Abstract
Objective
To evaluate the association between promoter DNA methylation and Discoidin domain receptor 1 (DDR1) gene expression in men with nonobstructive azoospermia (NOA).
Design
We compared fibroblalsts cultured from testicular biopsies using a high resolution Infinium 450K methylation array. We validated promoter methylation, mRNA and protein levels of the CpG sites identified from array.
Setting
Basic research laboratory.
Interventions
None
Patient(s)
Men with NOA (n = 16) and with normal spermatogenesis (n = 5).
Main outcome measure(s)
Bisulfite Clonal Sequencing was used for validation and quantification of CpG methylation of DDR1. Gene expression analysis of DDR1 with quantitative polymerase chain reaction and immunohistochemistry was performed to validate the array results at mRNA and protein levels.
Results
Differentially methylated CpG sites (~20K) were identified using an F-Test in the NOA samples. We identified 20 genes with >30% difference in DNA methylation within the promoter region of men with NOA and fertile controls. Of the aberrantly methylated genes, 10 were hypomethylated and 10 were hypermethylated genes. From the top 10 hypermethylated genes, six genes (MRI1, DCAF12L1, TMEM95, CECR2, DDR1, NPHS2) were selected for validation since they were shown to be expressed in the testis. Of the 6 genes expressed in the fibroblasts cultured from testis, DDR1 showed an abnormal gene expression pattern. Three patients (19%) out of the 16 NOA men for whom gene expression data available had lower DDR1 expression levels (1.8× fold decrease) than fertile men, whereas four (25%) men had higher expression levels (2× fold increase) of DDR1 compared to levels in fertile men. Quantitative analysis by bisulfite clonal sequencing showed that one of the CpG sites (cg13329862) of DDR1 promoter was hypermethylated in NOA patients compared with fertile controls (53% versus 15%). Immunohistochemical analysis suggests presence of DDR1 within cytoplasm of germ cells and peritubular connective tissue (in men with hypospermatogenesis) and decreased expression of the protein in men with Sertoli-cell only syndrome.
Conclusions
Abnormal gene expression of DDR1 is associated with NOA. The functional relevance of aberrant methylation of DDR1 to expression of DDR1 in men with NOA warrants further investigation.
Keywords: infertility, promoter, epigenetics
INTRODUCTION
Non-obstructive azoospermia (NOA), lack of sperm in the ejaculate, results from spermatogenic failure. Spermatogenesis requires coordinated regulation of testicular gene expression. DNA methylation, which is the stable, covalent addition of a methyl group to cytosine, has been shown to be a contributor to regulation of gene expression and alteration to DNA methylation can modify gene expression. Both human and animal studies indicate that abnormal sperm DNA methylation patterns, including aberrant methylation of both imprinted (1–5) and non-imprinted genes (6, 7) are associated with subfertility in oligospermic men.
Spermatogenesis is the transformation of spermatogonial cells into spermatozoa over an extended period of time involving constant cell proliferation and differentiation. Therefore genes involved in regulation of epithelial cell differentiation would be important for spermatogenesis. Discoidin domain receptor 1 (DDR1) is a member of a small subfamily of receptor tyrosine kinases that is involved in proliferation, apoptosis, cell morphogenesis and differentiation (8). DDR1 is expressed in human post-meiotic germ cells of testis (9). The ligands of the DDR1 receptor are collagen and Ecadherin(10). It is well recognized that collagen is essential for spermatogenesis (11) and E-cadherin plays an important role in primordial germ cell migration and development (12).
Sertoli and germ cells rely on structural support provided by the basement membrane at different stages of the seminiferous epithelial cycle. Hence, we hypothesized that abnormal DDR1 expression due to aberrant DNA methylation might trigger a mechanism that can compromise spermatogenesis in a subset of men with idiopathic NOA. In the present study, we utilized a high-density methylation array to investigate the pattern of DNA methylation that could be associated with abnormal gene expression. Of the candidate genes identified from the methylation microarray, we selected DDR1 and performed validation at promoter methylation, mRNA and protein levels.
PATIENTS AND METHODS
Sixteen azoospermic men and five men with proven fertility were recruited for this study with the approval and oversight of Baylor College of Medicine’s Institutional Review Board for Human Subjects. Subjects gave informed consent. Azoospermia in all patients was confirmed by analysis of at least two different centrifuged ejaculate specimens according to World Health Organization guidelines. On the day of surgery, another ejaculated sample was obtained and azoospermia confirmed via an extended sperm preparation. Karyotype analysis and Y chromosome microdeletion analysis was performed on all patients to confirm a normal karyotype and absence of Y-chromosome microdeletions. Men who were proven fertile reported a history of fathering a biological child.
We used the high resolution Infinium 450K methylation array and compared fibroblasts cultured from testicular biopsies of 16 NOA men and 5 fertile men (supplementary figure). Microarray data was analyzed using Minfi (R software package) and normalized utilizing subset-quantile within array normalization. Using an F-test, we generated a dataset of methylated CpG sites differentiating between fertile controls and men with NOA. We used a very stringent selection criterion that combined the following three factors: 1. Statistical significance (p-value) 2. Magnitude of effect (difference in % methylation between men with NOA and fertile men) and 3. Location of CpG sites (immediately upstream of the transcription start site of genes).
Primary testicular fibroblast culture
Testicular specimens were collected and divided into two portions. One tissue fragment was fixed in Bouin’s solution, paraffinized and stained for routine histological examination. A second portion of the sample (from the same biopsy) was cultured in vitro to establish a primary testicular fibroblast culture. To serve as controls, testis biopsies also were collected from men with proven fertility (done as part of routine clinical practice) undergoing vasectomy reversal. Primary cultures were maintained in Dulbecco’s modified Eagle medium, supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin (all provided by Gibco–Life Technologies; Grand Island, NY, USA). Peripheral blood was collected from each patient and used for DNA purification.
RNA Isolation and cDNA Synthesis
RNA was isolated from fibroblasts cultured from human testis biopsies using RNeasy Plus kit (Qiagen) according to the manufacturer's instructions. For reverse transcription, High Capacity cDNA Reverse Transcription kit (Cat#4368814, Applied Biosystems, USA) was used with a total concentration of 500 ng RNA (25ng/µl) in a total of 20 µl reaction mixture in accordance with the manufacturer’s instructions and incubated for 10 min at 25 °C followed by 30 min at 48 °C and 5 min at 95 °C. The cDNA was stored at −20°C until further use.
Clonal bisulfite sequencing
We used the cloning-based bisulfite sequencing (CBS) method for our quantitative examination of DDR1 CpG methylation in 8 NOA patients and 2 fertile controls chosen randomly from the study population included in the microarray. Genomic DNA (500 ng) was bisulfite-converted using EZ DNA methylation kit (Catalog # D5001, Zymo Research). Each of the DNA samples was amplified by PCR as follows: a PCR reaction mix containing ~2 µl of the bisulfite-treated DNA, 2 µL (2 µM) combined of forward and reverse primers (Forward TGATTTTGGGGTTGTTTGTTAGTA; Reverse AATACTTTTTCCCCACTCAACACTA), 1 × PCR buffer, 10mM dNTP mix, 1mM MgCl2, and ZymoTaq™ DNA Polymerase in a total volume of 20 µl. PCR cycle conditions were as follows: Initial denature at 95°C for 10 min, DNA was amplified in 35 cycles for 30s at 95°C, 40s at 59°C (for DDR1) and 1 min at 72°C followed by a final extension at 72°C for 10 min. Amplified products were directly cloned into the pCR®4- TOPO® vector using the TOPO Cloning kit (Catalog # K4575-02, Life technologies, USA). Individual clones were sent to Genewiz for DNA sequencing. Approximately 10 different clones from each PCR product were sequenced to analyze the methylation status of the CpG site (cg13329862). The methylation status was analyzed using BISMA (http://services.ibc.uni-stuttgart.de/BDPC/BISMA/index.php). BISMA counterchecks all sequences for existence of clonal amplifications from the same genomic template for CBS efficiency. Sequences with < 95% conversion efficiency were omitted by the software.
Quantitative RT-PCR (qPCR)
Quantification of DDR1 expression in fibroblasts cultured from human testicular biopsies was performed by qPCR using Taqman probe (Hs01058430_m1, Life technologies) designed for the amplification of DDR1 in 21 men (5 fertile controls and 16 men with NOA). Gene expression was normalized to an internal housekeeping gene GAPDH (Hs02758991_g1, Life technologies). Reactions were carried out in Quant Studio Plus 12K Flex Real-Time PCR System and efficiency of the reactions was determined for all primer sets using 1:10 dilutions of cDNA samples. Reaction mixtures consisted of TaqMan Gene expression master mix (Applied Biosystems) 1X, 1µL of Taqman probe, and 1 µL of cDNA in a final volume of 20 µL. All reactions were carried out in triplicate. Normalized expression values were calculated according to a published mathematical model proposed by Pfaffl (13).
Immunohistochemistry
Formalin-fixed specimens from the testes of 2 patients (1 patient with hypospermatogenesis and 1 Maturation arrest syndrome histology) were processed and mounted. Genitourinary pathologists at Baylor College of Medicine determined histologies. Samples were then heated in boiling water bath for antigen retrieval (10 mmol/L citrate buffer, pH 6, 20 min). The sections were allowed to cool in citrate buffer, washed thrice (de-ionized water, PBS, pH 7.4, 3 min each) and incubated in blocking solution for 30 min. Next, they were washed with PBS (3 × 10 min) and incubated overnight with primary rabbit polyclonal anti-DDR1 antibody (1:800 diluted, Santa-Cruz Biotechnology, sc-532) at 4 °C. After being washed with PBS (3 × 10 min), the sections were incubated with horseradish peroxidase conjugated goat anti-rabbit IgG for 1 h at room temperature. Finally, peroxidase activity was visualized using 0.05% diaminobenzidine (DAB; Sigma), in 0.05 mol/L Tris buffer, pH 7.6, containing 0.01% hydrogen peroxide.
Statistical Analyses
Aside from array normalization procedures, the R software environment (R Foundation for Statistical Computing, Vienna, Austria) was used for all statistical analysis for data from microarray. Gene expression values are expressed as mean ± SD, for comparison of the results between fertile control and men with NOA. All relative quantification was assessed using REST software 2009, RG mode, using the pair-wise fixed randomization test with 10,000 permutations (13). The significant level of differences was taken as P <0.05.
RESULTS
Differentially methylated CpG sites (~10K) were identified using an F-Test (p<0.05) in the NOA men. We identified 20 CpG sites within the promoter region of men with NOA with >30% difference in methylation as compared to fertile controls that had p<0.05. Of the 20 aberrantly methylated CpGs within promoter regions, 10 were hypomethylated and 10 were hypermethylated. From the top 10 hypermethylated CpG sites, six genes (MRI1, DCAF12L1, TMEM95, CECR2, DDR1, NPHS2) were selected for validation since they were shown to be expressed in the testis (Supplementary Table 1). Of the 6 genes, 4 genes were not expressed in the fibroblasts. MRI1 gene expression was similar in men with NOA and men with proven fertility.
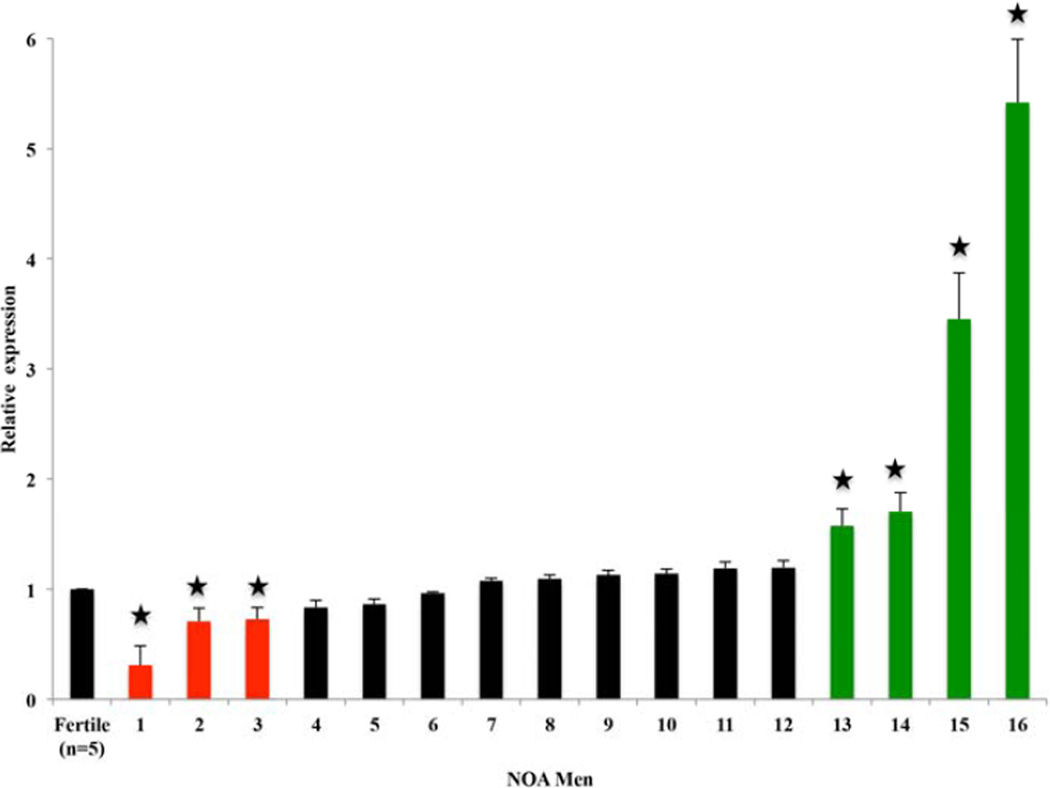
DDR1 showed an abnormal gene expression pattern in 16 men with NOA. The level of DDR1 mRNA in 3/16 men with NOA was approximately twofold lower than that in fertile controls (P < 0.05). Additionally, the expression level of DDR1 mRNA was approximately twofold higher in 4/16 NOA patients compared to fertile controls (P=0.012) (Figure 1).
Figure 1.
Relative expression of DDR1 mRNA in men with nonobstructive azoospermia compared to fertile controls. * p < 0.05
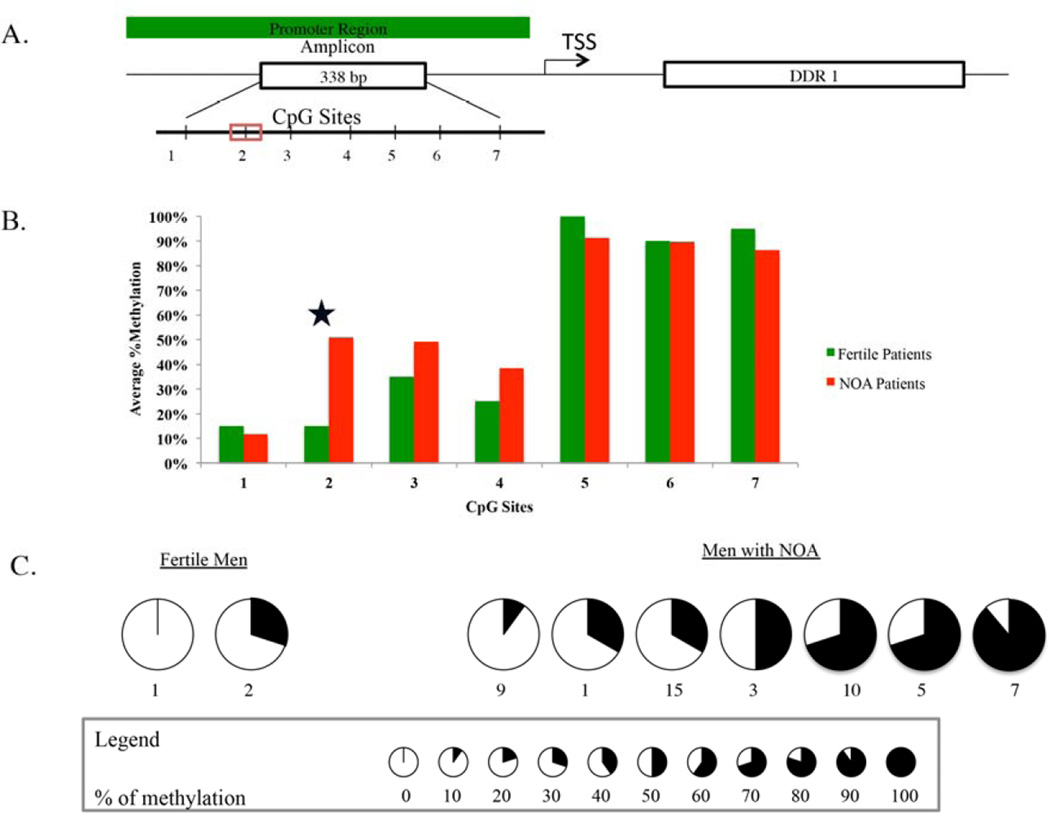
To further investigate the regulation of expression of DDR1 promoter and validate the microarray, we performed clonal bisulfite sequencing. Of the 7 CpG sites in the DDR1 amplicon (Figure 2A) (338–bp), we identified that the methylation of a single CpG site (cg13329862 identified from array) was differentially methylated between fertile and NOA men (Figure 2B and 2C). Fertile men had 15% methylation at the CpG site, whereas men with NOA had 50% methylation (p< 0.05) at the CpG site, cg13329862. Of the 3 men that had decreased gene expression, two men had genomic DNA available for CBS. One man had 30% and the other had 50% methylation of CpG site, cg13329862 (Supplementary Table 2).
Figure 2.
Map of the 338-bp CpG island around exon 1 in DDR1. Vertical bars indicate CpG sites (A). Clonal bisulfite sequencing was done to assess methylation status of CpG sites in the amplicon (B) and site # 2 (cg13329862) in men with NOA and fertile controls (c). Ten clones were sequenced from each patient. The percentage of methylation in each CCG site is denoted by pie charts, as indicated.
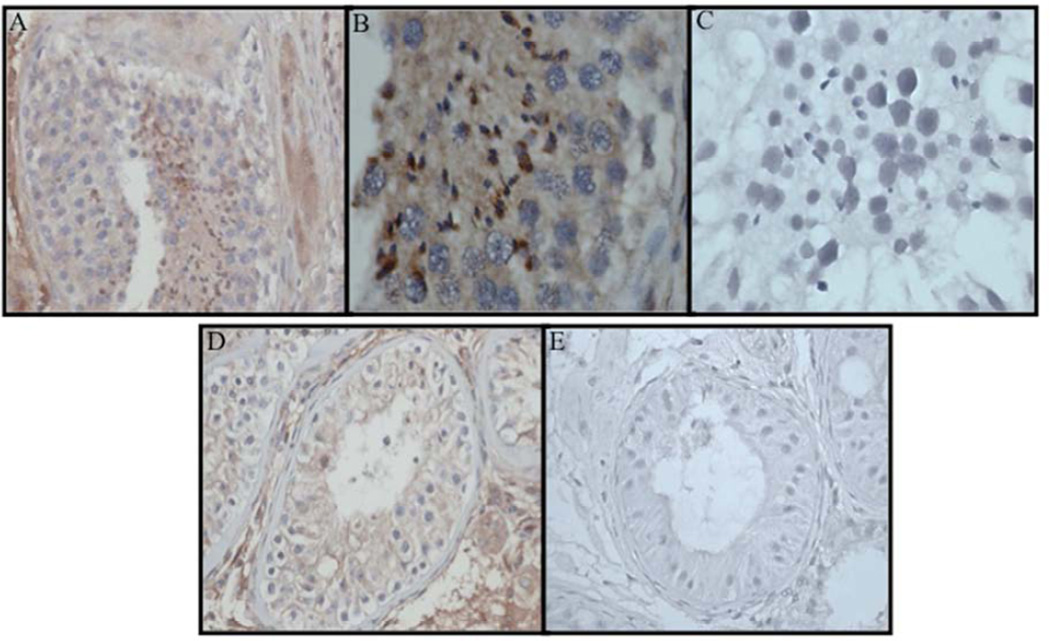
To identify DDR1 expression in men with hypospermatogenesis and Sertoli-cell only syndrome, DDR1 immunostaining signals were evaluated by qualitative IHC. DDR1 was expressed in the germ cells and peritubular connective tissue in men with hypospermatogenesis (Figure 3A – C) and expressed only in peritubular tissue men with maturation arrest (Figure 3D and 3E).
Figure 3.
Immunohistochemistry demonstrating DDR1 protein in germ cells and peritubular connective tissue from testis biopsy from man with NOA and hypospermatogenesis at low power 100x (A) and high power 400x (B) and decreased expression of DDR1 protein in man with Maturation arrest histology (D). Lack of DDR1 staining is demonstrated in negative controls (C and E).
DISCUSSION
In this study, we characterized the effect of methylation and the expression of DDR1 in men with NOA. Of the total of 16 men that had fibroblasts cultured from testicular biopsy samples from men with NOA, 3 patients had a relatively low (1.8×) DDR1 expression and 4 men had a higher DDR1 (2×) expression than did the 5 fertile men. DDR1 represents a small subfamily of receptor tyrosine kinases (RTKs) found predominantly in epithelial cells(14). RTKs are involved in proliferation, differentiation, and migration of cells, all processes that are essential for spermatogenesis. Ddr1 is expressed in postmeiotic germ cells of the rat testis (9). We demonstrated the presence of DDR1 protein in a man with spermatogenesis and the lack of DDR1 protein in a man with Sertoli-cell only syndrome. In one man, ~30% methylation of CpG site cg13329862 was associated with > 2X lower DDR1 gene expression and in another man ~50% methylation of CpG site cg13329862 was associated with 1.5X lower DDR1 expression (Supplementary Table 2). With a larger sample size, we could have demonstrated that the phenomenon of hypermethylation of the CpG site leads to decreased gene and protein expression in more men with NOA.
It is of great clinical importance to identify markers that can indicate the presence or absence of foci of spermatogenesis in a testicular biopsy sample, because it is widely accepted that the histological results of diagnostic testicular biopsies, the peripheral serum levels of FSH, serum levels of inhibin B, and even a testicular biopsy are not highly accurate markers (15). Of the 16 men with NOA that underwent testicular sperm extraction, we identified sperm in 9 men (43%). Unfortunately, there was no association between either DDR1 mRNA expression or methylation at CpG site of DDR1 promoter with finding sperm with testis biopsy.
Previously, CpG islands were considered to be almost entirely unmethylated except within imprinted regions and when on the inactive X chromosome (16).However, accumulating evidence suggests that differential methylation of tissue-specific regions is associated with modulated gene expression (17) and impaired spermatogenesis. Our observation of a low rate of abnormal methylation in NOA samples is consistent with the published data (1, 18). Due to the limited number of NOA samples analyzed, the nonsignificant difference in methylation at DDR1 in men with NOA needs to be confirmed in future studies. In addition, genomic DNA extracted from these testicular biopsies may not represent the methylation level in germ cells alone. These testicular biopsies contain many somatic cells, such as Leydig cells, Sertoli cells, etc. In the extreme case of Sertoli cell-only or tubular sclerosis, the DDR1 methylation rate indicated a high value as expected. Therefore, an aberrant methylation of DDR1 promoter could be one possible cause of idiopathic NOA.
The study has some limitations. Due to lack of germ cells in men with NOA, we used fibroblasts cultured from testis biopsies to evaluate both gene expression and methylation. Sertoli and Leydig cell could have been isolated and used as the primary target. Although DNA methylation in fibroblasts may not mimic the changes that can occur in germ cells, fibroblasts are the best surrogate for investigating potential constitutional epimutations. In addition, studying fibroblasts provides an adequate comparison between men with proven fertility and men with NOA because we studied the same cell type. We did not have gene expression or clonal bisulfite sequencing data on all the men in the microarray either due to lack of cells (from fibroblast culture) or were unable to extract RNA or genomic DNA for CBS. Ideally, we would have liked to correlate gene and protein expression. Unfortunately, we had access to only 2 / 16 patient blocks for IHC. We therefore used IHC as a qualitative measure to identify DDR1 within the seminiferous tubules in the 2 patients (one man with hypospermatogenesis and one man with maturation arrest histology). We used a man with NOA and hypospermatogenesis (instead of a fertile man) as an internal control to demonstrate that absence of DDR1 expression is dependent on the presence of germ cells and not due to other non-disease related mutations. Not all men who had hypermethylation of the CpG site had down regulation of gene expression. The lack of association between gene expression and methylation could be the result of aberrant methylation in other CpG sites (not identified from the array). Given the aberrant nature of gene expression and DNA methylation, it is possible that other factors such as histone deacetylation that could be responsible for altered gene expression. Although 7/16 men with NOA had statistically significant changes in gene expression, the biological significance remains unclear. Larger screening studies are necessary to validate the importance of DDR1 in men with NOA.
In summary, we believe that aberrant methylation of DDR1 promoter could modify gene expression. Altered DDR1 gene expression may therefore be a marker for men with idiopathic NOA.
Supplementary Material
Acknowledgments
Financial support: RR is supported by a Male Reproductive Health Research (MRHR) Career Development Physician-Scientist Award (K12, HD073917-01) from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) Program to DJL. This project was supported by the Pathology and Histology Core at Baylor College of Medicine with funding from the NIH (NCI P30-CA125123).
Abbreviations
- NOA
nonobstructive azoospermia
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors have no potential conflicts of interest to disclose
References
- 1.Hammoud SS, Purwar J, Pflueger C, Cairns BR, Carrell DT. Alterations in sperm DNA methylation patterns at imprinted loci in two classes of infertility. Fertility and sterility. 2010;94:1728–1733. doi: 10.1016/j.fertnstert.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 2.Houshdaran S, Cortessis VK, Siegmund K, Yang A, Laird PW, Sokol RZ. Widespread epigenetic abnormalities suggest a broad DNA methylation erasure defect in abnormal human sperm. PloS one. 2007;2:e1289. doi: 10.1371/journal.pone.0001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marques CJ, Carvalho F, Sousa M, Barros A. Genomic imprinting in disruptive spermatogenesis. Lancet. 2004;363:1700–1702. doi: 10.1016/S0140-6736(04)16256-9. [DOI] [PubMed] [Google Scholar]
- 4.Marques CJ, Costa P, Vaz B, Carvalho F, Fernandes S, Barros A, et al. Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Molecular human reproduction. 2008;14:67–74. doi: 10.1093/molehr/gam093. [DOI] [PubMed] [Google Scholar]
- 5.Lewejohann L, Damm OS, Luetjens CM, Hamalainen T, Simoni M, Nieschlag E, et al. Impaired recognition memory in male mice with a supernumerary X chromosome. Physiology & behavior. 2009;96:23–29. doi: 10.1016/j.physbeh.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 6.Wu W, Shen O, Qin Y, Niu X, Lu C, Xia Y, et al. Idiopathic male infertility is strongly associated with aberrant promoter methylation of methylenetetrahydrofolate reductase (MTHFR) PloS one. 2010;5:e13884. doi: 10.1371/journal.pone.0013884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Navarro-Costa P, Nogueira P, Carvalho M, Leal F, Cordeiro I, Calhaz-Jorge C, et al. Incorrect DNA methylation of the DAZL promoter CpG island associates with defective human sperm. Human reproduction. 2010;25:2647–2654. doi: 10.1093/humrep/deq200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeh YC, Wu CC, Wang YK, Tang MJ. DDR1 triggers epithelial cell differentiation by promoting cell adhesion through stabilization of E-cadherin. Molecular biology of the cell. 2011;22:940–953. doi: 10.1091/mbc.E10-08-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mullenbach E, Walter L, Dressel R. A novel discoidin domain receptor 1 (Ddr1) transcript is expressed in postmeiotic germ cells of the rat testis depending on the major histocompatibility complex haplotype. Gene. 2006;372:53–61. doi: 10.1016/j.gene.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 10.Borza CM, Pozzi A. Discoidin domain receptors in disease. Matrix biology : journal of the International Society for Matrix Biology. 2013 doi: 10.1016/j.matbio.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siu MK, Cheng CY. Extracellular matrix and its role in spermatogenesis. Advances in experimental medicine and biology. 2008;636:74–91. doi: 10.1007/978-0-387-09597-4_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Carlo A, De Felici M. A role for E-cadherin in mouse primordial germ cell development. Developmental biology. 2000;226:209–219. doi: 10.1006/dbio.2000.9861. [DOI] [PubMed] [Google Scholar]
- 13.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in realtime PCR. Nucleic acids research. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alves F, Vogel W, Mossie K, Millauer B, Hofler H, Ullrich A. Distinct structural characteristics of discoidin I subfamily receptor tyrosine kinases and complementary expression in human cancer. Oncogene. 1995;10:609–618. [PubMed] [Google Scholar]
- 15.Ramasamy R, Padilla WO, Osterberg EC, Srivastava A, Reifsnyder JE, Niederberger C, et al. A comparison of models for predicting sperm retrieval before microdissection testicular sperm extraction in men with nonobstructive azoospermia. The Journal of urology. 2013;189:638–642. doi: 10.1016/j.juro.2012.09.038. [DOI] [PubMed] [Google Scholar]
- 16.Rakyan VK, Down TA, Thorne NP, Flicek P, Kulesha E, Graf S, et al. An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs) Genome research. 2008;18:1518–1529. doi: 10.1101/gr.077479.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schilling E, Rehli M. Global, comparative analysis of tissue-specific promoter CpG methylation. Genomics. 2007;90:314–323. doi: 10.1016/j.ygeno.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 18.Hartmann S, Bergmann M, Bohle RM, Weidner W, Steger K. Genetic imprinting during impaired spermatogenesis. Molecular human reproduction. 2006;12:407–411. doi: 10.1093/molehr/gal040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.