Abstract
Next generation sequencing was used to identify Notch mutations in a large collection of diverse solid tumors. NOTCH1 and NOTCH2 rearrangements leading to constitutive receptor activation were confined to triple negative breast cancers (TNBC, 6 of 66 tumors). TNBC cell lines with NOTCH1 rearrangements associated with high levels of activated NOTCH1 (N1-ICD) were sensitive to the gamma-secretase inhibitor (GSI) MRK-003, both alone and in combination with pacitaxel, in vitro and in vivo, whereas cell lines with NOTCH2 rearrangements were resistant to GSI. Immunohistochemical staining of N1-ICD in TNBC xenografts correlated with responsiveness, and expression levels of the direct Notch target gene HES4 correlated with outcome in TNBC patients. Activating NOTCH1 point mutations were also identified in other solid tumors, including adenoid cystic carcinoma (ACC). Notably, ACC primary tumor xenografts with activating NOTCH1 mutations and high N1-ICD levels were sensitive to GSI, whereas N1-ICD-low tumors without NOTCH1 mutations were resistant.
Introduction
The Notch signaling pathway is an evolutionarily conserved regulator of cell fate, differentiation, and growth. In mammals, Notch signaling is mediated by four Notch receptors (NOTCH1–4) and at least four functional ligands [Delta-like-1 (DLL1), DLL3, DLL4 JAG1 and JAG2]. Canonical Notch signaling is initiated by ligand-binding to the Notch ectodomain. This triggers a series of proteolytic cleavage events, culminating in the release of the Notch intracellular domain (NICD) by gamma-secretase (GS). Upon GS cleavage, NICD translocates to the nucleus where it forms a Notch transcription complex with the DNA-binding factor CSL (also known as RBPJ) and co-activators of the MAML family (for review, see (1)).
Deregulated Notch signaling is oncogenic in specific cell types; for example, it is strongly associated with T-cell acute lymphoblastic leukemia (T-ALL), in which somatic activating mutations in NOTCH1 are present in >50% of cases (2). Most NOTCH1 mutations in human T-ALL fall into two classes: i) in-frame mutations or indels in exons 25–28 that disrupt an extracellular juxtamembrane negative regulatory region (NRR), leading to ligand-independent receptor proteolysis and release of the NOTCH1 ICD (N1-ICD); ii) and stop codons or frameshift mutations in exon 34 that result in deletion of a C-terminal PEST degron domain, stabilizing N1-ICD. Less commonly in human T-ALL NOTCH1 is the target of (7;9) translocations in which the 3’ end of NOTCH1 is fused to promoter/enhancer elements of TCRB (3). The rearranged NOTCH1 alleles in tumors with the t(7;9) drive expression of truncated mRNAs that initiate translation from a conserved methionine lying within the NOTCH1 transmembrane domain (4).
Oncogenic Notch signaling is also implicated in breast cancer. Recently, RNA-seq was used to identify abnormal Notch mRNAs in human breast carcinoma cell lines and primary tumors (5). The aberrant transcripts resulted from cytogenetically silent deletions involving either NOTCH1 or NOTCH2; although several of the rearrangements produce fusion genes, none encode chimeric proteins. Instead, the rearranged NOTCH1 genes drive expression of truncated mRNAs that initiate translation from the same internal methionine implicated in human T-ALLs with the t(7;9), while the rearranged NOTCH2 genes drive expression of truncated mRNAs that initiate translation from a methionine residue located within the intracellular domain of NOTCH2, internal to the GS cleavage. Because of this distinction, NOTCH1-rearranged breast cancers are predicted to be GS inhibitor (GSI)-sensitive, while NOTCH2-rearranged breast cancers are predicted to be GSI-resistant.
In addition to its oncogenic roles, genetic evidence suggests that Notch is a tumor suppressor in human squamous cell carcinoma (SCC). Whole exome deep sequencing identified likely loss-of-function mutations in a least one Notch signaling component in roughly 15–20% of head and neck SCC (6, 7); similarly, at least one putative loss-of-function mutation involving either NOTCH1 or NOTCH2 was identified in 19 out of 26 primary cutaneous SCC or derived cell lines (8). In additional, one trial of a GSI in Alzheimer patients reportedly led to an increase in skin cancers (9).
Despite concerns about the complications of long-term GSI treatment, preclinical studies in animals and clinical trials in cancer patients suggest that intermittent treatment with GSIs is well tolerated, and GSIs continue to hold promise as targeted therapy for malignancies in which Notch is an oncogenic driver (9). However, clinical responses to GSIs have been modest, possibly because GSI trials to date have not used biomarkers that predict responsiveness as a criterion for enrollment. To address the need for biomarkers, we first screened large collections of cell lines, primary tumors and metastases for Notch gene mutations, reasoning that tumors with gain-of-function mutations are most likely to be sensitive to GSI. Our studies show that triple negative breast cancers are uniquely enriched among tumors screened for activating NOTCH1 and NOTCH2 deletions. Using xenograft models, we demonstrate that the GSI-sensitivity of NOTCH1-rearranged breast cancer cell lines correlates with N1-ICD levels, and that NOTCH2-rearranged tumors are indeed GSI-resistant. We also identify other human solid tumors with novel activating point mutations in the NOTCH1 NRR, including a subset of adenoid cystic carcinoma cell lines and primary tumors. Like NOTCH1-rearranged breast cancers, xenografted NOTCH1-mutated adenoid cystic carcinomas are GSI-sensitive, while Notch wild type xenografts are not. Finally, we identify HES4, a known Notch target gene, as a gene whose expression is correlated with poor clinical outcome in triple negative breast cancer. Our findings suggest that assessment of Notch gene mutational status, activated Notch protein levels, and expression levels of particular target genes, such as HES4 in TNBC, will be useful in selecting patients for GSI trials.
Results
Whole exome sequencing identifies NOTCH gene rearrangements in breast cancer
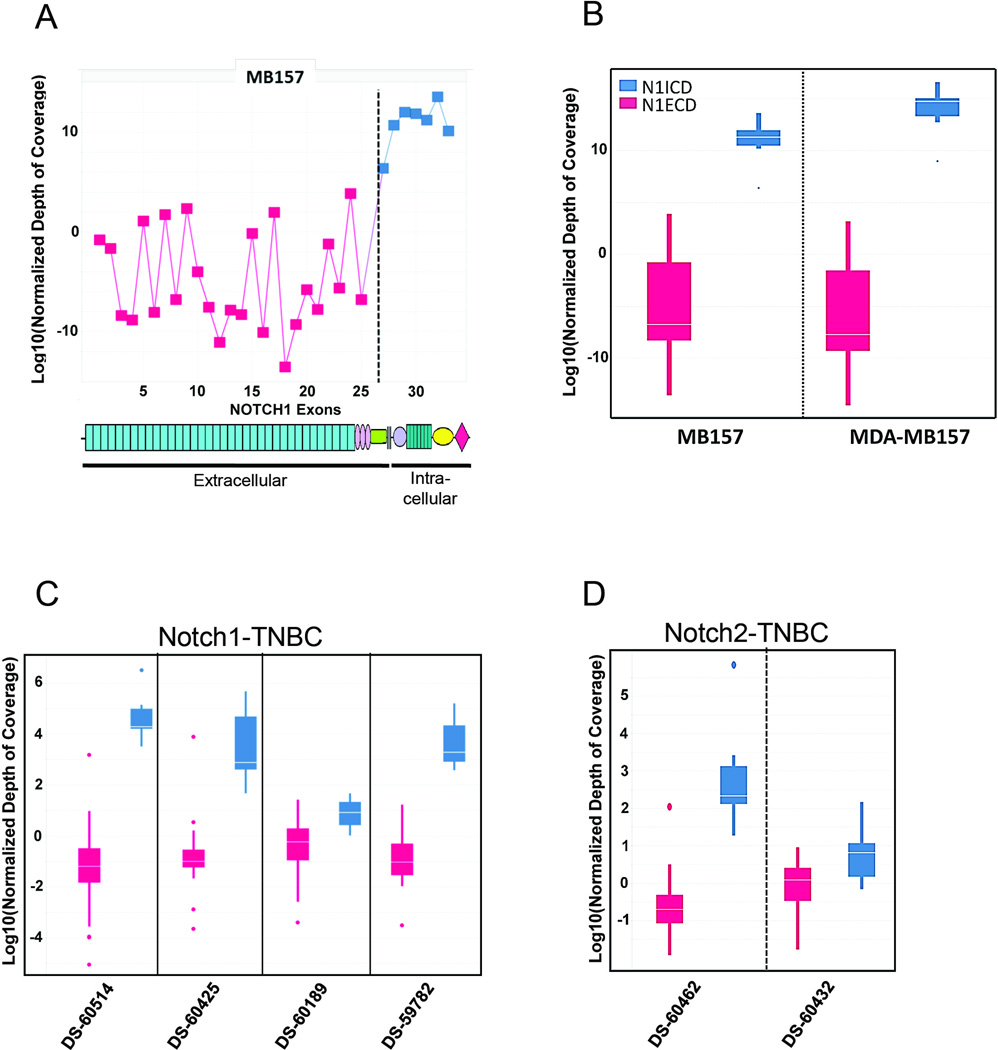
To detect Notch gene rearrangements, we used targeted exome sequencing (TES) data from human cancer cell lines and primary tumors to identify imbalances in exon coverage, which can be used to infer the presence of intragenic rearrangements, particularly deletions. Analysis of TES data from 608 cancer cell lines identified exon imbalances in NOTCH1 or NOTCH2 in 5 cell lines and in 1 cell line, respectively (Figure 1A and Supplemental Figure S1A; ). Exon imbalances were only found in triple negative breast cancer (TNBC) lines and stemmed from the deletion of exons encoding the Notch extracellular EGF repeats and negative regulatory region (NRR). For example, in the MB-157 and MDA-MB157 cell lines the read coverage of 5’ NOTCH1 exons was markedly lower than 3’ NOTCH1 exons (Figure 1A and 1B). A similar imbalance was observed in read coverage for 5’ and 3’ NOTCH2 exons in the HCC1187 cell line (Supplemental Fig S1A). Our data are consistent with previously reported Notch gene breakpoints in the HCC1187 and HCC2218 cell lines, which were originally suspected based on RNA sequencing data (5), and indicate that exon read imbalances in TES data can be used to identify tumors with NOTCH1 and NOTCH2 rearrangements. This capacity was confirmed by detection of novel NOTCH1 rearrangements in the MB-157 and MDA-MB-157 cell lines, both of which are derived from the same patient with triple negative breast cancer. Importantly, a fusion transcript consisting of noncoding RNA derived from SEC16A, a gene flanking NOTCH1 on chromosome 9q34.3, and exons 27–34 of NOTCH1 was identified by sequencing of RNA prepared from the MB-157 cell line (data not shown), consistent with the presence of an interstitial deletion that created a SEC16A-NOTCH1 fusion gene. Similarly, sequencing of RNA prepared from the HCC1599 cell line confirmed the presence of an aberrant transcript consisting of exon 2 joined out of frame to exon 27 of NOTCH1, consistent with the presence of an intragenic deletion involving NOTCH1, as described (5).
Figure 1. Identification of Notch gene rearrangements through analysis of DNA sequencing data.
(A) Relative coverage of NOTCH1 exons in MB157 cell line. Exons 27–34, which encode the NOTCH1 intracellular domain, are shown in blue, while exons 1–26, which encode the NOTCH1 extracellular domain, are shown in red. Significant (p<10−6) underrepresentation of reads mapping to exons 1–26 is evident. (B) Boxplots of read coverage of NOTCH1 exons 1–26 (red) and 27–34 (blue) in cell lines; the boxes span from the 25th to the 75th percentiles, while the white line is the median. MB157 and MDA-MB157 show significant (p<10−5) underrepresentation of reads mapping to exons 1–26. The NOTCH1 deletions in these cell lines are heterozygous, as confirmed by RT-PCR (data not shown). (C) Boxplots of read coverage of NOTCH1 exons 1–26 (red) and 27–34 (blue) in triple negative breast cancers. The boxes span from the 25th to the 75th percentile, while the white line is the median. Four out of 66 tumors show significant (p<10−5) underrepresentation of read coverage for NOTCH1 exons 1–26. (D) The same analysis as in C for NOTCH2. Two out of 66 tumors show significant (p<10−5) underrepresentation of read coverage for exons 1–26.
Next we analyzed WES data from 66 triple negative primary breast tumors. We identified NOTCH1 or NOTCH2 5’ deletions in 6 tumors, as indicated by significantly decreased read coverage (p < 10−5) in 5’ exons as compared to 3’ exons (Figure 1C and 1D, and Supplemental Figure S1B). An excess of transcripts containing 3’ Notch exons was confirmed by RT-qPCR (data not shown). Each of the inferred NOTCH1 deletions is predicted to create mutated alleles driving the expression of truncated transcripts encoding membrane-tethered NOTCH1 polypeptides that depend on GS for activity (Supplemental Figure S1C) (4, 5). By contrast, as with previously reported NOTCH2 deletions (5), the rearranged NOTCH2 alleles are predicted to drive expression of truncated transcripts encoding constitutively active, GS-independent NOTCH2 polypeptides (Supplemental Figure S1C).
TNBC is a molecularly heterogeneous tumor that can be classified based on patterns of gene expression into basal-like, immune, mesenchymal stem cell-like, and luminal androgen receptor subtypes (10). Consistent with previous reports, we observed the highest level of expression of NOTCH1 mRNA in the basal-like tumors (P<0.001; data not shown). Next we examine if the presence of NOTCH1 or NOTCH2 re-arrangements correlate with a particular subtype of TNBC. Of the 66 tumors analyzed, Notch gene rearrangements were observed in the basal and luminal androgen receptor subtypes (see Supplemental Table S1), but not in the immune or mesenchymal stem cell-like subtypes.
Notch mutations that disrupt the NRR coding region are associated with GSI sensitivity
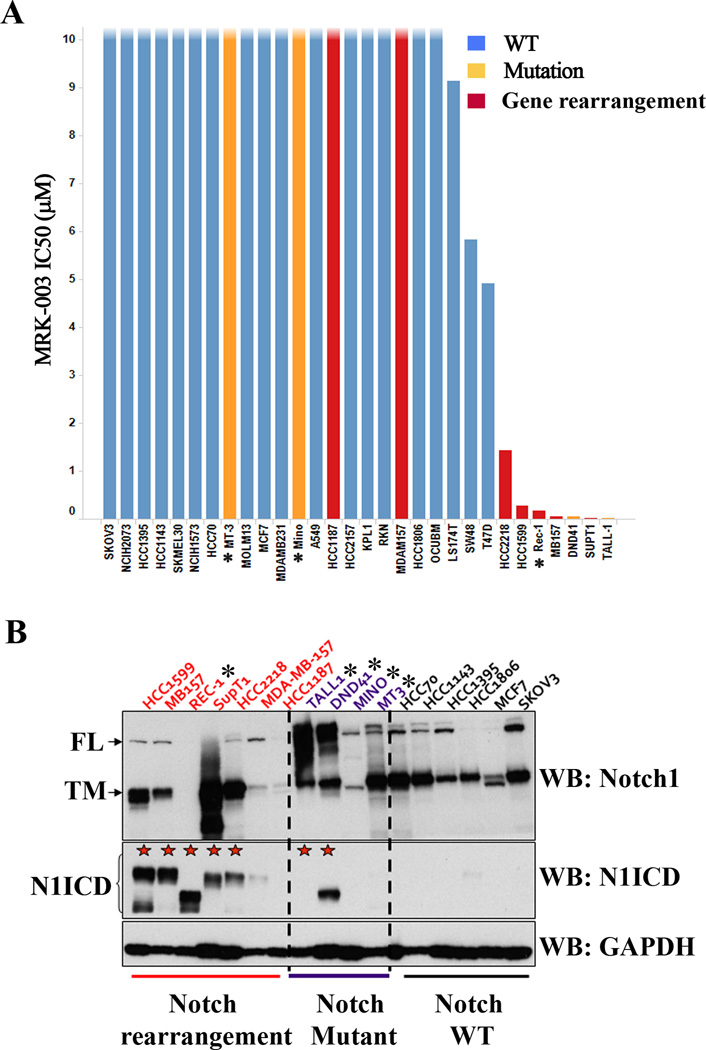
To determine the relationship between Notch gene mutational status and sensitivity to GSI we evaluated the anti-proliferative activity of MRK-003, a potent and selective GSI (11), in a panel of breast cancer cell lines. T-ALL and mantle cell lymphoma cell lines with mutations that disrupt the NOTCH1 NRR (2, 4) and TALL1, an additional GSI-sensitive T-ALL line with an activating NRR mutation in NOTCH3 (J.C.A., unpublished data), were included as positive controls. The IC50 for MRK-003 in the triple negative breast cancer cell lines HCC1599 and MB157 with NOTCH1 rearrangements was <1μM, similar to the sensitivity of lymphoid cell lines with NOTCH1 or NOTCH3 point mutations or gene rearrangements that disrupt the NRR coding region (Figure 2A, Supplemental Figure S2A). By contrast, the HER2+ breast cancer cell line HCC2218, which also harbors a SEC16A-NOTCH1 fusion gene (5), demonstrated intermediate sensitivity (Figure 2A, Supplemental Figure S2A). As a group, breast cancer cell lines with Notch gene rearrangements that disrupt the NRR coding region were significantly more sensitive to MRK-003 than cell lines with wild type Notch alleles, with two exceptions, the cell lines HCC1187 and MDA-MB-157. HCC1187, a breast cancer cell line harboring a SEC22-NOTCH2 fusion gene (5), was predicted to be GSI-resistant because the translation start site in aberrant NOTCH2 transcripts is internal to the site of GS cleavage (Supplemental Figure S1C). By contrast, the resistance of the cell line MDA-MB-157 (Supplemental Figure S2A) was unexpected, since this line has a deletion that disrupts the NOTCH1 NRR coding region. We also noted that the mantle cell lymphoma cell lines (Mino & MT3), which harbor NOTCH1 mutations leading to deletion of the C-terminal PEST domain, were also resistant to MRK-003. Thus, GSI sensitivity is confined to cell lines with mutations that disrupt the NRR, but this association is not absolute.
Figure 2. Notch mutational status determines pathway activity and sensitivity to MRK-003 in cancer cell lines.
(A) A panel of cancer cell lines was treated with increasing concentrations of MRK-003 for 72h and the IC50 was determined for cell proliferation as described in material and methods. Cell lines harboring NOTCH1 rearrangements (red) or mutations (orange) showed sensitivity to the gamma-secretase inhibitor MRK-003. ★ MT3, Mino: PEST mutations; Rec-1, DND41: dual ectodomain and PEST mutations; TALL-1: high N3ICD levels (B) Western blot analysis on selected cell lines harboring NOTCH1 rearrangements (red), point mutations (purple) or wild type (black) alleles. Blots were stained with antibodies specific for the intracellular domain of NOTCH1, which recognize the full-length NOTCH1 polypeptide (N1-FL) and the furin-cleaved N1-TM subunit, or an antibody specific for gamma-secretase cleaved, activated NOTCH1 (N1-ICD). Equal loading was confirmed by staining with an antibody against GAPDH.
MRK-003 sensitivity correlates with NICD levels
To assess the relationship between NOTCH1 mutational status, Notch signal strength, and GSI sensitivity, we evaluated N1-ICD protein levels. We observed high levels of N1-ICD in most cell lines with NOTCH1 gene rearrangements or mutations involving the NRR (Figure 2B). Of interest, the exception to this rule is the GSI-resistant MDA-MB157 cell line, which had low levels of N1-ICD despite the presence of a NOTCH1 rearrangement (Figure 2B). As expected, N1-ICD levels were low or absent in GSI-resistant NOTCH1 wild type cell lines, as well as in GSI-resistant mantle cell lymphoma lines (MT-3 and Mino), which harbor only PEST domain mutations (Figure 2B). This is consistent with studies showing that while PEST domain mutations stabilize NICD, they have no effect on NICD levels in the absence of NRR mutations or exposure to ligands (2). As a consequence, N1-ICD protein level was more highly correlated with GSI sensitivity than Notch gene mutational status (Supplemental Figures 2B and 2C).
We also compared the protein levels of all four Notch receptors in the breast cell line panel (n=8; Supplemental Figure S2D) using antibodies that recognize full-length Notch receptors and their furin-cleaved transmembrane subunits. We observed variable levels of all four receptors, expression of which did not correlate with sensitivity to MRK-003. For example, high NOTCH1 polypeptide levels were observed in both GSI-sensitive HCC1599 cells and GSI- resistant HCC70 and SKBR3 cells. Similarly, NOTCH2-4 polypeptide levels did not correlate with sensitivity to MRK-003 in this breast cancer cell line panel. These results suggest that N1-ICD level is more highly associated with sensitivity to MRK-003 treatment than expression of Notch receptors per se, with the important caveat that reagents to identify other forms of activated Notch receptors (N2-4-ICD) are not yet available.
HES4 expression correlates with Notch activation in multiple tumor types and with outcome in triple negative breast cancer
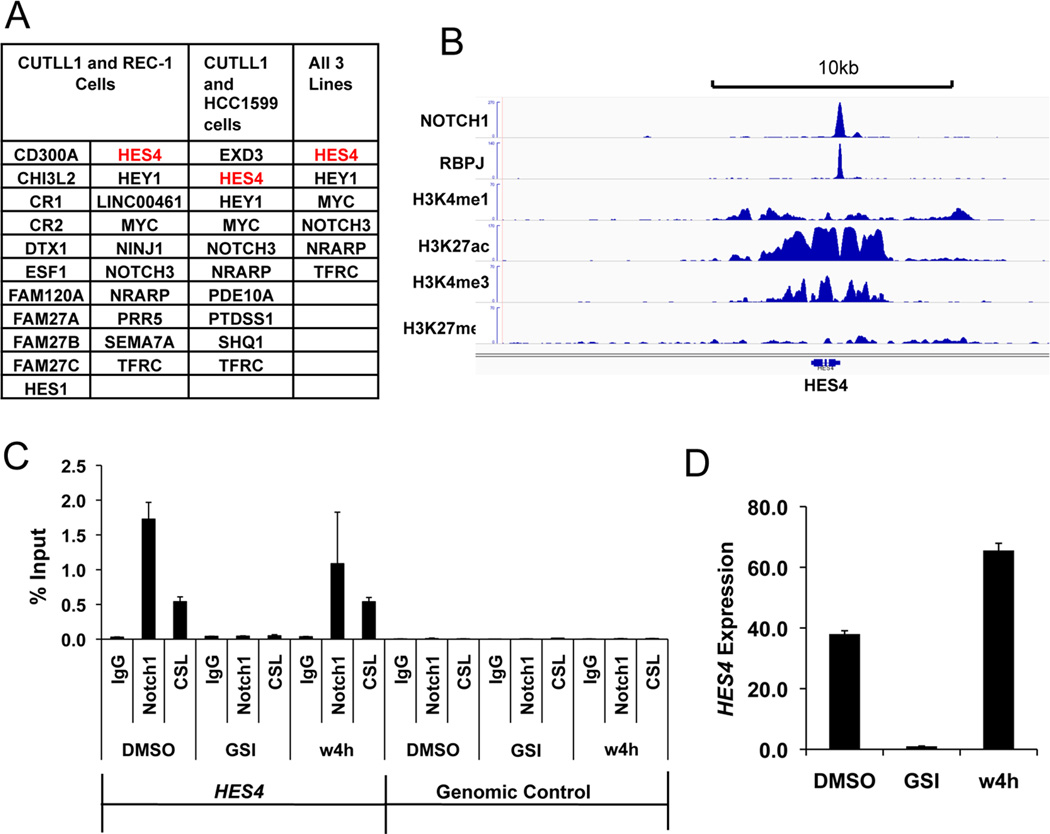
While NICD would appear to be a promising biomarker, because sensitive and specific antibodies only exist for detection of N1-ICD, there is a need for additional markers of Notch activation in diverse cellular lineages. To identify Notch target genes in Notch-addicted cancer cell lines in an unbiased fashion, we performed RNAseq on CUTLL1 T-ALL cells, REC-1 mantle cell lymphoma cells, and HCC1599 cells (Gene bank accession number GSE59810), all of which have mutations that disrupt the NOTCH1 NRR. RNAseq was performed in the Notch-off, GSI-treated state, and 4 hours following washout of GSI, a strategy that rapidly generates N1-ICD and upregulates NOTCH1 target genes in a temporally controlled fashion. Notably, although >80 genes were up-regulated upon GSI washout in each cell line (FDR <0.05), only 6 target genes were common to all three cell lines---HEY1, MYC, NOTCH3, TFRC, NRARP, and HES4 (Fig. 3A). ChIP-Seq analysis of CUTLL1 cells (12) showed that NOTCH1 and RBPJ bind to a site in the HES4 proximal promoter on chromosome 1 (Fig. 3B). To confirm that this site is highly dynamic, a feature that characterizes functional RBPJ/NOTCH1 binding sites (13), we performed local ChIP for RBPJ and NOTCH1 in REC-1 cells. This showed that GSI depleted NOTCH1 from the HES4 promoter, and that GSI washout resulted in rapid reloading of NOTCH1 onto this site (Fig. 3C). Finally, we confirmed that GSI markedly inhibited HES4 expression in HCC1599 cells, and that GSI washout rapidly restored expression (Fig. 3D).
Figure 3. HES4 is a direct Notch1 target gene in diverse Notch-addicted cancer cell lines.
(A) Summary of overlapping direct Notch1 target genes in the T-ALL cell line CUTLL1, the mantle cell lymphoma cell line REC-1, and the triple negative breast cancer cell line HCC1599. (B) Chromatin landscapes near HES4 in CUTLL1 cells. A gamma-secretase inhibitor-sensitive RBPJ/NOTCH1 binding site is present in the HES4 promoter. (C) NOTCH1/RBPJ complexes associate with the HES4 promoter site in REC-1 cells. Local ChIP for NOTCH1 and RBPJ was performed under steady state conditions (DMSO), in cells treated with the GSI compound E (1µM) for 72hr (the Notch-off state), and in cells treated for 72hr with GSI followed by 4hr of recovery following GSI washout (w4h). (D) HES4 expression in HCC1599 cells is Notch dependent. RT-PCR with HES4 specific primers was performed under steady state conditions (DMSO), in cells treated with the GSI compound E (1µM) for 72hr, and in cells treated for 72hr with GSI followed by 4hr of recovery following GSI washout (w4h).
To further study the relationship between activating NOTCH1 mutations and gene expression, we carried out Affymetrix based micro-array analysis in the panel of 608 cell lines (Gene bank accession number GSE59242) screened previously for Notch gene mutations using TES. We observed that high HES4 expression was significantly correlated (FDR <0.05) with the presence of activating mutations involving Notch genes in several cancer cell lines (Supplemental Figure S3 and Table S2). We next compared expression levels of various Notch target genes among primary TNBCs, and noted that HES4 was the gene whose expression was most strongly correlated with Notch gene-rearrangements in TNBC (Supplemental Table S3). High HES4 expression was also associated with poor patient outcome in TNBC (Log-Rank P<0.0001), as patients with metastatic disease in the HES4 high group had a median survival of 0.89 years [n=21; 95% CI=0.31], whereas patients with metastatic disease in the HES4 low group had a median survival of 2.97 years [n=139; 95% CI=2.18], (Supplemental Figure S4). Similarly, in patients without measurable metastatic disease, high HES4 expression in primary tumors also predicted a poor prognosis (Supplemental Figure S4). These results suggest that HES4 expression levels (and by extension, activated Notch levels) may be a useful prognostic biomarker in TNBC patients.
Notch inhibition by GSI is associated with induction of senescence and apoptosis and down-regulation of MYC in tumor models with Notch gene rearrangements
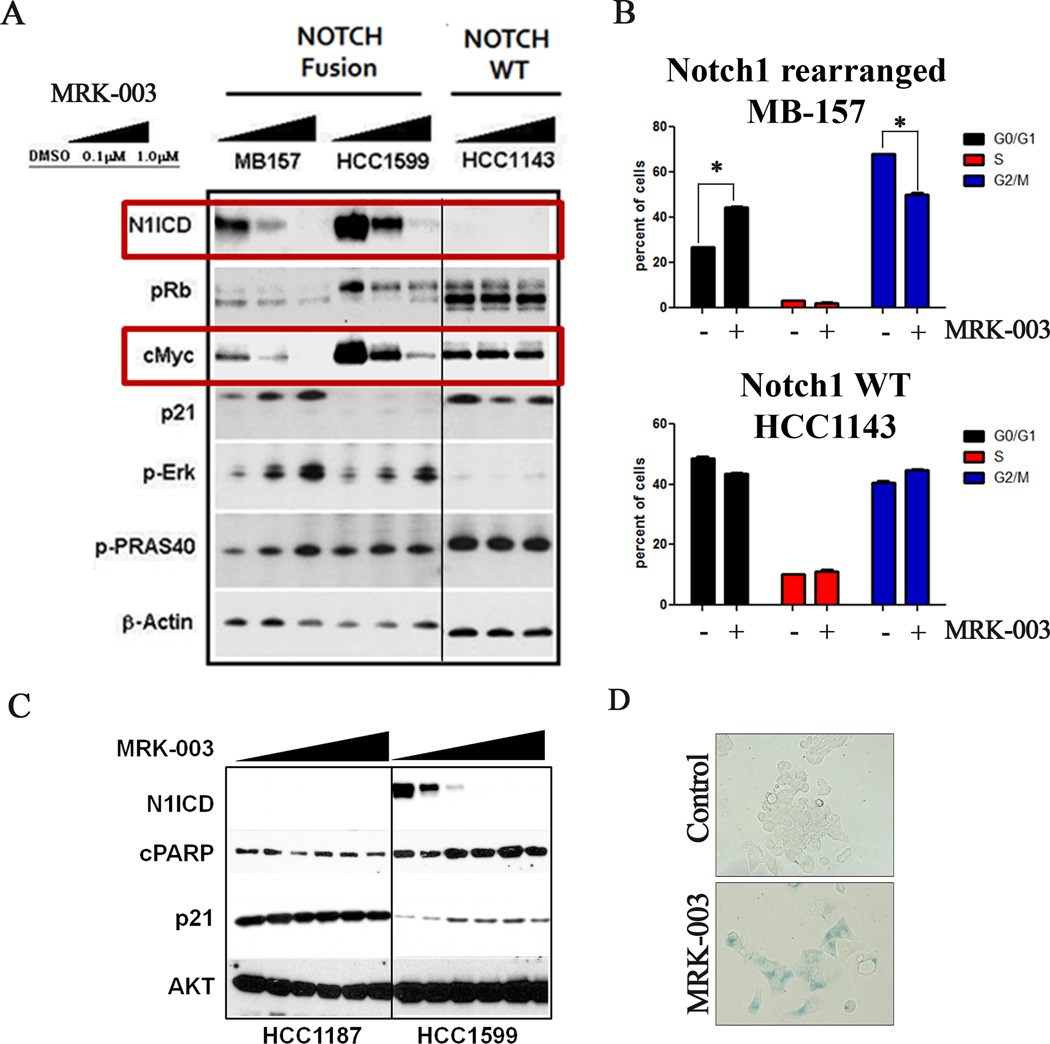
To investigate the how GSI affects cell growth in Notch-gene-rearranged breast cancer models, we evaluated the effect of GSI treatment on molecular pathways regulated by Notch. MRK-003 treatment resulted in a dose dependent decrease in levels of activated Notch as measured by N1-ICD in the NOTCH1 rearranged TNBC cell lines MB-157 and HCC1599 (Figure 4A). NOTCH1 inhibition was accompanied by decreases in MYC protein and RB phosphorylation and increases in the level of the CDK inhibitor p21 (Figure 4A). In addition, compensatory activation of the MAPK/PI3K pathway was observed, as indicated by GSI-dose-dependent increases in phosphorylation of ERK and PRAS40, an AKT substrate (Figure 4A). These results suggest the existence of a compensatory feedback loop between Notch and the PI3K/MAPK signaling pathways in Notch-dependent breast cancer cell lines. By contrast, MRK-003 treatment did not significantly affect any of these signaling events in the Notch wild type cell line HCC1143.
Figure 4. MRK-003 induces cell cycle arrest, apoptosis, and senescence in cancer cell lines harboring NOTCH1 gene rearrangements.
(A) Western blot analysis on lysates prepared from cell lines with or without NOTCH1 gene rearrangement following treatment with MRK-003 or vehicle (DMSO) for 24hr. Primary antibodies used were specific for activated NOTCH1 (N1-ICD), pRb, MYC, p21, p-ERK, p-PRAS40 and β-actin (loading control). (B) Cell cycle analysis performed following treatment of NOTCH1-rearranged MB157 cells and HCC1143 cells with wild type NOTCH1 alleles with 1µM MRK-003 for 72hr. (C) HCC1599 cells harboring a NOTCH1 rearrangement and HCC1187 cells harboring a NOTCH2 rearrangement were treated with increasing concentrations of MRK-003 for 24hr. Western blots were stained with antibodies specific for N1-ICD, cleaved PARP (c-PARP), p21, and total AKT (loading control). (D) Induction of senescence. β-galactosidase staining was performed after once weekly treatment of MB157 cells with 1uM MRK-003 or DMSO (vehicle) for four weeks. Over 95% of cells treated with MRK-003 were positive for β-galactosidase, versus 1% of the cells exposed to DMSO.
To investigate possible synergistic anti-tumor effects of NOTCH and PI3K/MAPK pathway co-inhibition we performed cell proliferation assays in the presence of MRK-003 and a selective ERK inhibitor (SCH772984; (14)). MRK-003 treatment produced dose-dependent growth inhibition of both MB-157 and HCC1599, but ERK inhibition failed to potentiate the effect of MRK-003 (See Supplemental Figure S5). Although these in vitro studies were negative, given the evidence of crosstalk between Notch and MAPK/PI3K signaling in breast cancer cells shown here and in other tumor cell types such as T-ALL cells (15), further evaluation of this combination in breast cancer may be warranted.
Next, we evaluated the effects of GSI treatment of Notch gene rearranged breast cancer cell lines on cell cycle progression and apoptosis. MRK-003 treatment resulted in a G1 cell cycle arrest and the appearance of a sub-G1 population, suggestive of apoptotic cell death, in MB-157 cells (Figure 4B). Similar effects were also observed in HCC1599 cells (data not shown). Furthermore, MRK-003 treatment resulted in a dose-dependent increase in cleaved PARP in HCC1599 cells (Figure 4C), whereas no significant change in cleaved PARP was observed in the NOTCH2-rearranged, GSI-resistant cell line HCC1187 (5). In the NOTCH1-rearranged MB-157 cell line, MRK-003 produced only a modest increase in apoptosis (data not shown), suggesting that GSI-mediated growth inhibition may stem from a different mechanism in the line. Consistent with this idea, we noted that prolonged MRK-003 treatment of MB-157 cells resulted in the induction of cellular senescence, as measured by β-galactosidase activity (Figure 4D). Furthermore, robust GSI-mediated up-regulation of p21 and down-regulation of MYC, events implicated in induction of cellular senescence (16), were also observed in MB-157 cells (Figure 4A).
The Notch pathway has also been suggested to be important in maintenance of stem-like cells with tumor-initiating activity (17) and to promote the growth of breast epithelial cells as mammospheres in culture, a phenotype linked to stem-like properties (5) (18). We evaluated the effect of MRK-003 treatment on stem-like cells using the markers CD44/CD24 and ALDH. When cultured in stem cell media, MB-157 cells contain a sub-population (~8%) of CD44+/CD24low stem-like cells that is depleted by MRK-003 treatment (Supplemental Figure S6A). Similarly, MRK-003 treatment of MB-157 cells also suppressed the ALDH+ cell population (Supplemental Figure S6B), another marker of cells with stem-like properties. Unlike the effects of MRK-003 on growth, which were restricted to breast cancer lines with NOTCH1 rearrangements, MRK-003 treatment also suppressed the ALDH+ cell population in NCIH226 and MFM223 cells (Supplemental Figure S6C), which have wild type Notch alleles. Thus, Notch gene rearrangements do not correlate with the effect of MRK-003 on breast cancer stem-like cells, possibly because the dose of Notch that is required to maintain stem-like cells is substantially lower than that required to drive cell growth. Taken together, Notch signaling in NOTCH1-rearranged TNBC cell lines appears to support cell growth through multiple mechanisms, including promotion of cell cycle progression, decreased apoptosis and cellular senescence, and increasing the fraction of stem-like cells.
In vivo efficacy of GSI therapy in TNBC models with Notch gene rearrangements
We next evaluated the effect of MRK-003 therapy on the growth of breast cancer xenograft models with NOTCH1 or NOTCH2 rearrangements (Table 1). MDA-MB-231, a TNBC cell line with wild type Notch alleles was used as a control model. To identify new primary tumor-derived xenograft models with NOTCH1 rearrangements, we used RT-PCR to screen for previously described NOTCH1 rearrangements (5). Of the 21 primary human tumor-derived TNBC xenograft models screened, two models, HBCx8 and HBCx14, yielded RT-PCR products consistent with the presence of NOTCH1 rearrangements. In HBCx8 cells, we observed a SEC16A-NOTCH1 fusion transcript identical to previously described rearrangements (data not shown). In HBCx14 cells, we were unable to detect RT-PCR products containing both exon 26 and exon 28 of NOTCH1 (data not shown); while consistent with the presence of a NOTCH1 rearrangement, we were unable to identify fusion junctions corresponding to previously described NOTCH1 rearrangements, suggesting that this line harbors a novel NOTCH1 rearrangement.
Table 1.
Summary of anti-tumor activity of MRK-003 alone or in combination with Paclitaxel in TNBC xenograft models.
| Cell line | NOTCH1 status | MRK-003 (300 mpk) |
Paclitaxel (15 mpk) |
MRK-003 + Paclitaxel |
IHC score |
|---|---|---|---|---|---|
| HCC1599 | Rearranged | 90% (TGI) | 1% (TGI) | 98% (TGI) | +++ |
| MB-157 | Rearranged | 22% (Reg) | 73% (TGI) | 72% (Reg) | ++ |
| HBCx-14 | Rearranged | 19% (TGI) | -19 (TGI) | 67% (TGI) | + |
| HBCx-8 | Rearranged | 46% (TGI) | 22% (TGI) | 75% (TGI) | + |
| HCC1187 | WT** | −34% (TGI) | 48% (TGI) | 76% (TGI) | Neg |
| MDA-MB-231 | WT | 27% (TGI) | 74% (TGI) | 86% (TGI) | Neg |
IHC score, N1-ICD staining intensity; TGI, tumor growth inhibition; Reg, tumor regression;
NOTCH2 translocation resulting in loss of GSI cleavage site; mgk, mg per kg.
To determine if NOTCH1 rearrangements correlate with NOTCH1 activation in vivo, we evaluated N1-ICD levels using a sensitive immunohistochemical staining method that is specific for N1-ICD (19). High levels of N1-ICD staining were detected in the HCC1599 and MB-157 models (Supplemental Figure S7A), a result that correlated well with the results of western blotting for N1-ICD (Supplemental Figure S7B). However, HBCx14 had lower levels of N1-ICD staining, and even less staining for N1-ICD levels was seen in the HBCx8 model, despite the presence of a SEC16A-NOTCH1 fusion gene. As expected, models with wild type NOTCH1 alleles were negative for N1-ICD staining. Next we evaluated sensitivity of these xenograft models to GSI treatment, given either alone or in combination with paclitaxel in vivo. We noted that sensitivity to MRK-003, alone or in combination with paclitaxel, was associated with N1-ICD levels; these results are summarized in Table1. For example, MRK-003 treatment produced dose-dependent growth cessation or regression of HCC1599 and MB-157 xenografts (Figure 5 A and Table1), but had only modest anti-tumor activity in the MDA-MB-231 xenograft model with wild type Notch genes (Supplemental Figure S7C, Table 1). Although previous studies have demonstrated significant anti-tumor activity with MRK-003 in MDA-MB-231 mode we would like to note that the dosing scheme utilized in these studies were not identical to our study (20). In this study we utilized a clinically tolerated dosing schedule (once a week dosing). Similarly, the HCC1187 xenograft model with a SEC-16-NOTCH2 fusion gene encoding NOTCH2 polypeptides that do not require gamma-secretase cleavage for activation was resistant to GSI therapy (Figure 5B). Monotherapy with MRK-003 had a modest anti-tumor activity in the primary human tumor derived xenografts HBCx8 and HBCx14 (Figure 5 & Table 1), which have low N1-ICD levels (Supplemental Figure S7A, B). These results indicate that N1-ICD levels can be substantially different in tumors with similar NOTCH1 rearrangements, and suggest that N1-ICD levels are more highly correlated with GSI responsiveness than Notch mutational status per se.
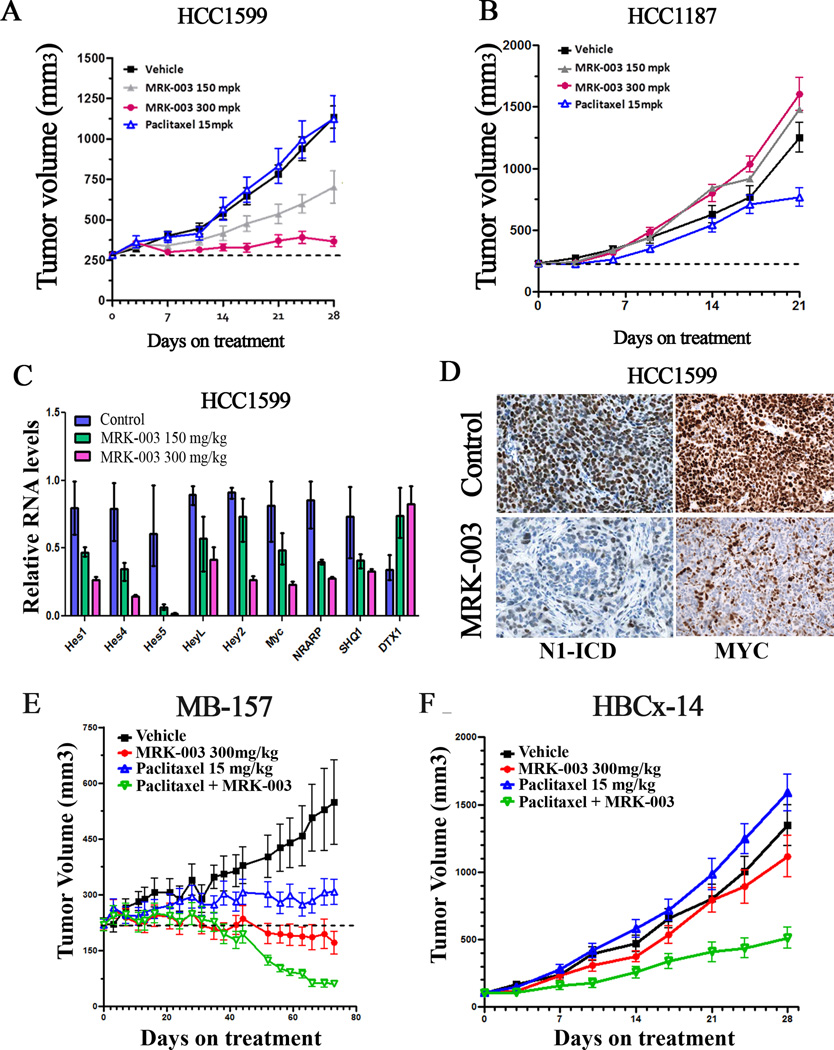
Figure 5. Treatment with MRK-003 leads to tumor regression in NOTCH1-rearranged TNBC xenograft models.
(A, B) Xenograft models of HCC1599 and HCC1187 cells were treated with 150mg/kg or 300mg/kg MRK-003 once a week, vehicle control, or 15mg/kg paclitaxel as indicated. A summary of tumor growth inhibition (TGI) is presented in Table 1. MRK-003 treatment at both doses (150mg/kg and 300mg/kg) resulted in significant tumor growth inhibition (p<0.001). (C) qRT-PCR analysis of tumor tissues from HCC1599 xenografts treated with 150mg/kg, 300mg/kg MRK-003 or vehicle control showing effects of Notch inhibition on a 9 gene signature 6hr after dosing. HES and HEY family members were significantly down-regulated together with MYC, NRARP and SHQ1, whereas DTX1 (which does not score as a target gene in breast cancer cells) was up-regulated. (D) Immunohistochemistry of formalin fixed paraffin embedded tumor sections from HCC1599 xenografts treated with 300mg/kg MRK-003 or vehicle control with a N1-ICD specific antibody, showing decreased nuclear levels in the MRK-003 treated mice. Tissue was harvested 6hr after dosing. (E, F) MB-157 and HBCx-14 xenograft models, treated with 150mg/kg or 300mg/kg MRK-003 by oral gavage once a week, vehicle control (methylcellulose) or 15mg/kg paclitaxel, alone or in combination with 300mg/kg MRK-003. The MB-157 model was treated for 70 days, while the HBCx-14 model was treated for 28 days. MRK-003 alone or in combination with Paclitaxel resulted in significant tumor growth inhibition (p<0.001) in the MB-157 model. In HBCx-14 model, only combination therapy with MRK-003 and paclitaxel was effective (p<0.001).
To further evaluate the association of the anti-tumor activity of MRK-003 with Notch pathway inhibition, we studied the effect of MRK-003 on Notch target gene expression in the HCC1599 xenograft model. Dose-dependent inhibition of multiple direct or indirect Notch target genes was observed in MRK-003-treated tumors (Figure 5C), which also showed depletion of N1-ICD, as judged by immunohistochemistry (Figure 5D). These data confirm that inhibition of the Notch pathway is correlated with the anti-tumor activity of MRK-003.
Multiple Notch targets may contribute to the anti-tumor activity of MRK-003 in vivo, but one that may be of special importance is MYC, which as previously noted (Figure 3) is one of the few common target genes across multiple types of Notch-“addicted” cancer cells in vitro. Treatment of HCC1599 xenografts with MRK-003 down-regulated MYC protein levels as judged by Western blotting and immunohistochemistry (Supplemental Figure S7D), an alteration that was accompanied by decreases in cyclin E, which is involved in G1/S phase progression. Regulation of MYC by Notch has been documented in T-ALL (21, 22) (23) (24) and in murine models of breast cancer (25), and our results suggest that targeting of the Notch-Myc signaling axis also underlies the responsiveness of human breast cancer models to MRK-003.
Combination therapy with GSI and Paclitaxel in breast cancer models
We next evaluated the effect of combination therapy with GSI and paclitaxel, an agent that is frequently used to treat TNBC. Modest anti-tumor activity was observed with single agent paclitaxel in the NOTCH1-rearranged breast tumor models. Although GSI monotherapy resulted in significant growth inhibition of HCC1599 or MB-157 cells, combination therapy with MRK-003 and paclitaxel significantly potentiated antitumor activity in both models and resulted in tumor regression in the MB-157 xenograft model (Figure 5E). In the primary human tumor-derived xenograft models with NOTCH1-rearrangements, HBCx8 and HBCx14, monotherapy with MRK-003 or paclitaxel was ineffective in blocking tumor growth, but combination treatment showed significant anti-tumor activity (Figure 5F and Table 1), whereas no potentiation was observed in a model with wild type Notch alleles. These results suggest that GSI/paclitaxel combination therapy may be effective in treatment of NOTCH1-rearranged tumors.
Characterization of novel NOTCH1 NRR mutations
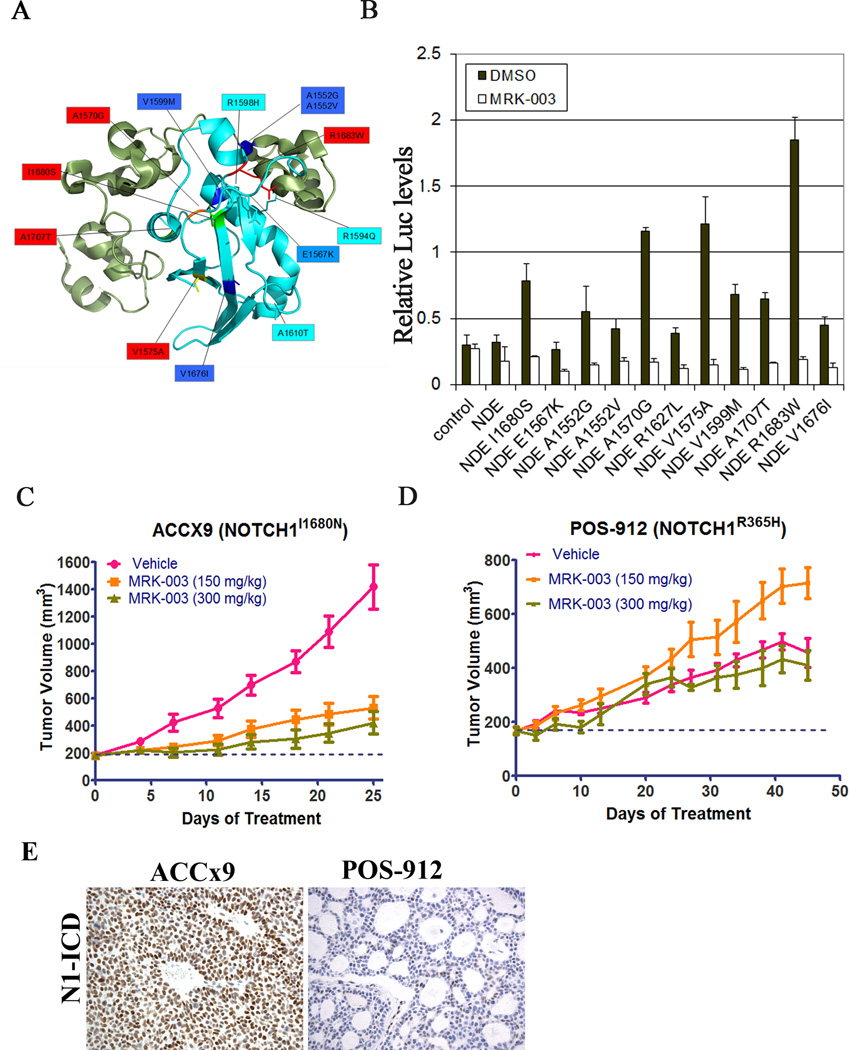
Targeted exome sequencing of over 4000 tumors identified several novel Notch gene mutations. As described previously, Notch mutations can be activating or inactivating depending on the location and nature of the substitution (26). To assess possible gain-of-function mutations, we focused on NOTCH1 mutations affecting the NRR region or the PEST domain (Table S4). Notch gene mutations in the NRR region and PEST domain were restricted to a small fraction of tumors (<5%), and most of these mutations mapped to the core of the NRR (Figure 6A), a region frequently involved by gain-of-function NOTCH1 mutations in human T-ALL (2). Tumor types in which recurrent Notch NRR and PEST domain mutations were identified are summarized in Table S5. Of note, NRR or PEST domain mutations were identified in 5 out of 105 TNBCs (Table S5). NRR mutations were also observed in 1 out of 3 adenoid cystic carcinomas (ACC), in line with recent sequencing studies that identified possible Notch gain-of-function mutations in a subset of tumors (27). To test whether these newly identified NOTCH1 mutations affect function, they were scored for their ability to activate a Notch-responsive luciferase reporter gene (Figure 6B). All mutations within the NOTCH1 NRR caused significant increases in MRK-003-sensitive luciferase activity, with amino acid substitutions at positions 1680, 1570, 1575 and 1683 having the greatest effect. By contrast, in accordance with previous studies, mutations in the PEST domain did not significantly increase luciferase reporter activity in the absence of ligand-mediated stimulation (data not shown).
Figure 6. Novel gain-of-function mutations in the NOTCH1 NRR are MRK-003 sensitive in vitro and in vivo.
(A) Ribbon diagram of the NOTCH1 NRR showing the position of the newly identified mutations. The NRR is composed of three LNR-A, B and C modules (green) and a heterodimerization (HD) domain (blue). All identified mutations localized to the HD domain. (B) HD domain mutations score as gain-of-function mutations in Notch-sensitive reporter gene assays. Mutations were introduced into a cDNA encoding a form of NOTCH1, ΔEGF, that lacks the NOTCH1 ligand-binding domain and cannot respond to ligand, but is sensitive to NRR mutations that trigger ligand-independent signaling. (C, D) Response of patient derived adenoid cystic carcinoma xenograft models to MRK-003. ACCx9, which harbors an activating NOTCH1 I1680N NRR mutation, and POS-912, which harbors a non-activating mutation in NOTCH1 EGF repeat region, were treated with 150mg/kg or 300mg/kg MRK-003 as described. MRK-003 treatment resulted in significant tumor growth inhibition in ACCx9 model (p<0.05). (E) Immunohistochemistry of formalin fixed paraffin embedded tumor sections from ACCx9 or POS-912 xenografts with a N1-ICD specific antibody, showing presence of nuclear levels in ACCx9 model.
To investigate the possible role of Notch in ACC, we sequenced NOTCH1 in 6 primary tumor derived xenograft models (see Supplemental Table S6). This led to the identification of NOTCH1 mutations in two adenoid cystic carcinoma models, both of which were tested for GSI sensitivity. MRK-003 treatment resulted in significant growth inhibition in the ACCx9 model (Figure 6C) that harbors a I1680N NOTCH1 NRR substitution, a mutation known to cause ligand-independent NOTCH1 activation (28). By contrast, MRK-003 treatment did not have any anti-tumor effect on the POS-912 model (Figure 6D), which harbors a NOTCH1 R365H point substitution in the EGF repeat region that is not expected to produce NOTCH1 gain-of-function. In line with these expectations, high levels of nuclear N1-ICD were observed in tumor cells in the ACCx9 model, while in the POS-912 model only a minor subset of tumor cells had detectable N1-ICD levels (Figure 6E). We also observed high levels of MYC positivity (>90% of tumor cells) in the ACCx9 model, while the POS-912 model showed much lower levels of MYC expression (see Supplemental Table S6), suggesting that MYC may be a target of Notch in Notch-mutated ACC. It should be noted, however, that high MYC expression was also observed in some ACCs with wild type NOTCH1 genes, such as the ACCx6 model, suggesting that MYC may sometimes be dysregulated in ACC through mechanisms unrelated to NOTCH1 mutations or NOTCH1 activation. In other analyses, we did not detect an association between Notch mutational status and HES4 expression in ACC models (data not shown), emphasizing the need to validate biomarkers of Notch activation in each cellular context of interest, and suggesting that the most reliable biomarker of Notch activation is direct assessment of NICD levels. Further evaluation of N1-ICD in primary ACC is needed to determine the prevalence of Notch pathway activation in this neoplasm, which appears to be a candidate for treatment with anti-Notch therapies such as GSIs.
Discussion
Aberrant Notch signaling has been implicated in numerous human diseases, including different types of cancers. Through unbiased sequencing of diverse cancer cell lines and primary tumors, we identified different types of activating Notch mutations in specific cancer subtypes. One striking finding is that deletions that remove the coding sequences of NOTCH1 and NOTCH2 ectodomains appear to be highly specific among human tumors for triple negative breast cancer. It is of interest to note that structurally similar notch1 deletions are common in murine T-ALL, where they are caused by DNA breakage at cryptic RAG recombinase sites (4). It is possible that underlying abnormalities of DNA repair make triple negative breast cancer cells susceptible to activating Notch gene deletions caused by random DNA breakage followed by non-homologous end joining (29). Recent studies by Shah et al (30) observed a high degree of clonal and mutational diversity in TNBC suggestive of genomic instability. By contrast, in human tumors with relatively small numbers of genetic changes, such as adenoid cystic carcinoma and human T-ALL, Notch gain-of-function mutations tend to consist mainly of point substitutions and small indels.
Several recent tumor genome sequencing studies have identified activating mutations in the Notch signaling pathway in a minority of ACCs (31–33). Ross et al reported genomic alterations in NOTCH1 in 11% (3 out of 28) of ACC, Ho et al., while reported alterations in NOTCH signaling pathway genes in 13% of samples. Stephens et al. reported activating mutations in NOTCH2 and loss of function mutations in SPEN, a gene encoding a transcriptional repressor that forms a complex with RBPJ and down-regulates Notch target genes. We did not observe mutations in SPEN in our primary tumor data set, and it remains to be determined if SPEN-mutated tumors and Notch-mutated tumors are comparable in terms of activation of downstream genes.
It might be anticipated that activating mutations in Notch genes would be robust predictors of tumor response to Notch pathway inhibitors such as GSIs, since recurrent mutation of oncogenes in particular types of tumors reliably identifies genes and pathways that are subject to selection during the initiation and progression of cancers. However, while GSIs have been proven safe(34), they are yet to be proven effective, and early experience with human T-ALL suggests that Notch gene mutational status, per se, is not highly correlated with response. Thus, additional biomarkers that are better predictors of response, preferably across a broad spectrum of cancers that are driven by Notch gain-of-function mutations, are highly desirable.
Based on the response of breast cancer and ACC xenografts to MRK-003, it appears that immunohistochemical staining for activated NOTCH1 (N1-ICD) may be one such biomarker. Xenografts with high levels of N1-ICD immunoreactivity showed excellent responses to GSI, alone and in combination with pacitaxel, whereas xenografts with low N1-ICD reactivity or GSI-refractory rearrangements in NOTCH2 show no response to GSI. It is notable in this regard that NOTCH1 mutations in human T-ALL were discovered through a cell line screen for GSI sensitivity in which the most sensitive cell lines were found to be those with dual NOTCH1 NRR and PEST domain mutations in cis, an alignment that produces high levels of N1-ICD (2). Our data suggest that it should be possible to select patients for clinical trials of GSI based on N1-ICD immunoreactivity in archival formalin-fixed paraffin-embedded tissue sections, which would enable rapid screening and identification of patients who are most likely to respond to GSI treatment. Of note, the neo-epitopes created by GS cleavage of NOTCH2, NOTCH3, and NOTCH4 are distinct from that created by GS cleavage of NOTCH1, and in principle it should be possible to extend this approach to other members of the NOTCH receptor family, pending development of additional, NICD isoform-specific antibody reagents.
Another biomarker emerging at the interface of expression profiling and genomic analysis is HES4, which is one of a small number of genes that are common to the Notch-driven gene signatures in breast cancer, T-ALL, and B cell lymphoma cell lines. HES4 expression is also well correlated with Notch mutational status in diverse cell lines, and identifies a group of patients with poor prognosis in triple negative breast cancer. These data suggest that HES4 is a potentially valuable biomarker in certain tumor types such as breast cancer, with the important caveat that the results shown here need to be validated in independent clinical cohorts.
Material and Methods
Detailed methods for cell growth assays, cell cycle analysis, stem cell marker analysis, luciferase reporter assays, whole exome sequencing, exome imbalance analysis and RNA-seq analysis are provided in supplementary methods.
Cell Lines
The T-ALL cell line CUTLL1 (kind gift of Adolfo Ferrando, Columbia University) was cultured in RPMI1640 containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin-glutamine. Other cell lines were purchased from cell line banks American Type Culture Collection (ATCC), Japanese Collection of Research Bioresources Cell Bank (RIKEN), or Deutsche Sammlung von Mikroorganismen und Zellkulturen (DKMZ), and were grown under culture conditions recommended by the vendors. The authenticity of the cell lines were verified by short tandem repeat (STR) profiling analysis or similar methodologies by the banks. In addition, the mutation and gene expression levels from the TES data were compared with the published mutation (COSMIC; Sanger data base) and gene expression data.
Western blotting
Cells were lysed in 50mM Tris, pH 8.0, containing 150mM NaCl and 1% NP40 supplemented with protease inhibitor cocktail (Thermo Scientific, Rockford, IL)). Protein amounts were determined using the Bio-Rad DCTM Protein Assay Kit II according to manufacturer’s protocol. Samples were mixed with Laemmli sample buffer (Bio-Rad, Hercules, CA) containing 5% β-mercaptoethanol, separated by 6% or 4–15% SDS-PAGE (Bio-Rad, Hercules, CA), and transferred onto a PVDF membrane using an iBlot dry transfer apparatus (Invitrogen, Grand Island, NY). The membrane was blocked with 5% non-fat dry milk (BioRad, Hercules, CA) or 3% BSA (Sigma, St. Louis, MO) in TBST (20mM Tris-HCl, 0.5M NaCl, 0.1% Tween 20) and incubated with a primary antibody overnight at 4°C. Following washes with TBST, the membrane was incubated with horseradish peroxidase-conjugated secondary antibody (Jackson Laboratories, West Grove, PA) and detected with ECL developing solution (Thermo Scientific, Rockford, IL). A list of primary antibodies used is provided in Supplemental Table S7.
Immunohistochemistry
Standard 5 micron paraffin-embedded tissue sections from xenografts were stained using an anti-N1-ICD rabbit monoclonal antibody (Cell Signaling Technology, clone D3B8, catalog #4147; final concentration, 17μg/mL) as described (19).
GSI washout assay
CUTLL1, REC-1 and HCC1599 cells were cultured for 3 days with GSI (compound E, 1uM) to establish a Notch-off state. Notch was the re-activated by GSI washout as described (22) and harvested for analysis 4hr later.
Quantitative Real-Time PCR
RNA was extracted from cultured cells or tumor xenografts using the RNeasy Mini Kit (Qiagen, Valencia, CA). cDNA was synthesized using SuperScript VILO MasterMix (Invitrogen, Grand Island, NY). Quantitative PCR was performed on an ABI 7900 using TaqMan® Gene Expression MasterMix (Invitrogen, Grand Island, NY) and their inventoried TaqMan probes/primers (Supplementary Table S8); the resulting qPCR data were analyzed using the ΔΔCt relative quantification protocol.
ChIP-qPCR
NOTCH1 and RBPJ ChIP were performed as described (8). REC-1 cells were cross-linked with 1% formaldehyde for 10 min at 37°C and sheared by sonication. Rabbit IgG (Jackson ImmunoResearch 011-000-003), NOTCH1 (35), and RBPJ (Cell Signaling Technology #5313) were added to the sonicate and incubated overnight at 4° C. DNA/protein complexes were captured with protein A-conjugated agarose beads, washed, and eluted. After reversal of cross-links, DNA was purified using the QIAquick PCR purification kit (QIAGEN). Input control was prepared in parallel without immunoprecipitation. Real-time PCR was performed in triplicates using primers specific for the HES4 promoter and genomic negative control. The primer sequences are as follows: HES4: forward 5’-GGTGTGTGAACCCGGCTCCG-3’, reverse 5’-CCGAGGCGTGACTGACAGCG-3’. Genomic negative control primers: forward 5’-AATGCTGGGCTTCCAAGGA-3’, reverse 5’-GACCTTGGTGACTGTTGAGGAAAC-3’.
Xenograft models
From 1–8×106 HCC1599, HCC1187, MB157, or MDA-MD-231 triple negative breast cancer cells were inoculated subcutaneously into the left flank of 4–6 week-old immunodeficient (nu/nu or NOD-SCID) female mice (Charles River Laboratories). Patient derived subcutaneous xenograft efficacy studies included the triple negative breast cancer models HBCx8 and HBCx-14 (Xentech, Evry, France) and the adenoid cystic carcinoma models CTG-0007 (ACCx9) and CTG-0009 (POS-912) (Champions Oncology Inc., Hackensack, NJ). Upon reaching an average tumor size of 150–250mm3, mice were randomized across control or treatment groups (n=10–12 mice per group). Tumor size was measured with calipers and body weight was recorded twice per week during the dosing phase. MRK-003 in 0.5% methylcellulose was given orally at the indicated dose/schedules, while paclitaxel in 0.9% NaCl was administered intraperitoneally at a 15mg/kg once per week. Mice were euthanized at the indicated time points and portions of the tumors were snap frozen in liquid nitrogen for biochemical analysis or fixed in 10% neutral buffered formalin for immunohistochemical analysis.
NOTCH gene analysis in human breast tumors
Detailed analysis of human tumor cell lines and breast tumors are provided in supplementary methods. A summary of the NOTCH gene coverage data from human tumors are provided (See Table S9; Cell line sequencing data can be accessed at Gene Bank accession number SRP044150). NOTCH gene signature analysis is provided in supplementary Table S10.
Supplementary Material
Significance.
NOTCH1 mutations, immunohistochemical staining for activated NOTCH1, and HES4 expression are biomarkers that can be used to identify solid tumors that are likely to respond to gamma-secretase inhibitor-based therapies.
Acknowledgments
Funding statement: Majority of the work was funded by Merck & Co Inc.; JCA was supported by NIH P01 grant (5P01CA119070-08); WSP was supported by Abramson Cancer Center Pilot Grant
Footnotes
CONFLICT OF INTEREST DISCLOSURES
AS; SL; AT; JY; CW; JM; PWG; JL; BBH; LZ; SF; GDG; TZ and SS all employees of Merck & Co and hold company stocks. JCA is a consultant for Cell Signaling Technologies and CytomX.
References
- 1.Kopan R. Notch signaling. Cold Spring Harb Perspect Biol. 2012;4(10) doi: 10.1101/cshperspect.a011213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weng AP, Ferrando AA, Lee W, Morris JP, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 3.Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66(4):649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 4.Ashworth TD, Pear WS, Chiang MY, Blacklow SC, Mastio J, Xu L, et al. Deletion-based mechanisms of Notch1 activation in T-ALL: key roles for RAG recombinase and a conserved internal translational start site in Notch1. Blood. 2010;116(25):5455–5464. doi: 10.1182/blood-2010-05-286328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson DR, Kalyana-Sundaram S, Wu YM, Shankar S, Cao X, Ateeq B, et al. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nature medicine. 2011;17(12):1646–1651. doi: 10.1038/nm.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333(6046):1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang NJ, Sanborn Z, Arnett KL, Bayston LJ, Liao W, Proby CM, et al. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc Natl Acad Sci U S A. 2011;108(43):17761–17766. doi: 10.1073/pnas.1114669108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aster JC, Blacklow SC. Targeting the notch pathway: twists and turns on the road to rational therapeutics. J Clin Oncol. 2012;30(19):2418–2420. doi: 10.1200/JCO.2012.42.0992. [DOI] [PubMed] [Google Scholar]
- 10.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. The Journal of clinical investigation. 121(7):2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis HD, Leveridge M, Strack PR, Haldon CD, O'Neil J, Kim H, et al. Apoptosis in T cell acute lymphoblastic leukemia cells after cell cycle arrest induced by pharmacological inhibition of notch signaling. Chem Biol. 2007;14(2):209–219. doi: 10.1016/j.chembiol.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 12.Palomero T, Barnes KC, Real PJ, Glade Bender JL, Sulis ML, Murty VV, et al. CUTLL1, a novel human T-cell lymphoma cell line with t(7;9) rearrangement, aberrant NOTCH1 activation and high sensitivity to gamma-secretase inhibitors. Leukemia. 2006;20(7):1279–1287. doi: 10.1038/sj.leu.2404258. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Zang C, Taing L, Arnett KL, Wong YJ, Pear WS, et al. NOTCH1-RBPJ complexes drive target gene expression through dynamic interactions with superenhancers. Proceedings of the National Academy of Sciences of the United States of America. 111(2):705–710. doi: 10.1073/pnas.1315023111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris EJ, Jha S, Restaino CR, Dayananth P, Zhu H, Cooper A, et al. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer discovery. 3(7):742–750. doi: 10.1158/2159-8290.CD-13-0070. [DOI] [PubMed] [Google Scholar]
- 15.Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nature medicine. 2007;13(10):1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu CH, van Riggelen J, Yetil A, Fan AC, Bachireddy P, Felsher DW. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(32):13028–13033. doi: 10.1073/pnas.0701953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGowan PM, Simedrea C, Ribot EJ, Foster PJ, Palmieri D, Steeg PS, et al. Notch1 inhibition alters the CD44hi/CD24lo population and reduces the formation of brain metastases from breast cancer. Mol Cancer Res. 9(7):834–844. doi: 10.1158/1541-7786.MCR-10-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazzone M, Selfors LM, Albeck J, Overholtzer M, Sale S, Carroll DL, et al. Dose-dependent induction of distinct phenotypic responses to Notch pathway activation in mammary epithelial cells. Proc Natl Acad Sci U S A. 2010;107(11):5012–5017. doi: 10.1073/pnas.1000896107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kluk MJ, Ashworth T, Wang H, Knoechel B, Mason EF, Morgan EA, et al. Gauging NOTCH1 Activation in Cancer Using Immunohistochemistry. PLoS ONE. 2013;8(6):e67306. doi: 10.1371/journal.pone.0067306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clementz AG, Rogowski A, Pandya K, Miele L, Osipo C. NOTCH-1 and NOTCH-4 are novel gene targets of PEA3 in breast cancer: novel therapeutic implications. Breast Cancer Res. 13(3):R63. doi: 10.1186/bcr2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao SS, O'Neil J, Liberator CD, Hardwick JS, Dai X, Zhang T, et al. Inhibition of NOTCH signaling by gamma secretase inhibitor engages the RB pathway and elicits cell cycle exit in T-cell acute lymphoblastic leukemia cells. Cancer Res. 2009;69(7):3060–3068. doi: 10.1158/0008-5472.CAN-08-4295. [DOI] [PubMed] [Google Scholar]
- 22.Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20(15):2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A. 2006;103(48):18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma VM, Calvo JA, Draheim KM, Cunningham LA, Hermance N, Beverly L, et al. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Molecular and cellular biology. 2006;26(21):8022–8031. doi: 10.1128/MCB.01091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klinakis A, Szabolcs M, Politi K, Kiaris H, Artavanis-Tsakonas S, Efstratiadis A. Myc is a Notch1 transcriptional target and a requisite for Notch1-induced mammary tumorigenesis in mice. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(24):9262–9267. doi: 10.1073/pnas.0603371103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.South AP, Cho RJ, Aster JC. The double-edged sword of Notch signaling in cancer. Semin Cell Dev Biol. 2012 doi: 10.1016/j.semcdb.2012.01.017. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephens PJ, Davies HR, Mitani Y, Van Loo P, Shlien A, Tarpey PS, et al. Whole exome sequencing of adenoid cystic carcinoma. The Journal of clinical investigation. 2013 doi: 10.1172/JCI67201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez-Irizarry C, Carpenter AC, Weng AP, Pear WS, Aster JC, Blacklow SC. Notch subunit heterodimerization and prevention of ligand-independent proteolytic activation depend, respectively, on a novel domain and the LNR repeats. Mol Cell Biol. 2004;24(21):9265–9273. doi: 10.1128/MCB.24.21.9265-9273.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amir E, Seruga B, Serrano R, Ocana A. Targeting DNA repair in breast cancer: a clinical and translational update. Cancer treatment reviews. 36(7):557–565. doi: 10.1016/j.ctrv.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 30.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 486(7403):395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho AS, Kannan K, Roy DM, Morris LG, Ganly I, Katabi N, et al. The mutational landscape of adenoid cystic carcinoma. Nature genetics. 45(7):791–798. doi: 10.1038/ng.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frierson HF, Jr, Moskaluk CA. Mutation signature of adenoid cystic carcinoma: evidence for transcriptional and epigenetic reprogramming. The Journal of clinical investigation. 2013:1–3. doi: 10.1172/JCI69070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ross JS, Wang K, Rand JV, Sheehan CE, Jennings TA, Al-Rohil RN, et al. Comprehensive genomic profiling of relapsed and metastatic adenoid cystic carcinomas by next-generation sequencing reveals potential new routes to targeted therapies. The American journal of surgical pathology. 38(2):235–238. doi: 10.1097/PAS.0000000000000102. [DOI] [PubMed] [Google Scholar]
- 34.Krop I, Demuth T, Guthrie T, Wen PY, Mason WP, Chinnaiyan P, et al. Phase I pharmacologic and pharmacodynamic study of the gamma secretase (Notch) inhibitor MK-0752 in adult patients with advanced solid tumors. J Clin Oncol. 30(19):2307–2313. doi: 10.1200/JCO.2011.39.1540. [DOI] [PubMed] [Google Scholar]
- 35.Wang H, Zou J, Zhao B, Johannsen E, Ashworth T, Wong H, et al. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc Natl Acad Sci U S A. 2011;108(36):14908–14913. doi: 10.1073/pnas.1109023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.