Abstract
BORIS/CTCFL is a member of cancer testis antigen family normally expressed in germ cells. In tumors, it is aberrantly expressed although its functions are not completely well-defined. To better understand the functions of BORIS in cancer, we selected the embryonic cancer cells as a model. Using a molecular beacon, which specifically targets BORIS mRNA, we demonstrated that BORIS positive cells are a small subpopulation of tumor cells (3–5% of total). The BORIS-positive cells isolated using BORIS-molecular beacon, expressed higher telomerase hTERT, stem cell (NANOG, OCT4, SOX2) and cancer stem cell marker genes (CD44 and ALDH1) compared to the BORIS-negative tumor cells. In order to define the functional role of BORIS, stable BORIS-depleted embryonic cancer cells were generated. BORIS silencing strongly down-regulated the expression of hTERT, stem cell and cancer stem cell marker genes. Moreover, the BORIS knockdown increased cellular senescence in embryonic cancer cells, revealing a putative role of BORIS in the senescence biological program. Our data indicate an association of BORIS expressing cells subpopulation with the expression of stemness genes, highlighting the critical role played by BORIS in embryonic neoplastic disease.
Introduction
Brother of the regulator of imprinting sites (BORIS) also designed as CTCFL, CCCTC-binding factor-like, is a DNA-binding protein with functions in cancer not fully understood. CTCF is a highly conserved, ubiquitously expressed, multifunctional chromatin factor that plays a role as a tumor suppressor gene [1]–[3]. BORIS is a mammalian paralog of CTCF, they share the same 11 zinc-finger domain but differ at N- and C- termini. Within this zinc-finger domain, BORIS and CTCF exhibit 70% of homology [4]. In normal tissues, BORIS expression is restricted to germ cells, where it is involved in epigenetic reprogramming [4], [5]. BORIS is expressed in spermatocytes during male germ line development, apparently in absence of CTCF [4]. In tumors, BORIS is aberrantly expressed and its transcription was detected at different levels in several cancer cell lines and in primary tumors [6]. Due to its restricted expression in normal germinal tissues and its re-expression in a wide variety of tumors, BORIS belongs to cancer testis antigen (CTA) family. It has been shown that BORIS induced expression of other CTA genes, as MAGE-A1, NY-ESO-1 [7], [8] and SPANX [9] but not in all tumors [10], [11]. In addition, we previously showed that BORIS activated hTERT expression by binding to the first exon of the hTERT telomerase gene in embryonic and ovarian tumor cells [12]. Furthermore, in studies of exogenous BORIS expression in normal BORIS-negative cells, we demonstrated that these transfected cells exhibited high levels of hTERT mRNA [12]. All these results revealed an important role of BORIS in the immortalization process during tumorigenesis. Interestingly, current reports show a correlation between hTERT expression and stem cell-like properties [13]–[17]. Further investigations regarding the correlation between BORIS functions and the main roles of hTERT in the immortalization and stemness properties have to be performed. Another question not yet clearly answered is how many cells, within a tumor cell line, express BORIS. In this paper, we address these questions. For this purpose, we selected to use the molecular beacon (MB) imaging technology, as it is an approved method to detect and also to visualize mRNA expression [18]. MBs are oligonucleotides structured as stem-loop hairpin with a fluorescence quencher at one end and, at the opposite end, a fluorescent dye also called fluorophore. Due to their specific structure, MBs emit fluorescence signals only when binding to their targets. To explore the frequency of BORIS-positive cells within tumor cell lines, we first designed a BORIS mRNA targeting MB, and then we analyzed BORIS expression in human embryonic and ovarian tumor cell lines, respectively NCCIT and OVCAR3. After verifying that BORIS-MB enable FACS sorting of BORIS-positive cells, we showed that the isolated BORIS-positive cells expressed higher mRNA level of hTERT and stemness genes compared to BORIS-negative and non-sorted NCCIT cells. We further confirmed this result by BORIS silencing studies. Moreover, we showed that BORIS protects from senescence process. Altogether, our data confirm a direct role of BORIS in embryonic neoplastic disease.
Materials and Methods
Cells
The human cell lines (BJ, foreskin fibroblast; HeLa, cervical adenocarcinoma; NCCIT, embryonic carcinoma; OVCAR3, ovary carcinoma) were purchased from the American Type Culture Collection (ATCC). The cells were cultured at 37°C with 5% CO2 either in Dulbecco's modified Eagle's medium (DMEM; Gibco, Invitrogen) for HeLa and BJ cells, or in RPMI-1640 medium (Gibco, Invitrogen) for NCCIT and OVCAR3, supplemented with 10% of heat inactivated fetal bovine serum (Invitrogen) and 1% of Penicillin-Streptomycin (Gibco, Invitrogen).
Molecular beacon (MB) design
Sequences of BORIS-MB1 and BORIS-MB2 were designed using Beacon Designer (Premier Biosoft). BORIS mRNA secondary structures were predicted using mFOLD software (mFOLD, http://www.bioinfo.rpi.edu/applications/mfold/) and specificity was determined by BLAST search (NCBI). The target sequence of BORIS-MB1 is located on exon 2 and that of BORIS-MB2 is located on exon 11 of BORIS mRNA. These locations were chosen since they are outside the zinc-finger domain and do not cross-hybridize with the CTCF homology regions. In addition, previous study has shown that the starting and the ending regions of mRNA are the more accessible for MBs hybridization [19]. The RANDOM-MB that was used as negative control does not match with any mammalian sequences [19]. Sequences were the following: BORIS-MB1 5′-CGCTGTCTCTGCACACTCCGTCTTCAGCG-3′; BORIS-MB2 5′-CAGCCATTCCTCTTTGACTCTGGCTG-3′ and RANDOM-MB 5′-CGACGCGACAAGCGCACCGATACGTCG-3′ (underlined bases indicating those complementary to the target sequences). A fluorophore (Cy3 or ATTO647) was 5′-conjugated and a Black Hole Quencher (BHQ-2) was linked to the 3′-end. The MBs were purchased from Sigma and they were purified by high-pressure liquid chromatography.
In vitro determination of MB specificity
Oligos were designed to be specific of the MBs targets (BORIS-MB1 specific target: 5′-AAGACGGAGTGTGCAGAGAGA-3′; BORIS-MB2 specific target: 5′-CAGCCAGAGTCAAAGAGGAA-3′ and RANDOM-MB specific target: 5′-TATCGGTGCGCTTGTCG-3′). A non-specific oligo was used for the in vitro test of the different MBs. This non-specific oligo, 5′-CGATGCCGAACCAATTCTCCAC-3′, corresponds to a transcript variant of CTCF. To test the specificity in solution, 200 nM of MB was mixed or not with 1 µM of oligo in 10 µL of Opti-MEM medium (Invitrogen).
The emission fluorescence profiles were obtained after heating the MB-target oligo mix to a progressive temperature elevation ranging from 15 to 80°C using 1°C steps. Fluorescence signal was acquired at the end of each increasing degree and detected on the Cy3 channel using a Rotor Gene 6000 Real-Time PCR system (Corbett Life Science).
MB delivery and cell fluorescence imaging
Cells were detached using 0.05% trypsin-EDTA (Invitrogen) and resuspended in serum-free DMEM medium at the concentration of 106 cells/ml. Firstly, Cy3-BORIS-MB or Cy3-RANDOM-MB (200 nM) was incubated at room temperature in presence of 1 µl/ml of Lipofectamine RNAiMAX siRNA transfection reagent (Invitrogen) using Opti-MEM medium. The Lipofectamine RNAiMAX reagent was used as delivery vehicle since in our conditions it gave less background compared to other reagents such as Streptolysin (data not shown). After 10 min, the transfection mix was added to the suspended cells and together incubated for 1 hour at 37°C. Hoechst 33342 (Invitrogen) was added at concentration of 5 µg/mL during the last 10 min of incubation. Then, cells were washed using Phosphate Buffered Saline (PBS, Invitrogen) and resuspended in PBS with 5 mM EDTA. Transfected cells were cytocentrifugated onto glass slide using a cytospin centrifuge and examined under fluorescent microscope (Axioplan2 Imaging, Zeiss). The fluorescence signal of Cy3-coniugated MB was analyzed using the red channel and Hoechst 33342 fluorescence emission was observed under blue channel.
FACS analysis and sorting using MB
For FACS analysis and cell sorting, we used MBs conjugated with ATTO 647, a dye characterized by its high photostability [20]. Cells were prepared and incubated with MBs as described above (except that Hoechst 33342 was not added) and were directly analyzed using Gallios flow cytometer (Beckman Coulter). At least 10,000 events were collected and analyzed by Kaluza Software. The BORIS-positive and BORIS-negative populations were sorted after exclusion of dead cells by Propidium Iodide (PI) staining using FACSAria I (Becton Dickinson) instrument at the Flow Cytometry Facility of UNIL (University of Lausanne, Switzerland). Ranges of 2×104–9×104 BORIS-positive cells and 2×105–9×105 BORIS-negative cells were sorted.
BORIS knockdown by inducible shRNA lentiviral system
Stable cell lines with inducible expressing shRNAs targeting human BORIS mRNA were generated using the doxycycline-inducible shRNA lentiviral system, pINDUCER [21]. The lentiviral vector pINDUCER11 constitutively expresses the eGFP fluorescent reporter protein, which enables to track cells transduced by the virus. This vector also contains a cassette with a doxycycline-inducible promoter that controls the transcription of a tRFP reporter gene together with the shRNA, which allows detection of cells with doxycycline activated shRNA transcription [21]. Four different shRNAmiR (shRNA) specifically targeting BORIS, and not its parolog CTCF (BORIS-sh1: 5′-ATTCACCAAGATCAAAGAACTC-3′, BORIS-sh2: 5′-GTTCTCACAGTTTCAAATTCAA-3′, BORIS-sh3: 5′-TTCATCCCGACTGTTTACAAAT-3′, BORIS-sh4: 5′TCCGACAGAAGCAACTTCTAAA-3′) and a control shRNA with scrambled sequence (CTR sh: 5′CAGAGCTAACTCAGATAGTACT3′) were synthetized (Sigma). They were PCR amplified and cloned into the pINDUCER11 backbone using EcoRI and XhoI restriction enzymes. The sequences of all constructs were verified by sequencing. Lentivirus were generated by co-transfection of the appropriate shRNA constructs along with the packaging vectors (pMD2G-VSVg, pCMV-dR8.74) into HEK-293T cells using FuGENE 6 reagent (Roche Diagnostics) according to the manufacturer's protocol. Viral supernatants were harvested 48 hours after transfection, filtered through a 0.45 µm pore filter, ultracentrifugated for 1.5 hours at 19,500 rpm in a Beckman SW28 rotor and resuspended in RPMI medium. The viral suspension combined with 8 µg/ml polybrene (Sigma) was used to infect target cells (NCCIT). Twenty-four hours post infection the medium was replaced and stably infected cells were eGFP-sorted using FACSAria I instrument (Becton Dickinson) at the Flow Cytometry Facility of UNIL. Induction of shRNA expression was obtained by addition to the medium of 2 µg/ml of doxycycline (Sigma). To maintain the knockdown, doxycycline-containing medium was refreshed every 3 days.
Ectopic BORIS expression in HeLa cells
The day prior transfection, HeLa cells were seeded at a density of 2×105 cells/well in 12-well/plates. Cells were transfected with 3 µg of the previously described pCMV-BORIS vector [12] using the Lipofectamine 2000 transfection reagent (Invitrogen) following the manufacturer's instructions. Cells were harvested 2 days post-transfection for cell fluorescence imaging.
Quantitative RT-PCR
Total RNA was isolated using the RNeasy mini kit (Qiagen) including on-column DNAse treatment according to the manufacturer's instructions. RNA concentration was determined using Nanodrop 2000 (Thermo Scientific) and Qubit Fluorescent Technology (Invitrogen).
A major limiting step was the low amount of total RNA isolated from cell sorting. To solve this technical limitation, we applied a method already described and validated [22], [23]. Firstly, 200 ng of total RNA were retrotranscribed using random hexamers and Superscript III reverse transcriptase (Invitrogen) in a final volume of 20 µl. Then 2 µl of cDNA were used for a preamplification reaction consisting on a multiplex PCR made with a mix of primers (Table S1) at 0.1 µM final concentration, 0.5 unit of Platinum Taq DNA Polymerase (Invitrogen), 1X PCR buffer, and 2 mM MgCl2 in a total volume of 25 µl.
For preamplification, PCR cycling conditions were: one denaturation step at 95°C for 5 min followed by 15 cycles of amplification (45 sec at 95°C, 30 sec at 60°C, 1 min at 72°C). Finally, for quantitative PCR, the preamplification reaction was 20-fold diluted and 2 µl of this dilution were used as template. Reaction was complemented with 0.5 units Platinum Taq DNA Polymerase (Invitrogen), 1X PCR buffer, 2.5 mM MgCl2, 2.5 µM SYTO9 green (Invitrogen) and 0.1 µM of each gene specific primer (Sigma) in a final volume of 20 µl. PCR conditions were: 95°C for 5 min followed by cycles (5 sec at 95°C, 30 sec at 60°C, 45 sec at 72°C). Melting curve analyses were performed at the end of the amplification to check the purity of the amplicons. The PCR products were also loaded on agarose gel to confirm the correct size of amplified products. Cycling and fluorescence acquisition were done in Rotor-Gene 6000 Real-Time PCR system (Corbett Life Science). Data analysis was performed using Rotor-Gene 6000 software. Relative expression levels were determined by ΔΔCt method with expression levels normalized to GAPDH level. The PCR efficiency ranged from 94 to 101%, with correlation coefficient (r2) ranging from 0.96 to 1.0, for all the amplified target genes. The Ct values of GAPDH were roughly the same (Ct = 10.07±0.25) for all the cells used in the quantitative RT-PCR experiments.
The human specific primers (Table S1) were designed using Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/) and OLIGO Primer Analysis software. Specificity was verified using BLAST search (NCBI). The designed primer pairs cross intron-exon boundaries to avoid genomic DNA contamination. BORIS primers were chosen (between exon 8 and 9) based on results previously obtained [24] in order to amplify the most abundant BORIS isoforms.
DNA methylation analysis
DNA was extracted using DNeasy kit (Qiagen). A range of 200 ng (from sorted cells) and 500 ng of DNA was used to bisulfite reaction using EpiTect Bisulfite kit (Qiagen) according to the manufacturer's instructions. The modified DNA was used to amplify a 123 bp fragment of the BORIS promoter.
The methylation assay was designed using the PyroMark Assay Design Software 1.0 (Qiagen). This assay allowed sequencing of 50 bp (from position −968 to −918) inside the promoter B of BORIS [25] and included 10 CpG. To perform sequencing, 3 µl of bisulfite treated DNA were first amplified by PCR. Sequences of the PCR primers were: BORIS-pyro Forward 5′ TGGTTTGTGGGTTTTGT 3′ and BORIS-pyro BIO Reverse 5′ CCCTTCACCCCCCCTCTTT 3′. PCR conditions were as follow: 95°C for 5 min; 45 cycles of 95°C for 30 s, 58°C for 15 s and 72°C for 1 min; and a final extension step at 72°C for 10 min. Then, purification and subsequent processing of the biotinylated single-stranded PCR fragment were performed according to the manufacturer's recommendations. Pyrosequencing of this PCR fragment was performed on a PyroMark Q24 instrument using Pyro Gold Q24 Reagents (Qiagen). The pyrosequencing primer (5′ GTGTTGTAGTTTATAGT 3′) was used at a final concentration of 0.3 µM. Resulting data were analyzed and quantified with the PyroMark Q24 software (Qiagen) which calculates the methylation percentage for each CpG site, allowing quantitative comparisons.
Cell proliferation assay
Cell proliferation was assessed by MTT assay. MTT (3-(4,5-dimethyl-2-thiazol)-2,5-diphenyltetrazolium bromide, Sigma) reagent was used according to the manufacturer's instructions. Briefly, stably infected cells were seeded at a density of 25×103 cells/well in 24-well/plates with doxycycline-containing medium. After 3 days, cells were incubated with MTT reagent (200 µg/ml final concentration) for 3 hours at 37°C. Then, cells were lysed adding isopropanol/HCl for 10 min and the plates were gently shaken for 5 min. Absorbance values were determined using a microplate reader (Synergy Mx, BioTek) at 570 nm. Each experiment was performed in triplicate and 3 independent experiments were conducted.
Apoptosis analysis
Apoptosis was measured in triplicates using Annexin V Apoptosis Detection Kit (BD bioscience) according to the manufacturer's protocols. Briefly, 5×104 cells/well were seeded in 12-well/plates and were grown in presence of doxycycline until confluence (5–7 days). The floating cells as well as trypsinized cells were collected, washed with PBS and resuspended in 100 µl Binding Buffer. Then, 5 µl of Annexin V-V500 and 5 µl of 7AAD were added and incubated with the cells for 30 min at room temperature. After addition of 400 µl of Binding Buffer the samples were immediately analyzed by Gallios flow cytometer (Beckman Coulter). At least 5×104 events were counted for all samples. The percentage of apoptotic cells was estimated after gating on eGFP and tRFP (transduced and doxycycline-induced, respectively) positive cells.
Western blot analysis
Whole cell lysates were obtained using RIPA buffer (Sigma) in presence of protease inhibitor cocktail (Sigma) and quantified using the BCA assay (Thermo Scientific). Thirty micrograms of protein were loaded on a 10% SDS-polyacrylamide gel, followed by blotting on a nitrocellulose membrane using a semi-dry transfer apparatus (BIO RAD). Non-specific binding was blocked by overnight incubation in 5% non-fat dried milk in TBS-T buffer (0.1% Tween 20 in tris-buffered saline, TBS) at 4°C. The membranes were then probed with monoclonal mouse anti-human BORIS/CTCFL antibody (produced and kindly provided by Dr Dmitri Loukinov, NIH/NIAD) used at 1:1000 dilution in 1% blocking buffer (1% low-fat dried milk in TBS-T buffer) and incubated at room temperature for 1.5 hours. As loading control, mouse anti-human β-actin antibody (Sigma) at 1:5000 dilution in 1% blocking buffer was used and incubated at room temperature for 45 min. The membranes were washed 3× with TBS-T and incubated at room temperature for 1 hour with horseradish peroxidase (HRP)-labeled rabbit anti-mouse IgG (Sigma) diluted at 1:5000 in 5% blocking buffer. After 3× washing with TBS-T, the membranes were developed using WesternBright Quantum (Advansta) and visualized with Fusion FX Chemiluminescence System (Vilber Lourmat).
Senescence-associated β-Galactosidase staining
Senescence-associated β-galactosidase (SA-β-gal) staining was performed using β-galactosidase staining kit (BioVision), according to the manufacturer's instructions. Briefly, 5×104 cells/well were seeded in 12-well/plates in presence of doxycycline and were grown until confluence (5–7 days). Then, cells were rinsed with PBS, fixed for 15 min and incubated with freshly prepared SA-β-Gal staining solution at 37°C for 24 hour. After washing with PBS, SA-β-gal activity was observed using inverted microscope (Nikon) by detection of blue stained cells. At least 10 separate fields were selected. For each field the number of blue stained cells and the number of total cells were counted. Results are expressed as percentage of SA-β-gal-positive cells calculated as: (number of blue cells/number of total cells) x 100.
Telomerase activity
Telomerase activity was measured using TRAPEZE RT telomerase detection kit (Millipore). This assay quantifies telomerase activity by SYBR Green real-time quantitative PCR [26]. Briefly, cells were lysed in 200 µl of CHAPS buffer and protein concentrations were determined with Nanodrop 2000. Aliquots of cell lysate (1.5 µg of protein/sample) were used. Inactivated samples, no-template reactions, and positive control were also assayed for quality control. A standard curve was prepared by serial dilution of TSR8 control template following manufacturer's instructions. Real-time amplifications were performed using Platinum Taq DNA Polymerase (2 unit/sample, Invitrogen). Cycling and fluorescence acquisition were done in Rotor Gene 6000 real-time PCR system (Corbett Life Science). Telomerase activity was calculated by comparing the average Ct values from each sample against the standard curve generated by the TSR8 control template.
Statistical analysis
Statistical significance was evaluated using two-tailed student t-test analysis. P-value <0.05 was considered statistically significant.
Results
In vitro validation of the molecular beacons (MBs)
Two different MBs specific to BORIS mRNA (BORIS-MB1 and BORIS-MB2) and a RANDOM-MB were used in this experiment. The hybridization temperatures and specificity of the MBs were first tested in solution. The fluorescence emission of these MBs was monitored at temperatures ranging from 25 to 80°C, under different conditions: MB alone, MB in presence of the specific target, in presence of a non-specific target or in presence of a plasmid containing BORIS cDNA (pCMV-BORIS). As shown in Figure S1, noticeable fluorescence signal was detected only when the MBs were mixed with their specific targets. Optimal fluorescence emission with acceptable signal-to-background ratio (>4) was observed below 40°C. The assay also showed that BORIS-MBs discriminate single and double stranded structures, since they do not emit fluorescence when incubated with the pCMV-BORIS plasmid. These results demonstrated that MBs specifically hybridize to their target sequences and strongly suggested that these MBs would be able to specifically bind mRNA and not genomic DNA.
Detection of BORIS mRNA using BORIS-MB
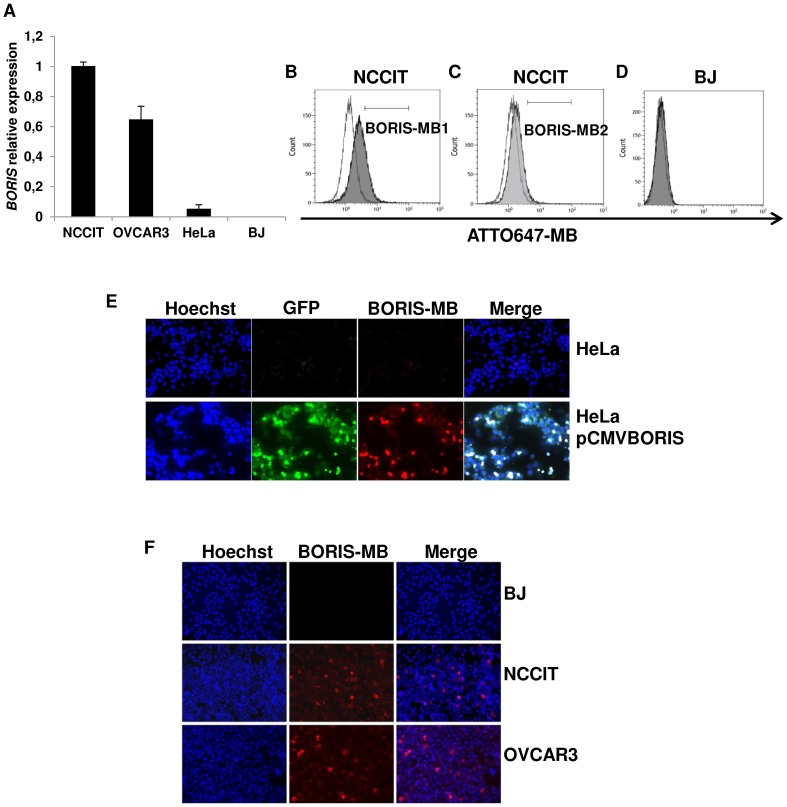
We first verified whether BORIS-MBs could be able to distinguish positive and negative BORIS expressing cells in living cells. Quantitative RT-PCR detected strong levels of BORIS mRNA in NCCIT and OVCAR3 cell lines, whereas this level was very low in HeLa cells and not detectable in BJ cells (Figure 1A), in accordance to previous studies [12]. Therefore, to study the specificity of MBs, NCCIT was used as a positive control cells and BJ as a negative control. NCCIT cells were transfected with BORIS-MB1 or BORIS-MB2 and fluorescence emissions were measured by flow cytometry. The mean fluorescent signals of the cells transfected with BORIS-MBs were higher compared to the mean fluorescent signal of the cells transfected with RANDOM-MB. An increase of 23.2 and 4.7 was observed for BORIS-MB1 and BORIS-MB2, respectively (Figure 1B–C). However, since BORIS-MB1 provided higher (5 fold) mean fluorescence signal compared to BORIS-MB2, BORIS-MB1 was selected for the subsequent analysis. From here onward BORIS-MB1 is referred as BORIS-MB. As expected, when BORIS negative BJ cells were transfected with BORIS-MB, they did not show any fluorescence signal (Figure 1D).
Figure 1. Detection of BORIS mRNA using BORIS-MB.
(A) BORIS expression in human cell lines. Total RNA were isolated from human tumoral cell lines: NCCIT (embryonic), OVCAR3 (ovarian), HeLa (cervical) and normal BJ (fibroblast) cells. mRNA levels of BORIS were analyzed by qRT-PCR. Results were normalized to GAPDH and related to NCCIT cells. BJ and NCCIT were considered as negative and positive controls, respectively. Error bars represent the mean ± SD of 3 independent experiments. (B) Fluorescent signals measured by flow cytometry of NCCIT cells transfected with ATTO647-BORIS-MB1 (dark grey peak) and with ATTO647-RANDOM-MB (white peak). (C) Fluorescent signals measured by flow cytometry of NCCIT cells transfected with ATTO647-BORIS-MB2 (weak grey peak) and with ATTO647-RANDOM-MB (white peak). (D) Fluorescent signals measured by flow cytometry of BJ cells treated with ATTO647-BORIS-MB1 (from here onward referred to BORIS-MB, grey peak) and with ATTO647-RANDOM-MB (white peak). (E) BORIS expression in HeLa cells using BORIS-MB. Representative images of HeLa cells transiently transfected with the BORIS expression vector, pCMV-BORIS (bottom) and non-transfected control cells (top), 20× magnification. (F) BORIS expression in human cell lines as detected using BORIS-MB. Representative images of BJ, NCCIT and OVCAR3 cells, 20× magnification. For fluorescence imaging, 1×106 cells were incubated at 37°C for 1 hour in serum-free DMEM medium with Cy3-BORIS-MB (200 nM). Hoechst 33342 5 µg/mL was added during the last 10 min of incubation. Slides were analyzed by fluorescence microscopy.
To further challenge the specificity of the BORIS-MB, HeLa cells that expressed BORIS mRNA at low level, were transiently transfected with the pCMV-BORIS expression plasmid, which contains the human BORIS cDNA fused to green fluorescent protein (GFP) gene. As expected, the transfected cells presented high fluorescence signals of both GFP and BORIS (Figure 1E). All together, these results confirmed the capacity of the BORIS-MB to reliably and specifically detect BORIS mRNA in living cells.
Hence, the cell lines were transfected with the BORIS-MB to visualize BORIS mRNA expression by fluorescence imaging. As can be seen from Figure 1F, the fluorescence signal of BORIS-MB was clearly visible in NCCIT and OVCAR3 cells, in which only a subset of cells were fluorescent. The percentage of BORIS positive cells in these cell lines was found to be between 3 and 5%. As expected, in BJ normal cell line no fluorescent cells were observed. In HeLa cells, the number of BORIS positive cells was extremely low, less than 0.1% (Figure 1E). Overall these experiments demonstrated that within tumor cell lines, BORIS mRNA is not present in all cells but rather occurs only in a subset of cells.
Isolation of cell population expressing BORIS mRNA using BORIS-MB
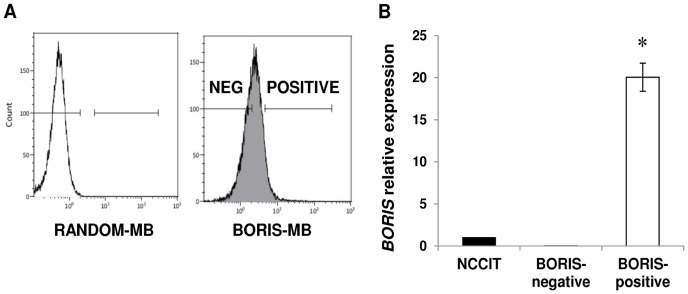
NCCIT cell line was used for the isolation of BORIS-positive cells. FACS sorting was performed after cell transfection with BORIS-MB. The BORIS-positive and BORIS-negative cells (8.4±1.5 and 41.5±7.2% of total, respectively; mean ± SD) were sorted (Figure 2A).
Figure 2. Isolation of BORIS-positive cells using BORIS-MB.
(A) NCCIT cells were transfected with ATTO647-RANDOM-MB (white peak) and ATTO647-BORIS-MB (grey peak). The two subpopulations, BORIS-negative and BORIS-positive cells were selected by comparing the fluorescent signal of RANDOM-MB to that of BORIS-MB. After exclusion of dead cells by PI staining, the two fractions were sorted. (B) BORIS expression of the isolated BORIS-negative and BORIS-positive fractions was analyzed by qRT-PCR. The results were normalized to GAPDH and related to non-sorted NCCIT cells. Error bars represent the mean ± SD of 3 independent experiments. Asterisk indicates statistically significant difference (p<0.05) between BORIS-positive and non-sorted cells.
BORIS expression analysis showed that BORIS mRNA level of sorted BORIS-positive cells was 20 fold higher compared to the non-sorted cells (Figure 2B). This result demonstrated the efficiency of the sorting method and the successful enrichment of a cell population that expressed BORIS mRNA.
The isolated BORIS-positive and BORIS-negative cells have similar methylation pattern of BORIS promoter B
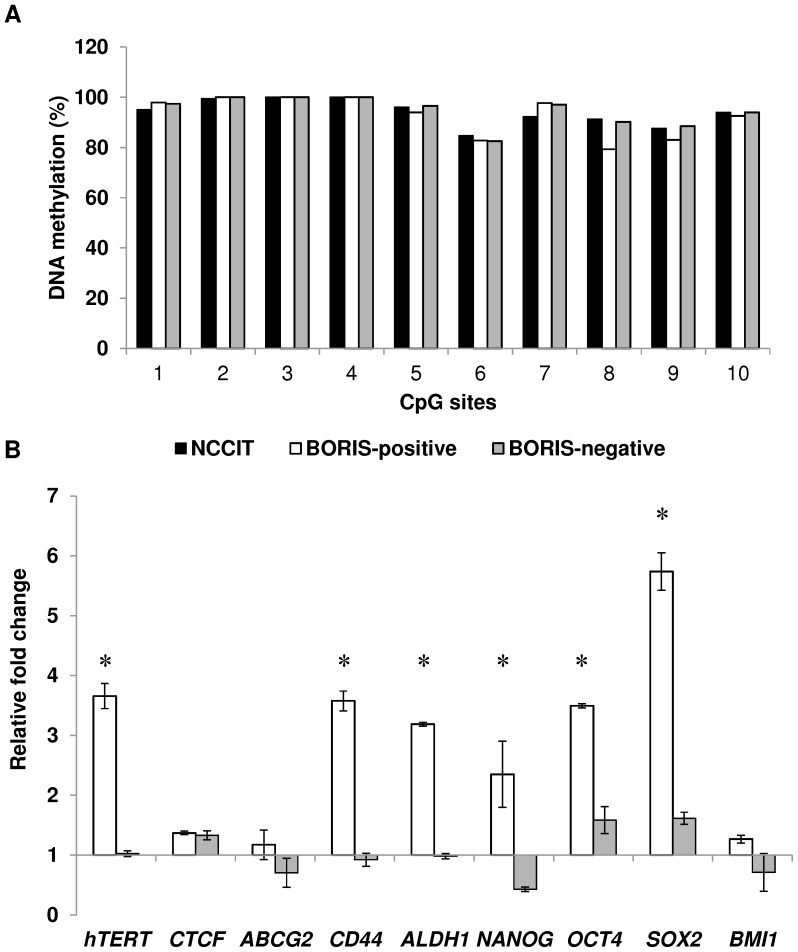
In a previous study, we have shown that BORIS expression is controlled by three alternative promoters, corresponding to transcription start sites at −1447, −899 and −658 bp upstream of the first ATG and designated promoters as A, B and C, respectively [25]. Interestingly, it has been observed that in tumors, demethylation of BORIS promoter B, is generally correlated with the expression of BORIS, which is not the case for promoters C and A [25]. Consequently, we interrogated the possible correlation between methylation of promoter B and BORIS expression in the sorted cells. BORIS methylation level of this promoter was measured by pyrosequencing, after bisulfite modification of DNA extracted from BORIS-positive and BORIS-negative cells. Pyrosequencing results indicated that in both fractions, the CpGs were heavily methylated (85–100% methylation) and no differences were detected (Figure 3A). This confirmed that in NCCIT cells, BORIS expression was not regulated by promoter B and suggested that the different expression of BORIS among the sorted fractions was not guided by DNA methylation status of promoter B but rather involved other levels of control.
Figure 3. BORIS promoter methylation and expression of hTERT, stem cell and CSC markers genes in BORIS-positive cells.
(A) Methylation analysis of 10 CpG islands within the BORIS promoter region (B promoter). The representative graphic shows the percentage of methylation of each CpG island for the isolated BORIS-positive (white), -negative (grey) and non-sorted (black) NCCIT cells. (B) Expression analysis of the isolated BORIS-positive (white) and BORIS-negative (grey) fractions. The indicated genes were analyzed by qRT-PCR. The results were normalized to GAPDH and related to non-sorted NCCIT cells. Graphic shown one representative experiment out of 3 independent experiments (the trend was similar in all independent experiments). Asterisks indicate statistically significant difference (p<0.05) between BORIS-positive and the non-sorted cells.
BORIS-positive cells express higher mRNA levels of hTERT as well as stem cell and cancer stem cell markers genes
We previously established that BORIS binds hTERT promoter at the same site of CTCF and can activate hTERT transcription in NCCIT cells [12]. To further investigate the correlation between BORIS, hTERT and CTCF expression, BORIS-positive and BORIS-negative populations were sorted as mentioned above and the expression of these genes was evaluated by qRT-PCR. Interestingly, three independent sorting experiments showed that hTERT mRNA level was significant higher (from 3.5 to 3.8 fold) in the BORIS-positive population compared to the BORIS-negative population. This result confirmed the positive correlation between BORIS and hTERT expression, whereas no difference was observed for CTCF (Figure 3B).
Expression of hTERT has been frequently associated with expression of stemness-related markers [13]-[17]. Consequently, considering our results, we also investigated if this correlation could exist with BORIS expression. To assess this, a panel of representative genes considered as stemness markers (NANOG, OCT4, SOX2 and BMI1) or as specific cancer stem cell (CSC) markers (ABCG2, CD44 and ALDH1) [27] were analyzed by qRT-PCR. Interestingly, it emerged that the BORIS-positive/hTERT-high population also expressed higher levels of stem cell-like markers. Indeed, mRNA levels of mostly all these genes were significantly higher in BORIS-positive population compared to BORIS-negative population and non-sorted cells (between 2 and 6 fold in average), at exception of BMI1 and ABCG2 genes (Figure 3B).
Knockdown of BORIS down-regulates expression of CTCF, hTERT, stem cell and CSC markers genes
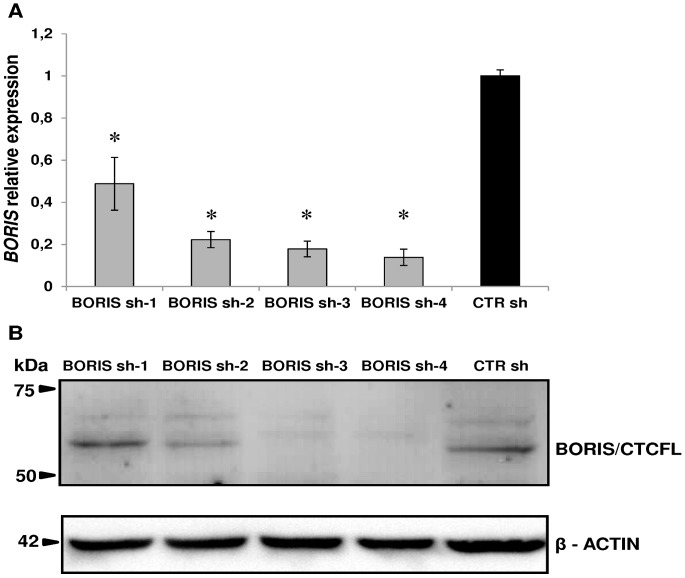
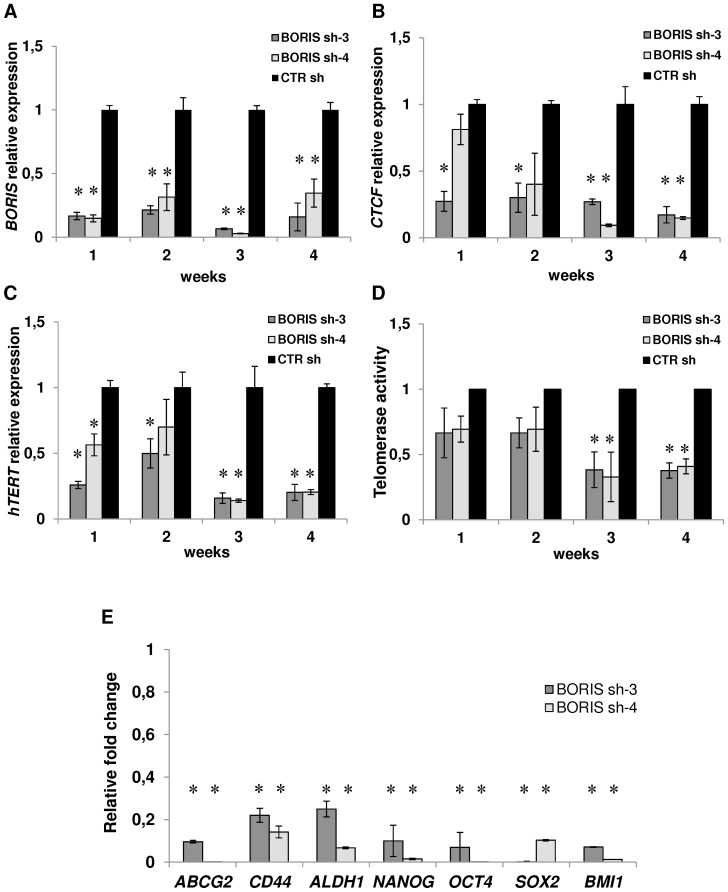
To investigate more directly the functions of BORIS, stable cell lines with inducible expressing shRNAs, targeting human BORIS mRNA, were generated using a lentiviral system. Four different BORIS shRNA lentivirus (BORIS sh-1, sh-2, sh-3 and sh-4) and a lentivirus carrying a scrambled sequence (CTR sh) were produced and tested. A significant reduction of BORIS mRNA was observed in all BORIS-depleted NCCIT-derived cells, compared to control cells (Figure 4A). The western-blot analysis indicated a remarkable decrease of BORIS protein level in BORIS sh-3 and sh-4 cells (Figure 4B). Therefore, all following knockdown experiments were performed using these BORIS sh-3 and sh-4 NCCIT-derived cells. The capacity of doxycycline-induction to maintain BORIS-knockdown overtime was also verified. As shown in figure 5A, BORIS mRNA levels were significantly reduced during 1 month. Interestingly, CTCF expression was dramatically decreased (Figure 5B). As expected, hTERT expression was down-regulated compared to control (Figure 5C) and this down-regulation was even stronger after 3 weeks. This observation was consistent with the correlation observed in the sorting experiments (Figure 3A) and was further confirmed by telomerase activity analysis. Indeed, we observed that telomerase activity was also decreased, especially after 3 and 4 weeks of BORIS silencing (Figure 5D). Notably, absence of BORIS triggered a dramatic down-regulation (between 75% and 99%) of the expression of stem cell and CSC marker genes (Figure 5E). This down-regulation was consistent up to the third week (data not shown).
Figure 4. BORIS-knockdown using lentivirus with doxycycline inducible BORIS-specific shRNA.
Four different BORIS shRNA lentiviral vectors (BORIS sh-1, sh-2, sh-3 and sh-4) and the vector carrying a scrambled sequence (CTR sh) were tested. NCCIT were transduced with the indicated lentivirus and sorted for eGFP marker expression. Then, cells were cultured with doxycycline-containing medium and after 3 days were analyzed. (A) BORIS mRNA levels were determined by qRT-PCR analysis, normalized to GAPDH and compared to that of CTR sh. Error bars represent the mean ± SD of 3 independent experiments. Asterisks indicate statistically significant difference (p<0.05) between BORIS sh-1, sh-2, sh-3 or sh-4 and CTR sh cells. (B) Representative western blot analysis. BORIS and β-actin (as a loading control) protein levels were determined by western blot. For both analyses (qRT-PCR and western blot) the knockdown was especially noticed with the BORIS sh-3 and sh-4 compared to the CTR shRNA.
Figure 5. Knockdown of BORIS resulted in decreasing expression of hTERT, stem cell and CSC markers genes.
NCCIT cells were engineered to stably exhibit knocked-down BORIS mRNA. BORIS sh-3, sh-4 and CTR sh (control with scrambled sequence) lentivirus were used to infect NCCIT cells. Each transduced cells were cultured with doxycycline to induce BORIS shRNA expression. Doxycycline-containing medium was replaced every 3 days. Each week over 1 month, mRNA levels of (A) BORIS, (B) CTCF and (C) hTERT were analyzed by qRT-PCR. Results were normalized with GAPDH and related to that of control cells (CTR sh) at each week. Error bars represent the mean ± SD of 3 independent experiments. (D) Telomerase activity was measured at each week by real-time quantitative PCR using TRAPEZE RT Telomerase Detection Kit. Values of telomerase activity of BORIS sh-3, sh-4 NCCIT-derived cells are related to that of control cells at each week. Error bars represent the mean ± SD of 3 independent experiments. (E) BORIS sh-3, sh-4 and CTR sh NCCIT-derived cells were cultured with doxycycline and after 7 days RNA was analyzed by qRT-PCR. mRNA levels of the indicated genes are related to that of control cells (CTR sh) after normalization with GAPDH. Error bars represent the mean ± SD of 3 independent experiments. Asterisks indicate statistically significant difference (p<0.05) between BORIS sh-3 or BORIS sh-4 and CTR sh cells.
All these results confirmed that BORIS regulates hTERT and strongly suggested that BORIS affects the transcription of stem cell and CSC marker genes, a pattern of genes expressed in CSCs.
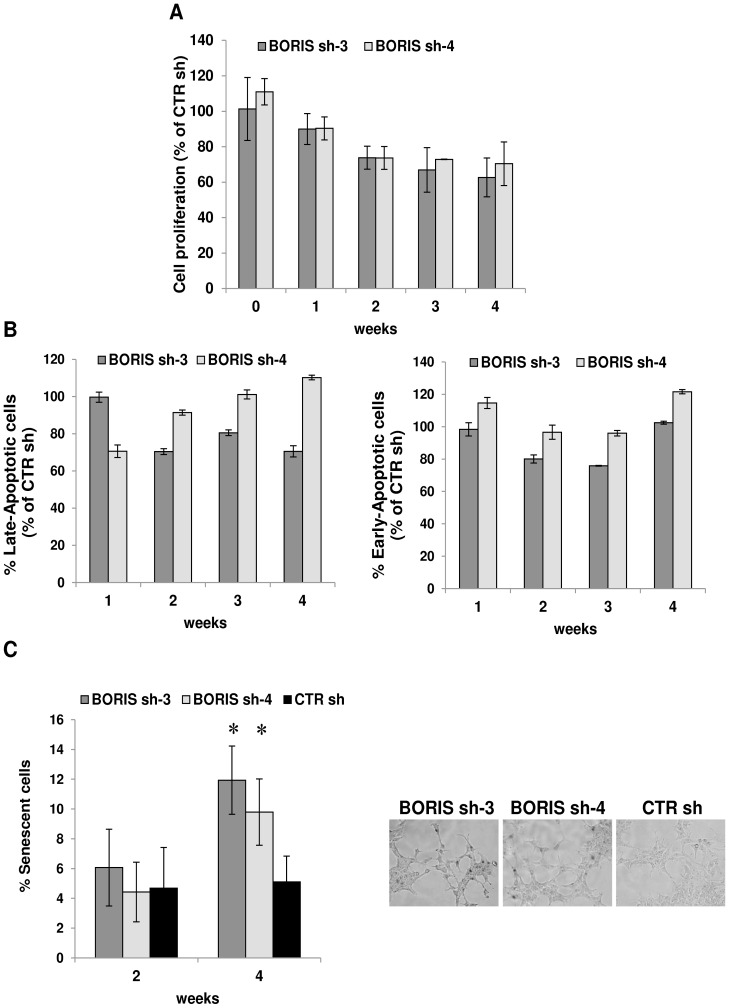
Knockdown of BORIS inhibits cell proliferation through cellular senescence
The impact of BORIS silencing on cell survival of embryonic tumor cells was also investigated. Cell proliferation was measured each week during 1 month by MTT assay (Figure 6A). An inhibition of cell growth was observed starting from the second week with 26% of reduction, to the fourth week with 40% of decrease compared to control. The analysis of apoptosis showed that the percentage of apoptotic cells (late apoptotic AnnexinV+/7AAD+ and early apoptotic AnnexinV+/7AAD−) was not significantly different between BORIS knockdown cells and control cells (Figure 6B). This result suggested that the decreasing of the observed cell growth is not due to cell apoptosis, therefore we further explored the causes of proliferation defects. As BORIS-knockdown led to a reduction of hTERT expression, a possible alteration of cellular senescence was investigated. Interestingly, analysis of senescence-associated β-galactosidase showed that, after 4 weeks of BORIS-knockdown, the percentage of senescent cells was 2 fold higher in BORIS silenced cells compared to control cells (Figure 6C). The percentage of senescent cells increased from 5.1 ± 1.7% in control cells to 11.9±2.3% and 9.8±2.2% in BORIS silenced with sh-3 and sh-4, respectively. These results indicated that BORIS knockdown led to the increase of cellular senescence in embryonic cancer cells.
Figure 6. BORIS knockdown impairs cell senescence but not apoptosis.
(A) Cell proliferation over 1 moth of dox-induced BORIS shRNA cells were analyzed by MTT assay. Results of the two specific BORIS-shRNA (BORIS sh-3 and sh-4) NCCIT-derived cells are indicated as a percentage compared to the cell proliferation of control cells (scrambled shRNA, CTR sh). Error bars represent the mean ± SD of 3 independent experiments. (B) After dox-induction of the BORIS specific shRNA in NCCIT cells, apoptosis was tested at each week using Annexin V Apoptosis Detection Kit. Results show the percentage of apoptotic cells (late apoptotic AnnexinV+/7AAD+ and early apoptotic AnnexinV+/7AAD−) of BORIS sh-3 and sh-4 cells compared to the control cells. Error bars represent the mean ± SD of 3 experiments. (C) The senescence-associated β-galactosidase (SA-β-gal) staining was performed using β-galactosidase staining kit. SA-β-gal was analyzed after 2 and 4 weeks of dox-induction of the BORIS specific shRNA in NCCIT cells. Results show the percentage of senescent cells of BORIS sh-3, BORIS sh4 and control cells. Error bars represent the mean ± SD of 3 experiments. Asterisks indicate statistically significant difference (p<0.05) between BORIS sh-3 or BORIS sh-4 and CTR sh cells. Representative images of NCCIT cells after 4 weeks of BORIS knockdown and stained with SA-β-gal were shown.
Discussion
Our study showed that in embryonic tumor cell lines, BORIS positive cells represent only a subset of the tumor cell population and the estimated frequency is approximately 3–5% of the total cells. This observation was confirmed by performing experiments with BORIS mRNA-targeting MB. Molecular beacon technology provides a powerful tool to discriminate target sequences, with very high specificity. Since their discovery [28], the MBs have been seen to distinguish target sequences differing only by a single nucleotide. Due to the characteristics of exceptional specificity and high sensibility, MBs have found a wide range of applications in biological sciences. MBs were used as taqman probes in real time quantitative PCR, for detection of mutations, SNP and allele, as indicators of contaminating infectious agents and also for in vivo detection of mRNA [29]. Furthermore, MBs are able to bind target sequences without modifying the functional and genomic characteristics of the cells. To date, there is no yet a validated antibody for in vivo detection of BORIS, therefore we used the MB technology to visualize BORIS mRNA positive cells. Recently, two publications demonstrated the use of MBs in FACS sorting [30], [31]. However, they showed different delivery systems of MBs to enter into the cells. One group described a delivery system with a cationic lipid vehicle [30] and the other group used electroporation with a dual-FRET MB [31]. Instead, we used the RNAiMAX Transfection Reagent, a cationic lipid formulation that was designed specifically for delivery single strand nucleotides (siRNA and miRNA). Additionally, we used for the sorting experiments, BORIS-MB conjugated with ATTO647, a fluorophore which confers a high photostability of fluorescence signal [20]. The analysis of BORIS expression demonstrated that we successfully enriched the cell population that expressed BORIS mRNA. In our experiments we used NCCIT cell line, classified as germ-cell tumor or embryonic cancer cells [32]. Due to its higher BORIS expression compared to the other tumor cell lines, the NCCIT cell line provides a good model for our studies, especially for the feasibility of isolation of BORIS positive cells by MB technology.
In a previous study, we have shown that BORIS modulates the transcription of hTERT telomerase gene in NCCIT and OVCAR3 cells, and in opposite manner compared to its paralog CTCF [12]. To further investigate the correlation between BORIS, hTERT and CTCF, we analyzed their expression in FACS-isolated BORIS-positive and BORIS-negative cells. The results confirmed the positive correlation between BORIS and hTERT, whereas no correlation was observed with CTCF. In the same previous work, it has been found that ectopic BORIS expression in normal BORIS negative cells allowed to expand the in vitro lifespan increasing cell passages. This finding could be explained by the expression of high levels of hTERT mRNA in these BORIS-transfected cells. Induction of hTERT expression and telomerase activity are well established as hallmarks of cancer and are prerequisite to cellular immortalization and malignant transformation [33]. Here, we confirm the role of BORIS in the immortalization process during tumorigenesis, since the isolated BORIS-positive cells expressed significantly higher levels of hTERT mRNA compared to the counterpart BORIS-negative cells.
In normal human cells, telomerase is generally absent in somatic cells but remains active in germ cells, progenitor cells and some adult stem cells [14]. It has been shown that telomerase is reactivated in a majority (approximately 90%) of tumors [34] and in clinical studies its reactivation is associated with poor outcomes of different tumors [35]–[38]. In addition, current literature supports the evidence that CSCs express telomerase and its inhibition suppresses the self-renewal of CSCs [39]–[41]. All these discoveries, together with our observations, indicate that BORIS could play an important and direct role in tumor malignancies by up-regulation of the hTERT telomerase gene. Consistent with this, we notably observed that the BORIS-positive/hTERT-high isolated cells expressed also high levels of the most important stem cell markers. The embryonic carcinoma cells that we investigated, indeed, provide a good model system to study the stem cell concept of cancer. They are stem cells derived from a teratocarcinoma and are also the malignant transformed embryonic stem cells [42]. They show gene expression profiles close to those of human embryonic stem cells [43]. Therefore, we analyzed the association of BORIS-positive/hTERT-high cells with the expression of the key-regulator genes of embryonic cells (NANOG, SOX2 and OCT4) and with some of the most known specific markers of CSCs [27]. CSCs are cancer cells characterized by stem cell properties and represent a small population of tumor cells that drives tumor development, progression, metastasis and drug resistance [27]. Interestingly, a correlation of the isolated BORIS-positive/hTERT-high cells with high expression of stem cell markers was observed. These findings were confirmed by BORIS silencing studies. Stable BORIS knockdown NCCIT-derived cells were generated by an efficient system of inducible-shRNA lentivirus [21]. After BORIS silencing, a significant decrease of hTERT expression was observed, as well as a down-regulation of telomerase activity, which is strictly regulated by hTERT gene transcription. CTCF expression was also decreased. This is consistent with our previous results [12] and confirms a role of BORIS in the transcriptional regulation of CTCF. This correlation between CTCF and BORIS was not observed in the expression analysis of the isolated BORIS-positive cells. This discrepancy could be due to the different experimental conditions. Indeed, in the expression analysis of the isolated BORIS-positive cells, CTCF was analyzed at steady state, while in BORIS silencing studies what we observed is the result of cellular and genetic modifications.
Importantly, after BORIS knockdown a decrease of expression of stem cell and CSC marker genes (NANOG, OCT4, SOX2, BMI1, ABCG2, CD44 and ALDH1) was also observed. These results highlight the importance of BORIS in malignant disease and its possible critical role on cancer growth and progression. Previous works have already showed a correlation of BORIS with stem cells. Monk et al. have observed the co-localization at the protein level of BORIS with ECSA, OCT4 and NANOG in cultured embryonic stem cells [44]. John et al. have detected BORIS expression in ECSA-expressing lung tumors [45]. In this paper, we additionally showed the molecular function of BORIS in embryonic cancer cells and all these data strongly suggested that BORIS affects the expression of stem cell genes.
Cell proliferation analysis revealed that the depletion of BORIS led to cell growth inhibition and an increase of cellular senescence in embryonic cancer cells. Cellular senescence is defined as the irreversible arrest of cell growth that is activated after alteration of telomeres or in response to different forms of stress [46]. Of note, cellular senescence is considered as a potent tumor suppressive mechanism, a protective barrier against neoplastic expansion [47]. Senescent cells cannot divide, even if they continue to be metabolically and synthetically active [48]. Senescent cells also show changes in chromatin organization and gene expression [49]. In our studies, the cellular senescence was measure by the most widely used senescence associated marker, the â-galactosidase activity [50]. The increasing of senescent cells after BORIS knockdown could be caused by the simultaneous telomerase inhibition. Consistent with the finding that inhibition of telomerase has been shown to initiate telomere shortening followed by cell senescence and cell death by apoptosis [51], [52].
In summary, the present study provides evidences that BORIS is only expressed in a small subset of tumor cells in embryonic tumors. As BORIS affects the expression of stemness-related genes, this subpopulation of BORIS positive cells might play an important role in cancer growth and progression. Our findings provide a rationale for investigating therapies that target BORIS/CTCFL in embryonic tumors.
Supporting Information
Fluorescence emission profiles of MBs. Representative fluorescence emission profiles of BORIS-MB1, BORIS-MB2 and RANDOM-MB. All the MBs were 5′-end Cy3-conjugated. All thermal profiles indicate the MBs (200 nM) alone (green dash line) and MBs mixed with specific target (red solid line), with non-specific target (black dash-dot line) and with plasmid (pCMV-BORIS, blue dot line). The targets were used at the final concentration of 1 µM. The samples were analyzed by Rotor Gene 6000 Real-Time PCR system and the fluorescence was measured at each temperature (from 25°C to 80°C) using filter for Cy3.
(DOCX)
Primer sequences for qRT-PCR analysis.
(DOCX)
Acknowledgments
We gratefully acknowledge Dr Dmitri Loukinov (Molecular Pathology Section, NIAID/NIH) for offering the monoclonal BORIS/CTCFL antibody.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by the Swiss National Science Foundation (grant number: 31003A-113505; www.snf.ch); and Emma Muschamp Foundation (www.s-a-v.org/-Fondations-associees-.html). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, et al. (2005) CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep 6: 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dunn KL, Davie JR (2003) The many roles of the transcriptional regulator CTCF. Biochem Cell Biol 81: 161–167. [DOI] [PubMed] [Google Scholar]
- 3. Herold M, Bartkuhn M, Renkawitz R (2012) CTCF: insights into insulator function during development. Development 139: 1045–1057. [DOI] [PubMed] [Google Scholar]
- 4. Loukinov DI, Pugacheva E, Vatolin S, Pack SD, Moon H, et al. (2002) BORIS, a novel male germ-line-specific protein associated with epigenetic reprogramming events, shares the same 11-zinc-finger domain with CTCF, the insulator protein involved in reading imprinting marks in the soma. Proc Natl Acad Sci USA 99: 6806–6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Klenova EM, Morse HC 3rd, Ohlsson R, Lobanenkov VV (2002) The novel BORIS + CTCF gene family is uniquely involved in the epigenetics of normal biology and cancer. Semin Cancer Biol 12: 399–414. [DOI] [PubMed] [Google Scholar]
- 6. de Necochea-Campion R, Ghochikyan A, Josephs SF, Zacharias S, Woods E, et al. (2011) Expression of the epigenetic factor BORIS (CTCFL) in the human genome. J Transl Med 9: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hong JA, Kang Y, Abdullaev Z, Flanagan PT, Pack SD, et al. (2005) Reciprocal binding of CTCF and BORIS to the NY-ESO-1 promoter coincides with derepression of this cancer-testis gene in lung cancer cells. Cancer Res 65: 7763–7774. [DOI] [PubMed] [Google Scholar]
- 8. Vatolin S, Abdullaev Z, Pack SD, Flanagan PT, Custer M, et al. (2005) Conditional expression of the CTCF-paralogous transcriptional factor BORIS in normal cells results in demethylation and derepression of MAGE-A1 and reactivation of other cancer-testis genes. Cancer Res 65: 7751–7762. [DOI] [PubMed] [Google Scholar]
- 9. Kouprina N, Noskov VN, Pavlicek A, Collins NK, Schoppee Bortz PD, et al. (2007) Evolutionary diversification of SPANX-N sperm protein gene structure and expression. PLoS One 2: e359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Woloszynska-Read A, James SR, Song C, Jin B, Odunsi K, et al. (2010) BORIS/CTCFL expression is insufficient for cancer-germline antigen gene expression and DNA hypomethylation in ovarian cell lines. Cancer Immun 10: 6. [PMC free article] [PubMed] [Google Scholar]
- 11. Kholmanskikh O, Loriot A, Brasseur F, De Plaen E, De Smet C (2008) Expression of BORIS in melanoma: lack of association with MAGE-A1 activation. Int J Cancer 122: 777–784. [DOI] [PubMed] [Google Scholar]
- 12. Renaud S, Loukinov D, Alberti L, Vostrov A, Kwon YW, et al. (2011) BORIS/CTCFL-mediated transcriptional regulation of the hTERT telomerase gene in testicular and ovarian tumor cells. Nucleic Acids Res 39: 862–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hiyama E, Hiyama K (2007) Telomere and telomerase in stem cells. Br J Cancer 96: 1020–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shay JW, Wright WE (2011) Role of telomeres and telomerase in cancer. Semin Cancer Biol 21: 349–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim H, Yoo JE, Cho JY, Oh BK, Yoon YS, et al. (2013) Telomere length, TERT and shelterin complex proteins in hepatocellular carcinomas expressing “stemness”-related markers. J Hepatol 59: 746–752. [DOI] [PubMed] [Google Scholar]
- 16. Paranjape AN, Mandal T, Mukherjee G, Kumar MV, Sengupta K, et al. (2012) Introduction of SV40ER and hTERT into mammospheres generates breast cancer cells with stem cell properties. Oncogene 31: 1896–1909. [DOI] [PubMed] [Google Scholar]
- 17. Schepers AG, Vries R, van den Born M, van de Wetering M, Clevers H (2011) Lgr5 intestinal stem cells have high telomerase activity and randomly segregate their chromosomes. EMBO J 30: 1104–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Monroy-Contreras R, Vaca L (2011) Molecular beacons: powerful tools for imaging RNA in living cells. J Nucleic Acids 2011: 741723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rhee WJ, Santangelo PJ, Jo H, Bao G (2008) Target accessibility and signal specificity in live-cell detection of BMP-4 mRNA using molecular beacons. Nucleic Acids Res 36: e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arden-Jacob J, Frantzeskos J, Kemnitzer NU, Zilles A, Drexhage KH (2001) New fluorescent markers for the red region. Spectrochim Acta A Mol Biomol Spectrosc 57: 2271–2283. [DOI] [PubMed] [Google Scholar]
- 21. Meerbrey KL, Hu G, Kessler JD, Roarty K, Li MZ, et al. (2011) The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc Natl Acad Sci USA 108: 3665–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Peixoto A, Monteiro M, Rocha B, Veiga-Fernandes H (2004) Quantification of multiple gene expression in individual cells. Genome Res 14: 1938–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Noutsias M, Rohde M, Block A, Klippert K, Lettau O, et al. (2008) Preamplification techniques for real-time RT-PCR analyses of endomyocardial biopsies. BMC Mol Biol 9: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pugacheva EM, Suzuki T, Pack SD, Kosaka-Suzuki N, Yoon J, et al. (2010) The structural complexity of the human BORIS gene in gametogenesis and cancer. PLoS One 5: e13872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Renaud S, Pugacheva EM, Delgado MD, Braunschweig R, Abdullaev Z, et al. (2007) Expression of the CTCF-paralogous cancer-testis gene, brother of the regulator of imprinted sites (BORIS), is regulated by three alternative promoters modulated by CpG methylation and by CTCF and p53 transcription factors. Nucleic Acids Res 35: 7372–7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wege H, Chui MS, Le HT, Tran JM, Zern MA (2003) SYBR Green real-time telomeric repeat amplification protocol for the rapid quantification of telomerase activity. Nucleic Acids Res 31: E3–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Medema JP (2013) Cancer stem cells: the challenges ahead. Nat Cell Biol 15: 338–344. [DOI] [PubMed] [Google Scholar]
- 28. Tyagi S, Kramer FR (1996) Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol 14: 303–308. [DOI] [PubMed] [Google Scholar]
- 29. Li Y, Zhou X, Ye D (2008) Molecular beacons: an optimal multifunctional biological probe. Biochem Biophys Res Commun 373: 457–461. [DOI] [PubMed] [Google Scholar]
- 30. Larsson HM, Lee ST, Roccio M, Velluto D, Lutolf MP, et al. (2012) Sorting live stem cells based on Sox2 mRNA expression. PLoS One 7: e49874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. King FW, Liszewski W, Ritner C, Bernstein HS (2011) High-throughput tracking of pluripotent human embryonic stem cells with dual fluorescence resonance energy transfer molecular beacons. Stem Cells Dev 20: 475–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Teshima S, Shimosato Y, Hirohashi S, Tome Y, Hayashi I, et al. (1988) Four new human germ cell tumor cell lines. Lab Invest 59: 328–336. [PubMed] [Google Scholar]
- 33. Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144: 646–674. [DOI] [PubMed] [Google Scholar]
- 34. Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, et al. (1994) Specific association of human telomerase activity with immortal cells and cancer. Science 266: 2011–2015. [DOI] [PubMed] [Google Scholar]
- 35. Domont J, Pawlik TM, Boige V, Rose M, Weber JC, et al. (2005) Catalytic subunit of human telomerase reverse transcriptase is an independent predictor of survival in patients undergoing curative resection of hepatic colorectal metastases: a multicenter analysis. J Clin Oncol 23: 3086–3093. [DOI] [PubMed] [Google Scholar]
- 36. Gertler R, Rosenberg R, Stricker D, Friederichs J, Hoos A, et al. (2004) Telomere length and human telomerase reverse transcriptase expression as markers for progression and prognosis of colorectal carcinoma. J Clin Oncol 22: 1807–1814. [DOI] [PubMed] [Google Scholar]
- 37. Terrin L, Rampazzo E, Pucciarelli S, Agostini M, Bertorelle R, et al. (2008) Relationship between tumor and plasma levels of hTERT mRNA in patients with colorectal cancer: implications for monitoring of neoplastic disease. Clin Cancer Res 14: 7444–7451. [DOI] [PubMed] [Google Scholar]
- 38. Tabori U, Ma J, Carter M, Zielenska M, Rutka J, et al. (2006) Human telomere reverse transcriptase expression predicts progression and survival in pediatric intracranial ependymoma. J Clin Oncol 24: 1522–1528. [DOI] [PubMed] [Google Scholar]
- 39. Vicente-Duenas C, Barajas-Diego M, Romero-Camarero I, Gonzalez-Herrero I, Flores T, et al. (2012) Essential role for telomerase in chronic myeloid leukemia induced by BCR-ABL in mice. Oncotarget 3: 261–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marian CO, Wright WE, Shay JW (2010) The effects of telomerase inhibition on prostate tumor-initiating cells. Int J Cancer 127: 321–331. [DOI] [PubMed] [Google Scholar]
- 41. Marian CO, Cho SK, McEllin BM, Maher EA, Hatanpaa KJ, et al. (2010) The telomerase antagonist, imetelstat, efficiently targets glioblastoma tumor-initiating cells leading to decreased proliferation and tumor growth. Clin Cancer Res 16: 154–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Przyborski SA, Christie VB, Hayman MW, Stewart R, Horrocks GM (2004) Human embryonal carcinoma stem cells: models of embryonic development in humans. Stem Cells Dev 13: 400–408. [DOI] [PubMed] [Google Scholar]
- 43. Sperger JM, Chen X, Draper JS, Antosiewicz JE, Chon CH, et al. (2003) Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc Natl Acad Sci USA 100: 13350–13355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Monk M, Hitchins M, Hawes S (2008) Differential expression of the embryo/cancer gene ECSA(DPPA2), the cancer/testis gene BORIS and the pluripotency structural gene OCT4, in human preimplantation development. Mol Hum Reprod 14: 347–355. [DOI] [PubMed] [Google Scholar]
- 45. John T, Caballero OL, Svobodova SJ, Kong A, Chua R, et al. (2008) ECSA/DPPA2 is an embryo-cancer antigen that is coexpressed with cancer-testis antigens in non-small cell lung cancer. Clin Cancer Res 14: 3291–3298. [DOI] [PubMed] [Google Scholar]
- 46. Campisi J, d'Adda di Fagagna F (2007) Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol 8: 729–740. [DOI] [PubMed] [Google Scholar]
- 47. Sager R (1991) Senescence as a mode of tumor suppression. Environ Health Perspect 93: 59–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Campisi J (2001) Cellular senescence as a tumor-suppressor mechanism. Trends Cell Biol 11: S27–31. [DOI] [PubMed] [Google Scholar]
- 49. Campisi J (2013) Aging, cellular senescence, and cancer. Annu Rev Physiol 75: 685–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dimri GP, Lee X, Basile G, Acosta M, Scott G, et al. (1995) A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA 92: 9363–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hahn WC, Stewart SA, Brooks MW, York SG, Eaton E, et al. (1999) Inhibition of telomerase limits the growth of human cancer cells. Nat Med 5: 1164–1170. [DOI] [PubMed] [Google Scholar]
- 52. Zhang X, Mar V, Zhou W, Harrington L, Robinson MO (1999) Telomere shortening and apoptosis in telomerase-inhibited human tumor cells. Genes Dev 13: 2388–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fluorescence emission profiles of MBs. Representative fluorescence emission profiles of BORIS-MB1, BORIS-MB2 and RANDOM-MB. All the MBs were 5′-end Cy3-conjugated. All thermal profiles indicate the MBs (200 nM) alone (green dash line) and MBs mixed with specific target (red solid line), with non-specific target (black dash-dot line) and with plasmid (pCMV-BORIS, blue dot line). The targets were used at the final concentration of 1 µM. The samples were analyzed by Rotor Gene 6000 Real-Time PCR system and the fluorescence was measured at each temperature (from 25°C to 80°C) using filter for Cy3.
(DOCX)
Primer sequences for qRT-PCR analysis.
(DOCX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.