Abstract
Purpose
Activated signal transducer and activator of transcription 3 (STAT3) regulates tumor growth, invasion, cell proliferation, angiogenesis, immune response and survival. Data regarding expression of phosphorylated (activated) STAT3 in diffuse large B-cell lymphoma (DLBCL) and the impact of phosphorylated STAT3 (pSTAT3) on prognosis are limited.
Experimental Design
We evaluated expression of pSTAT3 in de novo DLBCL using immunohistochemistry, gene expression profiling and gene set enrichment analysis. Results are analyzed in correlation with cell-of-origin, critical lymphoma biomarkers and genetic translocations.
Results
pSTAT3 expression was observed in 16% of DLBCL and was associated with advanced stage, multiple extranodal sites of involvement, activated B-cell-like (ABC) subtype, MYC expression and MYC/BCL2 expression. Expression of pSTAT3 predicted inferior overall survival (OS) and progression-free survival (PFS) in de novo DLBCL patients. When DLBCL cases were stratified according to cell-of-origin or MYC expression, pSTAT3 expression did not predict inferior outcome, respectively. Multivariate analysis showed that the prognostic predictability of pSTAT3 expression was due to its association with the ABC subtype, MYC expression and adverse clinical features. Gene expression profiling demonstrated up-regulation of genes, which can potentiate function of STAT3. Gene set enrichment analysis showed the JAK-STAT pathway to be enriched in pSTAT3+ DLBCL.
Conclusions
The results of this study provide a rationale for the ongoing successful clinical trials targeting the JAK-STAT pathway in DLBCL.
Keywords: phosphorylated STAT3, MYC, NF-κB, JAK/STAT, HDAC3, diffuse large B-cell lymphoma
INTRODUCTION
Diffuse large B-cell lymphoma (DLBCL) is the most common type of lymphoma and encompasses a heterogeneous group of tumors with several variants, subgroups and subtypes or entities (1). With the current standard treatment of rituximab, cyclophosphamide, doxorubicin, vincristine and prednisone (R-CHOP), 5-year overall survival for all patients with DLBCL ranges from 30 to 50% (2). Gene expression profiling (GEP) has identified distinct molecular subtypes, in particular the germinal center B-cell-like (GCB) and activated B-cell-like (ABC) DLBCL (3). The introduction of rituximab in the past decade has improved the survival of DLBCL patients (4), but patients with ABC-like DLBCL still display a worse outcome (5).
STAT3 is a member of the signal transducer and activator of transcription (STAT) family of transcription factors. In unstimulated cells, STAT3 is inactive and located in the cytoplasm. STAT3 is usually activated by growth factors and cytokines, such as IL-6 or IL-10 (6, 7). Activation of STAT3 is mediated by phosphorylation of a tyrosine residue (Tyr 705), which induces STAT3 dimerization and translocation to the nucleus (8). STAT3 transcriptional activity and DNA binding can be further modified by phosphorylation of Ser 727 (9) or p300-mediated acetylation at Lys 685 (10). Activated STAT3 regulates tumor growth, invasion, cell proliferation, angiogenesis, immune response and survival (11, 12). Constitutive activation of STAT3 has been shown in several solid tumors, including melanoma (13) and carcinomas of the lung (14), breast (15), ovary (16), colon (17), and prostate (18), while STAT3 mutation is not a frequent event in cancers with the exception of T-cell large granular lymphocytic leukemia (19).
Both in vitro and in vivo studies have shown that STAT3 is found to be preferentially activated in the ABC DLBCL (20, 21). In some earlier studies with a limited number of DLBCL patients treated with R-CHOP (22) or epratuzumab/R-CHOP (23), pSTAT3 expression did not show significant survival differences. However, in a very recent cohort study of DLBCL patients treated with R-CHOP, pSTAT3 expression predicted poorer outcome (24). In the present study, we evaluated the prevalence of pSTAT3 expression, its prognostic value and potential molecular insight in a large cohort (n=443) of patients with DLBCL receiving R-CHOP.
MATERIALS AND METHODS
Patient selection
A cohort of 443 patients with de novo DLBCL treated with the standard R-CHOP regimen was collected as part of The International DLBCL Rituximab-CHOP Consortium Program Study (25). All cases were centrally reviewed by a group of hematopathologists and classified according to the World Health Organization classification criteria (1). Cases that were excluded from this study included: large cell transformation of low-grade B-cell lymphoma, immunodeficiency-associated lymphoproliferative disorders (especially human immunodeficiency virus infection), primary mediastinal large B-cell lymphoma, primary cutaneous B-cell lymphoma, and primary central nervous system DLBCL. All patients had sufficient clinical and follow-up data. The present study was conducted in accord with the Declaration of Helsinki. Utilization of archived diagnostic tissue samples and therapy and outcome data procurement was approved by the Institutional Review Boards (IRBs) of all participating collaborative institutions. The overall study was approved by the IRB at The University of Texas MD Anderson Cancer Center in Houston, Texas, USA.
Tissue microarray immunohistochemical studies
Representative areas with the highest percentage of tumor cells were selected for tissue microarray (TMA) construction as described previously (5). Immunohistochemical (IHC) studies for various markers were performed; B-cell lymphoma 2 (BCL2), B-cell lymphoma 6 (BCL6), cyclin D1, CD10, CD30, Forkhead box protein P1 (FOXP1), Germinal Center B cell-expressed Transcript-1 (GCET1), Multiple Myeloma Oncogene 1 (MUM1), MYC, Nuclear factor-κB (NF-κB) components (p50, p52, p65, and c-Rel), p53, phosphorylated protein kinase B (pAKT), and phosphorylated signal transducer and activator of transcription 3 (pSTAT3). Receiver-operating characteristic (ROC) curves and X-tile analyses were utilized to assess a cutoff (5). When an optimal cutoff could not be determined by ROC curve and X-tile analyses, a conventional cutoff value for individual markers was decided based on previous reports in the literature. The cutoff scores for these markers were as follows: 10% for cyclin D1; 20% for CD30 and p53; 30% for CD10, and BCL6, 40% for MYC; 50% for pSTAT3; 60% for GCET1, MUM1 and FOXP1; and 70% for AKT and BCL2. All markers except cyclin D1 were determined by ROC curve. Any nuclear expression of each NF-κB component was considered positive.
Fluorescence in situ hybridization and TP53 sequencing
Fluorescence in situ hybridization (FISH) analysis was performed on formalin-fixed, paraffin-embedded tissue sections using BCL2 and BCL6 dual-color break-apart probes (Vysis, Downers Grove, IL), locus-specific IGH/MYC/CEP8 tricolor dual-fusion probes (Vysis, Downers Grove, IL) and a locus-specific MYC dual-color break-apart probe (Vysis, Downers Grove, IL) as reported (26). TP53 exon sequencing was performed as described previously (27). Sequencing data was compared with the TP53 reference sequence (NC_000017.10) in the Genbank database for data analysis.
Gene expression profiling and gene set enrichment analysis
Total RNA was extracted from formalin-fixed, paraffin-embedded tissue samples using the HighPure RNA Extraction Kit (Roche Applied Science, Indianapolis, IN) and subjected to gene expression profiling (GEP) as described previously (5). The robust multi-array analysis (RMA) algorithm was used for background correction (28), and then quantile normalization was conducted (29). Cell of origin (COO) classification was determined primarily based on GEP data and secondarily on immunohistochemical results using the Visco-Young algorithm as described previously (5).
Gene set enrichment analysis was performed with GSEA application (Broad Institute at MIT, Cambridge, MA) using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway gene sets (186 gene sets). GSEA results with false discovery rate (FDR) ≤ 0.3 were considered to be significant.
Statistical analysis
Comparison among categorical variables and numerical variables were carried out by using the Fisher’s exact test and Kruskal-Wallis test, respectively. OS and PFS were defined from the date of diagnosis to the date of last follow-up or death and from the date of diagnosis to the date of progression or death, respectively. Survival probability was determined using the Kaplan-Meier method, with difference compared by the log-rank test. A Cox proportional-hazards model was used for univariate and multivariate analysis. All variables with P<0.05 were considered to be statistically significant. SPSS Statistics V21 (IBM, Chicago, IL), GraphPad Prism V5 (GraphPad software, La Jolla, CA) and X-Tile version 3.6.1 (Yale University, New Haven, CT) were used for statistical analyses.
RESULTS
pSTAT3 expression is associated with poor outcome
The clinical features of the patient cohort are similar to that of our prior studies (30). Nuclear expression with or without cytoplasmic expression of pSTAT3 in large atypical lymphoid cells was regarded as positive in association with intensity (Figure S1). Expression of pSTAT3 in small, mature lymphocytes was disregarded. Clinical characteristics of patients with pSTAT3+ versus pSTAT3− DLBCL are summarized in Table 1. pSTAT3 was positive in 72 (16%) patients of which 49 were men and 23 women. pSTAT3+ DLBCL did not show distinct morphologic features compared to pSTAT3− DLBCL, having predominantly centroblastic morphology. Advanced stage (66% vs. 52%, p=0.036), involvement of multiple extranodal sites (33% vs. 20%, p=0.026) and the ABC subtype (67% vs. 45%, p=0.001) were more commonly observed in pSTAT3+ DLBCL compared with pSTAT3− DLBCL. Significant differences with respect to pSTAT3 expression were not observed for gender, age, B symptoms, ECOG score, serum LDH, IPI, tumor size and treatment response (p>0.05).
Table 1.
Clinical characteristics of 443 cases of de novo DLBCL with regard to phosphorylated STAT3 expression
| pSTAT3+ | pSTAT3− | ||
|---|---|---|---|
| N (%) | N (%) | P value | |
| Patients | 72 (16) | 371 (84) | |
| Gender | |||
| Male | 49 (68) | 211 (57) | |
| Female | 23 (32) | 160 (43) | 0.089 |
| Age | |||
| <60 | 26 (36) | 159 (43) | 0.300 |
| ≥60 | 46 (64) | 212 (57) | |
| B symptoms | |||
| Absent | 39 (60) | 223 (68) | 0.252 |
| Present | 26 (40) | 107 (32) | |
| ECOG | |||
| <2 | 53 (83) | 267 (85) | 0.706 |
| ≥2 | 11 (17) | 48 (15) | |
| Stage | |||
| I/II | 24 (34) | 172 (48) | 0.036 |
| III/IV | 46 (66) | 185 (52) | |
| Extranodal site | |||
| <2 | 47 (67) | 283 (80) | 0.026 |
| ≥2 | 23 (33) | 70 (20) | |
| LDH | |||
| Normal | 25 (37) | 116 (35) | 0.780 |
| Elevated | 42 (63) | 216 (65) | |
| IPI | |||
| 0–2 | 32 (44) | 210 (58) | 0.051 |
| 3–5 | 40 (56) | 154 (42) | |
| Tumor size, cm | |||
| <6 | 34 (63) | 194 (68) | 0.529 |
| ≥6 | 20 (37) | 92 (32) | |
| Treatment response | |||
| CR/PR | 63 (88) | 332 (90) | 0.678 |
| No response | 9 (12) | 39 (10) | |
| COO classification | |||
| GCB | 24 (33) | 204 (55) | 0.001 |
| ABC | 48 (67) | 166 (45) |
pSTAT3, phosphorylated STAT3; OS, overall survival; PFS, progression-free survival; ECOG, Eastern Cooperative Oncology Group; LDH, lactic dehydrogenase; IPI, International Prognostic Index; CR, complete remission; PR, partial remission; COO, cell-of-origin; GCB, germinal center B-cell-like; ABC, activated B-cell-like
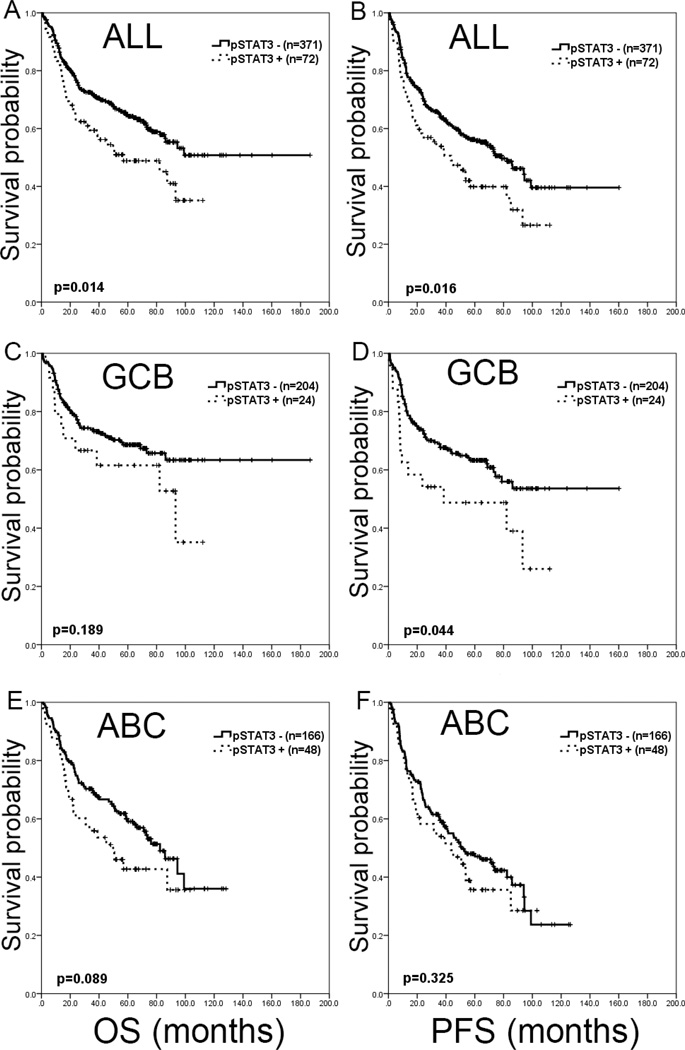
Patients with pSTAT3+ DLBCL had significantly worse OS and PFS compared with patients with pSTAT3− DLBCL (Figure 1-A and B). The 5-year OS was 48.8% for pSTAT3+ DLBCL and 64.7% for pSTAT3− DLBCL patients (p=0.014). The 5-year PFS was 39.9% for pSTAT3+ DLBCL and 56.2% for pSTAT3− DLBCL patients (p=0.016). The median OS was 56.9 months (95% CI, 18.1–95.7) for patients with pSTAT3+ DLBCL and was not reached for pSTAT3− DLBCL patients. The median PFS were 43.5 months (95% CI, 21.1–65.8) for pSTAT3+ DLBCL and 78.7 months (95% CI, 61.9–95.5) for pSTAT3− DLBCL patients.
Figure 1.
Survival impact of pSTAT3 in de novo DLBCL. A and B. Overall, pSTAT3 showed inferior overall survival (OS) and progression-free survival (PFS). C and D. In the germinal center B-cell-like (GCB) subtype, significant survival difference is only seen in PFS (p=0.044). E and F. In the activated B-cell-like (ABC) subtype, significant survival difference is not seen in either OS or PFS.
When stratified into GCB and ABC subtypes, patients with pSTAT3+ DLBCL showed a trend suggestive of inferior OS compared with pSTAT3− DLBCL patients in the GCB subgroup (p=0.189) and the ABC subgroup (p=0.089), respectively (Figure 1-C and E). Patients with pSTAT3+ DLBCL had worse PFS in the GCB subgroup (p=0.044), but only showed a trend in the ABC subgroup (p=0.325) (Figure 1-D and F). Separating stage I–II vs. III–IV, pSTAT3 retained prognostic value in the lower stage subgroup (p=0.008 for OS and p=0.021 for PFS), but not in the advanced stage subgroup (p=0.591 for OS and p=0.441 for PFS). In the patient subgroup with < 2 extranodal sites of involvement, pSTAT3 predicted poorer prognosis (p=0.029 for OS and p=0.028 for PFS), but not in patients with ≥ 2 extranodal sites (p=0.958 for OS and p=0.881 for PFS).
We also measured STAT3 mRNA levels and evaluated if STAT3 mRNA level correlated with pSTAT3 expression. To measure mRNA expression of the STAT3 gene, from the GEP dataset, we retrieved the intensity of four STAT3 probe-sets and used the average value as STAT3 expression at the mRNA level. Patients were divided into 3 groups for survival analysis according to the mean values of mRNA expression: low STAT3 mRNA (< mean – one standard deviation), high STAT3 mRNA (> mean + one standard deviation), and intermediate STAT3 mRNA levels (the remaining cases). The mean values of pSTAT3 expression in cases with low, intermediate and high STAT3 mRNA were 12%, 22% and 36%, respectively, showing correlation between STAT3 mRNA and pSTAT3 expression (p=0.0002) (Figure S2). However, STAT3 mRNA did not show significant difference with respect to OS in all cases (p=0.1487) and ABC DLBCL (p=0.6183). Although significant stratification was seen in GCB DLBCL, cases with high STAT3 mRNA and low mRNA showed the best and worst survival, respectively. Seven cases with high STAT3 mRNA in GCB DLBCL is insufficient number to achieve statistical power and could be an random event. The worse prognosis in cases with low STAT3 mRNA could be due to contamination from background inflammatory cells.
pSTAT3 expression is associated with MYC and MYC/BCL2 expression
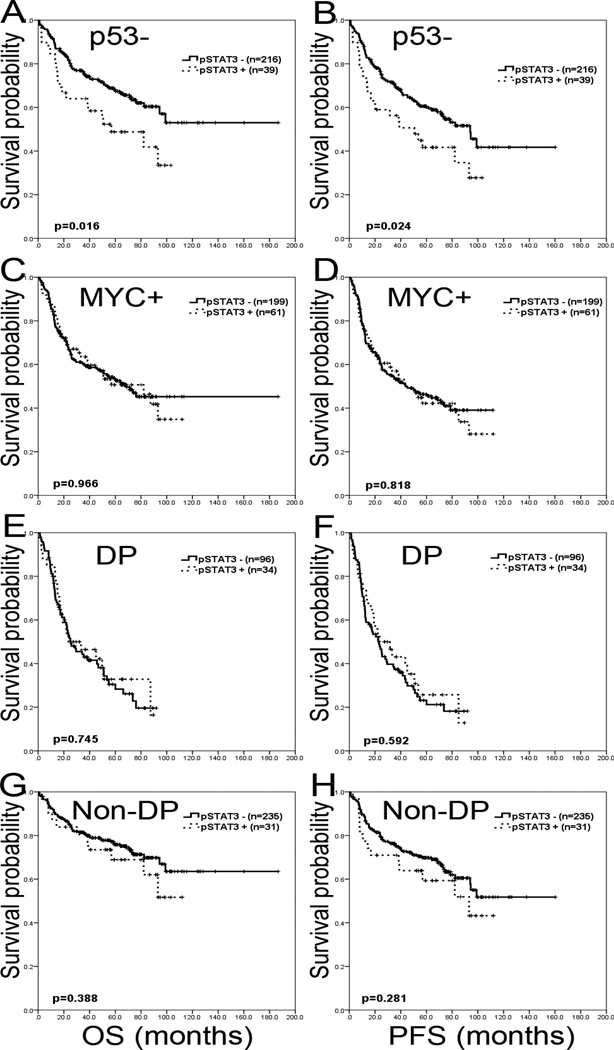
Immunohistochemical and genetic characteristics of pSTAT3+ and pSTAT3− DLBCL are summarized in Table 2. An association between STAT3 and the MYC and BCL2 genes has been reported previously by other investigators in limited patient series (31, 32). Therefore, in this study we comprehensively assessed MYC and BCL2 expression with respect to pSTAT3, in association with genetic aberrations. MYC and MYC/BCL2 double expression were more commonly observed in pSTAT3+ DLBCL cases (p<0.001 and p=0.001, respectively). However, BCL2 expression was not significantly different in DLBCL cases positive or negative for pSTAT3 expression (p=0.420). Rearrangements of BCL2, BCL6 or MYC detected by FISH did not show any significant differences between pSTAT3+ DLBCL and pSTAT3− DLBCL (p>0.05). One pSTAT3+ case (2%) showed rearrangements of both MYC and BCL6 genes and no case had coexistent MYC and BCL2 rearrangements. Regarding TP53 exon mutations detected by sequencing, TP53 gene deletions detected by FISH, or p53 expression, there were no significant differences between pSTAT3+ and pSTAT3− DLBCL cases (p>0.05). Stratifying our cohort according to p53 expression, pSTAT3+ DLBCL showed a significant worse outcome in OS (p=0.016) and PFS (p=0.024) in the p53 negative group (Figure 2-A and B). A total of 13 (19%) DLBCLs harbored TP53 mutations. All of the TP53 mutations were confined to exons 4 to 8, which are the most critical sites for DNA binding (33). Eleven cases were missense mutations and 2 cases were nonsense mutations.
Table 2.
Immunophenotypic and genetic characteristics of 433 cases of de novo DLBCL with regard to phosphorylated STAT3 expression
| Overall (%) | pSTAT3+ (%) | pSTAT3− (%) | P value | |
|---|---|---|---|---|
| MYC expression | 260/398 (65) | 61/65 (94) | 199/333 (60) | <0.001 |
| BCL2 expression | 192/398 (48) | 35/66 (53) | 157/332(47) | 0.420 |
| MYC/BCL2 co-expression | 130/396 (33) | 34/65 (52) | 96/331 (29) | 0.001 |
| BCL2 rearranged | 69/378 (18) | 7/59 (12) | 62/319 (19) | 0.201 |
| BCL6 rearranged | 107/321 (33) | 20/50 (40) | 87/271 (32) | 0.327 |
| MYC rearranged | 35/382 (9) | 4/62 (6) | 31/320 (10) | 0.630 |
| Double hit* | 13/287 (5) | 1/42 (2) | 12/245 (5) | 0.700 |
| TP53 deletion | 47/402 (12) | 5/67 (7) | 42/335 (13) | 0.300 |
| TP53 mutated | 91/408 (22) | 13/67 (19) | 78/341 (23) | 0.631 |
| p53 expression | 140/395 (36) | 27/66 (41) | 113/329 (34) | 0.326 |
| p50 expression | 154/428 (36) | 27/70 (39) | 127/358 (35) | 0.683 |
| p52 expression | 119/422 (28) | 27/71 (38) | 92/351 (26) | 0.050 |
| p65 expression | 144/428 (34) | 27/71 (38) | 117/357 (33) | 0.411 |
| cRel expression | 99/422 (24) | 13/67 (19) | 86/355 (24) | 0.436 |
| p50/p65 co-expression | 77/422 (18) | 14/70 (20) | 63/352 (18) | 0.735 |
| p50/cRel co-expression | 47/420 (11) | 6/67 (9) | 41/353 (12) | 0.674 |
| p52/p65 co-expression | 41/414 (10) | 11/70 (16) | 30/344 (9) | 0.081 |
| p52/cRel co-expression | 44/406 (11) | 6/66 (9) | 38/340 (11) | 0.829 |
| Classical NF-κB expression | 100/427 (23) | 15/70 (21) | 85/357 (24) | 0.758 |
| Alternative NF-κB expression | 71/417 (17) | 12/70 (17) | 59/347 (17) | 1.000 |
| NF-kB expression** | 134/417 (32) | 20/69 (29) | 114/348 (33) | 0.575 |
| CD30 expression | 72/439 (16) | 12/72 (17) | 60/367 (16) | 1.000 |
| pAKT expression | 76/435 (17) | 17/71 (24) | 59/364 (16) | 0.125 |
| Cyclin D1 expression | 10/432 (2) | 2/71 (3) | 8/361 (2) | 0.672 |
pSTAT3, phosphorylated STAT3
double hit, MYC and BCL2 or BCL6 rearrangements
Summation of the classical and alternative NF-κB pathway expression
Figure 2.
Survival impact of pSTAT3 in subsets of de novo DLBCL. A and B. In group of DLBCL without p53 expression by immunohistochemistry, pSTAT3 expression shows adverse effect in OS and PFS. C and D. In the group of MYC expression, pSTAT3 expression does not show different outcome in OS and PFS. E and F. In the group of MYC/BCL2 double protein expression (DP), pSTAT3 expression does not have prognostic impact. G and H. In the group without MYC/BCL2 double expression (non-DP), pSTAT3 expression does not show differences in OS and PFS..
Association of pSTAT3 expression with NF-kB components, cyclin D1 or pAKT
Cooperation of STAT3 and NF-κB is well known (21, 34). However, few studies have correlated expression of individual NF-κB components (p50, p52, p65 and cRel) with pSTAT3 expression. In this study, a significant trend that p52 single component and p52/p65 dimer were more commonly observed in pSTAT3+ DLBCL compared to pSTAT3− DLBCL (p=0.05 and p=0.081, respectively). Other NF-κB single components or combinations of NF-κB dimers (p50/p65, p50/cRel, and p52/cRel) were not more commonly expressed in pSTAT3+ DLBCL (p>0.05). Neither the classical NF-κB pathway (p50/p65 or p50/cRel), nor the alternative NF-κB pathway (p52/p65 or p52/cRel), nor both showed significant association with pSTAT3 expression (p>0.05).
Cyclin D1 is a known downstream target of pSTAT3 (35) and protein kinase B (AKT) can be activated via the JAK/STAT pathway (36). Therefore we assessed protein expression of cyclin D1 and pAKT in this study. Neither cyclin D1 nor pAKT protein expression was significantly different between pSTAT3+ DLBCL and pSTAT3− DLBCL (p>0.05).
Multivariate Analysis of Prognostic Parameters
Stratified according to MYC expression, the prognostic effect of pSTAT3 was abrogated in patients with MYC+ DLBCL (p=0.966 for OS and p=0.818 for PFS) (Figure 2-C and D). Survival analysis was not performed in MYC- group because only 4 MYC-/pSTAT3+ cases were present. Stratifying DLBCL cases into MYC/BCL2 protein double positive (DP) and non-double positive (non-DP) groups, pSTAT3 showed no effect on survival in the DP (p=0.745 for OS and p=0.592 for PFS) as well as the non-DP patient subgroups (p=0.388 for OS and p=0.281 for PFS) (Figure 2-E, F, G and H). When stratifying DP and non-DP subgroups into COO classification, no effect on survival was seen (Figure S3 and Table S1).
Univariate analysis identified advanced age, multiple extranodal sites of involvement, ABC subtype, pSTAT3 expression and MYC expression increased hazard in patients with de novo DLBCL (P<0.05 in all variables). Multivariate analysis using these 5 variables demonstrated that pSTAT3 is not an independent variable (p=0.460 for OS and p=0.523 for PFS). All the other variables were significant (Table 3).
Table 3.
Multivariate analysis of clinicopathologic variables in de novo DLBCL
| OS | PFS | |||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p value | HR | 95% CI | p value | |
| Stage III/IV | 2.182 | 1.464 to 3.252 | <0.001 | 1.758 | 1.235 to 2.503 | 0.002 |
| EN sites ≥2 | 1.560 | 1.066 to 2.284 | 0.022 | 1.662 | 1.161 to 2.379 | 0.005 |
| ABC subtype | 1.473 | 1.050 to 2.067 | 0.025 | 1.483 | 1.086 to 2.026 | 0.013 |
| pSTAT3 | 0.856 | 0.568 to 1.292 | 0.460 | 0.882 | 0.601 to 1.295 | 0.523 |
| MYC | 2.188 | 1.440 to 3.326 | <0.001 | 1.971 | 1.359 to 2.860 | <0.001 |
OS, overall survival; PFS, progression-free survival; EN sites, extranodal sites; ABC, activated B-cell-like
GEP and GSEA of pSTAT3+ DLBCL
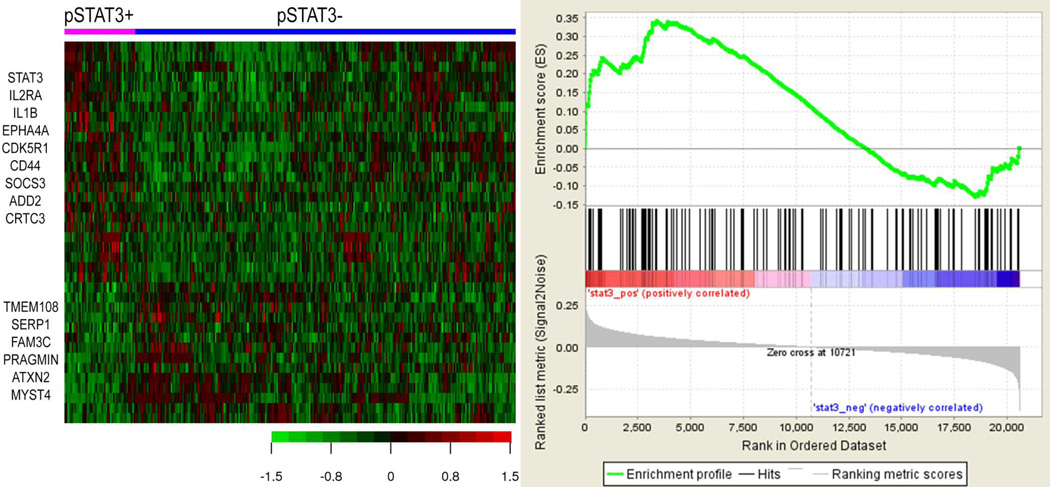
To further characterize pSTAT3+ DLBCL, we compared the GEP signatures of pSTAT3+ and pSTAT3− DLBCL using the Affymetrix HGU133plus2 platform (Affymetrix, Santa Clara, CA, USA) (Figure 3A). With a false discovery rate (FDR) threshold of 0.3, a total of 26 genes were differentially expressed between the two groups including 15 up-regulated genes and 11 down-regulated genes in pSTAT3+ DLBCL. Among the up-regulated genes in pSTAT3+ DLBCL, STAT3, IL2RA, CD44, EPHA4, and CDK5R1 were significant. Upregulation of the IL2RA gene suggests a positive feedback mechanism of pSTAT3 because IL-2 activates STAT3 via activating Janus kinase (JAK). CD44 can acetylate and dimerize STAT3 in a growth factor- and cytokine-independent manner and has been shown to be involved in cell migration, tumor invasion and metastasis, potentiating STAT3 function and partially explaining the adverse outcome in pSTAT3+ DLBCL (37). EPHA4 encodes ephrin receptor A4, which has receptor tyrosine kinase activity. CDK5R1 encodes CDK5/p35, which is a downstream target molecule of ephrin receptor A4 and it expression has been shown to predict inferior survival in patients with non-small cell lung cancer (38). Many of the down-regulated genes encode proteins of unknown function. Huang et al proposed a prognostically valuable 11-gene signature in PY-STAT3+ DLBCLs (24). The 11 genes were HSD17B4, MT1X, NAT8L, PCNX, RHEB, RNF149, SLA, SLC2A13, ZNF805, C15orf29 and ZNF420. However, few of these genes (RHEB, PCNX and ZNF420) were expressed with marginal significance for survival in our cohort and combination of the 11-genes showed overall significant trend (p=0.07). GSEA analysis showed trends for several gene sets, including the JAK-STAT pathway gene set, being enriched in pSTAT3+ DLCBCL (Figure 3-B).
Figure 3.
Gene expression profiling and gene set enrichment analysis. A. Gene expression profiling shows that pSTAT3+ DLBCL is genetically distinct from pSTAT3− DLBCL. Representative up-regulated and down-regulated genes in the pSTAT3+ DLBCL are illustrated at the upper end and lower end, respectively. B. Gene set enrichment analysis shows a trend that KEGG JAK-STAT pathway is enriched in the pSTAT3+ DLBCL.
DISCUSSION
In the present study, 16% of de novo DLBCL cases expressed pSTAT3. The prevalence of pSTAT3 expression in DLBCL is lower than that reported in earlier studies in which 30–40% of DLBCL expressed pSTAT3 (22, 24, 39, 40). Of note, pSTAT3 expression are 67%, 51%, 34%, 23%, 16%, and 11% if the pSTAT3 cutoffs are 10%, 20%, 30%, 40%, 50% and 60%, respectively (Table S2). With 30% cutoff, the prevalence of pSTAT3 expression is in similar range to prior data. However, our cutoff value for pSTAT3 was determined based on a ROC curve to achieve optimal sensitivity and specificity. Compared to survival analysis using 50% cutoff (Figure 1), 30% cutoff does not show any significant differences between pSTAT3+ DLBCL and pSTAT3− DLBCL (Figure S4). Furthermore, no clinical differences are observed between the two groups using 30% cutoff except COO classification (Table S3). We additionally conducted survival analysis in 4 separate quartile groups (Q1: <30%, Q2: ≥30% and <50%, Q3: ≥50% and <70%, and Q4: ≥70%, Table S4). Although overall quartile analysis showed significant stratification, Q3 and Q4 did not show significant difference (Figure S5, lower row). Merging Q3 and Q4, we conducted survival analysis in 3 groups (T1: <30%, T2: ≥30% and <50%, T3: ≥50%, Table S5). T2 and T3 showed significant difference, whereas T1 and T2 did not (Figure S5, upper row). Based on our analysis, we chosed 50% as an optimal cutoff for pSTAT3 expression. With 50% cutoff, we could minimize possible contamination of pSTAT3 expression in non-lymphoma cells. As has also been shown by others (22–24, 39), pSTAT3 was more commonly expressed in ABC-DLBCL. Such a finding can be explained by Ding et al who showed STAT3 gene transcription was negatively regulated by BCL6, hence providing a basis for STAT3 activation in ABC-DLBCL (20). We also showed that STAT3 mRNA level is quantitatively correlated with pSTAT3 expression. (Figure S2)
We showed that pSTAT3 expression in de novo DLBCL is associated with adverse outcome, confirming a similar observation by Huang et al and Meier et al (24, 40). We could not, however, show that pSTAT3 expression is an independent prognostic factor in DLBCL patients in multivariate analysis (Table 3). Furthermore, pSTAT3 expression is associated with MYC expression and MYC/BCL2 double expression. However, when MYC expression was controlled, pSTAT3 did not significantly predict prognosis (Figure 2). In cases with MYC/BCL2 DP or non-DP, significant difference was not observed, either. No survival difference was seen after stratified into GCB/ABC in DP and non-DP groups (Figure S4). Furthermore, multivariate analysis did not identify pSTAT3 as an independent hazard. Instead, other variables, such as stage, cell-of-origin classification and MYC expression, were shown to be independent prognosticators (Table 3). We additionally conducted bivariate analyses including each variable in the multivariate analysis and pSTAT3 expression to see if increased hazardous effect of pSTAT3 is observed (Table S6). pSTAT3 did not show increased hazard except when pSTAT3 co-analyzed with ABC phenotype. Therefore, inferior outcome of patients with pSTAT3+ DLBCL appeared to be a result of the associated poor prognostic indicators such as advanced stage disease and multiple extranodal sites of involvement. Interestingly, in patients with less aggressive or advanced disease such as limited stage (stage I or II), involvement of < 2 extranodal sites, absence of p53 expression, or GCB-type DLBCL, pSTAT3 expression showed a worse outcome. Our data therefore suggest that pSTAT3 expression predicts poor outcome in less aggressive/advanced DLBCL, but it does not provide additional survival impact in patients with advanced disease or with already known poor prognostic factors including MYC expression or BCL2/MYC co-expression (23, 30, 41).
The results of GEP analysis in this study showed that pSTAT3+ DLBCL is distinct from pSTAT3− DLBCL. Many of the up-regulated genes, such as STAT3, IL2RA, IL1B, EPHA4, and CDK5R1 in pSTAT3+ DLBCL contain several binding sites for STAT3 in their promoters and some of the up-regulated genes have been reported to correlate with aggressive features in carcinomas (37, 38). The results of GSEA also provide evidence that the JAK-STAT pathway is enriched in pSTAT3+ DLBCL, although the statistical significance is marginal. In a recent study by Huang et al (24), the authors suggested that an 11-gene signature was associated with poor OS in patients with STAT3+ DLBCL, treated with CHOP and R-CHOP, respectively, without achieving statistical significance. Some of these genes were marginally up-regulated in pSTAT3+ DLBCL by GEP in our study, partially in agreement with Huang et al (24).
Although our GEP results did not show that MYC was significantly up-regulated in pSTAT3+ DLBCL, it is not surprising to observe the correlation between MYC expression and pSTAT3 in this study because MYC is a known target of STAT3 (31, 42, 43). Kikuchi et al demonstrated that STAT3 was essential for MYC mRNA expression in a proB cell line (BAF/B03) (31). Barre et al showed that STAT3 was recruited to the MYC promoters upon IL-6 stimulation of glioblastoma cells using chromatin immunoprecipitation (ChIP) (42). Bowman et al reported that MYC expression was induced by STAT3 in fibroblasts (43). In primary DLBCL patient samples, Gupta et al showed a trend between pSTAT3 and MYC expression in a small study of 23 patients (23).
NF-κB and pSTAT3 are known to cooperate and four different modes of reciprocal interaction in transcriptional control have been suggested (34). 1) STAT3/p65 in the nucleus can recruit p300 histone acetyltransferase, which acetylates histone as well as p65. Acetylated p65 is less prone to nuclear export, hence persists longer in the nucleus where it is transcriptionally active. 2) In the nucleus, NF-κB/STAT3 can bind to particular DNA target sequences at sites where neither can bind alone. 3) Unphosphorylated STAT3 can bind with and displace IκB so that NF-κB can be activated without any upstream signals. 4) I-κBζ, which is induced by NF-κB, can bind to STAT3 and inhibit STAT3− DNA binding. We showed a significant trend that p52 single expression and p52/p65 dimer expression was more commonly seen in pSTAT3+ DLBCL, supporting a crosstalk between STAT3 and NF-κB (44, 45). Lam et al showed the induction of IL-6 and IL-10 by NF-κB pathway in ABC DLBCL cell lines (21). Considering constitutive activation of NF-κB and common pSTAT3 expression in ABC DLBCL, targeting either or both of them appears to be an attractive addition to the current R-CHOP regimen especially in this subtype.
Recently, STAT3 was found to be a substrate of histone deacetylase 3 (HDAC3) in ABC-DLBCL (46). Although the primary substrates of HDACs are histones, non-histone proteins including NF-κB transcription factors also could be targeted by HDACs (47). In the context of crosstalk between NF-κB and STAT3, targeting HDAC seems to be a reasonable approach. Gupta et al demonstrated that a HDAC inhibitor panobinostat (also known as LBH589) could dephosphorylate STAT3 in an ABC-DLBCL cell line, (Ly3), as well as DLBCL patient samples, in a dose-dependent manner (46). However, a recent phase I study using panobinostat with everolimus in patients with relapsed and refractory lymphoma did not show a response in patients with DLBCL (48). Lam et al also showed that blocking JAK/STAT3 activation using a JAK inhibitor in ABC-DLBCL cell lines reduces cell proliferation and survival (21). They also demonstrated a synergistic effect on tumor cell death by combining JAK/STAT3 and NF-κB inactivation in ABC-DLBCL cell lines. A phase II clinical trial using an oral JAK inhibitor (INCB18424) is in progress in patients with relapsed or refractory DLBCL or peripheral T-cell lymphoma (NCT01431209).
Scuto et al demonstrated that inhibition of STAT3 with short hairpin RNA was associated with apoptosis in a human ABC-like DLBCL cell line (Ly3), as well as tumor regression in NOD-SCID (nonobese diabetic/severe combined immunodeficient) mice injected by Ly3 cells (49). In their study, direct STAT3 inhibition induced significant reduction of STAT3 targets such as MYC and survivin at the protein level. Survivin is an anti-apoptotic protein and has been shown to be unfavorable prognostic marker in DLBCL (50). Furthermore, Scuto et al also showed that inhibition of STAT3 thwarted IL-10-dependent STAT3 activation, proving that STAT3 can be an attractive therapeutic target. Currently, a multi-institution phase I/II clinical trial using an antisense oligonucleotide inhibitor targeting STAT3 in DLBCL patients is in progress (NCT01563302).
In summary, pSTAT3 expression is more commonly seen in ABC-DLBCL and is associated with advanced stage, multiple extranodal sites of involvement, MYC expression and MYC/BCL2 double expression. Although pSTAT3 is not an independent prognostic marker, pSTAT3 expression in DLBCL predicts an unfavorable outcome. Gene expression profiling suggests that pSTAT3+ DLBCL is molecularly distinct from pSTAT3− DLBCL with a unique oncogenic pathway activation signature, which identify a group of patients with who might benefit from the use of molecularly targeted therapies. Ongoing successful clinical trials targeting the STAT3 pathway may shed new light on the significance of STAT3 expression, particularly in refractory/resistant DLBCL.
Supplementary Material
Translational Relevance.
Activated signal transducer and activator of transcription 3 (STAT3) plays a role in tumor invasion, cell proliferation, angiogenesis and immune response. Enhanced STAT3 activity has been found in many cancers, but its role in diffuse large B-cell lymphoma (DLBCL) has not been heavily studied. In this study, we showed that expression of phosphorylated STAT3 (pSTAT3) was seen in 16% of de novo DLBCL and was associated advanced stage, multiple extranodal sites of involvement, activated B-cell-like (ABC) subtype, MYC expression and MYC/BCL2 expression. Although multivariate analysis did not show expression of pSTAT3 was an independent prognostic marker, the expression of pSTAT3 showed inferior overall survival and progression-free survival in de novo DLBCL patients treated with R-CHOP. pSTAT3+ DLBCL up-regulates genes potentiating function of STAT3, that has been found to be an attractive therapeutic targets.
ACKNOWLEDGEMENTS
This work was supported by the Fellowship Award (C.Y.O); the Harold C. and Mary L. Daily Endowment Fellowships and Shannon Timmins Fellowship for Leukemia Research Award (Z.Y.X.-M.); The University of Texas MD Anderson Cancer Center Institutional Research Grant Award, an MD Anderson Lymphoma Specialized Programs of Research Excellence (SPORE) Research Development Program Award, an MD Anderson Myeloma SPORE Research Development Program Award, MD Anderson Collaborative Research Funds with High-Throughput Molecular Diagnostics, Gilead Pharmaceutical, Adaptive Biotechnologies, and Roche Molecular Systems (K.H.Y.). This work was also partially supported by National Cancer Institute and National Institutes of Health grants (R01CA138688, 1RC1CA146299, P50CA136411 and P50CA142509), and by the MD Anderson Cancer Center Support Grant CA016672.
Footnotes
Authorship
Contribution: C.Y.O and K.H.Y designed and conducted the research and performed the statistical analysis; C.Y.O., J.C., Z.Y.X.-M., A.T., G.C.M., L.L., C.V., S.M.M., K.D., A.C., A.O., Y.Z., G.B., K.L.R., E.D.H., W.W.L.C., J.H.K., J.H., X.Y.Z., M.P., A.J.M.F., M.B.M., F.B., J.P.F., M.A.P., J.N.W., L.J.M., and K.H.Y. contributed vital new reagents, resources, technology, and analytical tools; C.Y.O., J.C., L.L., A.T., C.V., S.M.M., K.D., A.C., A.O., Y.Z., G.B., K.L.R., E.D.H., W.W.L.C., J.H.K., J.H., X.Y.Z., M.P., A.J.M.F., M.B.M., J.P.F., M.A.P., J.N.W., and K.H.Y. collected clinical and follow-up data under approval by the institutional review boards and the material transfer agreement; C.Y.O, L.J.M., and K.H.Y. wrote the manuscript; and all authors contributed vital strategies, participated in discussions, and provided scientific input.
DISCLOSURE OF CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
References
- 1.Stein H, Warnke RA, Chan WC, Jaffe ES, Chan JKC, Gatter KC, et al., editors. Diffuse large B-cell lymphoma, not otherwise specified. 4th ed. Lyon: International Agency for Research on Cancer (IARC); 2008. [Google Scholar]
- 2.Cultrera JL, Dalia SM. Diffuse large B-cell lymphoma: current strategies and future directions. Cancer Control. 2012;19:204–213. doi: 10.1177/107327481201900305. [DOI] [PubMed] [Google Scholar]
- 3.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 4.Sehn LH, Donaldson J, Chhanabhai M, Fitzgerald C, Gill K, Klasa R, et al. Introduction of combined CHOP plus rituximab therapy dramatically improved outcome of diffuse large B-cell lymphoma in British Columbia. J Clin Oncol. 2005;23:5027–5033. doi: 10.1200/JCO.2005.09.137. [DOI] [PubMed] [Google Scholar]
- 5.Visco C, Li Y, Xu-Monette ZY, Miranda RN, Green TM, Tzankov A, et al. Comprehensive gene expression profiling and immunohistochemical studies support application of immunophenotypic algorithm for molecular subtype classification in diffuse large B-cell lymphoma: a report from the International DLBCL Rituximab-CHOP Consortium Program Study. Leukemia. 2012;26:2103–2113. doi: 10.1038/leu.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niemand C, Nimmesgern A, Haan S, Fischer P, Schaper F, Rossaint R, et al. Activation of STAT3 by IL-6 and IL-10 in primary human macrophages is differentially modulated by suppressor of cytokine signaling 3. J Immunol. 2003;170:3263–3272. doi: 10.4049/jimmunol.170.6.3263. [DOI] [PubMed] [Google Scholar]
- 7.Yu H, Jove R. The STATs of cancer--new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 8.Bhattacharya S, Schindler C. Regulation of Stat3 nuclear export. J Clin Invest. 2003;111:553–559. doi: 10.1172/JCI15372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wen Z, Zhong Z, Darnell JE., Jr Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 10.Wang R, Cherukuri P, Luo J. Activation of Stat3 sequence-specific DNA binding and transcription by p300/CREB-binding protein-mediated acetylation. J Biol Chem. 2005;280:11528–11534. doi: 10.1074/jbc.M413930200. [DOI] [PubMed] [Google Scholar]
- 11.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, et al. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 12.Calo V, Migliavacca M, Bazan V, Macaluso M, Buscemi M, Gebbia N, et al. STAT proteins: from normal control of cellular events to tumorigenesis. J Cell Physiol. 2003;197:157–168. doi: 10.1002/jcp.10364. [DOI] [PubMed] [Google Scholar]
- 13.Xie TX, Huang FJ, Aldape KD, Kang SH, Liu M, Gershenwald JE, et al. Activation of stat3 in human melanoma promotes brain metastasis. Cancer Res. 2006;66:3188–3196. doi: 10.1158/0008-5472.CAN-05-2674. [DOI] [PubMed] [Google Scholar]
- 14.Gao SP, Mark KG, Leslie K, Pao W, Motoi N, Gerald WL, et al. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest. 2007;117:3846–3856. doi: 10.1172/JCI31871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berishaj M, Gao SP, Ahmed S, Leslie K, Al-Ahmadie H, Gerald WL, et al. Stat3 is tyrosine-phosphorylated through the interleukin-6/glycoprotein 130/Janus kinase pathway in breast cancer. Breast Cancer Res. 2007;9:R32. doi: 10.1186/bcr1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang C, Lee H, Jove V, Deng J, Zhang W, Liu X, et al. Prognostic significance of B-cells and pSTAT3 in patients with ovarian cancer. PLoS One. 2013;8:e54029. doi: 10.1371/journal.pone.0054029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin L, Liu A, Peng Z, Lin HJ, Li PK, Li C, et al. STAT3 is necessary for proliferation and survival in colon cancer-initiating cells. Cancer Res. 2011;71:7226–7237. doi: 10.1158/0008-5472.CAN-10-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barton BE, Murphy TF, Shu P, Huang HF, Meyenhofer M, Barton A. Novel single-stranded oligonucleotides that inhibit signal transducer and activator of transcription 3 induce apoptosis in vitro and in vivo in prostate cancer cell lines. Mol Cancer Ther. 2004;3:1183–1191. [PubMed] [Google Scholar]
- 19.Koskela HL, Eldfors S, Ellonen P, van Adrichem AJ, Kuusanmaki H, Andersson EI, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med. 2012;366:1905–1913. doi: 10.1056/NEJMoa1114885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding BB, Yu JJ, Yu RY, Mendez LM, Shaknovich R, Zhang Y, et al. Constitutively activated STAT3 promotes cell proliferation and survival in the activated B-cell subtype of diffuse large B-cell lymphomas. Blood. 2008;111:1515–1523. doi: 10.1182/blood-2007-04-087734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lam LT, Wright G, Davis RE, Lenz G, Farinha P, Dang L, et al. Cooperative signaling through the signal transducer and activator of transcription 3 and nuclear factor-{kappa}B pathways in subtypes of diffuse large B-cell lymphoma. Blood. 2008;111:3701–3713. doi: 10.1182/blood-2007-09-111948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu ZL, Song YQ, Shi YF, Zhu J. High nuclear expression of STAT3 is associated with unfavorable prognosis in diffuse large B-cell lymphoma. J Hematol Oncol. 2011;4:31. doi: 10.1186/1756-8722-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta M, Maurer MJ, Wellik LE, Law ME, Han JJ, Ozsan N, et al. Expression of Myc, but not pSTAT3, is an adverse prognostic factor for diffuse large B-cell lymphoma treated with epratuzumab/R-CHOP. Blood. 2012;120:4400–4406. doi: 10.1182/blood-2012-05-428466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang X, Meng B, Iqbal J, Ding BB, Perry AM, Cao W, et al. Activation of the STAT3 Signaling Pathway Is Associated With Poor Survival in Diffuse Large B-Cell Lymphoma Treated With R-CHOP. J Clin Oncol. 2013;31:4520–4528. doi: 10.1200/JCO.2012.45.6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu-Monette ZY, Moller MB, Tzankov A, Montes-Moreno S, Hu W, Manyam GC, et al. MDM2 phenotypic and genotypic profiling, respective to TP53 genetic status, in diffuse large B-cell lymphoma patients treated with rituximab-CHOP immunochemotherapy: a report from the International DLBCL Rituximab-CHOP Consortium Program. Blood. 2013;122:2630–2640. doi: 10.1182/blood-2012-12-473702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tzankov A, Xu-Monette ZY, Gerhard M, Visco C, Dirnhofer S, Gisin N, et al. Rearrangements of MYC gene facilitate risk stratification in diffuse large B-cell lymphoma patients treated with rituximab-CHOP. Mod Pathol. 2013 Dec 13; doi: 10.1038/modpathol.2013.214. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 27.Xu-Monette ZY, Wu L, Visco C, Tai YC, Tzankov A, Liu WM, et al. Mutational profile and prognostic significance of TP53 in diffuse large B-cell lymphoma patients treated with R-CHOP: report from an International DLBCL Rituximab-CHOP Consortium Program Study. Blood. 2012;120:3986–3996. doi: 10.1182/blood-2012-05-433334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 29.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 30.Hu S, Xu-Monette ZY, Tzankov A, Green T, Wu L, Balasubramanyam A, et al. MYC/BCL2 protein coexpression contributes to the inferior survival of activated B-cell subtype of diffuse large B-cell lymphoma and demonstrates high-risk gene expression signatures: a report from The International DLBCL Rituximab-CHOP Consortium Program. Blood. 2013;121:4021–4031. doi: 10.1182/blood-2012-10-460063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiuchi N, Nakajima K, Ichiba M, Fukada T, Narimatsu M, Mizuno K, et al. STAT3 is required for the gp130-mediated full activation of the c-myc gene. J Exp Med. 1999;189:63–73. doi: 10.1084/jem.189.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fukada T, Hibi M, Yamanaka Y, Takahashi-Tezuka M, Fujitani Y, Yamaguchi T, et al. Two signals are necessary for cell proliferation induced by a cytokine receptor gp130: involvement of STAT3 in anti-apoptosis. Immunity. 1996;5:449–460. doi: 10.1016/s1074-7613(00)80501-4. [DOI] [PubMed] [Google Scholar]
- 33.Joerger AC, Ang HC, Fersht AR. Structural basis for understanding oncogenic p53 mutations and designing rescue drugs. Proc Natl Acad Sci U S A. 2006;103:15056–15061. doi: 10.1073/pnas.0607286103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grivennikov SI, Karin M. Dangerous liaisons: STAT3 and NF-kappaB collaboration and crosstalk in cancer. Cytokine Growth Factor Rev. 2010;21:11–19. doi: 10.1016/j.cytogfr.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7:41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 36.Wu K, Chang Q, Lu Y, Qiu P, Chen B, Thakur C, et al. Gefitinib resistance resulted from STAT3-mediated Akt activation in lung cancer cells. Oncotarget. 2013;4(12):2430–2438. doi: 10.18632/oncotarget.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.So JY, Smolarek AK, Salerno DM, Maehr H, Uskokovic M, Liu F, et al. Targeting CD44-STAT3 signaling by Gemini vitamin D analog leads to inhibition of invasion in basal-like breast cancer. PLoS One. 2013;8:e54020. doi: 10.1371/journal.pone.0054020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu JL, Wang XY, Huang BX, Zhu F, Zhang RG, Wu G. Expression of CDK5/p35 in resected patients with non-small cell lung cancer: relation to prognosis. Med Oncol. 2011;28:673–678. doi: 10.1007/s12032-010-9510-7. [DOI] [PubMed] [Google Scholar]
- 39.Stewart DA, Bahlis N, Mansoor A. pY-STAT3 and p53 expression predict outcome for poor prognosis diffuse large B-cell lymphoma treated with high dose chemotherapy and autologous stem cell transplantation. Leuk Lymphoma. 2009;50:1276–1282. doi: 10.1080/10428190903015628. [DOI] [PubMed] [Google Scholar]
- 40.Meier C, Hoeller S, Bourgau C, Hirschmann P, Schwaller J, Went P, et al. Recurrent numerical aberrations of JAK2 and deregulation of the JAK2-STAT cascade in lymphomas. Mod Pathol. 2009;22:476–487. doi: 10.1038/modpathol.2008.207. [DOI] [PubMed] [Google Scholar]
- 41.Valera A, Lopez-Guillermo A, Cardesa-Salzmann T, Climent F, Gonzalez-Barca E, Mercadal S, et al. MYC protein expression and genetic alterations have prognostic impact in patients with diffuse large B-cell lymphoma treated with immunochemotherapy. Haematologica. 2013;98:1554–1562. doi: 10.3324/haematol.2013.086173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barre B, Avril S, Coqueret O. Opposite regulation of myc and p21waf1 transcription by STAT3 proteins. J Biol Chem. 2003;278:2990–2996. doi: 10.1074/jbc.M210422200. [DOI] [PubMed] [Google Scholar]
- 43.Bowman T, Broome MA, Sinibaldi D, Wharton W, Pledger WJ, Sedivy JM, et al. Stat3-mediated Myc expression is required for Src transformation and PDGF-induced mitogenesis. Proc Natl Acad Sci U S A. 2001;98:7319–7324. doi: 10.1073/pnas.131568898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee H, Herrmann A, Deng JH, Kujawski M, Niu G, Li Z, et al. Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer Cell. 2009;15:283–293. doi: 10.1016/j.ccr.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang J, Liao X, Agarwal MK, Barnes L, Auron PE, Stark GR. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFkappaB. Genes Dev. 2007;21:1396–1408. doi: 10.1101/gad.1553707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gupta M, Han JJ, Stenson M, Wellik L, Witzig TE. Regulation of STAT3 by histone deacetylase-3 in diffuse large B-cell lymphoma: implications for therapy. Leukemia. 2012;26:1356–1364. doi: 10.1038/leu.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams SA, Chen LF, Kwon H, Ruiz-Jarabo CM, Verdin E, Greene WC. NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 2006;25:139–149. doi: 10.1038/sj.emboj.7600900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oki Y, Buglio D, Fanale M, Fayad L, Copeland A, Romaguera J, et al. Phase I Study of Panobinostat plus Everolimus in Patients with Relapsed or Refractory Lymphoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:6882–6890. doi: 10.1158/1078-0432.CCR-13-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scuto A, Kujawski M, Kowolik C, Krymskaya L, Wang L, Weiss LM, et al. STAT3 inhibition is a therapeutic strategy for ABC-like diffuse large B-cell lymphoma. Cancer Res. 2011;71:3182–318. doi: 10.1158/0008-5472.CAN-10-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adida C, Haioun C, Gaulard P, Lepage E, Morel P, Briere J, et al. Prognostic significance of survivin expression in diffuse large B-cell lymphomas. Blood. 2000;96:1921–1925. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.