Abstract
In this study the first complete sequence of the West Nile virus (WNV) lineage 2 strain currently circulating in Romania was determined. The virus was detected in a Hyalomma marginatum marginatum tick collected from a juvenile song thrush (Turdus philomelos) in the Romanian Danube Delta close to the city of Tulcea, end of August 2013. Our finding emphasizes the role of ticks in introduction and maintenance of WNV infections. Sequence analyses revealed close genetic relationship of the Romanian WNV strain to strain Reb_Volgograd_07_H, which was isolated from human brain tissue during an outbreak of West Nile neuroinvasive disease (WNND) in Russia in 2007. In 2010 the Eastern European lineage 2 WNV caused an outbreak of human WNND in Romania. Partial sequences from subsequent years demonstrated that this WNV strain became endemic in Eastern Europe and has been causing outbreaks of varying sizes in southern Russia since 2007 and in Romania since 2010.
Introduction
Mosquitoes, primarily of the genus Culex, are considered the main vectors of West Nile virus (WNV), a zoonotic member of the genus Flavivirus. Wild birds constitute the principal hosts of the virus amplifying it in a bird-mosquito cycle. Certain ‘bridge’ mosquito species have been determined to transmit the virus to humans and other mammals, which are regarded dead-end hosts [1], [2]. The role of ticks as WNV vectors had been poorly investigated to date [3].
Romania has a long-standing history of WNV infections, including severe outbreaks of human West Nile neuroinvasive disease (WNND) in 1996 with 393 confirmed cases [4], and in 2010 with 57 cases. Affected patients were distributed among 19 districts in the southern, western, central and eastern parts of the country [5]. The ‘2010’ WNV strain became endemic and has been the cause of outbreaks of varying sizes each following year (http://www.ecdc.europa.eu/en/healthtopics/west_nile_fever/Pages/epidemiological_updates.aspx; http://www.ecdc.europa.eu/en/healthtopics/west_nile_fever/West-Nile-fever-maps/Pages/historical-data.aspx). Nevertheless, the complete sequence of the main WNV strain circulating in Romania since 2010 has not been determined as yet. Therefore goals of this study were to determine the complete genomic sequence of the WNV strain currently circulating in Romania, assess its pathogenicity and neuroinvasive markers, investigate its phylogenetic relatedness to other WNV strains, and discuss the role of ticks in WNV introduction and maintenance.
Materials and Methods
A total of 32 ticks were found randomly on a total of 23 birds, which had been captured using mist nets [6] in the Danube Delta Biosphere Reserve, Romania. They were investigated for the presence of WNV within the framework of the European Union FP7 project EDENext. Specifically, the birds were captured at the following locations: Enisala (44°53′28.28″N/28°49′50.97″E), Gr. Lupilor (44°41′46.82″N/28°56′15.70″E), Salcioara (44°47′55.30″N/28°53′57.57″E), Maliuc Mila (45°10′34.61″N/29° 3′53.91″E), Rachitarie (45°11′34.74″N/29° 5′8.16″E), and Maliuc (45°10′35.96″N/29° 6′29.51″E). All ethical and research certifications, approvals and permits were issued by the responsible authorities, i.e. the Romanian Ornithological Centre, Danube Delta Biosphere Reserve Authority, Sanitary, Veterinary and Food Safety County Direction Tulcea, and the Scientific Council of the Danube Delta National Institute. The field studies did not involve endangered or protected species. The tick-infested birds were Passer montanus (n = 20), Acrocephalus arundinaceus (1), Turdus philomelos (1) and Oriolus oriolus (1). The latter was found to be most infested by ticks (n = 10), whereas merely a single tick was retrieved from each of the remaining birds. Ticks from Acrocephalus arundinaceus and Oriolus oriolus (n = 11) were collected in May 2012, from four Passer montanus in July 2012, and from sixteen further Passer montanus as well as one Turdus philomelos in August 2013. The ticks were removed using tweezers and stored at −80°C until further investigation.
Ticks were homogenized individually in a TissueLyser II (Qiagen, USA) using Tungsten Carbide Beads 3 mm (Qiagen, USA) and 150 µl nuclease free water (Promega, Madison, USA) for 3 min at 30 Hz. Both genomic DNA and total RNA were extracted from each suspension employing the QIAamp Viral RNA Mini Kit (Qiagen, USA) following the manufacturer's instructions.
For rapid detection of both WNV lineages 1 and 2 in the samples, a reliable real-time RT-qPCR method [WNV (lin.1+2) RT-qPCR] targeting the highly conserved 5′ non-coding region (NCR) was established. The sequences were as follows: WNV_8F: 5′-CGCCTGTGTGAGCTGACAAA-3′, WNV_118R 5′-GCCCTCCTGGTTTCTTAGACATC-3′, and WNV_67T: 5′-FAM-TGCGAGCTGTTTCTTAGCACGA-TAMRA-3′ for the forward primer, reverse primer, and TaqMan probe, respectively. The numbering refers to the WNV lineage 1 sequence AF196835.
Primers and probe for the WNV (lin.1+2) RT-qPCR were designed with the help of the AbiPrism Primer Express 2.0.0 software (Applied Biosystems, USA). The real-time RT-qPCR was performed in an Applied Biosystems 7300 Real-Time PCR System using a SuperScript III Platinum One-Step Quantitative RT-PCR System kit (Invitrogen, USA) following the manufacturer's instructions.
The positive sample was subsequently tested by various RT-PCRs using published universal flavivirus primer pairs, e.g., [7]–[9], primers specific for WNV lineage 2 [10] (online appendix: http://www.cdc.gov/ncidod/EID/vol12no04/05-1379_app.htm], and self-designed primers (Table 1)).
Table 1. In this table primer pairs which have been used in addition to primers described by Bakonyi et al. [10] are listed (available as online appendix at: http://www.cdc.gov/ncidod/EID/vol12no04/05-1379_app.htm).
| Sequence 5′–3′ (F, forward primer; R, reverse primer) | Primer position (refers to the sequence VLG_07, FJ425721) | Length of the PCR product (nt) |
| AGCACGAAGATCTCGATGTC(F)* | 49–68 | 593 |
| GTGCACCARCAGTCRATGTC(R)* | 641–622 | |
| CCGCGGATTGTCCTTGATAG(F) | 129–148 | 766 |
| CACGACGCGTTGCATYGTGT(R) | 894–875 | |
| CAATCTGTTGTGGCTCTAGG(F) | 1681–1700 | 845 |
| TCCATCCAGGCTTCCACATC(R) | 2525–2506 | |
| GCCGGAGCGATTCCTGTTGA(F) | 1732–1751 | 1340 |
| AGCTTCCARGTGTCGTTGAG(R) | 3071–3052 | |
| TCCTTGCAGTTGGAGGAGTT(F) | 2387–2406 | 826 |
| CCTGGTCTCCTGTTGTGATT(R) | 3212–3193 | |
| GGCACGCACAACCACTGAGA(F) | 3327–3346 | 642 |
| AGCAGCGGCACCACCACATT(R) | 3968–3949 | |
| GGCCTGCTACAGAAGTGATG(F) | 4190–4209 | 552 |
| CCTTAGTGGTGTGCCACAGT(R) | 4741–4722 | |
| GGTCTGGCAGAACTTGACAT(F) | 4243–4262 | 431 |
| CCAAGCAGACCTCGAGTCAT(R) | 4673–4654 | |
| TGCTGAGATCACAGGCTCTA(F) | 4380–4399 | 1021 |
| GTGTGGAGACATCAGCCTAT(R) | 5400–5381 | |
| AGATTGAGGACGGCTGTGCT(F) | 5224–5243 | 638 |
| TTGCGGCTGTCGATCACTCT(R) | 5861–5842 | |
| ATAGGCTGATGTCTCCACAC(F) | 5381–5400 | 591 |
| TTCTTCCTATGCGTCCTCT(R) | 5971–5953 | |
| CAGAGGCTCGCATCATGCTA(F) | 6047–6066 | 795 |
| ACACAGCGAGCTGGTTGTCA(R) | 6841–6822 | |
| CCTGAGCGCGAGAAGGTGTA(F) | 6112–6131 | 828 |
| GTGTCCTAGCAGGCTGCTAA(R) | 6939–6920 | |
| CCTGAGAACAGCTGACTTAC(F) | 6189–6208 | 519 |
| GTGGCAGCTCCTAAGATTAC(R) | 6707–6688 | |
| TGTTGGATGGCTGAAGTCTC(F) | 6715–6734 | 789 |
| TGCTGCTGCTGTAGTCAGAA(R) | 7503–7484 | |
| GACTCTGACCGTGACTGTGA(F) | 7203–7222 | 715 |
| ATAGCACCAGCCGCCTCTAC(R) | 7917–7898 | |
| AGCGGAAGCTATGCGATCTG(F) | 7275–7294 | 847 |
| TTCTACCTCGGCACTTGACG(R) | 8121–8102 | |
| GGCCATTACTGAAGTTGACC(F) | 7740–7759 | 1061 |
| ACAGCCAGTTCGTGGTCTCA(R) | 8800–8781 | |
| ACCGTCCGTGTCTTGGAGAT(F) | 8131–8150 | 448 |
| TTCCGTGGTAGTTCCAGGT(R) | 8578–8560 | |
| AGCTGACCTCGAGAATGAAG(F) | 9288–9307 | 990 |
| CGGACCTGATTRATTGCTAC(R) | 10277–10258 | |
| TAGCGCGGTCCATCATCGAG(F) | 9348–9367 | 1633 |
| GCGCACTGTGCCGTGTGGCT(R) | 10980–10961 | |
| GCCACCGGAAGTTGAGTAGA(F)* | 10515–10534 | 466 |
| CTGGTTGTGCAGAGCAGAG(R)* | 10962–10944 | |
| GCTGCGAGGTGATCCACGTA(F) | 10579–10598 | 398 |
| ACTGTGCCGTGTGGCTGGTT(R) | 10976–10957 |
Primers marked with stars are suitable for detection of both WNV lineages 1 and 2.
All primer pairs for the conventional RT-PCRs were developed with the help of Primer Designer program (Scientific & Educational Software). The RT-PCR assays were carried out using One Step RT-PCR Kit (Qiagen, USA).
Primer and probe synthesis as well as sequencing in both directions were carried out by Microsynth (Balgach, Switzerland).
The obtained WNV sequences were verified by BLAST search and compiled to one continuous sequence. The complete genome of the newly determined WNV was compared with 23 other WNV strains, representing complete lineage 2 sequences from different hosts, countries and years. Multiple alignments were performed using BioEdit Sequence Alignment Editor (version 7.0.9.0) and verified by Clustal X program (version 1.8).
Phylogenetic neighbor joining analysis was conducted with the help of the MEGA5 program. The evolutionary distances were computed using Maximum Composite Likelihood model [11]. Bootstrap resampling analysis with 1000 replicates was employed. Information about several sequences deposited atGenBank was obtained from [12]. Sequence translation was carried out using the program (http://www.expasy.org/genomics).
To explore the pathogenicity and neuroinvasiveness markers of the newly determined WNV strain, predicted N-glycosylation sites of all viral proteins were analyzed using the program NetNGlyc 1.0 (http://www.cbs.dtu.dk/services/NetNGlyc/) according to [13] (summarized in [14]).
In order to explore the hypothesis of the existence of a single Eastern European lineage 2 WNV cluster, partial E and partial NS5 gene sequences from Russia and Romania, available in GenBank (Table 2) were phylogenetically analyzed as described above for the complete genome sequences. Unfortunately the Russian and Romanian partial sequences targeted different WNV genes, thus additional two phylogenetic trees had to be established. Due to various lengths, the sequences had to be adjusted to 474 nt (E gene sequences) and 466 nt (NS5 gene sequences), respectively.
Table 2. Characteristics of WNV lineage 2 partial sequences additionally included in Figures 2 and 3.
| GenBank acc. no. | Designation | Length (nt) | Country | Area | Collection date | Host | Reference |
| Partial WNV E gene sequences additionally included in Figure 2 | |||||||
| FJ265699 | Isolate VLG-07-2657 | 1563 | Russia | Volgograd | 2007 | Human (blood) | Platonov et al., GenBank |
| FJ265700 | Isolate VLG-07-26428 | 1563 | Russia | Volgograd | 2007 | Human (blood) | Platonov et al., GenBank |
| FJ265701 | Isolate VLG-07-26571 | 1563 | Russia | Volgograd | 2007 | Human (blood) | Platonov et al., GenBank |
| FJ265702 | Isolate VLG-07-26298 | 1563 | Russia | Volgograd | 2007 | Human (blood) | Platonov et al., GenBank |
| FJ265703 | Isolate VLG-07-26304 | 1563 | Russia | Volgograd | 2007 | Human (blood) | Platonov et al., GenBank |
| FJ425729 | Isolate 57_VLG_07_M | 1563 | Russia | Volgograd | 2007 | Mosquito (Culex pipiens) | Platonov et al., GenBank |
| HQ237494 | Isolate ROS-2-2010-H | 774 | Russia | Rostov | 2010 | Human (blood) | Karan et al., GenBank |
| HQ237498 | Isolate VLG-609-2010-H | 774 | Russia | Volgograd | 2010 | Human (brain) | Karan et al., Genbank |
| JQ014116 | Isolate VOLGOGRAD-01/918-2011 | 474 (length-limiting sequence) | Russia | Volgograd | 2011 | Human (urine) | Antonov et al., GenBank |
| JX844662 | Isolate VOLGOGRAD-03/619-2012 | 492 | Russia | Volgograd | 2012 | Human (brain) | Antonov et al., GenBank |
| Partial WNV NS5 gene sequences additionally included in Figure 3 | |||||||
| HE984574 | Isolate 89-2011 | 1189 (with 100nt gap in the middle) | Romania | Danube Delta (Mila 26) | 2011 | Mosquito (Culex pipiens) | Panculescu-Gatej et al., GenBank |
| HG328830 | Isolate RO_mo151/2012 | 642 | Romania | Not available | 07-Sep-2012 | Mosquito (Culex pipiens) | Dinu et al., Genbank |
| HG328831 | Isolate RO_hu121351/2012 | 705 | Romania | Not available | 2012 | Human (serum) | Dinu et al., GenBank |
| HG514461 | Isolate RO_mo48/2012 | 618 | Romania | Danube Delta (Mila 26) | 26-Aug-2012 | Mosquito (Culex pipiens) | Dinu et al., GenBank |
| HG514462 | Isolate RO_mo98/2012 | 636 | Romania | Danube Delta (Mila 26) | 25-Aug-2012 | Mosquito (Culex pipiens) | Dinu et al., Genbank |
| HG514463 | Isolate RO_mo418/2012 | 614 | Romania | Danube Delta (Mila 26) | 30-Sep-2012 | Mosquito (Culex modestus) | Dinu et al., Genbank |
| HG514464 | Isolate RO_mo419-2012 | 585 (length-limiting sequence) | Romania | Danube Delta (Mila 26) | 30-Sep-2012 | Mosquito (Culex modestus) | Dinu et al., Genbank |
| HG514465 | Isolate RO_mo426/2012 | 633 | Romania | Danube Delta (Mila 26) | 30-Sep-2012 | Mosquito (Culex modestus) | Dinu et al., GenBank |
| HG514466 | Isolate RO_mo434-2012 | 638 | Romania | Danube Delta (Mila 26) | 29-Sep-2012 | Mosquito (Anopheles hyrcanus) | Dinu et al., GenBank |
| HG514467 | Isolate RO_mo444-2012 | 633 | Romania | Danube Delta (Mila 26) | 30-Sep-2012 | Mosquito (Culex pipiens) | Dinu et al., GenBank |
| HG514468 | Isolate RO_mo460/2012 | 639 | Romania | Danube Delta (Mila 26) | 28-Sep-2012 | Mosquito (Culex pipiens) | Dinu et al., GenBank |
| LK022077 | Isolate RO_mo292-2013 | 795 | Romania | Danube Delta (Mila 26) | 04-Aug-2013 | Mosquito (Culex pipiens) | Dinu et al., GenBank |
| LK022078 | Isolate RO_mo294/2013 | 664 | Romania | Danube Delta (Mila 26) | 04-Aug-2013 | Mosquito (Culex pipiens) | Dinu et al., GenBank |
| LK022079 | Isolate RO_mo313-2013 | 752 | Romania | Danube Delta (Mila 26) | 05-Aug-2013 | Mosquito (Culex pipiens) | Dinu et al., GenBank |
| LK022080 | Isolate RO_mo354/2013 | 673 | Romania | Danube Delta (Mila 26) | 02-Sep-2013 | Mosquito (Culex pipiens) | Dinu et al., GenBank |
| LK022081 | 1isolate RO_mo531/2013 | 728 | Romania | Bucharest | 25-Aug-2013 | Mosquito (Culex pipiens) | Dinu et al., GenBank |
| HG918026 | Isolate RO_hu149704/2013 | 600 | Romania | Bucharest | 02-Sep-2013 | Human (serum) | Dinu et al., GenBank |
| HG918027 | Isolate RO_hu149802/2013 | 562 (length-limiting sequence) | Romania | Fetesti | 02-Sep-2013 | Human (serum) | Dinu et al., GenBank |
| HG918029 | Isolate RO_mo10/2013 | 705 | Romania | Danube Delta (Mila 26) | 28-Aug-2013 | Mosquito (Coquillettidia richiardii) | Dinu et al., GenBank |
| HG918031 | Isolate RO_mo17/2013 | 681 | Romania | Danube Delta (Mila 26) | 28-Aug-2013 | Mosquito (Culex pipiens) | Dinu et al., GenBank |
| HG918033 | Isolate RO_mo34/2013 | 666 | Romania | Danube Delta (Mila 26) | 28-Aug-2013 | Mosquito (Coquillettidia richiardii) | Dinu et al., GenBank |
| HG918036 | Isolate RO_mo168/2013 | 614 | Romania | Danube Delta (Mila 26) | 02-Sep-2013 | Mosquito (Culex pipiens) | Dinu et al., GenBank |
| HG918037 | Isolate RO_mo233-2013 | 582 | Romania | Danube Delta (Mila 26) | 01-Aug-2013 | Mosquito (Culex pipiens) | Dinu et al., GenBank |
Molecular determination of the tick species was performed using a PCR assay targeting the mitochondrial 12S rDNA gene with primers recommended previously [15], [16]. For this purpose a Fast Cycling PCR Kit (Qiagen, USA) was applied.
Upon removal of the tick from the bird, oral and cloacal swabs as well as serum were collected from the bird, and later tested using the above-mentioned real-time WNV (lin.1+2) RT-qPCR. The serum sample was additionally investigated for the presence of antibodies against WNV by INGESIM West Nile Compac ELISA (Ingenasa, Madrid, Spain) following the manufacturer's instructions, and by PRNT [17], [18].
Results
One immature tick (nymph) was positive upon WNV (lin.1+2) RT-qPCR. This tick was genetically identified as Hyalomma marginatun marginatum (H. m. marginatum). It was found on a juvenile song thrush (Turdus philomelos) which had been captured in a mist-net in Enisala/Romanian Danube Delta on 27.08.2013. All other investigated ticks tested negative. All WNV-negative ticks were also identified as H. m. marginatum, except one tick which was identified as Haemaphyalis sp. (collected in August 2013 from a Passer montanus).
By application of several published and self-designed primer pairs a complete, 11,013 nt long WNV sequence was generated from the infected tick, encoding a 3,434 aa long polyprotein, which consists of all typical WNV proteins C, prM, M, E, NS1, NS2a, NS2b, NS3, NS4a, NS4b and NS5 with the corresponding lengths of 123, 92, 75, 501, 352, 231, 131, 619, 149, 256 and 905 aa, respectively.
The Romanian tick-derived WNV was most closely related to strain Reb_VLG_07_H (GenBank acc.no. FJ425721) with only 58 nucleotide differences (identity rate 99.45%).
The comparison of the polyprotein sequences of both WNV strains revealed only six amino acid substitutions (identity rate 99.83%): Thr to Ile (position 108 of the C gene), Ser to Gly (position 199 of the E gene), Met to Leu (pos. 90 of the NS2a gene), Ser to Pro (pos. 100 of the NS4a gene), as well as Tyr to His and Ala to Glu (positions 18 and 370 of the NS5 gene), of which two amino acid substitutions at positions 108 in C gene and 199 in E gene were unique for the Romanian tick WNV, compared to all complete WNV genomes investigated in this study.
The known pathogenicity and neuroinvasiveness markers could be identified in the Romanian WNV: N-glycosylation motif NYS at position 154 of the E protein as well as three potential N-glycosylation sites at positions 130, 175, and 207 in the NS1 gene, prolin at position 250 of the NS1 gene and histidine at position 249 of the NS3 gene.
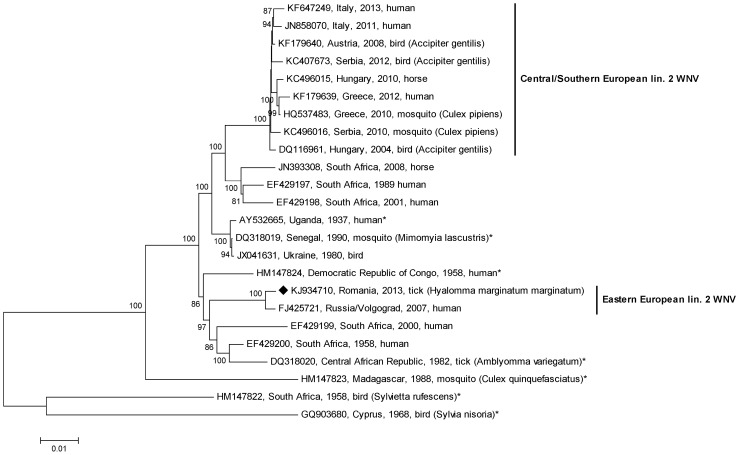
Phylogenetic analysis of 24 complete WNV lineage 2 sequences confirmed the close genetic relationship of the newly determined Romanian tick WNV with the Russian human-derived WNV from 2007 (Figure 1). All other viruses in this major cluster are of African origin, while the Central/Southern European lineage 2 viruses form an independent clade (Figure 1).
Figure 1. Phylogenetic tree of 24 representative WNV lineage 2 complete genomic sequences.
The WNV sequence derived from a Hyalomma marginatum marginatum tick collected from a song thrush in Romania (marked with a black diamond) is most closely related to the human-derived WNV strain VLG_07 from Russia. All other WNV strains related to this Russian/Romanian cluster originate from Central and South Africa, suggesting an introduction of this WNV lineage 2 variant from Africa to Europe. The cluster of another independent introduction of a WNV lineage 2 to Central Europe is also indicated. Black stars indicate sequences for which information was obtained from McMullen et al. [12]. The percentage of replicates in the bootstrap test (1000 replicates) is shown next to the branches. Values less than 70% are hidden.
Comparison of the WNV strain obtained in this study with partial human and mosquito-derived E gene sequences of 10 Russian WNVs obtained between 2007 and 2012 as well as with partial human- and mosquito-derived NS5 gene sequences of 23 Romanian WNVs obtained between 2011 and 2013 exhibited both 99–100% nucleotide identities.
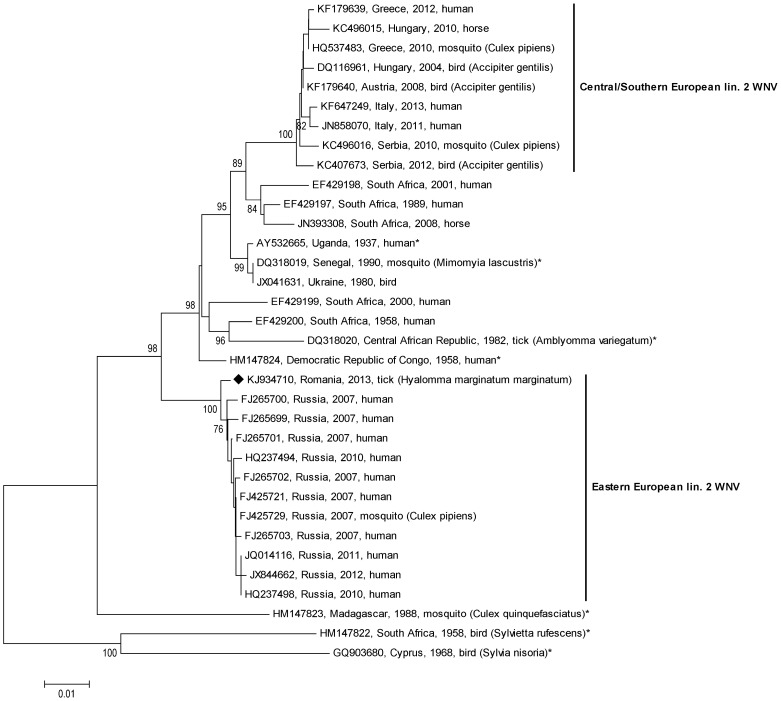
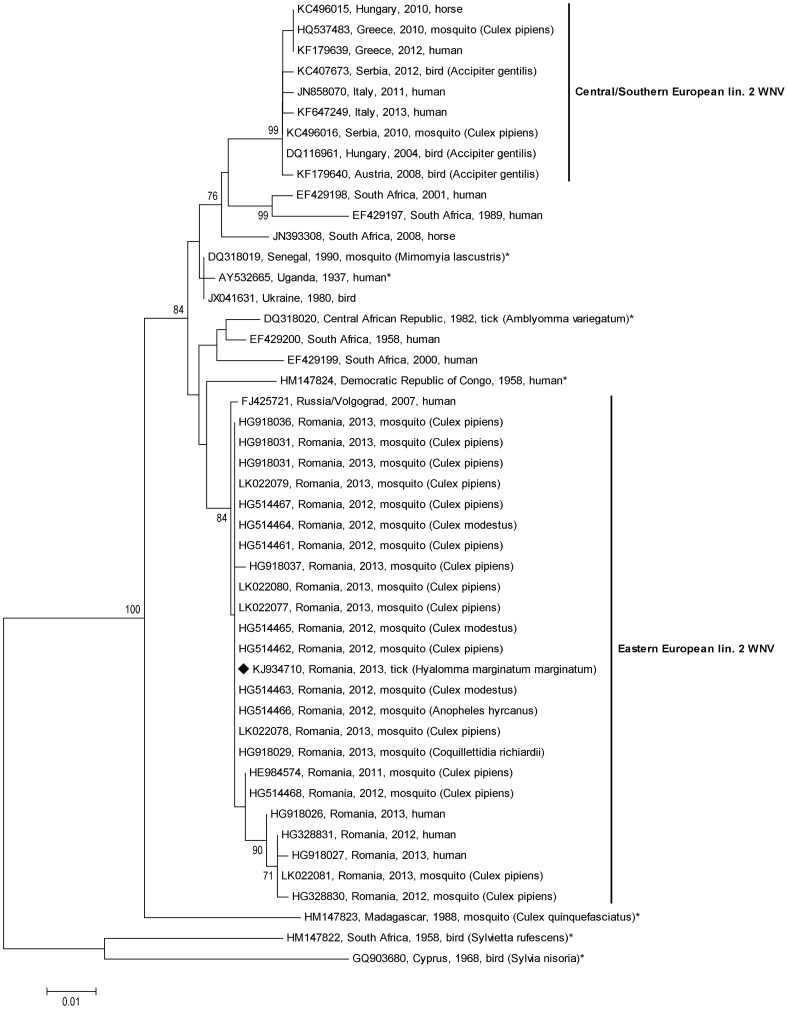
The phylogenetic analyses of the partial E gene sequences (Figure 2) and NS5 gene sequences (Figure 3), respectively, revealed a single Eastern European lineage 2 WNV cluster of closely related Russian and Romanian sequences from 2007 to 2013.
Figure 2. Phylogenetic tree of 474 nt long nucleic acid sequences (corresponding to nucleotide positions 1002–1475 of reference strain Reb_VLG_07_H, GenBank acc. no. FJ425721) within the E gene of the 24 WNV sequences included in Figure 1 and additional 10 WNVs isolated in Russia between 2007 and 2013.
Please note the distinct Eastern European lineage 2 WNV cluster consisting of Russian and Romanian sequences.
Figure 3. Phylogenetic tree of 466 nt long nucleic acid sequences (corresponding to nucleotide positions 9463–9928 of reference strain Reb_VLG_07_H, GenBank acc. no. FJ425721) within the NS5 gene of the 24 WNV sequences included in Figure 1 and additional 23 WNVs identified in Romania between 2011 and 2013.
Please note the distinct Eastern European lineage 2 WNV cluster consisting of Romanian and Russian sequences.
Nucleic acid extracts of both cloacal and oral swabs of the bird which carried the WNV positive tick as well as of its serum were negative upon the WNV (lin.1+2) RT-qPCR. The serum of this bird tested weakly-positive by WNV antibody ELISA. The confirmatory PRNT assay revealed a borderline positive result.
The complete sequence of the newly determined WNV strain, termed WNV lineage 2 strain Hyalomma/Romania/2013, is available from GenBank under accession number KJ934710. The 341bp long 12S rDNA gene sequence of the WNV-positive Hyalomma tick is available at GenBank under accession number KJ862057.
Discussion
The first major human WNV epidemic in Europe occurred in Romania in 1996, with a high rate of neurological symptoms [4]. A WNV lineage 1 was subsequently determined as the cause of this outbreak, and introduction of this virus by migrating birds from sub-Saharan Africa was suggested [19]. Interestingly, a closely related WNV was the etiologic agent of another large outbreak of WNND in 1999 in the Volgograd region of Russia with more than 800 hospitalized patients [20].
A decade later, a similar sequence of events was noticed, this time, however, the Volgograd outbreak (2007 [21]) occurred three years prior to the Romanian outbreak (2010 [5]). A newly introduced WNV lineage 2 strain was responsible for both outbreaks. The very close genetic relationship of the Romanian virus with the Russian Reb_Volgograd_07_H virus has been demonstrated in the present paper. Sirbu et al. [5] already reported that a 780 nt long WNV sequence determined from serum of an affected patient was 99.3% identical to strain Volgograd_07. In the following years several Romanian partial WNV sequences within the NS5 gene, obtained from patients and mosquitoes, were submitted to GenBank (Table 2). Our align analyses confirmed their close relationships to the above-mentioned Russian strain and revealed almost 100% nucleotide identity with the Hyalomma-derived strain determined in this study. Partial E gene sequences of Russian WNVs isolated between 2007 and 2012 (Table 2) confirmed the close relationship between Russian and Romanian lineage 2 WNV strains, too, and phylogenetic analyses of both partial E (Figure 2) and NS5 (Figure 3) gene sequences resulted in a distinct Russian/Romanian WNV lineage 2 genetic cluster, indicating local circulation and persistence of this WNV cluster in Eastern Europe.
The genetically most similar relatives of the Russian/Romanian lineage 2 WNV cluster are much older African viruses (Figures 1–3), suggesting an introduction of this virus from Africa, possibly via migrating birds. Interestingly, one of these old African WNV strains was isolated from a tick [12].
A slightly earlier independent introduction of a WNV lineage 2 from Africa to Europe occurred in or before 2004 [10]. This virus strain also managed toprevail and spread from Central Europe [22] via the Balkan states [23] to Southern European countries such as Greece [24] and Italy [25].
Ciccozzi et al. [26] suggested that the WNV lineage 2 introduction to Central Europe took place around 1999, followed by an independent introduction of another lineage 2 strain to Russia in the year 2000.
Bucharest and Volgograd are approximately 1,500 km apart, however, several flightpaths of certain species of birds, e.g. the song thrush, between these regions exist. It was not possible to determine whether the captured song thrush was a migrating or local bird. As a migrant, the song thrush breeds in most of Europe, and its migration to the Mediterranean starts in late August [27]. Its journey crosses Romania.
Migrating birds have been generally accepted as vehicles carrying viruses from Africa to Europe. Frequently, however, viremia lasts for merely a week in birds [2], a period which is considered too short to introduce exotic viruses to Europe.
Ticks are known carriers of viruses. In Israel, a total of 1.6% of Argas arboreus tick pools collected from wild and domestic birds and their nests proved WNV-positive, however all Hyalomma species tested negative [28]. WNV RNA and antigen were also detected in the tick species Ixodes pavlovskyi and I. persulcatus, which were collected from small mammals, lizards and birds in the region of Tomsk, Russia, at an average rate between 5.2 and 11.7% [29]. Depending on the tick species, WNV may persist for a very long time in ticks, e.g. at least 132 days, as demonstrated by [3].
Experimental infection with WNV performed on four ixodid tick species in the USA [30] and on H. marginatum ticks in Portugal [31] revealed that these tick species were able to acquire the virus from infected animals and to transmit it between various developmental stages. In case of H. marginatum nymphs and adults, subsequent virus transmission to uninfected hosts was observed [31]. H. marginatum is a ‘hard tick species’ occurring in southern and eastern Europe, South Asia and Africa. It is a common ectoparasite of – especially passerine – birds. Immature Hyalomma ticks may remain attached to their vertebrate hosts for up to four weeks, which enables their passive transport across continents (http://www.ecdc.europa.eu/en/healthtopics/vectors/ticks/Pages/hyalomma-marginatum-.aspx). As a two-host species moulting from larva to nymph on its first host and infesting the second host as an adult, H. marginatum ticks are able to infest a broad spectrum of vertebrate hosts including birds [32]–[34] and humans [35], [36], thereby disseminating WNV infection. Although H. marginatum usually prefers relatively dry and warm regions with low humidity, its import to Germany [37], the Netherlands [38], the United Kingdom [32], and Russia [39] has already been reported.
In the present study, the song thrush had cleared WNV, as evidenced by absence of viral RNA in samples of the bird and a low WNV antibody titer. In the attached tick, however, WNV persisted.
In the current study one out of 32 investigated ticks proved to be infected with WNV. However further research is necessary in order to draw general conclusions regarding the role of ticks in the introduction of WNV to new areas and as virus reservoir and bridge-vector.
Conclusions
Infected ticks on migrating birds may carry (new) pathogens to other areas much more efficiently than their avian hosts. The determination of the complete sequence of the currently in Romania circulating WNV strain revealed the most similar genetic relationship to the neuroinvasive Russian WNV strain Reb_Volgograd_07_H. Based on these sequences, future evolution of the Eastern European lineage 2 WNV cluster may be monitored.
Acknowledgments
The authors would like to thank Michael Kolodziejek, Dr. Karin Pachler, Dr. Tamas Bakonyi, Mag. Katharina Dimmel, Dr. Karin Sekulin, Nicholas Derby and Prof. Zdenek Hubalek for their technical assistance and general support, respectively, as well as Dr. James O. Rushton for a final language check.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.
Funding Statement
This study was partially supported by the European Union grants FP7-261504 EDENext (www.edenext.eu) to JK, MM, BK, VA, and NN as well as FP7-261391 EuroWestNile (www.eurowestnile.org) to JK and NN. Funder of both grants: European Commission. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hubálek Z, Halouzka J (1999) West Nile fever – a reemerging mosquito-borne viral disease in Europe. Emerg Infect Dis 5: 643–650 10.3201/eid0505.990505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reiter P (2010) West Nile virus in Europe: understanding the present to gauge the future. Euro Surveill 15: 19508. pii = 19508. [PubMed]
- 3. Lawrie CH, Uzcátegui NY, Gould EA, Nuttall PA (2004) Ixodid and argasid tick species and West Nile virus. Emerg Infect Dis 10: 653–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tsai TF, Popovici F, Cernescu C, Campbell GL, Nedelcu NI (1998) West Nile encephalitis epidemic in southeastern Romania. Lancet 352: 767–771. [DOI] [PubMed] [Google Scholar]
- 5.Sirbu A, Ceianu CS, Panculescu-Gatej RI, Vazquez A, Tenorio A, et al.. (2011) Outbreak of West Nile virus infection in humans, Romania, July to October 2010. Euro Surveill 16(2): pii = 19762. [PubMed]
- 6. Keyes BE, Grue CE (1982) Capturing birds with mist nets: a review. North American Bird Bander 7(1): 2–14. [Google Scholar]
- 7. Kuno G (1998) Universal diagnostic RT-PCR protocol for arboviruses. J Virol Methods 72(1): 27–41. [DOI] [PubMed] [Google Scholar]
- 8. Scaramozzino N, Crance JM, Jouan A, DeBriel DA, Stoll F, et al. (2001) Comparison of flavivirus universal primer pairs and development of a rapid, highly sensitive heminested reverse transcription-PCR assay for detection of flaviviruses targeted to a conserved region of the NS5 gene sequences. J Clin Microbiol 39: 1922–1927 10.1128/JCM.39.5.1922-1927.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weissenböck H, Kolodziejek J, Url A, Lussy H, Rebel-Bauder B, et al. (2002) Emergence of Usutu virus, an African Mosquito-Borne Flavivirus of the Japanese Encephalitis Virus Group, Central Europe. Emerg Infect Dis 8(7): 652–656 10.3201/eid0807.020094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bakonyi T, Ivanics E, Erdélyi K, Ursu K, Ferenczi E, et al. (2006) Lineage 1 and 2 strains of encephalitic West Nile virus, central Europe. Emerg Infect Dis 12: 618–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McMullen AR, Albayrak H, May FJ, Davis CT, Beasley DW, et al. (2013) Molecular evolution of lineage 2 West Nile virus. J Gen Virol 94: 318–325 10.1099/vir.0.046888-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brault AC, Huang CY, Langevin SA, Kinney RM, Bowen RA, et al. (2007) A single positively selected West Nile viral mutation confers increased virogenesis in American crows. Nat Genet 39: 1162–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kolodziejek J, Pachler K, Bin H, Mendelson E, Shulman L, et al. (2013) Barkedji virus, a novel mosquito-borne flavivirus identified in Culex perexiguus mosquitoes, Israel, 2011. J Gen Virol 94: 2449–2457 10.1099/vir.0.056200-0 [DOI] [PubMed] [Google Scholar]
- 15. Beati L, Keirans JE (2001) Analysis of the systematic relationships among ticks of the genera Rhipicephalus and Boophilus (Acari: Ixodidae) based on mitochondrial 12S ribosomal DNA gene sequences and morphological characters. J Parasitol 87(1): 32–48 doi: http://dx.doi.org/10.1645/0022-3395(2001)087 [];0032:AOTSRA2.0.CO2"> 10.1645/0022-3395(2001)087 10.1645/0022-3395(2001)087[];0032:AOTSRA2.0.CO2"> http://dx.doi.org/10.1645/0022-3395(2001)087 [0032:AOTSRA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 16. Toma L, Mancini F, Di Luca M, Cecere JG, Bianchi R, et al. (2014) Detection of microbial agents in ticks collected from migratory birds in central Italy. Vector Borne Zoonotic Dis 14: 199–205 10.1089/vbz.2013.1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sikutová S, Hornok S, Hubálek Z, Dolezálková I, Juricová Z, et al. (2009) Serological survey of domestic animals for tick-borne encephalitis and Bhanja viruses in northeastern Hungary. Vet Microbiol 135: 267–271 10.1016/j.vetmic.2008.09.082 [DOI] [PubMed] [Google Scholar]
- 18. Rushton JO, Lecollinet S, Hubálek Z, Svobodová P, Lussy H, et al. (2013) Tick-borne encephalitis virus in horses, Austria, 2011. Emerg Infect Dis 19(4): 635–637 10.3201/eid1904.121450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Savage HM, Ceianu C, Nicolescu G, Karabatsos N, Lanciotti R, et al. (1999) Entomologic and avian investigations of an epidemic of West Nile fever in Romania in 1996, with serologic and molecular characterization of a virus isolate from mosquitoes. Am J Trop Med Hyg 61: 600–611. [DOI] [PubMed] [Google Scholar]
- 20. Platonov AE, Shipulin GA, Shipulina OY, Tyutyunnik EN, Frolochkina TI, et al. (2001) Outbreak of West Nile virus infection, Volgograd Region, Russia, 1999. Emerg Infect Dis 7: 128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Platonov AE, Fedorova MV, Karan LS, Shopenskaya TA, Platonova OV, et al. (2008) Epidemiology of West Nile infection in Volgograd, Russia, in relation to climate change and mosquito (Diptera: Culicidae) bionomics. Parasitol Res 103 Suppl 1: S45–53 10.1007/s00436-008-1050-0 [DOI] [PubMed] [Google Scholar]
- 22. Bakonyi T, Ferenczi E, Erdélyi K, Kutasi O, Csörgő T, et al. (2013) Explosive spread of a neuroinvasive lineage 2 West Nile virus in Central Europe, 2008/2009. Vet Microbiol 165: 61–70 10.1016/j.vetmic.2013.03.005 [DOI] [PubMed] [Google Scholar]
- 23.Popović N, Milošević B, Urošević A, Poluga J, Lavadinović L, et al.. (2013) Outbreak of West Nile virus infection among humans in Serbia, August to October 2012. Euro Surveill 18(43). pii: 20613. [DOI] [PubMed]
- 24. Papa A, Bakonyi T, Xanthopoulou K, Vázquez A, Tenorio A, et al. (2011) Genetic characterization of West Nile virus lineage 2, Greece, 2010. Emerg Infect Dis 17: 920–922 10.3201/eid1705.101759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bagnarelli P, Marinelli K, Trotta D, Monachetti A, Tavio M, et al.. (2011) Human case of autochthonous West Nile virus lineage 2 infection in Italy, September 2011. Euro Surveill 16(43). pii: 20002. [PubMed]
- 26. Ciccozzi M, Peletto S, Cella E, Giovanetti M, Lai A, et al. (2013) Epidemiological history and phylogeography of West Nile virus lineage 2. Infect Genet Evol 17: 46–50 10.1016/j.meegid.2013.03.034 [DOI] [PubMed] [Google Scholar]
- 27.Clement P, Hathway R, Wilczur J. (2000) Thrushes (Helm Identification Guides). Christopher Helm Publishers Ltd. 392–395.
- 28. Mumcuoglu KY, Banet-Noach C, Malkinson M, Shalom U, Galun R (2005) Argasid ticks as possible vectors of West Nile virus in Israel. Vector Borne Zoonotic Dis 5: 65–71 10.1089/vbz.2005.5.65 [DOI] [PubMed] [Google Scholar]
- 29.Moskvitina NS, Romanenko VN, Ternovoi˘ VA, Ivanova NV, Protopopova EV, et al.. (2008) Detection of the West Nile virus and its genetic typing in ixodid ticks (Parasitiformes: Ixodidae) in Tomsk City and its suburbs. Parazitologiia 42: 210–225. [Article in Russian]. [PubMed]
- 30. Anderson JF, Main AJ, Andreadis TG, Wikel SK, Vossbrinck CR (2003) Transstadial transfer of West Nile virus by three species of ixodid ticks (Acari: Ixodidae). J Med Entomol 40: 528–533. [DOI] [PubMed] [Google Scholar]
- 31. Formosinho P, Santos-Silva MM (2006) Experimental infection of Hyalomma marginatum ticks with West Nile virus. Acta Virol 50: 175–180. [PubMed] [Google Scholar]
- 32. Jameson LJ, Morgan PJ, Medlock JM, Watola G, Vaux AG (2012) Importation of Hyalomma marginatum, vector of Crimean-Congo haemorrhagic fever virus, into the United Kingdom by migratory birds. Ticks Tick Borne Dis 3(2): 95–99 10.1016/j.ttbdis.2011.12.002 [DOI] [PubMed] [Google Scholar]
- 33. Toma L, Mancini F, Di Luca M, Cecere JG, Bianchi R, et al. (2014) Detection of microbial agents in ticks collected from migratory birds in central Italy. Vector Borne Zoonotic Dis 14(3): 199–205 10.1089/vbz.2013.1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hagman K, Barboutis C, Ehrenborg C, Fransson T, Jaenson TG, et al.. (2014) On the potential roles of ticks and migrating birds in the ecology of West Nile virus. Infect Ecol Epidemiol 15(4). doi: 10.3402/iee.v4.20943. [DOI] [PMC free article] [PubMed]
- 35. Papa A, Chaligiannis I, Xanthopoulou K, Papaioakim M, Papanastasiou S, et al. (2011) Ticks Parasitizing Humans in Greece. Vector-Borne and Zoonotic Dis 11(5): 539–542 10.1089/vbz.2010.0036 [DOI] [PubMed] [Google Scholar]
- 36. Bursali A, Tekin S, Keskin A, Ekici M, Dundar E (2011) Species diversity of ixodid ticks feeding on humans in Amasya, Turkey: seasonal abundance and presence of Crimean-Congo hemorrhagic fever virus. J Med Entomol 48(1): 85–93. [DOI] [PubMed] [Google Scholar]
- 37. Kampen H, Poltz W, Hartelt K, Wolfel R, Faulde M (2007) Detection of a questing Hyalomma marginatum marginatum adult female (Acari, Ixodidae) in southern Germany. Exp Appl Acarol. 2007 43(3): 227–231. [DOI] [PubMed] [Google Scholar]
- 38. Nijhof AM, Bodaan C, Postigo M, Nieuwenhuijs H, Opsteegh M, et al. (2007) Vector-Borne and Zoonotic Dis. 7(4): 585–596 10.1089/vbz.2007.0130 [DOI] [PubMed] [Google Scholar]
- 39. Movila A, Alekseev AN, Dubinina HV, Toderas I (2013 Detection of tick-borne pathogens in ticks from migratory birds in the Baltic region of Russia. Med Vet Entomol 27(1): 113–117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.