SUMMARY
Xylan is the third most abundant glycopolymer on earth after cellulose and chitin. As a major component of wood, grain and forage, this natural biopolymer has far-reaching impacts on human life. This highly acetylated cell wall polysaccharide is a vital component of the plant cell wall, which functions as a molecular scaffold, providing plants with mechanical strength and flexibility. Mutations that impair synthesis of the xylan backbone give rise to plants that fail to grow normally due to collapsed xylem cells in the vascular system. Phenotypic analysis of these mutants has implicated many proteins in xylan biosynthesis. However, the enzymes directly responsible for elongation and acetylation of the xylan backbone have not been unambiguously identified. Here we provide direct biochemical evidence that two Arabidopsis thaliana proteins, IRREGULAR XYLEM 10-L (IRX10-L) and ESKIMO1/ TRICOME BIREFRINGENCE 29 (ESK1/TBL29), catalyze these respective processes in vitro. By identifying the elusive xylan synthase and establishing ESK1/TBL29 as the archetypal plant polysaccharide O-acetyltransferase, we have resolved two long-standing questions in plant cell wall biochemistry. These findings shed light on integral steps in the molecular pathways utilized by plants to synthesize a major component of the world's biomass and expand our toolkit for producing glycopolymers with valuable properties.
Keywords: plant cell wall, glycosyltransferase, O-acetyltransferase, xylan, xylan synthase, Arabidopsis thaliana
INTRODUCTION
The survival of plants that dominate our environment and that are the ultimate source of most of our food depends on their ability to synthesize xylan-rich cell walls, which are required for the efficient transport of water from the soil to the leaves. Xylans are a major component of the cereals and vegetables we consume and constitute up to one third of the woody biomass we use to build our homes. As a major component of the plant cell walls in both monocots and dicots, these agronomically important natural biopolymers play a central role in human health and well-being. Xylans are being increasingly used as a renewable resource for the manufacture of valuable products such as biocompatible materials (Simkovic, 2013).
Like most polysaccharides, xylans are synthesized by glycosyltransferases (GTs) that use the chemical energy in donor molecules, such as nucleotide sugars, to drive the attachment of monosaccharides to the growing polymer (Fig. 1A) (Lairson et al., 2008). All xylan molecules produced by vascular plants have the same backbone, composed of 1,4-linked β-D-xylopyranosyl (Xyl) residues. However, the structures that decorate this backbone vary depending on the plant tissue and species (York and O'Neill, 2008). Several enzymes that diversify xylan structure by functionally modifying its backbone have been identified. For example, glucuronic acid (GlcA) substituents are added to glucuronoxylan in dicots such as Arabidopsis thaliana by members of the Carbohydrate Active enZyme (CAZy) family GT8 (Lairson et al., 2008)α-glucuronosyltransferases encoded by GLCA SUBSTITUTION OF XYLAN (GUX) genes (Rennie et al., 2012). We have recently shown that the GlcA residues are further modified by GLUCURONOXYLAN METHYLTRANSFERASE 1 (GXMT1) to form 4-O-methyl glucuronic acid (MeGlcA) (Urbanowicz et al., 2012). In dicots, the most abundant substituents of the backbone Xyl residues are O-acetates (Teleman et al., 2000). For example, the glucuronoxylan extracted from poplar wood and Arabidopsis thaliana stems (Teleman et al., 2000, Yuan et al., 2013) contains approximately six times as many O-acetyl substituents as (Me)GlcA substituents. Recently, analysis of Arabidopsis thaliana mutants with reduced xylan acetylation phenotypes implicated members of two protein families, REDUCED WALL ACETYLATION (RWA) and TRICHOME BIREFRINGENCE-LIKE (TBL), in xylan acetylation (Gille and Pauly, 2012, Rennie and Scheller, 2014). Phenotypic evidence suggests that RWA proteins are involved in the early steps of the acetylation process while members of the TBL family are specific polysaccharide O-acetyltransferases (Gille and Pauly, 2012, Rennie and Scheller, 2014). However, it has not been possible thus far to obtain direct biochemical evidence showing that any of the enzymes required for O-acetylation of xylan in vivo actually catalyze acetylation of the polysaccharide (Gille and Pauly, 2012).
Figure 1. IRX10-L catalyzed transfer of xylosyl residues from UDP-Xyl to a chemically labeled acceptor, Xyl6-2AB.
(a) In vitro xylosyltransferase reaction scheme.
(b) MALDI-TOF MS of the products after a 16-hour reaction in the absence and presence of IRX10-L. The series of annotated [M+H]+ ions are due to structures with a mass difference of 132 Da, consistent with the sequential addition of xylosyl residues to the Xyl6-2AB acceptor to generate oligosaccharides with DP 7-13.
(c) 1D 1H NMR analysis of products generated by incubating unlabeled xylohexaose for 120 hours with IRX10-L and UDP-Xyl. The increased signal intensities for internal β-Xyl (shaded) relative to those of terminal and reducing β-Xyl establishes that IRX10-L catalyzes the addition of β-(1,4)-linked Xyl residues to the chain.
(d) Kinetics of xylosyl transfer to xylohexaose (Xyl6) as determined by measuring the amounts of uridine triphosphate (UDP) formed upon transfer of the xylosyl residue to increasing amounts of Xyl6 (0–4 mM) in the presence of IRX10-L (15.8 pmol). Three technical replicates are shown. Kinetic constants Km (mM) and Vmax (pmol UDP min−1) were calculated by fitting the initial velocities (V, pmol UDP min−1) as a function of the acceptor substrate concentration to the Michaelis-Menten equation using nonlinear curve fitting.
(e) The effect of divalent metals on IRX10-L activity, determined by RP-HPLC analysis. Data are averages of two biological replicates. Error bars are SD.
It has been hypothesized (Richmond and Somerville, 2000, Sandhu et al., 2009) that the enzymes responsible for the synthesis of all plant β-1,4 glycans, including xylan, are members of CAZy family GT2 (Lairson et al., 2008). Due to their structural homology to cellulose synthase (CESA) (Pear et al., 1996), all of these proteins are classified as cellulose synthase-like (CSL) GTs (Richmond and Somerville, 2000). Enzymes in this family have been shown to catalyze β-1,4 glycan backbone synthesis for almost all other hemicellulosic polysaccharides, including mannan [CSLA (Dhugga et al., 2004)], xyloglucan [CSLC (Cocuron et al., 2007)] and 1,3;1,4-β-D-glucan [CSLF (Burton et al., 2006) and CSLH (Doblin et al., 2009)]. Interestingly, the β-1,4-galactosyltransferase responsible for galactan synthesis was recently identified as a GT92 enzyme, demonstrating that CSL proteins are not always responsible for β-1,4 glycan synthesis in plants (Liwanag et al., 2012). Although cell wall galactans are β-1,4 glycans, they are not configurationally homologous to cellulose, mannan, xyloglucan and xylan because the glycosidic oxygens in the main chain have an axial orientation relative to C-4 of the β-galactosyl residue. This has profound effects on the overall shape and physical properties of the polysaccharide, much the same way that a change from the β-anomeric configuration (equatorial glycosidic oxygen) to the α-anomeric configuration (axial glycosidic oxygen) causes in cellulose and starch to have completely different biophysical properties. Identification of the GT92 galactan synthase thus requires a modest refinement of the above hypothesis as follows: “The enzymes responsible for the synthesis of all plant β-1,4 glycans glycans with configurational homology to cellulose are members of CAZy family GT2.”
In contrast to this hypothesis, most of the enzymes so far implicated in xylan backbone synthesis are classified as GT43 or GT47 enzymes. The genes encoding these proteins were originally discerned by phenotypic analysis of Arabidopsis thaliana IRREGULAR XYLEM (IRX) mutants that produce xylan with reduced chain length (Brown et al., 2005, Peña et al., 2007, Brown et al., 2009, Wu et al., 2009, Scheller and Ulvskov, 2010). Three distinct complementation pairs were identified: IRX9/IRX9-L (GT43), IRX14/IRX14-L (GT43), and IRX10/IRX10-L (GT47) (Wu et al., 2009, Wu et al., 2010, Jensen et al., 2013, Rennie and Scheller, 2014,). The first gene of each pair (IRX9, IRX10 and IRX14) is the most highly expressed in secondary cell walls. Complementation with either gene in a pair is able to rescue double mutants in which both genes have been inactivated, indicating that each gene pair encodes two isoenzymes that have very similar or identical catalytic functions (Brown et al., 2009, Wu et al., 2009, Wu et al., 2010). Xylan with a reduced chain length has been isolated from the irx9 irx9-L double mutant. In contrast, no xylan was detected in irx14 irx14-L or irx10 irx10-L stem tissues (Wu et al., 2009, Wu et al., 2010), indicating that IRX10/IRX10-L and IRX14/IRX14-L play vital roles in the formation of this polymer. Based on the increased xylosyltransferase activity of microsomes from tobacco plants heterologously expressing both IRX9 and IRX14, Lee et al. proposed that these two GT43 enzymes are xylosyltransferases (Lee et al., 2012). However, no unambiguous biochemical evidence that establishes the precise molecular functions of any of these proteins in xylan backbone synthesis has been published. Recently, Jensen and colleagues performed transcriptional profiling of the heteroxylan rich psyllium (Plantago ovata Forsk) seed mucilaginous layer and showed that a transcript corresponding a homolog of Arabidopsis IRX10-L displayed the highest expression levels of all the putative glycosyltransferases surveyed (Jensen et al., 2013). However transcripts of GT43 (IRX9, IRX14) genes were found in very low abundance in the mucilage, but were present at levels comparable to IRX10 homologs in the stem tissue (Jensen et al., 2011, Jensen et al., 2013), indicating that IRX10/10-L may be sufficient for xylan backbone synthesis in select tissues.
RESULTS AND DISCUSSION
To determine whether IRX10-L from Arabidopsis thaliana is capable of elongating the β-(1,4)-xylan backbone, we expressed a recombinant tagged form this enzyme in human embroyonic kidney (HEK) 293 cells and purified it from the culture medium (Figure S1a). The catalytic activity of the enzyme was evaluated by means of a xylosyltransferase assay using uridine 5′-diphosphate-xylose (UDP-Xyl) as a sugar donor and 2-aminobenzamide β-(1,4)-xylohexaose (Xyl6-2AB) as a fluorescently labeled acceptor (Figure 1a). Matrix-assisted laser desorption-ionization time-of-flight mass spectrometry (MALDI-TOF MS) analysis of the reaction products indicated recombinant IRX10-L catalyzes the transfer of several xylosyl residues from UDP-Xyl to Xyl6-2AB (Figure 1b). This transfer cannot occur at the reducing end of the substrate, which is blocked by the fluorescent tag (2AB). Treatment of the synthesized polymer with a β-1,4-endoxylanase depolymerized the products (Figure S1b), consistent with the conclusion that IRX10-L catalyzes the addition of β-1,4-linked xylosyl residues to the xylan oligosaccharides. Having established a robust xylosyltransferase assay, we expressed and purified IRX9 and IRX14 as described above and also evaluated their ability to transfer Xyl to Xyl6-2AB. However, these proteins showed no discernible activity, alone or in combination, under these assay conditions (Figure S2). In order to obtain more rigorous characterization of the products of the reaction catalyzed by IRX10-L, we used MALDI-TOF MS (Figure S3a) and nuclear magnetic resonance (NMR) spectroscopy to fully characterize the products of a scaled-up reaction using unlabeled β-(1, 4)-xylohexaose as the acceptor. The NMR spectra of these products (Figures 1c, S3b, and S4a,b) showed an increase in the relative intensity of signals assigned to internal β-(1→4) linked xylose compared to those assigned to terminal β-Xyl and reducing α- and β-Xyl. This is consistent with the fact that all plant GT47 glycosyltransferases characterized to date employ an inverting catalytic mechanism (Geshi et al., 2010), which in the case of IRX10-L transfers xylose from an α-linked donor (UDP-Xyl) to form a β-linked product (β-1,4-xylan). These data unambiguously establish that IRX10-L is a polymerizing GT that forms β-(1→4) linkages by transferring multiple xylosyl residues from UDP-Xyl to the non-reducing end of xylooligosaccharide acceptors.
Having established that IRX10-L catalyzes xylan backbone synthesis in vitro, we next examined the biochemical properties of this enzyme using a bioluminescent assay to detect and quantify xylosyltransferase activity. This approach measures uridine diphosphate (UDP), which is released as a product of the transfer of Xyl from UDP-Xyl onto the elongating chain (Figure 1a). Xylosyltransferase activity was shown to be enzyme concentration dependent in the presence of UDP-Xyl as a donor and Xyl6 as an acceptor, with no observable release of UDP when IRX10-L was reacted with UDP-Xyl alone (Figure S1c). Further, we performed experiments using various acceptors, including xylose, β-(1→4) xylooligosaccharides with degrees of polymerization (DP) ranging from 2 to 6 and cellopentaose, a β-(1→4) glucooligosaccharide. The data indicated that IRX10-L requires an exogenously added acceptor with at least two sugar residues and does not use cellopentaose as an acceptor (Figure S1d). The Km for IRX10-L activity using Xyl6 as an acceptor was determined to be 1.17 mM (Figure 1d), which is similar to that reported for crude wheat microsomal membrane preparations using xylopentaose (Xyl5) as an acceptor (2.9 mM) (Kuroyama and Tsumuraya, 2001). Using the MALDI-TOF MS based assay, we also observed that activity was optimal at pH 6.7-6.8 (Figure S5), also consistent with previous studies using microsomal membranes (Kuroyama and Tsumuraya, 2001). We tested whether IRX10-L can utilize other UDP-sugars, including UDP-arabinopyranose, UDP-galactose, UDP-galacturonic acid, UDP-glucose and UDP-glucuronic acid, as sugar donors. No glycosyltransferase activity was detected with any of these nucleotide-sugars except for very low activity when UDP-arabinopyranose was added. We attribute this activity to the presence of UDP-Xyl, which is a known contaminant (approximately 0.35%) of the UDP-arabinopyranose preparation (CarboSource Services, personal communication).
GTs that utilize nucleotide sugar donor substrates adopt two main types of three-dimensional (3-D) folds. GT-A enzymes are typically divalent metal dependent and often have a cation binding Asp-X-Asp (DXD) signature motif, while most GT-B proteins are metal ion-independent (Lairson et al., 2008). Although no 3-D structures are yet available to place GT47 enzymes such as IRX10-L into one of the GT superfamilies, they have been predicted to adopt a GT-B type fold ( Liu and Mushegian, 2003, Lairson et al., 2008). This is consistent with our data showing that purified IRX10-L is a nucleotide sugar-dependent GT that retains full catalytic activity after ion chelation with EDTA (Figure 1e). The reported increase in xylan synthase activity in wheat microsomes observed when metal ions are added in the absence of exogenous acceptors (Porchia and Scheller, 2000) suggests that other factors that contribute to xylan synthesis in vivo (Rennie and Scheller, 2014) may be metal-dependent.
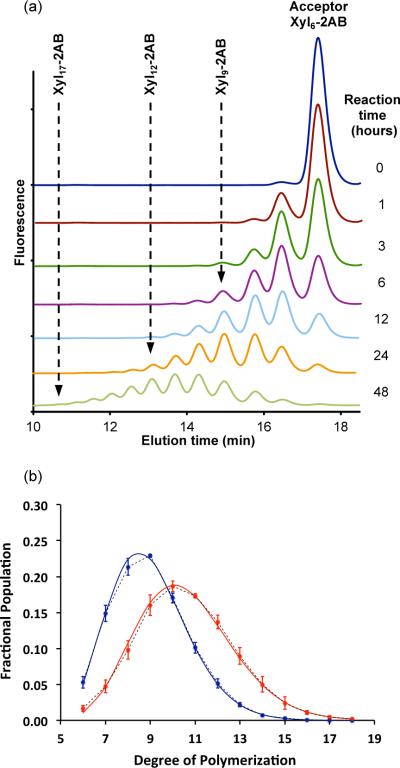
Polymerizing GTs catalyze glycosyl chain elongation by either a distributive mechanism, where the acceptor is released after addition of a single residue, or processive mechanism, where the acceptor is released after addition of several residues (May et al., 2009). A distributive mechanism generates products corresponding to the addition of one or two residues during the initial stages of the reaction followed by a Poisson distribution of larger products as the reaction progresses. In contrast, a processive mode of action produces larger oligosaccharides during the initial stages of the reaction. To distinguish these two possibilities, we analyzed and quantified the reaction progress by reversed-phase high-performance liquid chromatography (RP-HPLC) (Figure 2a). These analyses demonstrate that IRX10-L catalyzes the time-dependent polymerization of xylan to produce large xylooligosaccharides after 48 hours (Figure 2a). The remarkable agreement between the observed and theoretical abundances is consistent with a distributive catalytic mechanism of action for the enzyme (Figure 2b). Taken together, our data indicates that IRX10-L, unaided by other enzymes, uses a distributive mechanism to catalyze xylan synthesis in vitro by repeated incorporation of xylosyl residues to the non-reducing end of the acceptor.
Figure 2. Characteristics of recombinant IRX10-L.
(a) RP-HPLC of the products formed by incubating IRX10-L (0.25 μg/ μL) with UDP-Xyl (2 mM), Xyl6-2AB (0.25 mM) and MgCl2 (1mM) in HEPES-HCl (75 mM) pH 6.8 for the indicated times.
(b) The time-dependent abundance distributions of oligosaccharide products generated as described in (a) and quantified by integration of the peaks in the RP-HPLC elution profiles. Data are averages of three reactions with purified protein from two biological replicates. Observed product distributions are shown for 24 hours (blue) and 48 hours (red) as fractional populations (dotted lines with data point symbols); theoretical abundances (solid lines) are Poisson probabilities calculated as described in Experimental Procedures.
O-Acetyl groups are the most abundant backbone substituents of xylan in dicotyledonous plants. Despite the high degree of acetyl substitution in xylan and many other plant glycopolymers, very little is known about the process of polysaccharide O-acetylation in plants (Gille and Pauly, 2012). Current data is consistent with the notion that plant polysaccharide O-acetylation may occur through a mechanism similar to that proposed for the O-acetylation of peptidoglycan by Gram-negative bacteria. For example, phenotypic evidence suggests that Neisseria gonorrhoeae employs a two-component system where an acetyl moiety is first translocated from a cytoplasmic acetyl-donor, assumed to be acetyl-CoA, to the periplasm by an integral membrane protein, peptidoglycan O-acetyltransferase (PatA). O-acetylated PatA is proposed to then act as the acetyl-donor for PatB in vivo, which carries out the transacetylation reaction (Moynihan and Clarke, 2010). Recently, in depth biochemical analysis of PatB has shown that the enzyme can use several simple acyl donors in vitro, indicating a great deal of promiscuity in terms of donor utilization (Moynihan and Clarke, 2013, Moynihan and Clarke, 2014). In plants, the integral membrane proteins from the RWA family are predicted to function in the acetylation pathway prior to TBL proteins. Recent chemotypic analysis of mutants lacking one or more RWA proteins show a global reduction in acetylation of several cell wall polysaccharides with varying effects on individual polymers (Manabe et al., 2013). This data suggests that they may function either as acetyl translocating proteins, similar to the mechanism proposed for PatA, as Golgi acetyl-CoA transporters, or in some other capacity such as components of a larger protein complex. In cases where in depth chemical analysis has been performed on hemicellulosic polysaccharides from TBL mutants, the phenotypes are manifested as reduced acetylation in one or two specific glycopolymers (Gille et al., 2011, Xiong et al., 2013, Yuan et al., 2013). Accordingly, phenotypic analysis has revealed that mutating a member of the plant specific TBL gene family (Gille and Pauly, 2012) TRICHOME BIREFRINGENCE-LIKE 29/ESKIMO1 (TBL29/ESK1) results in plants with a chemotype distinguished by xylan and mannan with a reduced degree of acetylation, indicating that TBL29 is a xylan or mannan O-acetyltransferase (Xiong et al., 2013, Yuan et al., 2013). However, no biochemical activity data has been published for any plant polysaccharide O-acetyltransferase, and the identity of the acetyl donor and the ability of this enzyme to directly acetylate xylan or mannan have not been established.
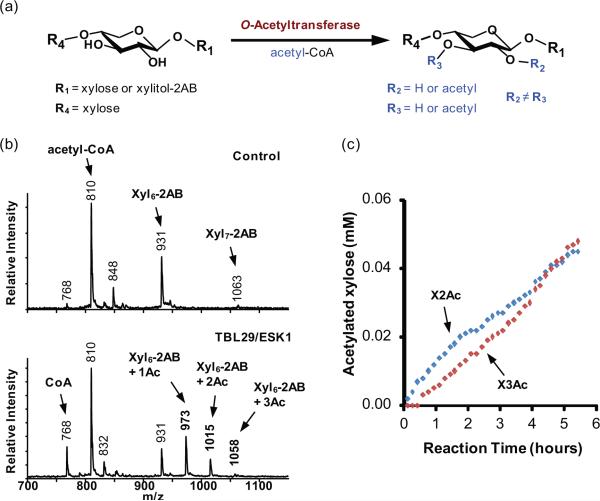
To determine whether TBL29/ESK1 is capable of functioning as an O-acetyltransferase, we expressed and purified a recombinant tagged form of the enzyme (Figure S6a), and evaluated its capacity to transfer O-acetyl groups to xylan or mannan oligomers. We developed an in vitro assay based on the same principles as the xylosyltransferase assay, using acetyl-coenzyme A (acetyl-CoA) as the acetyl donor and Xyl6-2AB or Man5-2AB as the acceptor (Figure 3a). MALDI-TOF MS analysis of the reaction products indicated that this enzyme is able to transfer up to three acetyl groups to Xyl6-2AB, generating coenzyme A (CoA) as a by-product (Figures 3b and S6b). However, no acetylation of Man5-2AB was observed under the same reaction conditions (Figure S6c), indicating that the enzyme is a xylan O-acetyltransferase. To confirm the structure of the O-acetylated xylooligosaccharides, we performed 2D 1H NMR analysis of the products of scaled-up reactions using unlabeled xylohexaose as the acceptor substrate. The resulting spectra contained signals diagnostic for monosubstituted (2-O-acetyl and 3-O-acetyl) xylosyl residues, but no signals corresponding to disubstituted xylosyl residues (Figure S7). Thus, the in vitro activity of recombinant TBL29/ESK1 is consistent with the reduced xylan acetylation chemotype of esk1 mutants, which produce xylan with fewer monoacetylated xylosyl residues but no apparent change in disubstituted xylosyl residues (Yuan et al., 2013). To establish the regiospecificity of TBL29/ESK1, we performed real-time NMR analysis of the acetylation reaction. Signals corresponding to monoacetylation at O-2 dominated during the initial stages of the reaction and signals corresponding to monoacetylation at O-3 were observed only after a lag period of approximately 30 minutes (Figures 3c and S8). This is consistent with enzyme-catalyzed transfer of O-acetyl groups to O-2 and spontaneous migration of the acetates between O-2 to O-3, approaching an equilibrium state in which acetylation at O-3 ultimately dominates. Together, our data establish that TBL29/ESK1 catalyzes the transfer of O-acetyl groups from acetyl-CoA to O-2 of xylosyl residues of β-(1→4) xylooligosaccharides in vitro, without the addition of any cofactors or divalent metals (Figure S9). The observation that TBL29/ESK1 is a metal independent enzyme that can use acetyl-CoA as a donor is comparable to biochemical analysis of PatB, which has also been shown to be a metal independent O-acetyltransferase, capable of using a variety of simple acetyl donors in vitro (Moynihan and Clarke, 2014). The availability of the recombinant enzymes and robust biochemical assays reported here will enable research aimed at resolving the complex molecular mechanisms that plants use for polysaccharide O-acetylation in vivo.
Figure 3. TBL29/ESK1 catalyzed acetylation of xylooligosaccharides using acetyl-CoA as the donor.
(a) In vitro O-acetyltransferase reaction scheme.
(b) MALDI-TOF MS of the products after incubating TBL29/ESK1 for 16 hours with Xyl6-2AB and acetyl-CoA. Each transfer of an O-acetyl (Ac) group increases the mass of the Xyl6-2AB acceptor by 42 Da to generate products corresponding to the annotated [M+H]+ ions.
(c) Amounts of acetylated xylosyl residues at O-2 (X2Ac) or O-3 (X3Ac) formed during the first 6 hours of the TBL29/ESK-catalyzed reaction. The product concentrations were determined by real time 1H NMR analysis.
Our data establishes the biochemical function of two crucial enzymes in the synthesis and substitution of one of the world most abundant glycopolymers, xylan. Xylan biosynthesis synthesis, like that of other hemicellulosic polymers, occurs in the Golgi apparatus. Accordingly, membrane topology studies indicate that the catalytic domain of IRX10-L is localized in the Golgi lumen (Sogaard et al., 2012), and TBL29/ESK1 is predicted to be a type II membrane protein also localized in the Golgi apparatus (Yuan et al., 2013). This led us to hypothesize that we could synthesize decorated polymers in vitro through the sequential or combined catalytic activity of these two proteins. To test this approach we first generated large (up to DP 21) 2AB-labeled xylooligosaccharides in vitro using IRX10-L (Figure 4a), then subjected them to TBL29/ESK1-catalyzed acetylation. MALDI-TOF MS analysis of the products demonstrated that TBL29/ESK1 adds up to five acetyl substituents to xylooligosaccharides with DP≥13 (Figure 4b). These findings led us to investigate whether IRX10-L and TBL29/ESK1 may act synergistically in vitro, increasing the solubility of nascent polymeric xylan through O-acetylation to produce polymers that are larger than those made by IRX10-L alone. However, contrary to our expectation, prior introduction of acetyl groups limited the ability of IRX10-L to extend the acceptor in vitro (Figure S10). This effect does not necessarily hinder xylan elongation in vivo, where, for example, the addition of each xylosyl residue to the polymer may be temporally or spatially isolated from its acetylation.
Figure 4. MALDI-TOF MS spectra of xylan synthesized and acetylated in vitro.
(a) Products formed after incubating IRX10-L for 96 hours with Xyl6-2AB and UDP-Xyl under standard assays conditions. Annotated [M+H]+ ions correspond to xylooligosaccharides with 6-21 residues.
(b) Products of the reaction described in (a) were incubated with TBL29/ESK1 and acetyl-CoA for 16 hours. The number of acetates introduced to each oligosaccharide is indicated by asterisks above its [M+H]+ ion.
In summary, we have demonstrated that IRX10-L has UDP-Xyl: β-(1→4) xylosyl transferase activity, and on this basis we propose the name XYLAN SYNTHASE-1 (XYS1) for this enzyme. Further, we have shown TBL29/ESK1 catalyzes the subsequent addition of O-acetyl groups from acetyl-CoA to the 2-position of xylosyl backbone residues, making it the only plant polysaccharide O-acetyl transferase that has been biochemically characterized to date. We thus propose the name XYLAN O-ACETYLTRANSFERASE-1 (XOAT1) for this enzyme. Our data make it unlikely but do not rule out the possibility (Lee et al., 2012) that the GT43 proteins IRX9 and IRX14 also catalyze the transfer of xylosyl residues from a donor to the elongating xylan chain. Rather, we propose that these proteins increase xylan synthesis in vivo either by playing a role in chain initiation or through a structural role such as formation of a multimeric xylan synthase complex. Notably, IRX10 and IRX10-L are both predicted to contain signal peptides rather than transmembrane domains (Wu et al., 2009). Hence, the possibility cannot be discounted that IRX9 and/or IRX14 actually play structural rather than catalytic roles in xylan synthesis, akin to the role that the catalytically inactive, membrane-anchored GT8 protein GAUT7 plays by tethering the α-1,4-galacturonosyltransferase GAUT1to the Golgi membrane, forming a core complex for pectin biosynthesis (Atmodjo et al., 2011). Notably, Arabidopsis IRX9 and its orthologs in rice and poplar lack a conserved DXD motif (Chiniquy et al., 2013).
Several lines of evidence suggests that there may be variations in the makeup of the xylan synthase complexes dependent on the xylan structures produced in different tissues and species ( York and O'Neill, 2008, Faik et al., 2014, Rennie and Schelle, 2014,). Our data does not preclude the participation of other enzymes or cofactors that may be required for xylan backbone synthesis in vivo. However, the characterization of XYS1 and XOAT1 described here are key advances toward understanding the complex mechanisms utilized by plants to synthesize xylans and other plant polysaccharides. Our findings also set the stage for the generation of advanced renewable biomaterials developed and produced using natural or engineered catalysts based on these biosynthetic enzymes.
EXPERIMENTAL PROCEDURES
Generation and purification of fusion proteins
Truncated coding region sequences of AtIRX10-L (amino acids 22-415), AtIRX9 (amino acids 49-351), AtIRX14 (amino acids 89-525) and AtTBL29/AtESK1 (amino acids 57-486) were amplified from cDNA prepared from inflorescence stems of wild-type Arabidopsis thaliana (Col-0) as a template (Urbanowicz et al., 2012). All reactions were carried out using Phusion High-Fidelity DNA Polymerase (Thermo Scientific). To create Gateway entry clones, attB-PCR products were generated using two-step adapter PCR as described in the Gateway Technology Manual (Life Technologies). The following primer pairs were used for gene-specific amplification of AtIRX10-L, AtIRX9, AtIRX14 and AtTBL29/AtESK1 sequences:
IRX10L_pDONR_GS-F, 5’- AACTTGTACTTTCAAGGCTTCCGTCTGAGTCGGAGTC-3’/
IRX10L_pDONR_GS-R, 5’- ACAAGAAAGCTGGGTCCTACCAAGGTTTCAGATCAGC-3’;
IRX14_pDONR_GS-F, 5’- AACTTGTACTTTCAAGGCGATGTTCACACAATCGG-3’ /
IRX14_pDONR_GS-R, 5’- ACAAGAAAGCTGGGTCCTAGTTTCTTTCTTGATGCTT-3’;
IRX9_pDONR_GS_F, 5’- AACTTGTACTTTCAAGGCTCTTATACTTCTACAAAATC-3’ /
IRX9_pDONR_GS_R, 5’- ACAAGAAAGCTGGGTCCTAGGTGCTTAAACGTGTTCTTG-3’; ESK1_pDONR_GS_F, 5’- AACTTGTACTTTCAAGGCTCAAAACCTCACGACGTC-3’ /
ESK1_pDONR_GS_R, 5’- ACAAGAAAGCTGGGTCCTAACGGGAAATGATACGTGT-3’. Underlined sequences denote the partial attB adapter sequences appended to primers used in the first round of PCR amplification. A second set of universal primers, attB_Adapter-F, 5’-GGGGACAAGTTTGTACAAAAAAGCAGGCTCTGAAAACTTGTACTTTCAAGGC-3’ / attB_Adapter-R, 5’-GGGGACCACTTTGTACAAGAAAGCTGGGTC-3’ was used to insert a tobacco etch virus (TEV) protease cleavage site and complete the attB recombination region. The completed attB-PCR products were gel purified using the AxyPrep DNA Gel Extraction Kit (Corning) and cloned into the pDONR221 plasmid vector (Life Technologies) using Gateway BP Clonase II Enzyme Mix (Life Technologies) according to the manufacturer's instructions. The transformants were confirmed by sequence analysis using M13 universal primers, M13 (-20) Forward and M13 Reverse. To generate expression clones, the entry clones were recombined into a Gateway adapted version of the pGEn2 mammalian expression vector (pGEn2-DEST)(Meng et al. 2013), using Gateway LR Clonase II Enzyme Mix (Life Technologies) and confirmed by sequence analysis. The resulting expression construct encodes a fusion protein comprised of an NH2-terminal signal sequence, an 8xHis tag, an AviTag recognition site, the “superfolder” GFP coding region, the 7 amino acid recognition sequence of the tobacco etch virus (TEV) protease, followed by the corresponding enzyme catalytic domain. For transfection, plasmids were purified using the PureLink HiPure Plasmid Filter Maxiprep Kit (Life Technologies).
Recombinant enzyme expression was accomplished by transient transfection of HEK293 cells as previously described (Meng et al., 2013). Briefly, suspension culture HEK293f cells (FreeStyle 293-F cells, Life Technologies, Grand Island, NY) were maintained in serum free Freestyle 293 expression medium (Life Technologies). Transfections were initiated at cell densities of 2.5×106 by addition of 3 μg/ml of the respective expression plasmid and 9 μg/ml polyethylenimine (linear 25 kDa PEI, Polysciences, Inc, Warrington, PA) to the culture. After 24 hours, the cultures were diluted 1:1 with Freestyle 293 expression medium containing valproic acid (2.2 mM final) and protein production was continued for a further 4-5 days at 37°C.
All chromatography experiments were carried out on an ÄKTA FPLC System (GE Healthcare). Purification of His8-GFP tagged enzymes secreted into the culture medium by HEK293 cells was performed using His TALON Superflow Cartriges (Clontech Laboratories) or HisTrap HP columns (GE Healthcare) according to the manufacturer's instruction. The purity of the proteins was confirmed by SDS-PAGE, Coomassie Brilliant Blue R-250 (Bio-Rad) staining and in-gel GFP fluorescence. For activity assays, proteins were buffer exchanged into HEPES sodium salt-HCl (75 mM), pH 6.8, using either a PD-10 gel filtration column (GE Healthcare) or dialysis (3500 molecular weight cut-off) and then concentrated using a 30 kDa molecular weight cut-off Amicon Ultra centrifugal filter device (Millipore). To examine the divalent metal-dependence of IRX10-L and TBL29/ESK1, metal-depleted enzyme was prepared by dialysis against HEPES sodium salt-HCl (75 mM), pH 6.8 buffer containing 5 g/L Chelex-100 (Bio-Rad) at 4°C for a minimum of 6 hours. For real-time NMR experiments, purified proteins were buffer exchanged into potassium bicarbonate (50 mM), pH 6.8, prepared with D2O (99.98%, Cambrige Isotope Laboratories) by diafiltration using a 30 kDa molecular weight cut-off Amicon Ultra centrifugal filter device (Millipore). Protein concentration was determined by the Bio-Rad protein assay (Bio-Rad).
Determination of xylosyltransferase activity
The 1,4-β-D-xylooligosaccharides, 1,4-β-D-mannooligosaccharides, 1,4-β-D - glucooligosaccharides and the endo-1,4-β-Xylanase M1 (Trichoderma viride) were obtained from Megazyme International (Wicklow, Ireland). UDP-xylose, UDP-arabinopyranose and UDP-galacturonic acid were purchased from CarboSource Services (Athens, GA) and UDP-galactose, UDP-glucose, UDP-glucuronic acid and acetyl-coenzyme A were purchased from Sigma-Aldrich. β-(1→4)-xylohexaose and β-(1→4)-mannopentaose were labeled at the reducing terminus with 2-aminobenzamide (2AB) as described previously (Ishii et al., 2002). Xylosyltransferase activity was routinely assayed by using an activated sugar donor (UDP-Xyl) and the chemically labeled acceptor 2AB-β-(1→4)-xylohexaose (Xyl6-2AB). The standard xylosyltransferase assay (15 μL) was performed in HEPES sodium salt-HCl (75 mM), pH 6.8 containing 3.8 μg of purified protein (IRX10-L, IRX9, IRX14), UDP-Xyl (2 mM) as donor, Xyl6-2AB (0.25 mM ) as acceptor. The standard assays were performed in the presence of MnCl2 (1 mM) and MgCl2 (1 mM) or only MgCl2 (1 mM). Reactions were incubated at 25 °C for the indicated times. For determination of the optimal pH, standard reactions were carried out at pH 5.5-6.7 in MES-KOH (50 mM) and at pH 6.8-8.0 in HEPES buffer (50 mM). To determine the effect of divalent cations on IRX10-L activity, metal depleted enzyme and donor substrate were incubated for 30 min at 25 °C in the presence of EDTA, MgCl2, MnCl2, CaCl2, CoCl2 (5 mM) or without additives. The reactions were then initiated by the addition of the acceptor substrate (Xyl6-2AB) and allowed to proceed for 16 hours. To determine if IRX10-L can utilize other UDP-sugars as sugar donors, IRX10-L and Xyl6-2AB (0.25 mM) were incubated for 16 h in the presence of each of the following activated nucleotide sugars: 2 mM of UDP-arabinopyranose, UDP-galactose, UDP-galacturonic acid, UDP-glucose or UDP-glucuronic acid. Reaction products were analyzed by MALDI-TOF MS and quantified by RP-HPLC as described below.
MALDI-TOF MS analysis
The reaction products were analyzed by MALDI-TOF MS using a LT Bruker LT Microflex spectrometer. Aliquots (5 μL) of the reaction mixture were incubated with 1 μL of a suspension of Dowex-50 cation exchanger resin in water for 1 hour. After centrifugation, 1 μL of the supernatants were mixed with an equal volume of matrix solution (20 mg/mL 2,5-dihydroxbenzoic acid in aqueous 50% methanol) on the target plate. The positive-ion spectra were recorded and at least 200 laser shots were summed to generate each spectrum.
RP-HPLC analysis and quantification
The xylosyltransferase activity was measured using RP-HPLC analysis of the fluorescent 2AB-labeled oligosaccharides generated by IRX10-L. HPLC was performed with an Agilent 1100 series LC system and a Shimadzu RF-10AXL fluorescence detector. The reaction products were separated using a Luna C18 column (250 mm long, 4.6 mm i.d.). The column was eluted at 0.5 ml/min with aqueous 20% methanol for 30 min followed by a 5 min gradient to 80% methanol, which was held for 5 min. The column was re-equilibrated by a 5 min gradient to return to the initial conditions and washed for an additional 5 min in these conditions. The elution of fluorescent oligosaccharides was monitored using a fluorescence detector (330 nm excitation and 420 nm emission). The molar concentrations of products were quantified assuming that the molar fluorescence response was the same for all labeled substrates and products and that the total molar concentration of these labeled molecules was equal to the initial concentration of the Xyl6-2AB acceptor. Each detected oligosaccharide was assigned an index i, which corresponds to the DP of the oligosaccharide. The signal si of each oligosaccharide peak was integrated, and used to calculate molar concentrations ci by the following equation: where and [acceptor] is the initial molar concentration of the Xyl6-2AB acceptor. The molar amount ai (nmol) of each product produced per μg of protein was calcluated by dividing its concentration by the concentration of protein (0.25 μg / μL) in the reaction: where [enzyme] is the concentration of the enzyme. The total amount x of xylosyl residues (nmol) transferred per μg of protein was quantified by multiplying the molar amount ai of each product per μg of protein by its extension number ni = i - 6, where 6 is the DP of the acceptor substrate, and summing the result: . Theoretical product population calculations were also performed for each time point. First, the observed abundances ai of the oligosaccharides were converted to fractional populations where . The Poisson expectation value λ was calculated by multiplying each fractional population by the extension number ni and summing the result. That is, . The value of λ corresponds to the average number of xylosyl residues transferred per oligosaccharide acceptor molecule in the reaction. The theoretical fractional population of each oligosaccharide pi was calculated as a Poisson probability P(ni λ) for each oligosaccharide in the standard way: . The observed and theoretical fractional populations for the products of the reaction catalyzed by IRX10-L were remarkably similar.
Structural characterization of xylooligosaccharides synthesized in vitro
NMR spectroscopy was used to structurally characterize the in vitro synthesized β-(1→4)-xylooligosaccharides. Scaled up enzyme-catalyzed reactions were carried out in potassium bicarbonate (50 mM), pH 6.8 in D2O containing unlabeled xylohexaose (100 μg) and UDP-Xyl (2 mM) as donor. The course of the reaction was monitored by real-time 1D 1H NMR. After 120 hours, the products of the scaled-up reaction were fractionated by SEC on a Superdex-75 HR10/30 SEC column (GE Healthcare Bio-Sciences Corp. Piscataway, NJ) eluted with water. The fractions containing the xylo-oligosaccharides were lyophilized, dissolved in D2O (0.3 mL, 99.9%; Cambridge Isotope Laboratories) and analyzed by 1D and 2D NMR spectroscopy. Data were recorded at 298 K with an Agilent-NMR spectrometer operating at 600 MHz equipped with a 5-mm NMR cold probe. The 2D homonuclear and hetereonuclear experiments were recorded using standard Varian pulse programs. Chemical shifts were measured relative to internal acetone (δ 1H 2.225; δ 13C 30.89). Data were processed using MestReNova software (Universidad de Santiago de Compostela, Spain). Long-range scalar couplings in the gHMBC spectrum were used to establish the glycosidic linkage positions between the xylosyl residues in the reaction products.
The products of the xylosyltransferase reaction catalyzed by IRX10-L for 16 h, using Xyl6-2AB as acceptor and under the same conditions described for the standard xylosyltransferase assay were treated for 1h with the endo-1,4-β-Xylanase M1. Aliquots of the reaction with and without the endoxylanase treatment were analyzed by RP-HPLC as was described above.
Kinetics analysis and determination of the acceptor DP preferences of IRX10-L
The amount of UDP formed as a by-product of the xylosyltransferase reactions was determined using the UDP-Glo™ Glycosyltransferase Assay (Promega) according to the manufacturer's instructions. For determination of kinetic parameters, xylosyltransferase reactions (5 μl) comprising HEPES sodium salt-HCl (75 mM), pH 6.8, and purified IRX10-L (1.25 μg) were carried out using UDP-xylose (800 μm, CarboSource Services) as a donor substrate and several different concentrations of xylohexaose (0-4.0 mM) as an acceptor. Reactions were performed at 26 °C for up to 120 min and were stopped at the indicated times by flash freezing in liquid nitrogen. For determination of protein concentration dependence, xylosyltransferase reactions were carried out with UDP-xylose (800 μm) as a donor, xylohexaose (1.0 mM) as an acceptor and increasing concentrations of purified IRX10-L (0-1.25 μg) for 60 min at 26 °C. For detection of UDP, reactions were mixed with an equal volume of UDP-Glo Detection Reagent (5 μl) in white polystyrene, low-volume, 384-well assay plates (Corning Incorporated) and incubated for 60 min at 23°C. After incubation, luminescence measurements were performed using a multifunctional microplate reader (POLARstar OPTIMA, BMG Labtech). A standard curve was used for quantification of UDP produced, and the detection limit of the luminescent assay was 40 nM UDP with a linear response up to 20 μM. The steady state parameters Km and Vmax were calculated by fitting the initial velocities to the Michaelis-Menten equation using nonlinear curve fitting in GraphPad Prism 6 (GraphPad Software, La Jolla, CA).
To investigate xylosyltransferase activity on xylooligomer acceptors of different degrees of polymerization, reactions were performed with the following unlabeled acceptor substrates: xylose (Xyl), xylobiose (Xyl2), xylotriose (Xyl3), xylotetraose (Xyl4), xylopentaose (Xyl5) xylohexose (Xyl6) and cellopentaose (Glc5). Each reaction (5 μl) was carried out for 12 hours at 25 °C and contained 0.4 μg of enzyme, UDP-Xyl (100 μM) donor and the indicated acceptor substrate (65 μM). Glycosyltransferase activity was measured using the bioluminescent UDPGlo™ Glycosyltransferase Assay (Promega), as described above.
Determination of xylan O-acetyltransferase activity
Xylan O-acetlytransferase activity was routinely assayed by using acetyl-coenzyme A (acetyl-CoA) as a donor and 2AB-Xyl6 or xylohexaose as acceptor substrates. The standard assay (15 μL) was performed in HEPES sodium salt-HCl (75 mM), pH 6.8 containing purified protein (4 μM), acetyl-CoA (1 mM) donor and of Xyl6-2AB (0.25 mM) acceptor unless indicated. Mannan O-acetlytransferase activity was assayed as described above using 2AB-β-(1→4)-mannopentaose (Man5-2AB) as an acceptor. The reaction products were analyzed by MALDI-TOF MS as described above. The optimal acetyl-CoA concentration and the effect of divalent cations on TBL29/ESK1 activity were determined using the same experimental procedures described for IRX10-L. For structural analysis of the reaction products, unlabeled xylohexaose (100 μg) and acetyl-CoA (2 mM) were incubated with TBL29/ESK1 in 50 mM potassium bicarbonate (50 mM), pH 6.8 in D2O for 48 hours. The high resolution gCOSY was recorded as previously described (Jia et al. 2005). Resonances were assigned to glycosyl residues based on our analysis of acetylated xylooligosaccharides released from Arabidopsis stem cell walls by the endo-1,4-β-xylanase M1 and published data (Kabel et al., 2003, Teleman et al., 2000).
To investigate the positional specificity of TBL29/ESK1, the reaction catalyzed by TBL29/ESK1 in vitro was monitored using real-time 1H NMR. The assay was performed at 25 °C in potassium bicarbonate (50 mM), pH 6.8 in D2O containing unlabeled xylohexaose (100 μg), acetyl-CoA (2 mM) and 50 μg of purified TBL29/ESK1. Data acquisition was initiated 5 min after the reaction components were mixed. 1D 1H spectra consisted of 64 transients that were acquired with water presaturation every 10 min over a 12 hour time period, generating a spectral array representing the progress of the reaction. The resonance peaks corresponding to the methyl protons of the acetyl groups attached to O-2 or O-3 of internal xylosyl residues were identified in the arrayed spectra and their areas were determined by integration or by deconvolution in the case of overlapping peaks. The concentrations of the acetylated products were calculated based on the initial concentration of acetyl-CoA added to the reaction. After monitoring the reaction for 12 hours, the products were analyzed by 1D and 2D NMR as described above. All data processing was performed using MestReNova software (Universidad de Santiago de Compostela, Spain).
Supplementary Material
ACKNOWLEDGMENTS
We thank M.A. O'Neill, Peter J. Smith and J. Backe for technical support and the Center for Plant and Microbial Complex Carbohydrates (DEFG0296ER20097) for equipment support. B.R.U., M.J.P. and W.S.Y. were funded by The BioEnergy Science Center (BESC), a U.S. Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science. H.A.M. and K.W.M. were supported by the National Institutes of Health (grant P41GM103390). The nucleotide sugars obtained from Carbosource Services, which is supported in part by the DOE funded Center For Plant and Microbial Complex Carbohydrates (grant DE-FG02-93ER20097). All authors contributed to the intellectual development and/or writing of the manuscript. All authors designed research and B.R.U., M.J.P., and H.A.M performed experiments.
Footnotes
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- Atmodjo MA, Sakuragi Y, Zhu X, Burrell AJ, Mohanty SS, Atwood JA, III, Orlando R, Scheller HV, Mohnen D. Galacturonosyltransferase (GAUT)1 and GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan: galacturonosyltransferase complex. Proc. Natl. Acad. Sci. USA. 2011;108:20225–20230. doi: 10.1073/pnas.1112816108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DM, Zeef LAH, Ellis J, Goodacre R, Turner SR. Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell. 2005;17:2281–2295. doi: 10.1105/tpc.105.031542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DM, Zhang Z, Stephens E, Dupree P, Turner SR. Characterization of IRX10 and IRX10-like reveals an essential role in glucuronoxylan biosynthesis in Arabidopsis. Plant J. 2009;57:732–746. doi: 10.1111/j.1365-313X.2008.03729.x. [DOI] [PubMed] [Google Scholar]
- Burton RA, Wilson SM, Hrmova M, Harvey AJ, Shirley NJ, Stone BA, Newbigin EJ, Bacic A, Fincher GB. Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-beta-D-glucans. Science. 2006;311:1940–1942. doi: 10.1126/science.1122975. [DOI] [PubMed] [Google Scholar]
- Chiniquy D, Varanasi P, Oh T, Harholt J, Katnelson J, Singh S, Auer M, Simmons B, Adams PD, Scheller HV, Ronald PC. Three novel rice genes closely related to the Arabidopsis IRX9, IRX9L, and IRX14 genes and their roles in xylan biosynthesis. Front. Plant Sci. 2013;4:83. doi: 10.3389/fpls.2013.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocuron J-C, Lerouxel O, Drakakaki G, Alonso AP, Liepman AH, Keegstra K, Raikhel N, Wilkerson CG. A gene from the cellulose synthase-like C family encodes a beta-1,4 glucan synthase. Proc. Natl. Acad. Sci. USA. 2007;104:8550–8555. doi: 10.1073/pnas.0703133104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhugga KS, Barreiro R, Whitten B, Stecca K, Hazebroek J, Randhawa GS, Dolan M, Kinney AJ, Tomes D, Nichols S, Anderson P. Guar seed beta-mannan synthase is a member of the cellulose synthase super gene family. Science. 2004;303:363–366. doi: 10.1126/science.1090908. [DOI] [PubMed] [Google Scholar]
- Doblin MS, Pettolino FA, Wilson SM, Campbell R, Burton RA, Fincher GB, Newbigin E, Bacic A. A barley cellulose synthase-like CSLH gene mediates (1,3;1,4)-beta-D-glucan synthesis in transgenic Arabidopsis. Proc. Natl. Acad. Sci. USA. 2009;106:5996–6001. doi: 10.1073/pnas.0902019106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faik A, Jiang N, Held M. Plants and BioEnergy. Springer; 2014. Xylan Biosynthesis in Plants, Simply Complex. pp. 153–181. [Google Scholar]
- Geshi N, Harholt J, Sakuragi Y, Jensen JK, Scheller HV. Glycosyltransferases of the GT47 Family. Annual Plant Reviews: Plant Polysaccharides, Biosynthesis and Bioengineering. 2010;41:265–283. [Google Scholar]
- Gille S, de Souza A, Xiong G, Benz M, Cheng K, Schultink A, Reca I-B, Pauly M. O-Acetylation of Arabidopsis Hemicellulose Xyloglucan Requires AXY4 or AXY4L, Proteins with a TBL and DUF231 Domain. Plant Cell. 2011;23:4041–4053. doi: 10.1105/tpc.111.091728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille S, Pauly M. O-acetylation of plant cell wall polysaccharides. Front. Plant Sci. 2012;3:12. doi: 10.3389/fpls.2012.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T, Ichita J, Matsue H, Ono H, Maeda I. Fluorescent labeling of pectic oligosaccharides with 2-aminobenzamide and enzyme assay for pectin. Carbohydr. Res. 2002;337:1023–1032. doi: 10.1016/s0008-6215(02)00087-3. [DOI] [PubMed] [Google Scholar]
- Jensen JK, Johnson N, Wilkerson CG. Discovery of diversity in xylan biosynthetic genes by transcriptional profiling of a heteroxylan containing mucilaginous tissue. Front. Plant Sci. 2013;4:183. doi: 10.3389/fpls.2013.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JK, Kim H, Cocuron J-C, Orler R, Ralph J, Wilkerson CG. The DUF579 domain containing proteins IRX15 and IRX15-L affect xylan synthesis in Arabidopsis. Plant J. 2011;66:387–400. doi: 10.1111/j.1365-313X.2010.04475.x. [DOI] [PubMed] [Google Scholar]
- Jia Z, Cash M, Darvill AG, York WS. NMR characterization of endogenously O-acetylated oligosaccharides isolated from tomato (Lycopersicon esculentum) xyloglucan. Carbohydr. Res. 2005;340:1818–1825. doi: 10.1016/j.carres.2005.04.015. [DOI] [PubMed] [Google Scholar]
- Kabel MA, de Waard P, Schols HA, Voragen AGJ. Location of O-acetyl substituents in xylo-oligosaccharides obtained from hydrothermally treated Eucalyptus wood. Carbohydr. Res. 2003;338:69–77. doi: 10.1016/s0008-6215(02)00351-8. [DOI] [PubMed] [Google Scholar]
- Kuroyama H, Tsumuraya Y. A xylosyltransferase that synthesizes beta-(1 -> 4)-xylans in wheat (Triticum aestivum L.) seedlings. Planta. 2001;213:231–240. doi: 10.1007/s004250000499. [DOI] [PubMed] [Google Scholar]
- Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: Structures, functions, and mechanisms. Annu. Rev. Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- Lee C, Zhong R, Ye Z-H. Arabidopsis Family GT43 Members are Xylan Xylosyltransferases Required for the Elongation of the Xylan Backbone. Plant Cell Physiol. 2012;53:135–143. doi: 10.1093/pcp/pcr158. [DOI] [PubMed] [Google Scholar]
- Liu J, Mushegian A. Three monophyletic superfamilies account for the majority of the known glycosyltransferases. Protein Sci. 2003;12:1418–1431. doi: 10.1110/ps.0302103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liwanag AJM, Ebert B, Verhertbruggen Y, Rennie EA, Rautengarten C, Oikawa A, Andersen MCF, Clausen MH, Scheller HV. Pectin Biosynthesis: GALS1 in Arabidopsis thaliana Is a beta-1,4-Galactan beta-1,4-Galactosyltransferase. Plant Cell. 2012;24:5024–5036. doi: 10.1105/tpc.112.106625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manabe Y, Verhertbruggen Y, Gille S, Harholt J, Chong S-L, Pawar PM-A, Mellerowicz EJ, Tenkanen M, Cheng K, Pauly M, Scheller HV. Reduced Wall Acetylation Proteins Play Vital and Distinct Roles in Cell Wall OAcetylation in Arabidopsis. Plant Physiol. 2013;163:1107–1117. doi: 10.1104/pp.113.225193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May JF, Splain RA, Brotschi C, Kiessling LL. A tethering mechanism for length control in a processive carbohydrate polymerization. Proc. Natl. Acad. Sci. USA. 2009;106:11851–11856. doi: 10.1073/pnas.0901407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Forouhar F, Thieker D, Gao Z, Ramiah A, Moniz H, Xiang Y, Seetharaman J, Milaninia S, Su M, Bridger R, Veillon L, Azadi P, Kornhaber G, Wells L, Montelione GT, Woods RJ, Tong L, Moremen KW. Enzymatic Basis for N-Glycan Sialylation: Structure of rat alpha2,6-sialyltransferase (ST6GAL1) reveals conserved and unique features for glycan sialylation. J. Biol. Chem. 2013;288:34680–34698. doi: 10.1074/jbc.M113.519041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynihan PJ, Clarke AJ. O-Acetylation of Peptidoglycan in Gram-negative bacteria identification and characterization of peptidoglycan o-acetyltransferase in Neisseria gonorrhoeae. J. Biol. Chem. 2010;285:13264–13273. doi: 10.1074/jbc.M110.107086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynihan PJ, Clarke AJ. Assay for peptidoglycan O-acetyltransferase: A potential new antibacterial target. Anal. Biochem. 2013;439:73–79. doi: 10.1016/j.ab.2013.04.022. [DOI] [PubMed] [Google Scholar]
- Moynihan PJ, Clarke AJ. Substrate specificity and kinetic characterization of peptidoglycan O-acetyltransferase B from Neisseria gonorrhoeae. J. Biol. Chem. 2014;289:16748–16760. doi: 10.1074/jbc.M114.567388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc. Natl. Acad. Sci. USA. 1996;93:12637–12642. doi: 10.1073/pnas.93.22.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña MJ, Zhong R, Zhou G-K, Richardson EA, O'Neill MA, Darvill AG, York WS, Ye Z-H. Arabidopsis irregular xylem8 and irregular xylem9: Implications for the complexity of glucuronoxylan biosynthesis. Plant Cell. 2007;19:549–563. doi: 10.1105/tpc.106.049320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porchia AC, Scheller HV. Arabinoxylan biosynthesis: Identification and partial characterization of beta-1,4-xylosyltransferase from wheat. Physiol. Plantarum. 2000;110:350–356. [Google Scholar]
- Rennie EA, Hansen SF, Baidoo EEK, Hadi MZ, Keasling JD, Scheller HV. Three Members of the Arabidopsis Glycosyltransferase Family 8 Are Xylan Glucuronosyltransferases. Plant Physiol. 2012;159:1408–1417. doi: 10.1104/pp.112.200964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennie EA, Scheller HV. Xylan biosynthesis. Curr. Opin. Biotech. 2014;26:100–107. doi: 10.1016/j.copbio.2013.11.013. [DOI] [PubMed] [Google Scholar]
- Richmond TA, Somerville CR. The cellulose synthase superfamily. Plant Physiol. 2000;124:495–498. doi: 10.1104/pp.124.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu APS, Randhawa GS, Dhugga KS. Plant Cell Wall Matrix Polysaccharide Biosynthesis. Mol. Plant. 2009;2:840–850. doi: 10.1093/mp/ssp056. [DOI] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P. Hemicelluloses. Annu. Rev. Plant Biol. 2010;61:263–289. doi: 10.1146/annurev-arplant-042809-112315. [DOI] [PubMed] [Google Scholar]
- Simkovic I. Unexplored possibilities of all-polysaccharide composites. Carbohydr. Polym. 2013;95:697–715. doi: 10.1016/j.carbpol.2013.03.040. [DOI] [PubMed] [Google Scholar]
- Søgaard C, Stenbaek A, Bernard S, Hadi M, Driouich A, Scheller HV, Sakuragi Y. GO-PROMTO Illuminates Protein Membrane Topologies of Glycan Biosynthetic Enzymes in the Golgi Apparatus of Living Tissues. Plos One. 2012;7:e31324. doi: 10.1371/journal.pone.0031324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman A, Lundqvist J, Tjerneld F, Stalbrand H, Dahlman O. Characterization of acetylated 4-O-methylglucuronoxylan isolated from aspen employing 1H and 13C NMR spectroscopy. Carbohydr. Res. 2000;329:807–815. doi: 10.1016/s0008-6215(00)00249-4. [DOI] [PubMed] [Google Scholar]
- Urbanowicz BR, Pena MJ, Ratnaparkhe S, Avci U, Backe J, Steet HF, Foston M, Li H, O'Neill MA, Ragauskas AJ, Darvill AG, Wyman C, Gilbert HJ, York WS. 4-O-methylation of glucuronic acid in Arabidopsis glucuronoxylan is catalyzed by a domain of unknown function family 579 protein. Proc. Natl. Acad. Sci. USA. 2012;109:14253–14258. doi: 10.1073/pnas.1208097109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A-M, Hornblad E, Voxeur A, Gerber L, Rihouey C, Lerouge P, Marchant A. Analysis of the Arabidopsis IRX9/IRX9-L and IRX14/IRX14-L Pairs of Glycosyltransferase Genes Reveals Critical Contributions to Biosynthesis of the Hemicellulose Glucuronoxylan. Plant Physiol. 2010;153:542–554. doi: 10.1104/pp.110.154971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A-M, Rihouey C, Seveno M, Hornblad E, Singh SK, Matsunaga T, Ishii T, Lerouge P, Marchant A. The Arabidopsis IRX10 and IRX10-LIKE glycosyltransferases are critical for glucuronoxylan biosynthesis during secondary cell wall formation. Plant J. 2009;57:718–731. doi: 10.1111/j.1365-313X.2008.03724.x. [DOI] [PubMed] [Google Scholar]
- Xiong G, Cheng K, Pauly M. Xylan O-Acetylation Impacts Xylem Development and Enzymatic Recalcitrance as Indicated by the Arabidopsis Mutant tbl29. Mol. Plant. 2013;6:1373–1375. doi: 10.1093/mp/sst014. [DOI] [PubMed] [Google Scholar]
- York WS, O'Neill MA. Biochemical control of xylan biosynthesis - which end is up? Curr. Opin. Plant Biol. 2008;11:258–265. doi: 10.1016/j.pbi.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Teng Q, Zhong R, Ye Z-H. The Arabidopsis DUF231 Domain-Containing Protein ESK1 Mediates 2-O- and 3-O-Acetylation of Xylosyl Residues in Xylan. Plant Cell Physiol. 2013;54:1186–1199. doi: 10.1093/pcp/pct070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.