Abstract
Purpose
To examine whether GNAQ and GNA11 somatic mutations previously identified in uveal melanomas of Caucasians are associated with uveal melanomas in Chinese patients.
Methods
Uveal melanomas treated by primary enucleation in Chinese patients underwent a mutation analysis of GNAQ and GNA11 with sequencing of exon 5 and exon 4.
Results
The study included 50 patients with uveal melanoma and with a mean age of 47.6±13.0 years. During the follow-up of at least 3 years, 20 (40%) patients developed extraocular metastases. The frequencies of GNAQ and GNA11 somatic mutations in uveal melanoma were 18% (9/50) and 20% (10/50), respectively. The mutations occurred exclusively in codon 209 of exon 5. No mutations were detected in exon 4. Mutations affecting codon 209 in GNAQ were c.626A>C(Q209P) (78%) and c.626A>T(Q209L) (22%). Mutations affecting codon 209 in GNA11 were exclusively c.626A>T(Q209L) (100%). In none of the tumors, mutations of BRAF and NRAS were detected. GNAQ/11 mutations were marginally (P = 0.045) associated with optic disc involvement. In Kaplan-Meier analysis, metastasis-free survival was not significantly (P = 0.94) associated with GNAQ/11 mutations.
Conclusions
Mutations of GNAQ and GNA11 can be found in Chinese patients as in Caucasian patients with uveal melanoma, with a higher frequency reported for Caucasian patients.
Introduction
Uveal melanoma is the most common intraocular malignant tumor in adults, which arises from melanocytes of the choroid, ciliary body, and iris. It accounts for approximately 5% of all melanomas and represents the second most common form of melanoma [1], [2]. Uveal melanoma has a strong tendency to metastasize to the liver and there have been no effective treatments for the metastases. The survival time after the detection of metastases is usually less than 5 to 8 months [3], [4].
An important signaling pathway in the tumor genesis of uveal melanomas is the mitogen-activated protein kinase (MAPK) pathway incorporating RAS, RAF, MEK, and ERK [5], [6]. Previous studies have revealed that cutaneous melanomas showed oncogenic mutations in some components of the MAPKinase cascade, particularly in BRAF and NRAS [7]–[9]. In contrast, uveal melanomas did not exhibit mutations in BRAF and NRAS [10]–[11]. Interestingly, the downstream effectors of BRAF and NRAS, MEK, ERK and ELK, were constitutively activated in uveal melanomas [11]. These mutations remained undetected in uveal melanomas until recently GNAQ and GNA11 mutations in uveal melanoma were identified [12]–[14].
GNAQ and GNA11 encode Gαq, a member of the q class of G-protein alpha-subunits which are involved in mediating signals between G-protein-coupled receptors (GPCRs) and downstream effectors [15], [16]. Recently, oncogenic mutations in the G-protein α-subunit q class were found in about 83% of uveal melanomas with constitutive an activation of downstream MAPK signaling [13]. Glu209 and Arg183 are conserved in the guanosine triphosphate (GTP) binding site of Gα subunits and essential in GTP hydrolysis. The substitutions of glutamine or arginine in GNAQ or GNA11 mutations cause constitutive G-protein activation due to a reduced GTP hydrolysis [17], [18]. Metthew et al. reported that R183 mutated oncoproteins were less potent than that of Q209 [19]. GNAQ or GNA11 mutations lead to an upregulating activation of the MAPKinase pathway and appear to be major contributors to the development of uveal melanomas [12], [13].
The landmark studies on the association of uveal melanoma with GNAQ and GNA11 mutations were conducted in Caucasian populations while the status of GNAQ and GNA11 mutations in uveal melanomas of Chinese has not been investigated yet. Considering the genetic variations and their role in tumor genesis in different ethnic groups, we investigated the status of GNAQ and GNA11 mutations in uveal melanoma of Chinese patients to decrypt potential oncogenic differences between Caucasian and Chinese populations. It may promote the understanding of molecular mechanisms and of the pathogenesis of uveal melanomas in patients of different ethnic background.
Methods
The study included globes which were enucleated for uveal melanomas arising either from the ciliary body or choroid. The study was approved by the medical ethics committee of Beijing Tongren Hospital and was conducted according to the Declaration of Helsinki Principles. All participants gave their written informed consent. The diagnosis of uveal melanoma was substantialized by histological examination of slides stained by hematoxylin and eosin, and by immunohistochemistry showing the melanoma markers of HMB-45, melanin-A and S-100. Clinical and pathologic data including age, gender, largest basal tumor diameter, tumor thickness, histopathologic cell type, ciliary body involvement, optic disc involvement and presence of metastasis were collected. Optic disc involvement was defined as the occurrence of tumor cells at the peripapillary border tissue of Elschnig and Jacoby. For the purpose of our study, the presence of detected metastasis was used as the primary end point. All metastasis-free patients had to have a follow-up of at least 3 years.
The DNA preparation and screening for mutations were performed in several steps. For each tumor, five 5-µm thick, formalin-fixed, paraffin-embedded histological sections were prepared for micro-dissection. Genomic DNA was extracted from the dissected tissues using a QIAamp DNA FFPE Tissue Kit (Qiagen Co., Hilden Germany). To detect hotspot mutations, exon 4 and exon 5 of both GNAQ and GNA11 were amplified by polymerase chain reaction (PCR) in at least two separate preparations of genomic DNA. The technique was described in detail previously [21]. The primer sequences are listed in Table 1. The PCR products were purified with QIAquick (Qiagen Co., Hilden Germany) and sequenced directly.
Table 1. Used Primer Sequences.
| GNAQexon 5F: | 5′-GACTTGGATGATCATCGTCATT-3′ |
| GNAQexon 5R: | 5′-AAGAAAGCAAAGAAGTAAGTTCAC-3′ |
| GNA11exon 5F: | 5′-AGCGTCCTTGCCCGTTCTA-3′ |
| GNA11exon 5R: | 5′-AGGGCCCACCTCGTTGTC-3′ |
| GNAQexon4F: | 5′-TGTCCTTCCCTTTCCGTAGA-3′ |
| GNAQexon4R: | 5′-TGGGAAATAGGTTTCATGGACT-3′ |
| GNA11exon4F: | 5′-GCTGGTTTGGGTGCTGTGT-3′ |
| GNA11exon4R: | 5′-GGCAAATGAGCCTCTCAGTG-3′ |
The statistical analysis was performed using a commercially available statistical software (SPSS 21.0, IBM-SPSS, Chicago, USA). The associations between the clinical or pathologic parameters and the mutation types (mutated GNAQ, mutated GNA11, or neither mutated) were evaluated and the mutational status was categorized into 2 categories (mutated GNAQ/11 or neither mutated). The Student’s t-test, analysis of variance and Fisher’s exact test were applied. The Kaplan-Meier method and the log-rank test were used for survival analyses. The primary end point for metastasis-free survival was defined as the time to the development of metastatic disease, whereby death due to other causes was treated as censored. P-values were based on two-sided tests and, if less than 0.05, were regarded as statistically significant.
Results
Our study included 50 globes with uveal melanoma treated by primary enucleation. The mean age was 47.6±13.0 years (Table 2). During the follow-up, 10 patients developed extraocular metastases and 10 patients had died from metastases. There were 4 patients as censors with follow-up data before the reference time horizon. All the metastasis-free patients had a follow-up of longer than 3 years. In detail, the metastasis free survival was 94% for 1-year, 90% for 2-years and 87% for 3-years follow-up in the wild type group, 89% for 1-year, 78% for 2-years and 78% for 3-years follow-up in the GNAQ mutation group, and 90% for 1-year, 80% for 2-years and 80% for 3-years follow-up in the GNA11 mutation group.
Table 2. Clinical and Pathological Parameters of 50 Patients With Uveal Melanoma.
| Clinical-Pathological Parameters | Mean ± Standard deviation (Range) or n (%) |
| Age (Years) | 47.6±13.0 (23–78) |
| Men/Women | 20 (40%)/30 (60%) |
| Largest Basal Tumor Diameter (mm) | 14.7±3.3 (6–20) |
| Tumor Thickness (mm) | 9.5±3.2 (4–20) |
| Histopathological Cell Type: | |
| Spindle Cell Type | 42 (84%) |
| Mixed Cell Type | 7 (14%) |
| Epitheloid Cell Type | 1 (2%) |
| Scleral Invasion | 32 (64%) |
| Ciliary Body Involvement | 9 (18%) |
| Optic Disc Involvement | 12 (24%) |
| Metastasis | 10 (20%) |
| Interval from Diagnosis to End Point (Months) | 52.3±22.2 |
To investigate mutations within GNAQ and GNA11, we amplified mutation hotspot regions (exon 4 and 5) of GNAQ and GNA11 and sequenced the purified PCR products directly. Sequencing of GNAQ and GNA11 was successful in all 50 tumors. Among the 50 samples screened for GNAQ and GNA11 mutations, the overall mutation frequency was 38% (19/50), with 18% (9/50) for GNAQ and 20% (10/50)for GNA11, respectively. These figures were lower than mutation frequencies reported in Caucasian patients [12]–[14]. In all samples, the GNAQ or GNA11 mutations were mutually exclusive and occurred exclusively at codon 209. Mutations affecting codon 209 in GNAQ were c.626A>C(Q209P) (78%) and c.626A>T(Q209L) (22%). These mutations predicted substitution by proline (Q209P) in 78% of samples that were analyzed and by leucine (Q209L) in 22% of the samples. Mutations affecting codon 209 in GNA11 were exclusively c.626A>T(Q209L) (100%) (Tables 3). These mutations predicted substitution by leucine (Q209L) in all samples analyzed. In addition, all of these tumors had been sequenced for BRAF and NRAS hotspots, but no mutations were found, which were consistent with previous reports.
Table 3. Mutations Found in GNAQ and GNA11 in Chinese Patients with Uveal Melanoma.
| Gene | Mutation | n | Total (%) |
| GNAQ exon 5 | c.626A>C (Q209P) | 7 | 77.8% |
| c.626A>T (Q209L) | 2 | 22.2% | |
| GNAQ exon 4 | – | 0 | 0 |
| GNA11 exon 5 | c.626A>T (Q209L) | 10 | 100% |
| GN11Q exon 4 | – | 0 | 0 |
Examining correlations between GNAQ mutations or GNA11 mutations and the clinical-pathologic features in univariate analysis revealed a statistically association between the presence of GNAQ/11 mutations and optic disc involvement (P = 0.045) and a statistically weak association between male gender and presence of GNAQ/11 mutations (P = 0.04) (Table 4). In a logistic multivariate analysis with GNAQ/11 mutations as the dependent variable and gender and optic disc involvement as independent variables showed that GNAQ/11 mutations remained to be significantly (P = 0.034; Odds ratio (OR): 4.76; 95% Confidence Interval (CI): 1.12, 20.2) associated with an optic disc involvement while gender was no longer significantly associated (P = 0.07; OR: 3.26; 95%CI: 0.92, 11.5).
Table 4. Associations of GNAQ and GNA11 Mutations with Clinical-Pathologic Features of Uveal Melanoma in Chinese Patients.
| Factors | Total n | GNAQ mut, n (%) | GNA11 mut, N (%) | WT, N (%) | P-Value | GNAQ/11 mut, N (%) | WT, N (%) | P-Value |
| 50 (100%) | 9 (18%) | 10 (20%) | 31 (62%) | 19 (38%) | 31 (62%) | |||
| Gender | ||||||||
| Men, n (%) | 20 (40%) | 6 (12%) | 5 (10%) | 9 (18%) | 0.11 | 11 (22%) | 9 (18%) | 0.043 |
| Women, n (%) | 30 (60%) | 3 (6%) | 5 (10%) | 22 (44%) | 8 (16%) | 22 (44%) | ||
| Age (Years) | ||||||||
| Mean (range) | 47.6 (23–78) | 43.8 (32–58) | 47.6 (25–68) | 48.7 (23–78) | 0.62 | 45.8 (25–68) | 48.7 (23–78) | 0.45 |
| <50, n (%) | 27 (54%) | 7 (14%) | 5 (10%) | 15 (30%) | 0.34 | 12 (24%) | 15 (30%) | 0.31 |
| ≥50, n (%) | 23 (46%) | 2 (4%) | 5 (10%) | 16 (32%) | 7 (14%) | 16 (32%) | ||
| Largest Basal Tumor Diameter (mm) | ||||||||
| Mean (Range) | 14.7 (6––20) | 13.8 (9–20) | 15.5 (7–20) | 14.8 (6–20) | 0.53 | 14.7 (7–20) | 14.8 (6–20) | 0.93 |
| <16, n (%) | 32 (64%) | 7 (14%) | 4 (8%) | 21 (42%) | 0.22 | 11 (22%) | 21 (42%) | 0.48 |
| ≥16, n (%) | 18 (36%) | 2 (4%) | 6 (12%) | 10 (20%) | 8 (16%) | 10 (20%) | ||
| Tumor Thickness (mm) | ||||||||
| Mean (range) | 9.5 (4–20) | 9.6 (6–14) | 10.7 (5–20) | 9.1 (4–16) | 0.39 | 10.2 (5–20) | 9.1 (4–16) | 0.26 |
| <10, n (%) | 26 (52%) | 4 (8%) | 4 (8%) | 18 (36%) | 0.61 | 8 (16%) | 18 (36%) | 0.27 |
| ≥10, n (%) | 24 (48%) | 5 (10%) | 6 (12%) | 13 (36%) | 11 (22%) | 13 (26%) | ||
| Epithelioid Cell Type | ||||||||
| Yes, n (%) | 8 (16%) | 1 (2%) | 3 (6%) | 4 (8%) | 0.54 | 4 (8%) | 4 (8%) | 0.72 |
| No, n (%) | 42 (84%) | 8 (16%) | 7 (14%) | 27 (54%) | 15 (30%) | 27 (54%) | ||
| Scleral Invasion | ||||||||
| Yes, n (%) | 32 (64%) | 4 (8%) | 7 (14%) | 21 (42%) | 0.41 | 11 (22%) | 21 (42%) | 0.48 |
| No, n (%) | 18 (36%) | 5 (10%) | 3 (6%) | 10 (20%) | 8 (16%) | 10 (20%) | ||
| Ciliary Body Involvement | ||||||||
| Yes, n (%) | 9 (18%) | 0 (0%) | 3 (6%) | 6 (12%) | 0.25 | 3 (6%) | 6 (12%) | 1.00 |
| No, n (%) | 41 (82%) | 9 (18%) | 7 (14%) | 25 (50%) | 16 (32%) | 25 (50%) | ||
| Optic Disc Involvement | ||||||||
| Yes, n (%) | 12 (24%) | 4 (8%) | 4 (8%) | 4 (8%) | 0.045 | 8 (16%) | 4 (8%) | 0.045 |
| No, n (%) | 38 (76%) | 5 (10%) | 6 (12%) | 27 (54%) | 11 (22%) | 27 (54%) | ||
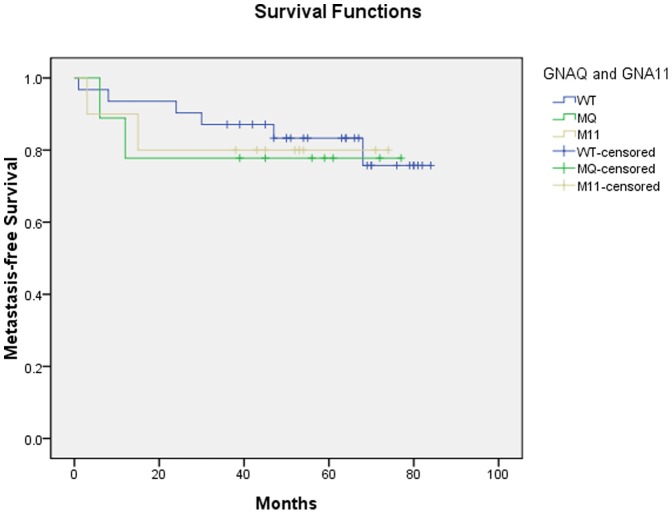
In the Kaplan-Meier analysis, the difference in metastasis-free survival among patients with GNAQ or GNA11 mutations and patients without the mutations was not statistically significant (P = 0.94), indicating that GNAQ and GNA11 mutations were not significantly associated with metastasis (Fig. 1). Due to the relatively small number of patients, a multivariable survival analysis was not performed.
Figure 1. Kaplan-Meier Estimate of Metastasis-Free Survival in Chinese Patients with Uveal Melanomas Harboring either a GNAQ or GNA11 Mutation Compared with Patients with Tumors without These Mutations.
WT: Wild type; MQ: GNAQ mutation; M11: GNA11 mutation.
Discussion
Our hospital-based study showed that mutation frequency of GNAQ and GNA11 in Chinese patients with uveal melanomas was 18% (9/50) for GNAQ and 20% (10/50) for GNA11 with an overall frequency of 38% (19/50). These results on a relatively low percentage of uveal melanomas showing mutations in GNAQ and GNA11 are different from the findings obtained in previous studies on Caucasian patients. Van Raamsdonk and her colleagues found somatic mutations in GNAQ or GNA11 in 83% of the globes with uveal melanomas [13]. The study sample of our investigation was relatively small and was not obtained in a population-based manner to allow a direct comparison of the frequency of GNAQ and GNA11 mutations between different studies. In Caucasian patients with uveal melanomas, the reported GNAQ mutation frequency varied between 36% and 53%. Van Raamsdonk and her colleagues detected GNAQ mutations in 46% of uveal melanomas [12]. Onken and his colleagues reported that 49% of the uveal melanomas harbored activating mutations in GNAQ [14]. In our study, the GNAQ mutation frequency was 18%, which was lower than the frequency in studies on Caucasian patients. It has remained unclear however, whether the difference in the reported frequencies of GNAQ mutations between the study populations allowed conclusions on inter-ethnic differences.
Interestingly with respect to cutaneous melanomas, the frequency of BRAF mutations was 66% in Caucasian patients with cutaneous melanomas while the frequency was only 25.5% in Chinese patients with cutaneous melanomas [9], [20]. Curtin et al. reported that mutations of KIT gene were detected in 29% of Caucasian patients with cutaneous melanomas while the frequency was only 11% in a Chinese cohort with cutaneous melanomas [21], [22]. These data may suggest that Caucasians and Chinese may differ in the genetic background of cutaneous melanomas. It has remained unclear whether any analogies between cutaneous melanomas and uveal melanomas can be drawn. It has also remained elusive whether the difference in iris and skin pigmentation between Caucasians and Chinese may play a role in the development of uveal melanoma, with fair complexion and light irides being generally considered risk factors for uveal melanoma [23].
Most of the reported GNAQ and GNA11 mutations occurred at codon 209, which was located in the activation domain of this kinase. Van Raamsdonk and her colleagues reported that most of GNAQ and GNA11 mutations occurred at codon 209, and few others occurred at codon 183 [12], [13]. In a similar manner, Onken and his colleagues found that 49% of primary uveal melanomas harbored activating mutations in GNAQ at codon 209 [14]. Consistent with previous studies, our data confirmed that GNAQ and GNA11 mutations in exon 5 occurred exclusively in codon 209. Although we did not detect mutations in exon 4, the relatively small number of patients included into our study may not allow concluding that mutations did generally not occur in exon 4 in a study population as ours. Interestingly, mutations in exon 4 appear also in Caucasians in a very low frequency [13].
In previous studies, associations between GNAQ and GNA11 mutations and clinical-pathologic features and prognosis were examined intensively. Van Raamsdonk and her colleagues reported that GNA11 mutations were more common in locally advanced primary tumors and in melanomas originating from the peripheral choroid or ciliary body [13]. But these associations were not statistically significant. In the study of Onken and colleagues, GNAQ mutations were not significantly associated with any clinical or histopathological parameter nor correlated with tumor progression [14]. Onken and coworkers concluded that the GNAQ mutation may be an early or initiating event in the tumorgenesis. Koopmans and his colleagues reported that patient survival in uveal melanoma was not correlated with oncogenic mutations in GNAQ and GNA11 [24]. It is in agreement with our study in which GNAQ/11 mutations were not significantly associated with age, largest tumor basis diameter, tumor thickness, tumor cell type, scleral invasion or ciliary body involvement, with these parameters being associated with a poor prognosis of uveal melanomas. Consequently, we also did not find statistically significant associations of GNAQ/11 mutations with metastasis.
Interestingly, GNAQ/11 mutations were associated with uveal melanomas involving optic disc in our study. Future studies may address whether this finding could be due to site-specificity. Specific windows for GNAQ signaling in terms of location, cell type and developmental time were previously reported, such as that GNAQ mutations induced melanocytic proliferations spared epithelial structures [16]. Our study did not allow addressing whether the association between GNAQ/11 mutations and uveal melanomas involving optic disc would be a parallel to the previous observation of two choroidal melanocytomas harboring mutations in GNAQ with one of them transforming into uveal melanoma [25].
Potential limitations of our study should be mentioned. First, it was a hospital-based study with the inherent risk of a referral bias. Second, the study sample was relatively small, so that the power of the statistical analysis was limited. Strength of the study was that it was the first study to investigate the frequency of GNAQ/11 mutations in uveal melanomas of Chinese patients.
In conclusion, mutations of GNAQ and GNA11 can be found in Chinese patients as in Caucasian patients with uveal melanoma, with the reported frequency being higher in Caucasian patients. Future studies may address whether as suggested by our study, the frequency of these mutations is indeed lower in Chinese than in Caucasians.
Acknowledgments
The authors thank Dr. Jian Jing and Prof. Xiang Jin for critically reading earlier versions of this article.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are included within the paper and its Supporting Information files.
Funding Statement
The study was supported by National Natural Science Foundation of China (Grant No. 81272981) and Beijing Nova Program (Grant No. 2008B46). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Egan KM, Seddon JM, Glynn RJ, Gragoudas ES, Albert DM (1988) Epidemiologic aspects of uveal melanoma. Surv Ophthalmol 32: 239–251. [DOI] [PubMed] [Google Scholar]
- 2. Singh AD, Bergman L, Seregard S (2005) Uveal melanoma: epidemiologic aspects. Ophthalmol Clin North Am 18: 75–84. [DOI] [PubMed] [Google Scholar]
- 3. Singh AD, Topham A (2003) Survival rates with uveal melanoma in the United States: 1973–1997. Ophthalmology 110: 962–965. [DOI] [PubMed] [Google Scholar]
- 4. Rietschel P, Panageas KS, Hanlon C, Patel A, Abramson DH, et al. (2005) Variates of survival in metastatic uveal melanoma. J Clin Oncol 23: 8076–8080. [DOI] [PubMed] [Google Scholar]
- 5. Dunn KL, Espino PS, Drobic B, He S, Davie JR (2005) The Ras-MAPK signal transduction pathway, cancer and chromatin remodelling. Biochem Cell Biol 83: 1–14. [DOI] [PubMed] [Google Scholar]
- 6. McArthur GA (2012) The coming age of ERK. The coming of age of MEK. Lancet Oncol 13: 744–745. [DOI] [PubMed] [Google Scholar]
- 7. Davies H, Bignell GR, Cox C, Stephens P, Edkins S, et al. (2002) Mutations of the BRAF gene in human cancer. Nature 417: 949–954. [DOI] [PubMed] [Google Scholar]
- 8. Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, et al. (2002) BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res 62: 6997–7000. [PubMed] [Google Scholar]
- 9. Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, et al. (2005) Distinct sets of genetic alterations in melanoma. N Engl J Med 353: 2135–2147. [DOI] [PubMed] [Google Scholar]
- 10. Cruz F, Rubin BP, Wilson D, Town A, Schroeder A, et al. (2003) Absence of BRAF and NRAS mutations in uveal melanoma. Cancer Res 63: 5761–5766. [PubMed] [Google Scholar]
- 11. Zuidervaart W, van Nieuwpoort F, Stark M, Dijkman R, Packer L, et al. (2005) Activation of the MAPK pathway is a common event in uveal melanomas although it rarely occurs through mutation of BRAF or RAS. Br J Cancer 92: 2032–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, et al. (2009) Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 457: 599–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, et al. (2010) Mutations in GNA11 in uveal melanoma. N Engl J Med 363: 2191–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Onken MD, Worley LA, Long MD, Duan S, Council ML, et al. (2008) Oncogenic mutations in GNAQ occur early in uveal melanoma.Invest Ophthalmol Vis Sci. 49: 5230–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neves SR, Ram PT, Iyengar R. G protein pathways. Science. 296: 1636–9. [DOI] [PubMed] [Google Scholar]
- 16.Van Raamsdonk CD, Fitch KR, Fuchs H, de Angelis MH, Barsh GS (2004) Effects of G-protein mutations on skin color. Nature Genet 36,961–968. [DOI] [PMC free article] [PubMed]
- 17. Sondek J, Lambright DG, Noel JP, Hamm HE, Sigler PB (1994) GTPase mechanism of G proteins from the 1.7-A crystal structure of transducin alpha-GDP-AIF-4. Nature 372: 276–279. [DOI] [PubMed] [Google Scholar]
- 18. Kalinec G, Nazarali AJ, Hermouet S, Xu N, Gutkind JS (1992) Mutated alpha subunit of the Gq protein induces malignant transformation in NIH 3T3 cells. Mol Cell Biol 12: 4687–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shirley MD, Tang H, Gallione CJ, Baugher JD, Frelin LP, et al. (2013) Sturge-Weber syndrome and port-wine stains caused by somatic mutation in GNAQ. N Engl J Med 368: 1971–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Si L, Kong Y, Xu X, Flaherty KT, Sheng X, et al. (2012) Prevalence of BRAF V600E mutation in Chinese melanoma patients: large scale analysis of BRAF and NRAS mutations in a 432-case cohort. Eur J Cancer 48: 94–100. [DOI] [PubMed] [Google Scholar]
- 21. Curtin JA, Busam K, Pinkel D, Bastian BC (2006) Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol 24: 4340–4346. [DOI] [PubMed] [Google Scholar]
- 22. Kong Y, Si L, Zhu Y, Xu X, Corless CL, et al. (2011) Large-scale analysis of KIT aberrations in Chinese patients with melanoma. Clin Cancer Res 17: 1684–1691. [DOI] [PubMed] [Google Scholar]
- 23. Mudhar HS, Parsons MA, Sisley K, Rundle P, Singh A, et al. (2004) A critical appraisal of the prognostic and predictive factors for uveal malignant melanoma. Histopathology 45: 1–12. [DOI] [PubMed] [Google Scholar]
- 24. Koopmans AE, Vaarwater J, Paridaens D, Naus NC, Kilic E, et al. (2013) Rotterdam Ocular Melanoma Study group. Patient survival in uveal melanoma is not affected by oncogenic mutations in GNAQ and GNA11. Br J Cancer 109: 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mudhar HS, Doherty R, Salawu A, Sisley K, Rennie IG (2013) Immunohistochemical and molecular pathology of ocular uvealmelanocytoma: evidence for somatic GNAQ mutations. Br J Ophthalmol 97: 924–928. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are included within the paper and its Supporting Information files.