expression in the myeloid cell lineage (Grivennikov et al., 2005). Remarkably, the conditional ablation of TNFα resulted in significant growth retardation of BARFV600E-4434 melanoma allografts (Figure 3F). When we analyzed the tumors for the presence of macrophages using a pan-macrophage anti-CD68 antibody, we found that the presence of CD68 positive cells within the tumor was significantly reduced in LysMcre/TNFαF/F mice, which was accompanied by significantly decreased TNFα expression (Figure 3G).
Macrophages can protect against MEK inhibitor induced apoptosis in a TNFα dependent manner
We have previously shown that elevated MITF expression provides resistance to MEK inhibitor (MEKi) induced cell death (6). Because TNFα induces MITF expression it was not surprising to see that it suppressed MEKi induced caspase3 cleavage (Figure 4A). Importantly, this response was dependent on MITF, because MITF depletion through RNAi resulted in loss of the TNFα mediated protective effect (Figure 4A).
Figure 4. Macrophages protect against MEKi induced apoptosis.
A. Western blot of WM266-4 cells transfected with scrambled control or MITF specific siRNAs, and treated with 2μM PD184 for 48hours in the absence or presence of 50ng TNFα. B. Schematic of co-culture assay of melanoma cells and differentiated macrophages. C. Survival assay of A375 melanoma cells. The cells were treated for 48hr with 2μM AZD6244 (MEKi) in presence of the indicated macrophages. D. TNFα production detected by ELISA from conditioned media from undifferentiated and differentiated macrophages cultured in the presence or absence of MEKi (AZD6244). E. Survival assay of A375 melanoma cells assessed by toluidine blue staining. The cells were treated for 48hr with 2μM AZD6244 in presence of the indicated macrophages with or without the addition of 5ng TNFα blocking antibody.
We then assessed whether macrophages can protect melanoma cells from MEKi induced apoptosis. For this, we co-cultured melanoma cells with macrophages using a trans-well technique (Figure 4B). This approach enabled the isolation of both melanoma cells and macrophages for separate analysis, and also excluded any macrophage-derived phagocytic activity. We found that the presence of either M1 or M2 macrophages significantly protected melanoma cells from MEKi exposure (Figure 4C, Supplementary Figure S5A). Analysis of TNFα expression and secretion by the macrophages showed no change when treated with MEKi (Figure 4D, Supplementary Figure S5B) confirming that TNFα was available for the melanoma cells under these conditions. Moreover, MITF expression in the melanoma cells was elevated in the presence of macrophages even in the presence of MEKi (Supplementary Figure S5C). Most importantly however, when a TNFα-blocking antibody was added during drug treatment, the protective function of macrophages towards MEKi-induced cell death was lost (Figure 4E).
BRAF and MEK inhibitor treatment increases the number of tumour-associated macrophages in vivo
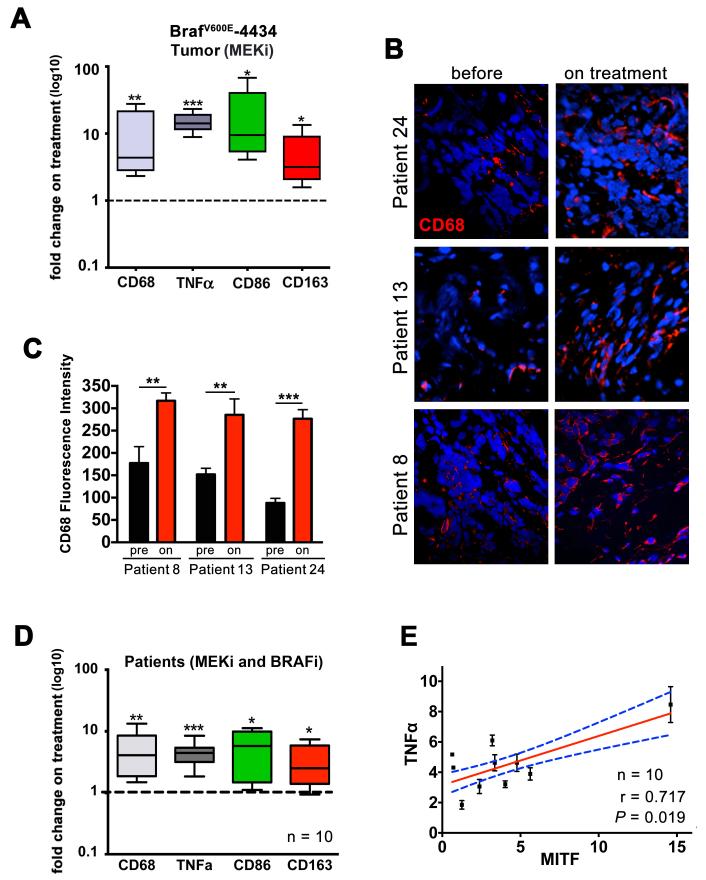
We next wanted to assess the effect of MEKi treatment on the presence of macrophages in the tumor environment in vivo. Histological sections of BRafV600E-4434 melanoma allografts from MEKi treated immunocompetent mice, showed increased staining for the pan macrophage marker CD68 when compared to tumors from vehicle treated mice (see Figure 7C). This increase in CD68 expression was confirmed at mRNA level (Figure 5A, Supplementary Figure S6A), indicating that MEKi treatment enhances macrophage accumulation within the tumor microenvironment. We also found significantly increased expression of the monocyte and macrophage marker F4/80 (not shown), the M1 macrophage marker CD86, the M2 marker CD163 and TNFα in tumors from MEKi treated over vehicle treated mice (Figure 5A, Supplementary Figure S6A).
Figure 7. IKKi and MEKi treatment synergizes in vivo and suppresses TNFα and MITF expression.
A. Growth of BRafV600E-4434 melanomas in C57J/B6 mice treated with 25 mg/kg/day PD184352 and 40 mg/kg/day BMS-345541 either alone or in combination. B. Real time qPCR analysis of cDNA isolated from BRafV600E-4434 tumors from PD184352 and BMS-345541 treated mice showing the fold change in expression from control mice. C. Immunofluorescence staining for CD68 in BRafV600E-4434 allografts from mice treated for 3 weeks as described in A. D. Relative immunofluorescence intensity of positive CD68 cells present in tissue samples. E. Quantitative real time PCR analysis of TNFα expression in differentiated macrophages either untreated or treated with 0.5μM BMS-345541 for 48hrs. F. Real time qPCR analysis of MITF in BRafV600E-4434 allografts (n=5) treated as indicated showing the fold change in expression from control mice (n=5). G. Model of MITF regulation through macrophage-derived TNFα at various treatment conditions.
Figure 5. Tumour-associated macrophage numbers increase during BRAFi and MEKi treatment.
A. Quantitative real time PCR analysis of cDNA isolated from PD184352 treated BRafV600E-4434 allografts showing the fold change in expression from control mice (n=7). B. CD68 histology of a melanoma sample from patients undergoing treatment with BRAF inhibitor or BRAF/MEK inhibitor combination. C. Mean CD68 immunofluorescence intensity of the tumor samples shown in B. (n = 8 fields for each tumor). D. Real-time qPCR analysis of CD68, TNFα, CD89 and CD163 expression in patients undergoing treatment with BRAF inhibitor or BRAF/MEK inhibitor combination (n=10). Data indicate the mean±s.e.m. E. Correlation of fold change in MITF and TNFα mRNA expression in patients undergoing treatment with BRAF inhibitor alone or BRAF and MEK inhibitor (n=10). Pearson correlation, r = 0.717, P = 0.019.
In order to validate the relevance of these findings for targeted therapy in melanoma, we examined paired BRAFV600E positive tumor biopsies from 11 patients prior to treatment and after 10-14 days of treatment with either BRAF inhibitor alone or a BRAF/MEK inhibitor combination (detailed patient data, see Table S1).
We found a significant increase in the macrophage marker CD68 (Figure 5B, C and D), as well as the M1 marker CD86 and the M2 marker CD163 in all patients in response to the treatment with BRAF and MEK inhibitors (Figure 5D, Supplementary Figure S6B), indicating an accumulation of M1 and M2 polarized macrophages. Moreover, TNFα expression was upregulated in response to treatment (Figure 5D, Supplementary Figure S6B), and the increase in TNFα expression significantly correlated with enhanced MITF expression in the tumors (Pearson correlation: r = 0.717, P = 0.019, Figure 5E), supporting the idea that that TNFα contributes to MITF expression in these patients during drug treatment. Importantly, there was no difference between patients on BRAFi mono-compared to patients on BRAFi/MEKi combination therapy (Supplementary Figure S6C), suggesting that inhibition of MEK does not alter the effect of BRAF inhibition on macrophage accumulation in vivo.
IKK and MEK inhibition synergise in vitro
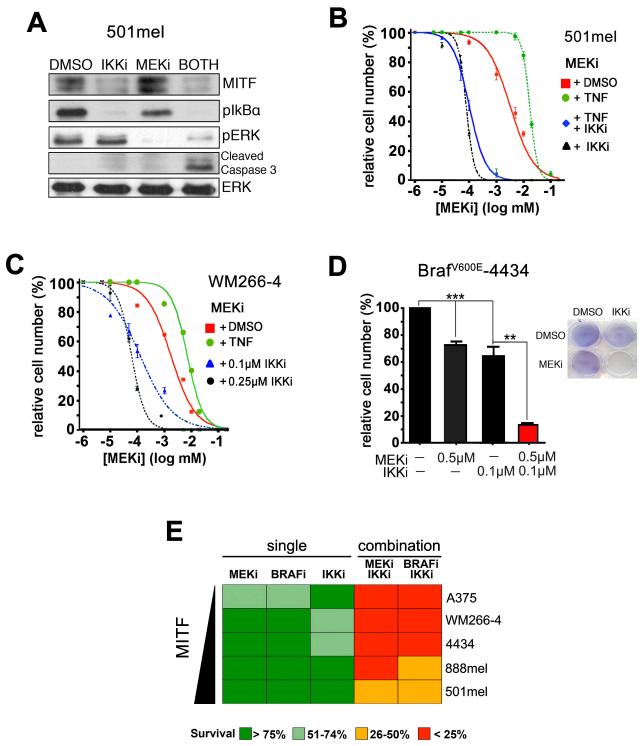
Our findings suggest that inhibition of IKK represents a possible strategy to overcome the TNFα/MITF mediated survival signals that protect melanoma cells from MEK inhibition. We therefore assessed whether IKKi mediated reduction in MITF expression can synergize with MEKi to induce cell death in melanoma cells. Indeed, at conditions where neither IKKi nor MEKi treatment alone was able to elicit apoptosis in 501mel cells, the combination of both inhibitors was able to induce cleavage of caspase3 (Figure 6A). In line with this, IKK inhibition using BMS345541 in the presence of MEKi reduced the EC50 ~26 fold (from 3.45uM to 132nM) (Figure 6B). Furthermore, as expected, TNFα protected 501mel melanoma cells from MEKi induced cell death, but IKK inhibition was able to counteract the protective effect of TNFα by reducing the EC50 ~51 fold (from 11.2μM to 214nM) (Figure 6B). Similar results were found with another cell line, WM266-4, where the combined treatment led to a dose dependent reduction in the EC50 of ~14 fold at 0.1μM and ~24 fold at 0.25μM IKKi, respectively (Figure 6C).
Figure 6. Inhibition of IKK and MEK synergize in vitro.
A. Western blot of 501mel human melanoma cells treated with 1μM AZD6244 (MEKi) and 0.5μM BMS-345541 (IKKi) either alone or in combination for 48hrs for MITF, pIKBα, cleaved caspase-3, pERK and ERK2. B. Drug dose response analysis of 501mel human melanoma cell survival in response to MEKi AZD6244, in combination of either 50ng TNFα or 0.1μM BMS-345541 for 72hrs. C. Drug dose response analysis of WM266-4 melanoma cell survival in response to MEKi PD184352, in combination of either 50ng TNFα or 0.1μM or 0.25μM BMS-345541 for 72hrs. D. BRafV600E-4434 melanoma cells were treated with 0.5μM AZD6244 (MEKi) and 0.1μM BMS-345541 (IKKi) either alone or in combination for 48hrs. E. Summary of drug treatments in the indicated cell lines.
The potentiating effect of IKK inhibition on the efficacy of the MEKi was further confirmed in other melanoma cell lines including BRafV600E-4434 melanoma cells (Figure 6D, Supplementary Figure S7A-C). Moreover, when we analyzed the effect of IKK inhibition on the efficacy of the BRAFi vemurafenib, we found that IKK inhibition significantly synergized with vemurafenib in cell killing (Figure 6E, Supplementary Figure S8A-B). According to MITF’s protective function, there was a trend of a more efficient response to the inhibitors when MITF expression was lower (Figure 6E, Supplementary Figure S8C).
IKK inhibition suppresses TNFα production in macrophages and enhances the efficacy of MEK inhibitors in vivo
To test whether our findings regarding the combination of MEKi and IKKi in vitro apply to the in vivo situation, we treated BRafV600E-4434 allograft-bearing immunocompetent mice with MEKi either alone or in combination with the IKKi BMS345541. Tumors from mice treated with the inhibitor combination showed significantly reduced growth when compared to either the single MEKi or IKKi treatment (Figure 7A), which is in agreement with the effects of the MEKi/IKKi combination on 4434 cells in vitro (see Figure 6D) and clearly demonstrates that inhibition of IKK sensitizes melanoma cells to MEK inhibition also in vivo.
To assess the consequences of the MEKi/IKKi combination treatment on the tumor microenvironment, we analyzed the tumors for macrophage markers. This analysis revealed a reduction in the expression of not only the pan macrophage marker CD68, but also the M1 and M2 macrophage markers CD86 and CD163, in the tumors from mice treated with the MEKi/IKKi combination when compared to the MEKi and IKKi only treatment (compare Figure 7B to Figure 5A and Supplementary Figure S9). This finding suggested that the MEKi induced effect on macrophage numbers is inhibited by the IKKi, which was confirmed when we assessed the presence of CD68 positive cells within the tumors (Figure 7C,D).
Most strikingly, the expression of TNFα in the tumors was reduced below basal level (=1) when the IKKi was present (Figure 7B), suggesting that in addition to decreasing macrophage numbers, IKK inhibition directly impacts on TNFα expression in the microenvironment, most probably the macrophages. Indeed, when we analyzed the effect of IKK inhibition on TNFα expression in isolated macrophages in vitro, we observed a strong suppression in response to the inhibitor (Figure 7E). Finally, in correlation with the severely reduced TNFα expression in the MEKi/IKKi treated tumors (Figure 7B), MITF mRNA levels had also dropped (Figure 7F). Thus, inhibition of IKK signaling suppresses not only stromal derived TNFα levels, but also MITF expression in melanoma cells, which creates an advantageous environment to increase the efficacy of MEK inhibitor activity (Figure 7G).
Discussion
Targeting the MAPK pathway has become a powerful therapy approach in melanoma. Nevertheless, the inevitable development of resistance demands for further improvement, which could come from combination therapies that tackle the mechanisms contributing to resistance. Furthermore, the combination of targeted approaches with recently developed immune therapies is considered an attractive novel strategy. However, initial attempts indicate that we are yet to completely understand the interplay of the immune-microenvironment and targeted therapy in melanoma (23). The full impact of targeted therapy on the immune response is not clear, which challenges the ability to predict how interfering with both simultaneously will affect the overall treatment outcome.
We found that MAPK pathway inhibition directly impacts on the tumor immune-microenvironment by increasing the number of macrophages, and that this can create a source for resistance to BRAF and MEK inhibitors. We identify TNFα as a potentially crucial factor in this resistance due to its ability to enhance the expression of the melanoma survival factor MITF (Figure 7G).
Although originally identified as an anti-tumorigenic factor, TNFα and its downstream effectors IKK and NF-κB are now well-accepted players in inflammation driven tumorigenesis (18, 24). As such, in mice TNFα is required for skin or liver carcinogenesis (25, 26), and IKK activity is essential for colitis-associated cancer (27). Moreover the depletion of the Ikk subunit Ikkβ protects from oncogenic H-Ras induced melanoma development in mice (28, 29). We now demonstrate a clear dependence of BRafV600E-driven melanoma growth on TNFα in vivo, and we show that MITF contributes to survival signaling downstream of TNFα in both mouse and human BRAF mutant melanoma cells.
Downstream of TNFα, IKK activity is required for the expression of MITF and its target genes CDK2, CDK4 or BCL2. This regulation appears to occur also in vivo, because reduced expression of these target genes is seen in Ras-transformed melanocytes of mice with conditional deletion of Ikkβ (29). Thus although NF-κB can regulate many important cell cycle and survival genes directly, in melanoma MITF appears to contribute to this regulation, thereby acting downstream of TNFα.
It is clear that IKKβ and NFKB are activated in cancer cells, and in melanoma enhanced NF-κB signaling has been correlated with progression (16). The source of TNFα to stimulate this signaling can be the cancer cells themselves, leading to autocrine TNFα signaling (18). Approximately 50% of the melanoma cells we analyzed displayed increased TNFα expression compared to melanocytes and this might contribute to enhanced basal NF-κB activation in these cells. On the other hand, paracrine signaling derived from the micro-environment clearly plays also an important role in IKK/NF-κB activation in cancer cells, and TNFα produced by myeloid cells, particularly macrophages can promote tumor growth in vivo and stimulate tumor cell invasion in vitro (26, 30, 31). We found that TNFα produced by myeloid cells was crucial for melanoma growth in vivo. While we could recapitulate the growth promoting effect of macrophages in vitro, this also revealed that TNFα acts in conjunction with other macrophage-derived factors and it is the overall balance of tumor-promoting and tumor-inhibiting factors that will produce the net effect of growth.
Our in vitro data suggest that TNFα directly acts as growth and survival factor in melanoma cells, but the reduced number of macrophages within the tumors grown in LysMcre/TNFαF/F mice indicates that TNFα also impacts on immune cell recruitment. Such a role for TNFα has been described previously (18) and reduced immune cell recruitment would result in a tumor microenvironment containing fewer tumor promoting cytokines and hence a less favorable milieu for tumor growth. Importantly we observe a significant effect on macrophage numbers when we inhibit IKKs, which as we show reduces TNFα dramatically. In line with our observations Ikkβ deletion from myeloid cells using LysMcre mice in a colitis-associated cancer model results in reduced expression in paracrine acting cytokines and reduced tumor growth (27).
We found that differentiated macrophages are able to protect melanoma cells from MEK inhibitor induced apoptosis in vitro, and that this protection is dependent on TNFα and MITF. We and others have demonstrated the relevance of MITF in resistance to MAPK pathway inhibitor treatment i.e. BRAF and MEK inhibitors in single and combination therapies (6, 8, 9). This MITF dependent increased survival is probably due to its central role regulating multiple anti-apoptotic genes such as BCL2, BCL2A1 and ML-IAP (8, 32, 33). We now show that targeting IKKs acts on the cell autonomous resistance by diminishing MITF expression in melanoma cells, thereby rendering them more sensitive to MAPK pathway inhibition. Moreover, the advantage of targeting IKKs lies in the concomitant inhibition of the external activation of the TNFα pathway stimulated by the stroma. Unfortunately, so far pre-clinical data using IKK inhibitors have not successfully been translated into the clinic due to toxicity issues (34), but our data suggest that when used in combination therapies, lower and therefore less toxic doses of IKK inhibitors could produce synergistic effects. In an approach to target TNFα directly, we had trialed a combination treatment with Enbrel™ (etanercept) and MEKi, but we did not observe any synergy (not shown). Besides scheduling and drug penetrance issues, we think that a reason for this observation could be that directly blocking TNFα action will have a broader impact, because it will inhibit all routes of signaling downstream of TNFR (including MKK7/JNK and MKK3 signaling). At the same time contrary to IKK inhibition, etanercept will not target the TNFα independent basal IKK/NF-κB activation found in melanoma cells.
An important finding of our study is that the number of macrophages within the tumor is increased in patients in response to BRAF and MEK inhibitor treatment and this is correlated with a significant increase in TNFα expression in the tumor microenvironment. However, despite this increase in cytokine production, for the development of novel strategies combining MAPK pathway targeted therapy with adoptive immunotherapy it will be crucial to fully understand the impact of BRAF and MEK inhibition on cytokine function. Interestingly, an increase in serum TNFα in patients during MAPK pathway inhibitor treatment has also been described in a study that showed that overall immunity is not perturbed during treatment (35). Furthermore, T-cell infiltration and clonality is enhanced in patients on MAPK pathway targeted therapy and BRAF inhibition enhances adoptive T-cell transfer therapy in mice (36-38). Although, in contrast to BRAF inhibition, MEK inhibition can affect viability and function of dendritic cells in vitro (39, 40), in patients T-cell recruitment and clonality is still increased in the presence of MEK inhibitor (36, 41-43). The exact impact of combined BRAF and MEK inhibition on the activity of the individual immune cell populations within the tumor remains to be investigated, but we find that in vitro macrophages protect melanoma cells in the presence of MEK inhibitor, and the inhibitor does not impact on the expression of TNFα or the ability of macrophages to stimulate MITF expression in melanoma cells (Supplementary Figure S5). Moreover the majority of patients in our study who displayed increased macrophage numbers had been on BRAFi/MEKi combination therapy.
Our finding of possible immune promoted resistance to MAPK pathway inhibitors has important implications for clinical strategies, because it means that we have to consider all components of the immune microenvironment in combination therapies. Targeting myeloid cell infiltration appears to be an attractive option, and indeed the CSF-1R inhibitor PLX3397 has been shown to reduce myeloid cell infiltration and enhance adoptive cell transfer immunotherapy in BrafV600E driven melanomagenesis in mice (44). Not surprisingly, a clinical trial combining PLX3397 with vemurafenib in melanoma has recently been initiated. In summary, our data suggest that using drug combinations that impact on both the tumor cells and tumor-microenvironment derived survival signals will increase the responsiveness to MAPK pathway inhibitors in melanoma and may have a greater chance of creating more durable responses.
Methods
Cell culture and survival assays
A375, WM266-4, SKMel28 and SKMel2 cells were bought from the American Type Culture Collection and 501mel and 888mel cells were a gift from Steve Rosenberg (NCI, MD); all were obtained in 2008. Additional cell lines in the panel used for RNA extraction were a gift from Adam Hurlstone (University of Manchester). All cell lines were authenticated in house by short tandem repeat profiling before and during the study; the last authentication was carried out in 2014. These cell lines were grown in DMEM/10% FCS (PAA, Yeovil, UK). 4434 melanoma cells were isolated from a BRafV600E mouse (45) and were grown in RPMI/10%FCS. THP1 cells were a gift from Adam Hurlstone (University of Manchester) and grown in RPMI/10% FCS (PAA, Yeovil, UK). Cell survival was measured as the optical density at 540nm (OD540) of solubilized toluidine blue from formalin fixed cells. Anoikis assays were performed by culturing 10,000 cells in non-adhesive plates in DMEM/2% FSC for 48hrs. Viable cells were assessed by trypan blue exclusion.
Inhibitors and cytokines
PD184352 was from Axon Medchem (Groningen, The Netherlands), selumetinib (AZD6244) from Selleck Chemicals (Newmarket, UK), BMS-345541 and SC-514 from Sigma, UK. Human recombinant TNFα, IL4 and IL13 as well as mouse recombinant TNFα were from PreproTech, London, UK.
In vivo melanoma models
All procedures involving animals were approved by the Animal Ethics Committees of the Institute of Cancer Research and The Cancer Research UK Manchester Institute in accordance with National Home Office regulations under the Animals (Scientific Procedures) Act 1986 and according to the guidelines of the Committee of the National Cancer Research Institute. C57J/B6 mice were purchased from Charles River, (Margate, UK), LysMcre mice (B6.129P2-Lyz2tm1(cre)Ifo/J) were from Jackson Laboratories. TNFα−/−, LysMcre and TNFαF/F have been described previously (46-48). For long-term survival tests BrafV600E;Tyr::CreERT2 and BrafV600E;Tnfα−/−;Tyr::CreERT2 mice were treated with tamoxifen as described previously (17). Controls were either Ethanol-treated BrafV600E;Tnfα−/−;Tyr::CreERT2 mice or tamoxifen-treated Tyr::CreERT2 mice. For allografts 5 × 106 4434 melanoma cells were injected subcutaneously into the flank of immunocompetent mice and tumor growth was monitored. For drug treatments, the tumors were allowed to establish and mice were dosed daily by oral gavage with vehicle (5% DMSO), PD184352 (25 mg/kg/day), BMS-345541 (40 mg/kg/day) or PD184352 (25 mg/kg/day) plus BMS-345541 (40 mg/kg/day)). Tumor size was determined by caliper measurements of tumor length, width, and depth, and volume was calculated as volume = 0.5236 × length × width × depth (mm).
Patient samples
Patients with mutant V600BRAF positive metastatic melanoma were treated with either a BRAF inhibitor (BRAFi), or a combination of BRAF and MEK inhibitors (BRAFi + MEKi) (for patient characteristics see Table S1). All patients were consented for tissue acquisition per an IRB-approved protocol. Tumor biopsies were obtained before treatment (day 0), at 10-14 days on treatment, and/or at time of progression if applicable. Patient cDNA samples were pre-amplified using the PreAmp Master Mix Kit (Applied Biosystems, Carlsbad, CA) according to the manufacturer’s instructions. Real time qPCR conditions and primer sequences are described in Supplementary data.
Histology
Cryo-sections of mouse or human tumors were permeabilized in a solution of 0.1% Trition and 1% saponin in PBS for 15mins. Sections were blocked in 10% BSA at 37°C for 30mins and incubated overnight at 4°C with primary CD68 antibody in 10% BSA PBS. The anti-mouse CD68 antibody (FA-11) was from Abcam Cambridge, UK. The anti-human CD68 monoclonal antibody (KP1) was from DAKO Cambridge, UK. Stained sections were washed in PBS and then incubated with secondary antibody for 2hrs at RT and mounted using vectashield.
Cell lysis and antibodies
Cells were lysed in SDS sample buffer and analysed by standard Western-blotting protocols. The antibodies used were as follows: MITF clone C5 from Neomarkers, Lab Vision, Runcorn, UK; CDK2 (D-12), CDK4 (H-22) and ERK2 (C-14) from Santa-Cruz Biotechnolgy, Santa Cruz, CA, USA. Antibodies against p65, cleaved caspase3 and pIκBα were from Cell Signaling, Boston, MA and against BCL2 and p27 from BD Biosciences, Oxford, UK. Anti-phospho-ERK was from Sigma, UK.
RNA isolation and qPCR analysis
RNA from cell lines was isolated with TRIZOL® and selected genes were amplified by quantitative real time PCR using SYBR green (Qiagen, Valencia, CA, USA). RNA was similar isolated from frozen sections of mouse tumor left in TRIZOL® for 2hrs.
Primers used for qPCR analysis
Primers used in the qPCR gene expression analyses were for human sequences: MITF: CCGTCTCTCACTGGATTGGT and TACTTGGTGGGGTTTTCGAG; GAPDH: CAATGACCCCTTCATTGACC and GACAAGCTTCCCGTTCTCAG; βACTIN: GCAAGCAGGAGTATGACGAG and CAAATAAAGCCATGCCAATC; primers for mouse sequences were: CD68: GCTACATGGCGGTGGAGTACAA and ATGATGAGAGGCAGCAAGATGG; CD86: TGCTCATCTATACACGGTTAC and TTTCTTGGTCTGTTCACTCTC; CD163: ACATAGATCATGCATCTGTCATTTG and CATTCTCCTTGGAATCTCACTTCTA; TNFα: GACGTGGAAGTGGCAGAAGAG and TGCCACAAGCAGGAATGAGA; GAPDH: TCTCCCTCACAATTTCCATCCCAG and GGGTGCAGCGAACTTTATTGATGG. Qiagen QuantiTect primers were used for TNFα:QT0002916; IL1β: QT00021385 and YM1: QT00068446;
TNFα ELISA
Conditioned medium was collected from macrophages and analyzed using a TNFα ELISA kit from PreproTech (London, UK) according to manufactures instructions. The TNFα blocking antibody (Ab6671, Abcam) was used at a concentration of 5ng/ml.
RNAi
Small interfering RNAs (siRNAs) were transfected using INTERFERin siRNA-transfection reagent (Polyplus, Illkirch, France) according to the manufacturer’s instructions. MITF target sequences are: MI#1: GAACGAAGAAGAAGAUUUAUU, MI#2: AAAGCAGUACCUUUCUACCAC, MI#3: GACCUAACCUGUACAACAAUU.
Colony formation assay
1 × 105 melanoma cells were plated into 10cm dishes and allowed to adhere o/n. The next day the medium was replaced with control medium or medium derived from M1 or M2 polarised macrophages containing no or a TNFα blocking antibody (5ng/ml). The medium was exchanged every 48h for 3 weeks after which cells were fixed, stained and quantified.
Gene expression analysis
Publically available ONCOMINE datasets used in this paper were the Haqq Melanoma dataset (21) containing 37 samples: 3 skin, 8 non-neoplastic nevi and 25 melanomas (6 primary, 19 metastases), and the Riker Melanoma dataset (Accession:GSE7553) (22) containing 72 samples: 4 skin samples, 1 normal epidermal melanocyte culture, 2 melanoma in situ, 14 primary and 40 metastatic melanomas as deposited in Oncomine. The datasets were analysed in Oncomine and the results were exported and further analysed using GraphPad Prism. Alternatively heat-maps were exported as publication-quality graphic (SVG).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation assays using control IgG (Santa Cruz) or antibodies specific for p65 (Ab7970 Abcam) were performed as described previously (49). Primers for the M-MITF promotor were: ACTGTCTGTGTTGTCAGGCA and ACATTCCCTTGGAGATAGCCT; for the negative control (MITF coding region):ACCACATACAGCAAGCCCAA and TCCCTCTTTTTCACAGTTGGAGT; for the positive control (GROα): CGTCGCCTTCCTTCCGGACTCG and GCTCTCCGAGATCCGCGAACCC.
Luciferase reporter assay
Cells were transfected with plasmid DNA using Attractene (Qiagen, Valencia, CA) and analyzed for luciferase activity 24hr after treatment with forskolin or TNFα using a RLB based luciferase assay kit (Promega). Data were normalized to renilla luciferase activity. The −2.3 kb M-MITF promoter fragment (−2293bps to +120bps) and the truncated promoter (−333) cloned into pGL2 (Promega) were described previously (49). The −1.8 kb M-MITF promoter fragment was created by deleting a 5′ KpnI/AvrII fragment from the −2.3 kb construct. The NF-kB mutation (see Supplementary Figure S2B) was created by site directed mutagenesis.
Image acquisition and processing
For immunofluorescence, a Zeiss Axioskop2 plus equipped with epifluorescence was used; images were taken at room temperature by a Photometrics Cool Snap HQ CCD camera driven by Metamorph software (Universal Imaging). Image analysis was performed using ImageJ software. All Western blots were processed using Photoshop CS5.1.
THP1-macrophage differentiation and trans-well co-culture assay
THP1 cells were differentiated in trans-well inserts (BD Biosciences, Oxford, UK). To differentiate THP1 cells into M2 activated macrophages they were treated with 10ng/ml TPA for 24h and subsequently with 20ng/ml IL4 and 20ng/ml IL13 for 48h. Alternatively TPA treated THP1 cells were stimulated with 15ng/ml LPS to differentiate them into activated M1 macrophages. After differentiation, the inserts were washed in RPMI three times before being placed in wells with pre-plated melanoma cells. Experiments using drugs and/or blocking antibodies were performed for 48h by adding the respective reagents to the wells, so that both cell populations were exposed to the same conditions.
Peripheral blood monocytes isolation and differentiation
Peripheral blood mononuclear cells (PBMCs) were isolated from leucocyte cones obtained from healthy donors (provided by NHS Blood and Transplant, Manchester, UK) by density gradient centrifugation using Ficoll Paque Plus (GE healthcare) for 50 min at 400g. PBMCs were transferred to flasks in serum-free RPMI 1640 Glutamax media (Life Technologies) to allow enrichment for monocytes (PBMs) by adherence to the tissue culture plastic for 1h at 37°C. Following thorough washing, adhered monocytes were incubated for 6 days in RPMI/10%FCS and 1% penicillin/streptomycin solution (Sigma) supplemented with 100ng/ml human M-CSF (Peprotech) to stimulate macrophage differentiation. Macrophages were washed and primed by incubating with RPMI media supplemented with 100ng/ml IFN-γ (Peprotech) or 100ng/ml IL4 and IL13 (Peprotech) for 24h to drive M1 or M2 polarization respectively. Unprimed macrophages were incubated with non-supplemented RPMI media. By adding 20ng/ml LPS to media containing priming stimuli for a further 24h, M1 and M2 macrophages were activated. Cells were thoroughly washed in PBS before incubating for a further 24h in non-supplemented RPMI media to produce conditioned media to be used in subsequent in vitro assays.
Statistical Analysis
If not indicated otherwise, data represent the results for assays performed in triplicate, with error bars to represent standard deviations or errors from the mean. Statistics used were: predominately Student t-test and One-way ANOVA with Tukeys’s post hoc test performed using GraphPad Prism version 4.00 for Mac OS, GraphPad Software, San Diego California USA, www.graphpad.com. Pearson correlation was used to analyze associated gene expression.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
This study identifies the immune-microenvironment as a source of resistance to MAPK pathway inhibitors through macrophage-derived TNFα, and reveals that in patients on treatment this source becomes reinforced. Inhibiting IκB-kinase enhances the efficacy of MAPK pathway inhibitors, which identifies this approach as a potential novel strategy to improve targeted therapy in melanoma.
Acknowledgements
We thank Adam Hurlstone for helping with the THP1 system, and Imanol Arozarena for providing melanoma cell lines.
Financial Support: This work was supported through funding of CW by Cancer Research UK (C11591/A16416), a Wellcome Trust Institutional Strategic Support Fund (ISSF) award [097820/Z/11/B] to the University of Manchester and a NCI/NIH U54CA163125 grant to JAW and KTF.
Footnotes
Potential conflict of interest: JAW received honoraria from DAVA Oncology
References
- 1.Wellbrock C, Hurlstone A. BRAF as therapeutic target in melanoma. Biochem Pharmacol. 2010 doi: 10.1016/j.bcp.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 2.Belden S, Flaherty KT. MEK and RAF inhibitors for BRAF-mutated cancers. Expert Rev Mol Med. 2012;14:e17. doi: 10.1017/erm.2012.11. [DOI] [PubMed] [Google Scholar]
- 3.Lito P, Rosen N, Solit DB. Tumor adaptation and resistance to RAF inhibitors. Nature medicine. 2013;19:1401–9. doi: 10.1038/nm.3392. [DOI] [PubMed] [Google Scholar]
- 4.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. The New England journal of medicine. 2012;367:1694–703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greger JG, Eastman SD, Zhang V, Bleam MR, Hughes AM, Smitheman KN, et al. Combinations of BRAF, MEK, and PI3K/mTOR inhibitors overcome acquired resistance to the BRAF inhibitor GSK2118436 dabrafenib, mediated by NRAS or MEK mutations. Mol Cancer Ther. 2012;11:909–20. doi: 10.1158/1535-7163.MCT-11-0989. [DOI] [PubMed] [Google Scholar]
- 6.Smith MP, Ferguson J, Arozarena I, Hayward R, Marais R, Chapman A, et al. Effect of SMURF2 targeting on susceptibility to MEK inhibitors in melanoma. J Natl Cancer Inst. 2013;105:33–46. doi: 10.1093/jnci/djs471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med. 2006;12:406–14. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 8.Haq R, Yokoyama S, Hawryluk EB, Jonsson GB, Frederick DT, McHenry K, et al. BCL2A1 is a lineage-specific antiapoptotic melanoma oncogene that confers resistance to BRAF inhibition. Proc Natl Acad Sci U S A. 2013;110:4321–6. doi: 10.1073/pnas.1205575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johannessen CM, Johnson LA, Piccioni F, Townes A, Frederick DT, Donahue MK, et al. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature. 2013 doi: 10.1038/nature12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–4. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray-Schopfer VC, Karasarides M, Hayward R, Marais R. Tumor necrosis factor-alpha blocks apoptosis in melanoma cells when BRAF signaling is inhibited. Cancer Res. 2007;67:122–9. doi: 10.1158/0008-5472.CAN-06-1880. [DOI] [PubMed] [Google Scholar]
- 12.Katerinaki E, Evans GS, Lorigan PC, MacNeil S. TNF-alpha increases human melanoma cell invasion and migration in vitro: the role of proteolytic enzymes. Br J Cancer. 2003;89:1123–9. doi: 10.1038/sj.bjc.6601257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torisu H, Ono M, Kiryu H, Furue M, Ohmoto Y, Nakayama J, et al. Macrophage infiltration correlates with tumor stage and angiogenesis in human malignant melanoma: possible involvement of TNFalpha and IL-1alpha. Int J Cancer. 2000;85:182–8. [PubMed] [Google Scholar]
- 14.Kashani-Sabet M, Shaikh L, Miller JR, 3rd, Nosrati M, Ferreira CM, Debs RJ, et al. NF-kappa B in the vascular progression of melanoma. J Clin Oncol. 2004;22:617–23. doi: 10.1200/JCO.2004.06.047. [DOI] [PubMed] [Google Scholar]
- 15.McNulty SE, del Rosario R, Cen D, Meyskens FL, Jr., Yang S. Comparative expression of NFkappaB proteins in melanocytes of normal skin vs. benign intradermal naevus and human metastatic melanoma biopsies. Pigment Cell Res. 2004;17:173–80. doi: 10.1111/j.1600-0749.2004.00128.x. [DOI] [PubMed] [Google Scholar]
- 16.Ueda Y, Richmond A. NF-kappaB activation in melanoma. Pigment Cell Res. 2006;19:112–24. doi: 10.1111/j.1600-0749.2006.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dhomen N, Reis-Filho JS, da Rocha Dias S, Hayward R, Savage K, Delmas V, et al. Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer Cell. 2009;15:294–303. doi: 10.1016/j.ccr.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 18.Balkwill F. Tumour necrosis factor and cancer. Nat Rev Cancer. 2009;9:361–71. doi: 10.1038/nrc2628. [DOI] [PubMed] [Google Scholar]
- 19.Liu G, Yang H. Modulation of macrophage activation and programming in immunity. J Cell Physiol. 2013;228:502–12. doi: 10.1002/jcp.24157. [DOI] [PubMed] [Google Scholar]
- 20.Zaidi MR, Davis S, Noonan FP, Graff-Cherry C, Hawley TS, Walker RL, et al. Interferon-gamma links ultraviolet radiation to melanomagenesis in mice. Nature. 2011;469:548–53. doi: 10.1038/nature09666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haqq C, Nosrati M, Sudilovsky D, Crothers J, Khodabakhsh D, Pulliam BL, et al. The gene expression signatures of melanoma progression. Proc Natl Acad Sci U S A. 2005;102:6092–7. doi: 10.1073/pnas.0501564102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riker AI, Enkemann SA, Fodstad O, Liu S, Ren S, Morris C, et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med Genomics. 2008;1:13. doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ribas A, Hodi FS, Callahan M, Konto C, Wolchok J. Hepatotoxicity with combination of vemurafenib and ipilimumab. The New England journal of medicine. 2013;368:1365–6. doi: 10.1056/NEJMc1302338. [DOI] [PubMed] [Google Scholar]
- 24.Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5:749–59. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 25.Moore RJ, Owens DM, Stamp G, Arnott C, Burke F, East N, et al. Mice deficient in tumor necrosis factor-alpha are resistant to skin carcinogenesis. Nature medicine. 1999;5:828–31. doi: 10.1038/10552. [DOI] [PubMed] [Google Scholar]
- 26.Pikarsky E, Porat RM, Stein I, Abramovitch R, Amit S, Kasem S, et al. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431:461–6. doi: 10.1038/nature02924. [DOI] [PubMed] [Google Scholar]
- 27.Greten FR, Eckmann L, Greten TF, Park JM, Li ZW, Egan LJ, et al. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–96. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 28.Yang J, Pan WH, Clawson GA, Richmond A. Systemic targeting inhibitor of kappaB kinase inhibits melanoma tumor growth. Cancer Res. 2007;67:3127–34. doi: 10.1158/0008-5472.CAN-06-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J, Splittgerber R, Yull FE, Kantrow S, Ayers GD, Karin M, et al. Conditional ablation of Ikkb inhibits melanoma tumor development in mice. The Journal of clinical investigation. 2010;120:2563–74. doi: 10.1172/JCI42358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hagemann T, Wilson J, Kulbe H, Li NF, Leinster DA, Charles K, et al. Macrophages induce invasiveness of epithelial cancer cells via NF-kappa B and JNK. J Immunol. 2005;175:1197–205. doi: 10.4049/jimmunol.175.2.1197. [DOI] [PubMed] [Google Scholar]
- 31.Oguma K, Oshima H, Aoki M, Uchio R, Naka K, Nakamura S, et al. Activated macrophages promote Wnt signalling through tumour necrosis factor-alpha in gastric tumour cells. EMBO J. 2008;27:1671–81. doi: 10.1038/emboj.2008.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dynek JN, Chan SM, Liu J, Zha J, Fairbrother WJ, Vucic D. Microphthalmia-associated transcription factor is a critical transcriptional regulator of melanoma inhibitor of apoptosis in melanomas. Cancer Res. 2008;68:3124–32. doi: 10.1158/0008-5472.CAN-07-6622. [DOI] [PubMed] [Google Scholar]
- 33.McGill GG, Horstmann M, Widlund HR, Du J, Motyckova G, Nishimura EK, et al. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell. 2002;109:707–18. doi: 10.1016/s0092-8674(02)00762-6. [DOI] [PubMed] [Google Scholar]
- 34.Liu F, Xia Y, Parker AS, Verma IM. IKK biology. Immunol Rev. 2012;246:239–53. doi: 10.1111/j.1600-065X.2012.01107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hong DS, Vence L, Falchook G, Radvanyi LG, Liu C, Goodman V, et al. BRAF(V600) inhibitor GSK2118436 targeted inhibition of mutant BRAF in cancer patients does not impair overall immune competency. Clinical cancer research. 2012;18:2326–35. doi: 10.1158/1078-0432.CCR-11-2515. [DOI] [PubMed] [Google Scholar]
- 36.Frederick DT, Piris A, Cogdill AP, Cooper ZA, Lezcano C, Ferrone CR, et al. BRAF inhibition is associated with enhanced melanoma antigen expression and a more favorable tumor microenvironment in patients with metastatic melanoma. Clinical cancer research. 2013;19:1225–31. doi: 10.1158/1078-0432.CCR-12-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koya RC, Mok S, Otte N, Blacketor KJ, Comin-Anduix B, Tumeh PC, et al. BRAF inhibitor vemurafenib improves the antitumor activity of adoptive cell immunotherapy. Cancer Res. 2012;72:3928–37. doi: 10.1158/0008-5472.CAN-11-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu C, Peng W, Xu C, Lou Y, Zhang M, Wargo JA, et al. BRAF inhibition increases tumor infiltration by T cells and enhances the antitumor activity of adoptive immunotherapy in mice. Clinical cancer research. 2013;19:393–403. doi: 10.1158/1078-0432.CCR-12-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boni A, Cogdill AP, Dang P, Udayakumar D, Njauw CN, Sloss CM, et al. Selective BRAFV600E inhibition enhances T-cell recognition of melanoma without affecting lymphocyte function. Cancer Res. 2010;70:5213–9. doi: 10.1158/0008-5472.CAN-10-0118. [DOI] [PubMed] [Google Scholar]
- 40.Ott PA, Henry T, Baranda SJ, Frleta D, Manches O, Bogunovic D, et al. Inhibition of both BRAF and MEK in BRAF(V600E) mutant melanoma restores compromised dendritic cell (DC) function while having differential direct effects on DC properties. Cancer Immunol Immunother. 2013;62:811–22. doi: 10.1007/s00262-012-1389-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cooper ZA, Frederick DT, Juneja VR, Sullivan RJ, Lawrence DP, Piris A, et al. BRAF inhibition is associated with increased clonality in tumor-infiltrating lymphocytes. Oncoimmunology. 2013;2:e26615. doi: 10.4161/onci.26615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knight DA, Ngiow SF, Li M, Parmenter T, Mok S, Cass A, et al. Host immunity contributes to the anti-melanoma activity of BRAF inhibitors. The Journal of clinical investigation. 2013;123:1371–81. doi: 10.1172/JCI66236. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Wilmott JS, Long GV, Howle JR, Haydu LE, Sharma RN, Thompson JF, et al. Selective BRAF inhibitors induce marked T-cell infiltration into human metastatic melanoma. Clinical cancer research. 2012;18:1386–94. doi: 10.1158/1078-0432.CCR-11-2479. [DOI] [PubMed] [Google Scholar]
- 44.Mok S, Koya RC, Tsui C, Xu J, Robert L, Wu L, et al. Inhibition of CSF1 Receptor Improves the Anti-tumor Efficacy of Adoptive Cell Transfer Immunotherapy. Cancer Res. 2013 doi: 10.1158/0008-5472.CAN-13-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dhomen N, Marais R. BRAF signaling and targeted therapies in melanoma. Hematol Oncol Clin North Am. 2009;23:529–45. doi: 10.1016/j.hoc.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 46.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–77. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 47.Grivennikov SI, Tumanov AV, Liepinsh DJ, Kruglov AA, Marakusha BI, Shakhov AN, et al. Distinct and nonredundant in vivo functions of TNF produced by t cells and macrophages/neutrophils: protective and deleterious effects. Immunity. 2005;22:93–104. doi: 10.1016/j.immuni.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 48.Marino MW, Dunn A, Grail D, Inglese M, Noguchi Y, Richards E, et al. Characterization of tumor necrosis factor-deficient mice. Proc Natl Acad Sci U S A. 1997;94:8093–8. doi: 10.1073/pnas.94.15.8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wellbrock C, Rana S, Paterson H, Pickersgill H, Brummelkamp T, Marais R. Oncogenic BRAF regulates melanoma proliferation through the lineage specific factor MITF. PLoS One. 2008;3:e2734. doi: 10.1371/journal.pone.0002734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Issa R, Xie S, Lee KY, Stanbridge RD, Bhavsar P, Sukkar MB, et al. GRO-alpha regulation in airway smooth muscle by IL-1beta and TNF-alpha: role of NF-kappaB and MAP kinases. American journal of physiology Lung cellular and molecular physiology. 2006;291:L66–74. doi: 10.1152/ajplung.00384.2005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.