SUMMARY
Nanog facilitates ESC self-renewal and iPSC generation during the final stage of reprogramming. From a genome-wide siRNA screen using a Nanog-GFP reporter line we discovered opposing effects of Snai1 and Snai2 depletion on Nanog promoter activity. We further discovered mutually repressive expression profiles and opposing functions of Snai1 and Snai2 during the Nanog-driven final stage of reprogramming. We found that Snai1, but not Snai2, is both a transcriptional target and protein partner of Nanog in reprogramming. Ectopic expression of Snai1 or depletion of Snai2 greatly facilitate the Nanog-driven reprogramming. Snai1 (but not Snai2) and Nanog co-bind to and transcriptionally activate pluripotency-associated genes including Lin28 and miRNA-290-295. Ectopic expression of miRNA-290-295 cluster genes partially rescues reprogramming inefficiency caused by Snai1 depletion. Our studies thus uncover a novel interplay between Nanog and mesenchymal transcription factors Snai1 and Snai2 in the transcriptional regulation of pluripotency-associated genes and miRNAs during the Nanog-driven reprogramming process.
Keywords: RNAi, Nanog, Snai1, Snai2, miRNA, pluripotency, reprogramming
INTRODUCTION
Embryonic or induced pluripotent stem cells (ESCs and iPSCs, respectively) have attracted great attention because of their potential for the study and possible treatment of human diseases. Somatic cells can acquire pluripotency through nuclear reprogramming by ectopic expression of the transcriptions factors Oct4, Sox2, Klf4 and c-Myc (Takahashi and Yamanaka, 2006). The acquisition of induced pluripotency is a multi-step process requiring the interplay of many transcription factors, epigenetic regulators and the cell-signaling network. In particular, the cellular changes during the final stage of reprogramming towards a fully reprogrammed stem cell comprise the final hurdle for achieving the pluripotent ground state. Nanog dependency of the transition to the ground state has been well characterized (Silva et al., 2009), although low-efficiency transition may happen in its absence (Carter et al., 2014; Schwarz et al., 2014). While our proteomic studies have identified the transcriptional repressor Zfp281 as a reprogramming barrier (Fidalgo et al., 2012) and DNA hydroxylases Tet1 and Tet2 as facilitators (Costa et al., 2013) of Nanog function in establishing ground state pluripotency, the full repertoire of pluripotency factors and epigenetic regulators that contribute to this process remains to be identified.
Several genome-wide RNAi studies have been performed with Oct4 (Chia et al., 2010; Ding et al., 2009; Hu et al., 2009) and Rex1 (Yang et al., 2012) reporter systems leading to the identification of many novel regulators of ESC identity. In contrast, studies that utilize a Nanog reporter system have thus far only been performed with small siRNA libraries targeting limited number of chromatin regulators (e.g., Mbd3, SWI/SNF) (Rais et al., 2013; Schaniel et al., 2009; Schaniel et al., 2010) or posttranslational modifiers (e.g., ubiquitin-proteasome system or UPS) (Buckley et al., 2012). In this study, we performed a genome-wide RNAi screen using a Nanog-GFP reporter line (Schaniel et al., 2009) under mild differentiation conditions for bidirectional alteration of GFP activity (see Table S1 and Figure 1). We identified genes that are required for maintaining (downregulation of Nanog-GFP upon knockdown) or exiting (upregulation of Nanog-GFP upon knockdown) the pluripotent state, providing a rich resource for further dissection of the molecular mechanisms underlying pluripotency and reprogramming.
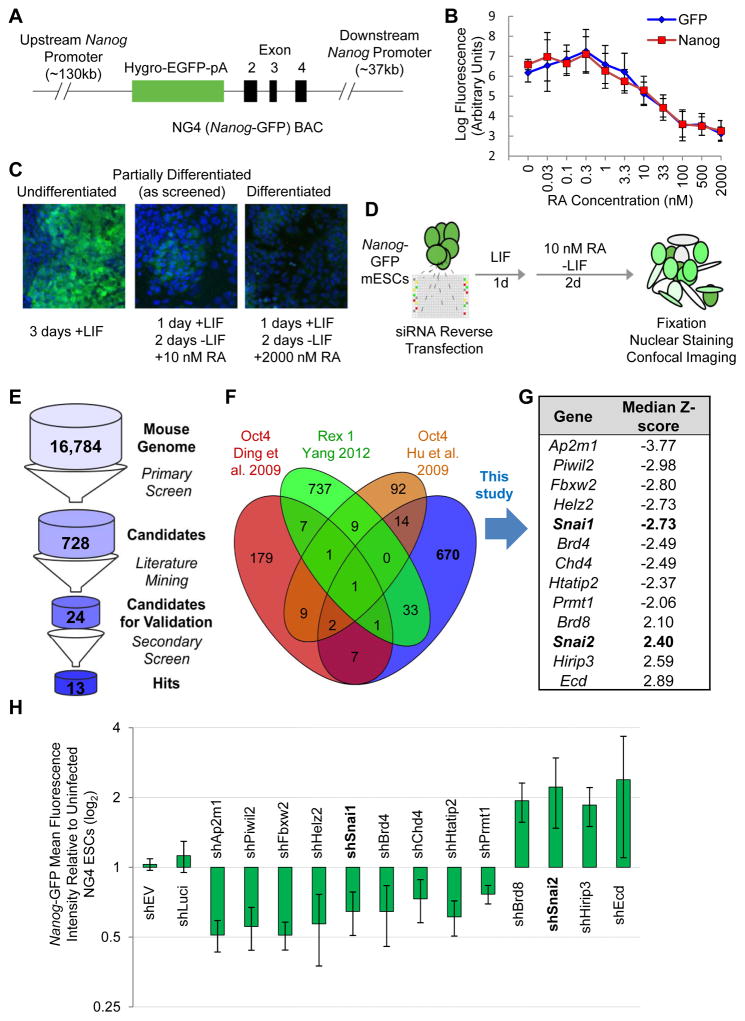
Figure 1. Genome-Wide RNAi Screening Strategy.
(A) NG4 line vector. BAC-based GFP reporter expression is driven by the Nanog promoter.
(B) Nanog reporter fluorescence correlates with Nanog protein levels during RA-mediated differentiation. Nanog reporter cells were treated with varying concentrations of RA as indicated and stained for immunofluorescence. Mean fluorescence per cell was calculated.
(C) Confocal fluorescence of reporter expression following RA-mediated differentiation. Nuclei are stained with Hoechst 33342 and shown in blue. Cytoplasmic GFP reflecting Nanog promoter activity is in green.
(D) Screening conditions. NG4 cells are reverse transfected and cultured for 3 days, the last 2 under mild RA-mediated differentiation conditions. Cells are fixed, nuclei stained and plates imaged at cell-level resolution.
(E) Screen analysis pipeline. Candidates among outlier genes from the primary screen are further narrowed down by removing known pluripotency regulators through literature mining and by a secondary screen with individual siRNAs.
(F) Comparison of our RNAi screen dataset with published RNAi screen datasets.
(G) List of the final candidate genes with |Median Z-scores|>2 validated in the secondary screen.
(H) Nanog-GFP mean fluorescence intensity for hits from RNAi screen. NG4 cells were infected with two independent shRNAs against each candidate gene and cultured under the same RA conditions as in the screen. Values are normalized to empty vector (shEV). Error bars indicate average ± SD. See also Figure S1.
We exploited the Nanog-dependency of establishing the naïve ground state to test the functional significance of candidate genes in positive or negative regulation of the reprogramming process. In particular, we uncovered mutually repressive expression and opposing roles of the mesenchymal transcription factors Snai1 and Snai2 (also known as Slug) in the establishment of ground state pluripotency. We found Snai1, but not Snai2, is a direct target of and transcriptionally activated by Nanog during the last stage of reprogramming. Finally, we discovered a novel partership between Nanog and Snai1 and an unexpected functional antagonism of Snai1 and Snai2 in promoting the final transition of partially reprogrammed cells into fully pluripotent cells.
RESULTS
A Genome-Wide RNAi Screen Strategy to Identify Modulators of Nanog Promoter Activity
We performed a genome-wide siRNA screen under mild retinoic acid (RA)-induced differentiation conditions in the previously characterized Nanog-GFP reporter ESC line (NG4) (Figure 1A) (Schaniel et al., 2009) to identify potential direct and indirect regulators of Nanog gene expression that would presumably play roles in pluripotency and reprogramming. Pluripotency, measured by GFP fluorescence levels, is progressively lost with increasing amounts of RA treatment (Figure 1B). We determined an optimal intermediate dose of RA (10 nM) to partially differentiate the ESC population over 2 days (Figure 1B–C) for bidirectional screening for candidates whose knockdown would either increase or decrease GFP levels (Figure 1D) (see Extended Experimental Procedures for details).
As expected, we found that siRNA targeting GFP dramatically reduced fluorescence (median Z-score = −3.16) and that siRNAs to retinoid X receptors effectively blocked the loss of fluorescence and produced median Z-scores of 3.20, 1.61 and 1.97 for RXRα, RXRβ and RXRγ, respectively (Figure S1A). In addition, a number of genes were identified, including known pluripotency genes (e.g., Sox2, Esrrb) and lineage specification markers (e.g., Zeb1, Fgf2), whose depletion decreased and increased, respectively, the median GFP fluorescence (Figure S1A). Gene ontology (GO) analyses of candidate hits (Table S2) with the most dramatic positive (429) or negative (299) effects on GFP expression indicated high enrichment of genes with DNA/protein-binding and nuclear transcriptional regulatory activities (Figure S1A). Collectively, these results establish a robust RNAi platform that is distinct from other RNAi screen studies (see detailed comparison in Table S1) for discovery of novel self-renewal and pluripotency regulators with nuclear functions.
Because we aimed to identify outliers with effects comparable to or stronger than retinoic acid receptor and GFP knockdown, we applied a |Z-score|>2 threshold. Using this approach, we found that 2,926 of the 50,339 non-control conditions, corresponding to 728 genes with consistent effects from a total of 16,784 targets included in the screen, were more than 2 standard deviations in median cell fluorescence from the null-effect (Figure 1E and Table S2).
To identify novel factors to further our understanding of pluripotency, we excluded candidates with established roles in pluripotency (e.g., Esrrb, Sox2, Sall4) or candidates already identified by other published RNAi screens (Figure 1F and Table S1), and selected candidates in transcription factor, chromatin-modifying and signaling GO categories that have been implicated but not directly studied in the context of pluripotency control, thus narrowing the list to 24 candidates (Figure 1E). In contrast to our primary screen with pooled siRNAs, we re-screened these 24 candidate genes in technical triplicate with their 4 component siRNAs in separate wells. 13 of the 24 candidates had repeatable effects on GFP levels (4 up- and 9 down-regulation) by multiple individual siRNAs targeting the same mRNA (Figure S1B–C). We further confirmed the regulatory effects of these 13 candidates on Nanog promoter activity by flow cytometry analyses of GFP in NG4 cells infected with two independent shRNAs against each of candidate genes under the screen condition (Figure 1H and S1D).
In summary, the unique genome-wide RNAi screening platform (Table S1) for Nanog-GFP reporter activity allowed us to identify novel pluripotency regulators that positively or negatively regulate the Nanog promoter activity.
Snai1 and Snai2 Differentially Regulate Pluripotency Genes in the Late Stage of Reprogramming
Although dispensable for stem cell pluripotency (Chambers et al., 2007) and for mouse embryonic fibroblast (MEF) reprogramming when supplemented with vitamin C (Schwarz et al., 2014), Nanog is required for efficient transition of partially reprogrammed pre-iPSCs into fully pluripotent iPSCs (Carter et al., 2014; Silva et al., 2009). We sought to understand how the two mesenchymal transcription factors Snai1 and Snai2 with opposing effects on Nanog promoter activity (Figure 1G–H) may influence this Nanog-driven critical stage of reprogramming. We therefore tested effects of Snai1/2 knockdown in the well-established pre-iPSC reprogramming system as described previously (Silva et al., 2008). Briefly, the pre-iPSCs were generated by retroviral transduction of neural stem (NS) cells harboring an Oct4-GFP reporter with Oct4, Klf4 and c-Myc (rOKM) followed by introduction of shRNAs of interest and establishment of stable pre-iPSC lines with appropriate drug selection. Reprogramming to pluripotency was initiated by a medium switch from serum+LIF to 2i+LIF (2i refers to the two chemical inhibitors for ERK and GSK signaling pathways (Ying et al., 2008)) and assessed by scoring total numbers of Oct4-GFP+ colonies (Figures S2A and S2C).
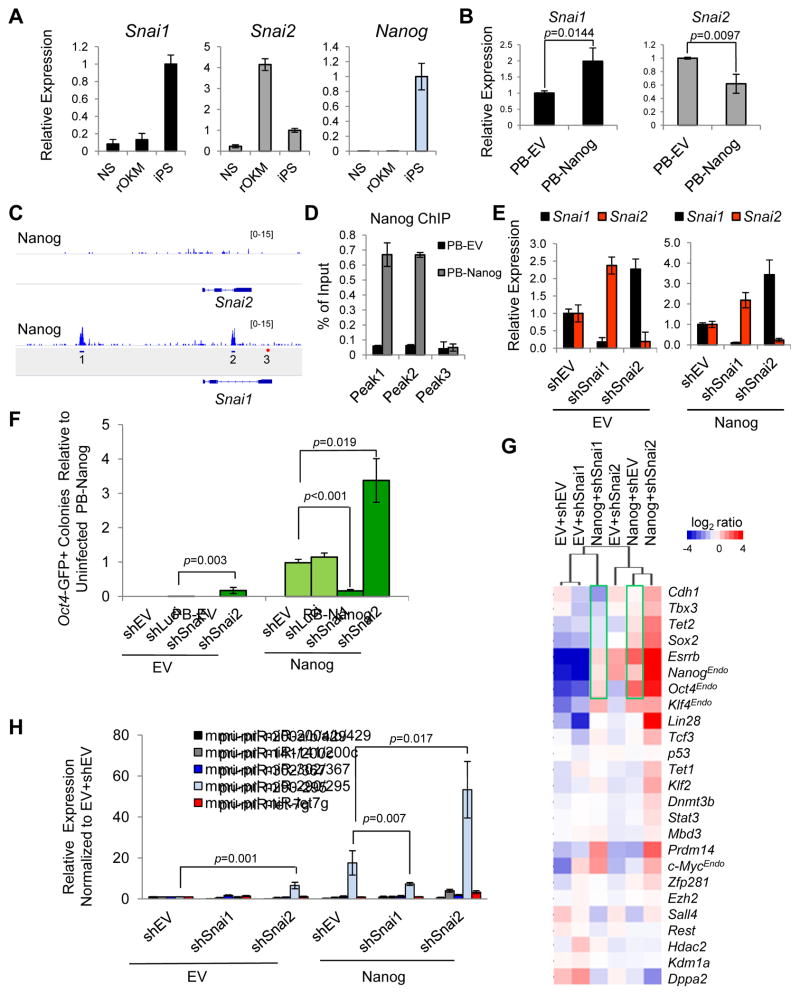
We found a differential expression pattern of Snai1 and Snai2 during the pre-iPSC to iPSC transition amid predicted downregulation of mesenchymal genes and upregulation of epithelial and pluripotency genes during Nanog-driven pre-iPSC reprogramming (Figure S2B). Notably, we observed high expression levels of Snai2 in pre-iPSCs (rOKM d0) that declined with Nanog overexpression (rOKM+N) to a minimal level at day 10 (Figure S2B) and in final iPSCs (Figure 2A). In contrast, Snai1 expression remained low in pre-iPSCs but peaked dramatically during reprogramming with ectopic Nanog at day 10 (Figure S2B) and in final iPSCs (Figure 2A). In addition, we found ectopic Nanog expression led to upregulation of Snai1 and downregulation of Snai2 in pre-iPSCs (Figure 2B), raising the possibility that Nanog may directly control Snai1/2 expression during the reprogramming process. Indeed, we confirmed that Snai1, but not Snai2, is a direct target gene of Nanog based on a previous ChIP-seq study (Marson et al., 2008) (Figure 2C) and our ChIP-qPCR analysis (Figure 2D). Furthermore, we observed a negative feedback control between Snai1 and Snai2 in pre-iPSCs (Figure 2E), suggesting that Nanog’s inhibition of Snai2 expression (Figure 2B) may be mediated by upregulated Snai1 levels. More importantly, we observed an antagonistic function of Snai1 and Snai2 in pre-iPSC reprogramming with shSnai1 compromising and shSnai2 enhancing Nanog-dependent pre-iPSC reprogramming assessed by both counting Oct4-GFP+ colonies (Figure 2F and Figure S2C–D) and by alkaline phosphatase (AP) staining (Figure S2E–F). Similar effects of Snai1 and Snai2 depletion on reprogramming were also observed in MEF (harboring a Nanog-GFP reporter)-derived pre-iPSC reprogramming (Figure S2G–I). We confirmed that the observed differential reprogramming effects cannot be explained by cell cycle alterations upon Snai1 and Snai2 depletion (Figure S2J).
Figure 2. Opposing Effects of Snai1 and Snai2 Depletion on pre-iPSC reprogramming.
(A) Relative expression levels of Snai1, Snai2, and Nanog in NS, rOKM, and iPSC cells.
(B) Ectopic expression of Nanog upregulates Snai1 and downregulates Snai2 in rOKM pre-iPSCs. Error bars indicate average ± SD (n=3). p values were calculated with the unpaired t-test.
(C) Nanog binding peaks are found on the Snai1, but not Snai2 locus (Marson et al., 2008). Amplicons 1 and 2 are at the Nanog-bound Snai1 locus. Amplicon 3 is a negative control.
(D) ChIP-qPCR validation of Nanog binding to the Snai1 locus. Amplicons are noted in Figure 2C.
(E) Mutually repressive expression of Snai1 and Snai2 in pre-iPSCs measured by qRT-PCR. Data are normalized to empty vector (shEV) and Gapdh. Error bars indicate average ± SD of two independent shRNAs each against Snai1 and Snai2 (n=3).
(F) Opposing effects of shSnai1 and shSnai2 on Nanog-dependent pre-iPSC reprogramming. Error bars indicate average ± SD (n=3) using two independent shRNAs against each Snai1 and Snai2. p values were calculated with the unpaired t-test.
(G) Relative expression of pluripotency genes measured by qRT-PCR in clonal pre-iPSCs cultured in serum+LIF at day 0 (before the medium switch).
(H) Relative expression of primary miR transcripts (pri-miRs) measured by qRT-PCR in pre-iPSCs transduced with indicated shRNAs against Snai1 and Snai2 under serum+LIF conditions. Values are normalized to Gapdh and snU6 for each condition. Error bars indicate average ± SD (n=3) using two independent shRNAs each against Snai1 and Snai2. p values were calculated with the unpaired t-test. See also Figure S2.
To elucidate how Snai1 and Snai2 may contribute to their opposing functions in reprogramming, we profiled expression of pluripotency-associated genes in pre-iPSCs upon Snai1 or Snai2 depletion in the presence or absence of ectopic Nanog expression (Figure 2G). These genes are known to be reactivated during the late phase of reprogramming (Buganim et al., 2012; Costa et al., 2013; Fidalgo et al., 2012; Polo et al., 2012). Gene expression analyses at day 0 (before initiation of reprogramming) revealed that shSnai1 samples (±Nanog) clustered with pre-iPSCs alone while shSnai2 samples (±Nanog) clustered closely with pre-iPSCs ectopically expressing Nanog (Figure 2G). Moreover, relative to pre-iPSCs alone that are infected with empty vectors (EV+shEV), depletion of Snai2 (EV+shSnai2), ectopic expression of Nanog (Nanog+shEV) or both (Nanog+shSnai2) upregulated many of these genes, among them Sox2, Esrrb, Cdh1 (or E-cadherin), Lin28, and endogenous Nanog (NanogEndo). In contrast, depletion of Snai1 (EV+shSnai1) had minimal impact on these genes (Figure 2G). Importantly, we found that Snai1 depletion led to decreased expression of pluripotency genes that are activated by ectopic Nanog (Figure 2G, compare the two green rectangles), which may explain the effect of shSnai1 in compromising Nanog reprogramming efficiency.
Taken together, our results suggest that the opposing effects of Snai1 and Snai2 on Nanog-driven reprogramming may be explained by their differential transcriptional control of key pluripotency genes.
Ectopic Expression of Snai1 and Snai2 Have Opposing Functions in Modulating Nanog-Driven pre-iPSC to iPSC Transition
We also assessed the direct effects of ectopic Snai1 and Snai2 expression on pre-iPSC reprogramming (Figures 3A and S3A). As expected, Nanog overexpression facilitated the transition of pre-iPSCs to Oct4-GFP+ and AP+ iPSCs (Figures 3B and S3B). Strikingly, while no colonies were observed after ectopic expression of Snai1 or Snai2 alone, Nanog+Snai1 samples yielded more than twice the number of Oct4-GFP+ colonies as Nanog alone while Snai2 overexpression had an inhibitory effect (Figures 3B and S3B). This differential reprogramming efficiency cannot be explained by cell cycle effects caused by Snai1 or Snai2 ectopic expression (Figure S3C). To confirm iPSC pluripotency, we established stable Oct4-GFP+ iPS colonies free of transgenes (Figure S3D) by PBase treatment to remove ectopic Nanog and Snai1 transgenes. The bona fide pluripotency of Nanog iPSCs upon transgene removal has been proven in previous studies (Costa et al., 2013; Silva et al., 2009; Theunissen et al., 2011). We confirmed the comparable pluripotency status of transgene-free, Nanog+Snai1 iPSCs and Nanog iPSCs by reduced H3K27me3 foci in female cells (indicative of X-chromosome reactivation), normal expression of pluripotency surface markers and genes by both immunostaining and mRNA profiling in iPSCs (Figure S3E–G) and multi-lineage differentiation propensities under three independent differentiation protocols (Figure S3H).
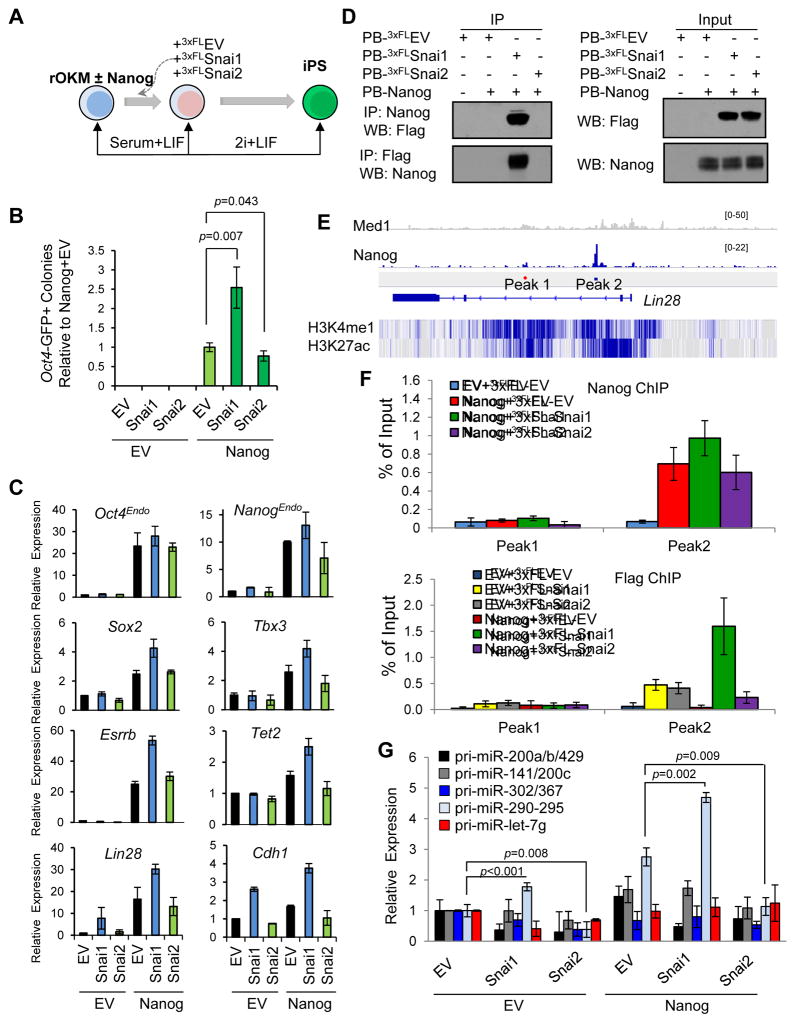
Figure 3. Ectopic Snai1 and Snai2 Have Opposite Effects on Nanog-Dependent pre-iPSC reprogramming.
(A) Reprogramming assay overview.
(B) Relative reprogramming efficiencies from the data shown in Figure S3B (presented as average ± SD (n=6)). p values were calculated with the unpaired t-test.
(C) Relative expression of pluripotency genes measured by qRT-PCR in clonal pre-iPSCs cultured in serum+LIF. Error bars indicate average ± SD (n=3).
(D) Snai1, but not Snai2, physically interacts with Nanog in pre-iPSCs.
(E) Depiction of Nanog binding peaks at the Lin28 locus. Peak 1 is a negative control. H3K4 mono methylation and H3K27 acetylation patterns are displayed below in blue.
(F) Cooperative binding of Nanog and Snai1 to the Lin28 locus. Data are from two independent ChIP experiments and presented as average ± SD (n=2).
(G) Relative expression of primary miR transcripts (pri-miRs) measured by qRT-PCR in pre-iPSCs transduced with Snai1 and Snai2 under serum+LIF conditions (normalized to Gapdh and snU6 for each condition). Error bars indicate average ± SD (n=3). p values were calculated with the unpaired t-test. See also Figure S3.
Quantitative (q) RT-PCR analysis further revealed that Snai1, acting together with Nanog, upregulated critical pluripotency genes such as Oct4Endo, NanogEndo, Sox2, Tbx3, Esrrb, Tet2, and Lin28, as well as the epithelial marker Cdh1 in pre-iPSCs (Figure 3C). In contrast, Snai2 had no effects on some (e.g., Oct4Endo, Sox2, and Esrrb) or slightly downregulated other (e.g., NanogEndo, Tbx3, Tet2, Lin28, and Chd1) genes (Figure 3C). Finally, we also confirmed the facilitating and hindering functions of ectopic Snai1 and Snai2, respectively, on Nanog-driven reprogramming of MEF-derived pre-iPSCs (Figure S3I–J), excluding the possibility of artifacts associated with NS-derived pre-iPSCs producing the differential effects of Snai1 and Snai2 on reprogramming.
To elucidate the molecular mechanism by which Nanog and ectopic Snai1/2 differentially regulate pluripotency-associated genes during reprogramming, we chose the Lin28 gene due to its pronounced expression change in Nanog+shSnai2 (Figure 2G) and Nanog+Snai1 (Figure 3C) and its established role in final stage of reprogramming (Tanabe et al., 2013). Lin28 is known to be a Nanog binding target (Marson et al., 2008). We first asked if Snai1 and Snai2 could impact Lin28 expression via interactions with Nanog. Interestingly, co-immunoprecipitation (co-IP) of ectopically expressed proteins in heterologous 293T cells indicated that Snai1 but not Snai2 interacts with Nanog (Figure S3K). More importantly, such a specific Nanog-Snai1 partnership was also preserved in pre-iPSCs (Figure 3D). These results suggest that Snai1 and Nanog participate in a protein complex to transcriptionally activate Lin28, and that Snai2 may indirectly and antagonistically modulate Lin28 expression via the negative feedback control of Snai1 (Figure 2E). To directly examine the contribution of differential Nanog and Snai1/2 partnership to the Lin28 gene regulation, we performed ChIP-qPCR analyses on all Nanog binding loci within the regulatory region of Lin28 as previously reported (Marson et al., 2008) (Figure 3E). Interestingly, we observed a slight and reproducible increase of Nanog binding to “Peak2” upon Snai1, but not Snai2 expression (compare green and purple bars with the red one in Figure 3F top panel). As expected, there was no Nanog binding to the negative control “Peak1” region irrespective of ectopic Snai1 or Snai2 expression (Figure 3F top panel). More importantly, we observed a dramatic Nanog-facilitated Snai1 binding to the regulatory region of Lin28 (green versus yellow/red bars in Figure 3F bottom panel) but no such cooperative binding between Nanog and Snai2 (purple versus grey/red bars in Figure 3F bottom panel).
Collectively, our results indicate that a novel Nanog-Snai1 partnership leads to direct transcriptional activation of pluripotency genes during the late stage of reprogramming.
Snai1 and Snai2 Differentially Regulate miRNA Genes during the Last Phase of Reprogramming
Snai1 and Snai2 have been shown to modulate cell proliferation and miRNA (miR) regulation in a cellular context-dependent manner (Zheng and Kang, 2013). However, cell cycle profiling of pre-iPSCs with loss or gain of Snai1 or Snai2 expression cannot explain their opposing functions in reprogramming (Figures S2J and S3C). We thus turned our attention to potential EMT/MET-implicated miRNAs such as the miR-200 cluster (Siemens et al., 2011; Wellner et al., 2009) as potential targets of Snai1 and Snai2 proteins during reprogramming as well as the miRNA genes regulated by Nanog (Marson et al., 2008) that play positive roles in reprogramming (Leonardo et al., 2012).
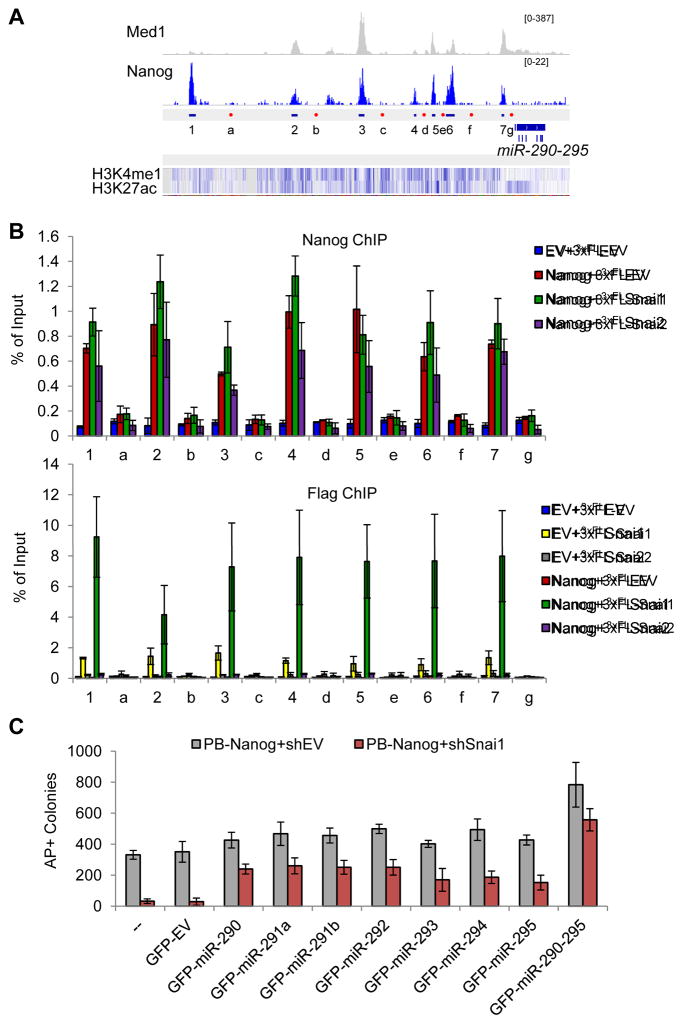
We found that both knockdown and ectopic expression of Snai1 or Snai2 alone have minimal effects on expression of these miRNA genes in pre-iPSCs in the absence of ectopic Nanog expression (EV panels in Figures 2H and 3G). We observed a slight upregulation of miR-290-295 upon depletion of Snai2 (EV+shSnai2) (Figure 2H) or ectopic expression of Snai1 (EV+Snai1) (Figure 3G). In contrast, ectopic Nanog greatly upregulated the expression of the miR-290-295 cluster (Nanog+shEV and Nanog+EV samples in Figures 2H and 3G). More importantly, knockdown of Snai1 reduced and ectopic Snai1 further enhanced expression of miR-290-295, whereas knockdown and ectopic expression of Snai2 had the opposite effects compared with the same treatment of Snai1 on miR-290-295 expression (Nanog panels in Figures 2H and 3G).
It is well established that miR-290-295 plays positive roles in reprogramming (Judson et al., 2009), although how it was regulated during the reprogramming process is not known. We decided to address whether the novel Snai1-Nanog partnership (Figure 3D and S3K) may contribute to direct transcriptional activation of this miR. We therefore performed ChIP-qPCR analyses on all Nanog binding loci (1–7) within the promoter/enhancer region of miR-290-295 as previously defined (Marson et al., 2008) (Figure 4A). While we confirmed Nanog binding to these loci (red bars in Figure 4B top), we also observed a slight but reproducible effect of ectopic Snai1 facilitating Nanog binding to 6 of 7 loci (compare green with red bars in Figure 4B top panel). In contrast, ectopic expression of Snai2 had no or slight downregulating effects on Nanog binding to these loci (compare purple with red bars in Figure 4B top panel). Interestingly, we also observed enrichment of Snai1, but not Snai2, at these Nanog-binding loci (yellow bars in Figure 4B bottom panel), which was strikingly enhanced by ectopic Nanog expression (Figure 4B bottom panel, compare the green bars with the yellow ones). However, there was no detectable binding of Snai2 to any of these loci irrespective of Nanog expression (grey and purple bars in Figure 4B bottom panel). To investigate the functional significance of the Nanog and Snai1 activated miR-290-295 cluster, we performed individual miR rescue of Nanog+shSnai1 reprogramming (Figure 4C). We retrovirally introduced GFP-marked individual miRs into pre-iPSCs stably expressing PB-Nanog+shEV or PB-Nanog+shSnai1. Two days later, we switched the culture from serum+LIF to 2i+LIF. Reprogramming was assessed by counting AP+ colonies after 10 days in 2i+LIF culture (GFP cannot be used due to its presence in the individual miRs (Chen et al., 2012)). Strikingly, we found that each member of the miRNA-290-295 can partly rescue the reprogramming defects of PB-Nanog+shSnai1 (compare red and grey bars between rescue and control samples), with the most pronounced rescue when the whole cluster is expressed (Figure 4C). These data conclusively establish the novel partnership of Nanog and Snai1 and its functional contribution to the final reprogramming stage by direct transcriptional activation of miR-290-295. The incomplete rescue also suggests that direct transcriptional activation of other key pluripotency genes such as endogenous Nanog, Esrrb and Lin28 (Figures 2G and 3C) likely also contribute to the enhanced reprogramming by Nanog and Snai1.
Figure 4. The Nanog and Snai1 Partnership in Transcriptional Regulation of Pluripotency-Associated miRNAs.
(A) Depiction of ChIP-seq peaks within the enhancer region of miR-290-295 that are bound by mediator 1 (Med1) and Nanog. “1–7” denote amplicons for the peaks, and “a–g” denote amplicons for negative controls. H3K4 mono methylation and H3K27 acetylation patterns are noted beneath the peaks.
(B) Nanog facilitated binding of Snai1, but not Snai2, to the enhancer of miR-290-295 in pre-iPSCs ectopically expressing the indicated factors. “1–7” are positive binding peaks, and “a–g” are negative control regions. Error bars indicate average ± SD from two independent ChIP studies.
(C) Ectopic expression of miR-290 family members partially rescues shSnai1 effect on Nanog-mediated reprogramming. Error bars indicate average ± SD (n=3). See also Figure S4.
While let-7 family miRs and miR-290-295 are known to have opposing effects on ESC self-renewal (Melton et al., 2010), how such an antagonistic effect is controlled is not known. We observed minimal expression changes of pri-miR-let-7g in pre-iPSCs expressing Nanog±shSnai1/2 or Nanog±Snai1/2 (Figures 2H and 3G). However, the marked upregulation of Lin28, an inhibitor of pri-miR-let-7 processing, in pre-iPSCs expressing Nanog+shSnai2 (Figure 2G) or Nanog+Snai1 (Figure 3C), and direct cooperative binding of Nanog and Snai1 to the Lin28 locus (Figure 3E–F) suggests that mature let-7 is kept minimal in these cells by the combined action of Nanog and ectopic Snai1 expression (or Snai2 depletion). Our study thus provides insights into the molecular control mechanism for these two opposing groups of miRNAs during the reprogramming process.
To investigate how miR-290-295 and let-7 family miRs may contribute to the differential function of Snai1 and Snai2 in reprogramming, we re-analyzed published microarray data comparing mRNA gene expression profiles of pre-iPSCs and iPSCs (Sridharan et al., 2009). We found that nearly 50% of differentially expressed mRNAs are predicted targets of miR-290-295 (Figure S4A left). These so-called “pre-iPSC genes” are greatly downregulated in Nanog+shSnai2 but not Nanog+shSnai1 pre-iPSCs (Figure S4B left). When analyzing let-7 targets in the same dataset (Sridharan et al., 2009), we found ~35% of differentially expressed mRNAs to be potential targets (Figure S4A right). These so-called “iPSC genes” include many pluripotency genes including Lin28 that are upregulated during the process of attaining full pluripotency (Figure S4B right). Therefore, miR-290-295 upregulation and let-7 downregulation are likely to be involved in the functional antagonism of Snai1 and Snai2 in the Nanog-driven final stage of reprogramming.
Snai2 Does Not Compete with Snai1 in Binding to the Nanog Sites at the Regulatory Regions of Lin28 and miR-290-295 Genes
Snai1 and Snai2 are known to bind the E-box motif “CANNTG” and transcriptionally regulate target genes (Wu and Zhou, 2010). The physical association of Nanog and Snai1 (Figures 3D and S3K) and opposing effects of Snai1 and Snai2 on Nanog function (Figures 2–3 and S2–3) in reprogramming prompted us to address whether the E-box motif is enriched in Nanog binding loci and whether Snai1 and Snai2 may compete in binding to the E-box motif within those Nanog binding loci. We searched the canonical E-box motif “CANNTG” among those published Nanog peaks and found that 78% and 69% of global Nanog binding peaks from the Marson et al. (Marson et al., 2008) and Chen et al. (Chen et al., 2008) studies contained the E-box motif, with an average of 1.67 and 1.29 E-box sites per Nanog peak, respectively, from the two Nanog ChIP-seq datasets (Figures 5 and S5). These results suggest a significant juxtaposition of Nanog and Snai1/2 binding loci in the genome, consistent with the physical association and functional cooperation between Nanog and Snai1 we identified in this study.
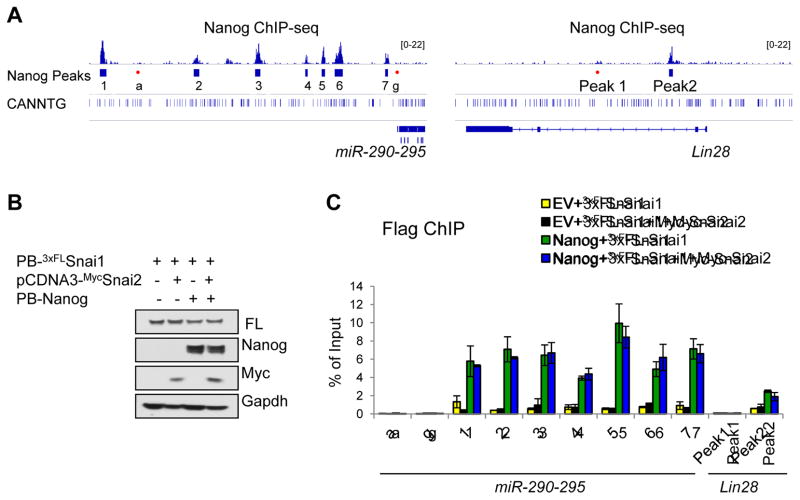
Figure 5. Snai2 Does Not Compete with Snai1 in Binding to the Nanog Sites at the Lin28 and miR-290-295 Loci.
(A) Depiction of ChIP-seq peaks at regulatory regions of both miR-290-295 and Lin28 genes. The E-box motif is also shown where present.
(B) Various combinations of Nanog, Snai1, and Snai2 were used to generate stable pre-iPSC clones. Western blots confirm transgenic Nanog, 3xFLAGSnai1 or Myc-Snai2 protein expression in pre-iPSCs cultured in serum+LIF.
(C) FLAG-ChIP and qPCR analyses of Snai1 binding to the Nanog sites at the miR-290-295 and Lin28a loci in pre-iPSCs. Error bars indicate average ± SD (n=2). See also Figure S5.
Bioinformatic analyses on the miR-290-295 and Lin28 genes revealed many E-box motifs at both Nanog binding and non-binding loci (Figure 5A). Snai1, but not Snai2, binds to these loci and its binding is specifically enriched in Nanog binding loci (#1–7) but not other non-binding loci (#a–g) (Figure 4B bottom panel), suggesting a Snai1/2 preference for certain internal dinucleotides (Figure S5A). Such sequence-specific motif binding preference has been reported for several Snai1 homologues (Kataoka et al., 2000). To test whether Snai2 could compete with Snai1 in binding to the miR-290-295 and Lin28 gene loci, we established control (EV) and Nanog pre-iPSC lines stably expressing ectopic 3xFLSnai1 in the presence and absence of Myc-Snai2 (Figure 5B) and performed 3xFLAG (for Snai1) ChIP, asking whether the overexpression of Myc-Snai2 would reduce Snai1 binding. As shown in Figure 5C, we did not detect significant change of Snai1 binding to these Nanog/Snai1 co-binding loci (yellow bars). More importantly, we confirmed that both Nanog-facilitated binding of Snai1 and direct Nanog binding to these loci were not affected by the overexpression of Myc-Snai2 (compare green with blue bars in Figures 5C and S5B). Together, our results confirm cooperative Snai1 and Nanog binding to regulatory regions of Lin28 and miR-290-295 genes that is more likely controlled by a differential interaction of Snai1/2 with Nanog but not a competitive binding to the E-box motifs within the Nanog sites.
DISCUSSION
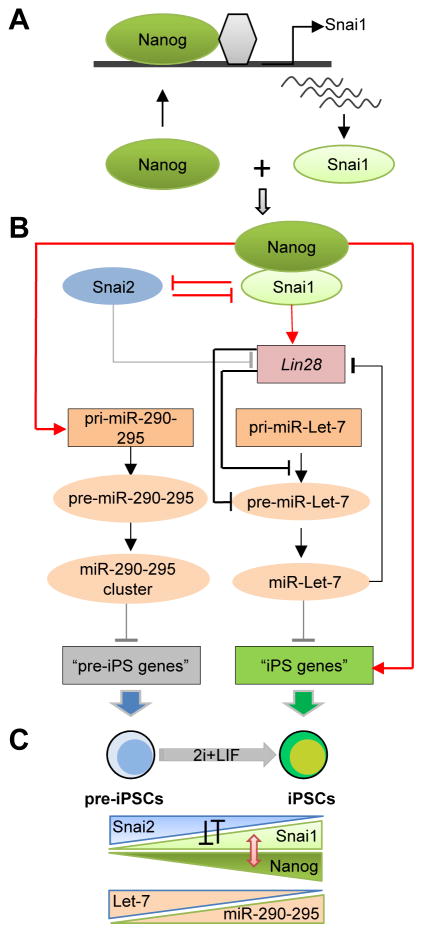
As noted previously (Subramanian et al., 2009), design differences contribute to the limited concordance between RNAi datasets (Figure 1F and Table S1). However, all these loss-of-function screening strategies have uncovered many previously unknown and complementary regulators of pluripotency. We were able to uncover 728 candidate hits with potential functional significance in pluripotency and reprogramming from our primary screen (Figure 1E and Table S2). We not only rediscovered many well-known self-renewal and pluripotency genes from our screen but, more importantly, identified several that have not previously been studied. 13 of the 24 total candidates in our secondary screen had a validated impact on Nanog promoter activity, 9 positively and 4 negatively (Figure 1G–H). We did not find Mbd3 in the candidate list as the median Z-score for this gene was 0.69. This is in contrast with its prominent status in the recent RNAi screen using Nanog-GFP reporter EpiSCs (Rais et al., 2013) but consistent with the minimal changes of Nanog expression in Mbd3 null ESCs (Kaji et al., 2006; Reynolds et al., 2012). It is likely that Mdb3 depletion would not impact Nanog expression during loss of pluripotency within the 2-day RA differentiation time window in our screen. It is also worth pointing out that the deterministic reprogramming of Mbd3-depleted somatic cells reported by Rais et al. (Rais et al., 2013) has been challenged by a more recent study demonstrating a requirement of Mbd3/NuRD function for efficient reprogramming (Dos Santos et al., 2014). Our screen platform is unique in that it allows for simultaneous identification of candidates with increased and decreased pluripotency reporter activity in the same screen (Table S1), which led to the identification of the antagonistic roles of Snai1 and Snai2 proteins in controlling pluripotency and reprogramming (Figure 6A–C).
Figure 6. A Model for the Opposing Function of Snai1 and Snai2 In Reprogramming.
(A) Transcriptional activation of Snai1, but not Snai2, by Nanog during pre-iPSC reprogramming.
(B) Summary of regulatory loops controlled by the Nanog-Snai1 complex and mutual inhibition of Snai1 and Snai2 in transcriptional control of pluripotency-associated genes and miRNAs. Red colored signs denote the novel regulatory controls defined in this study. Grey lines denote the regulatory controls indicated but not yet defined in this study. Black lines are the regulatory controls already established from the published literature.
(C) Summary of the salient features of functional antagonisms of Snai1 versus Snai2 (indicated by mutual inhibition) and Let7 versus miR-290-295, as well as the physical and functional cooperation of Nanog and Snai1 during the pre-iPSC to iPSC transition. The red two-pointed arrow denotes the protein-protein interaction between Nanog and Snai1. See also Figure S6.
Because the limited overlap between our candidates and those from additional screens (Figure 1F) may be due to distinct differentiation conditions, we checked whether our candidates were differentially regulated (>2-fold either up or down) relative to the day 0 time-point following 2 days of either LIF withdrawal differentiation (Hailesellasse Sene et al., 2007) or 2 μM RA differentiation (Ivanova et al., 2006). Restricting our analysis to the genes probed in both microarrays as well as in our RNAi screen (Figure S1E, left), we found that our candidates were not overrepresented in the differentially regulated genes under either LIF withdrawal or RA conditions (Figure S1E, right, and S1F). Thus, although the effect of candidate depletion on Nanog promoter activity may be dependent on the differentiation conditions, our candidates are not themselves preferentially regulated by them.
We focused on Snai1 and Snai2 for detailed mechanistic studies for several reasons. First, it is well known that both Snai1 and Snai2 maintain the mesenchymal phenotype by directly repressing epithelial gene expression (Thiery et al., 2009), thus functioning as a barrier in inhibiting the requisite MET process during early reprogramming (Li et al., 2010; Samavarchi-Tehrani et al., 2010). The mutually repressive expression pattern and opposing reprogramming effects during the late stage of reprogramming are totally unexpected. Second, it was recently demonstrated that highly coordinated proteome dynamics during reprogramming involves an EMT feature in the late stage of the reprogramming process (Hansson et al., 2012), although the role of Snai1 and Snai2, if any, was not clear. Third, a reprogramming protocol (Liu et al., 2013) noted the association of optimal reprogramming efficiency with sequential EMT-MET during the early stage of reprogramming but had not attempted to study the late reprogramming process. It was found that the reprogramming factor Oct4 activates early EMT through Snai2 regulation (Liu et al., 2013). Interestingly, our study found that Nanog activates Snai1, but not Snai2, during the last stage of reprogramming (Figure 2A–D).
We recognize that Snai1 can also function outside of EMT control (Wu and Zhou, 2010). Upregulation of both Snai1 and Snai2 together with downregulation of Cdh1/E-cadherin are the hallmarks of EMT. However, our findings that Snai1 and Snai2 are mutually repressive (Figure 2E) and that Cdh1/E-cadherin is upregulated upon ectopic expression of Snai1 in pre-iPSCs (Figure 3C) preclude the EMT process from being considered the defining feature of Snai1 and Snai2 action during the critical transition stage of pre-iPSC reprogramming. Rather, the Snai1-Nanog partnership in cooperative and direct binding to and transcriptional activation of the pluripotency-associated genes (e.g., Lin28 and miR-290-295) suggests a more pronounced activator function than the well-established MET repressor function of Snai1 in early stage of reprogramming (Polo and Hochedlinger, 2010) and in cancer (Peinado et al., 2004). Supporting this, Snai1+/E-cadherin+ populations have also been reported during the reprogramming process using single-cell expression analyses (Buganim et al., 2012). The positive regulation of Cdh1/E-cadherin by a Snai1 counterpart in Drosophila was previously reported (Tanaka-Matakatsu et al., 1996), although the molecular mechanism underlying such a positive control is not clear. We suspect that Snai1 cooperates with Nanog forming a multi-protein complex that transcriptionally activates the pluripotency-associated genes and miRNAs (Figure 6A–B) much like other pluripotency factors that operate via such combinatorial binding in the enhancers for target gene activation (Chen et al., 2008; Kim et al., 2008). In this regard, it is noteworthy that the interrogated Nanog/Snai1 co-binding loci are also bound by Med1 and marked with H3K4me1 and H3K27ac as active enhancers (Figures 3E and 4A). Interestingly, the Snai1 regulatory region was one of the retroviral integration (presumably activation) sites during reprogramming (Aoi et al., 2008).
Snai1 protein was recently shown to be expressed in conventionally cultured (serum/LIF) ESCs and its mRNA upregulated in ground state pluripotency 2i culture (Lin et al., 2014), suggesting its role in controlling stem cell pluripotency. However, Snai1 is dispensable for self-renewal and ESC maintenance, and rather, it is required for EMT during early differentiation and subsequently epiblast stem cell exit and mesoderm commitment (Lin et al., 2014). The upregulation of both Nanog (Leitch et al., 2013) and Snai1 (Lin et al., 2014) under 2i/LIF relative to serum/LIF culture supports their physical and functional relationship during reprogramming under the same 2i/LIF condition as defined in our study. However, neither overexpression nor depletion of Snai1 affects Nanog expression and its function in controlling self-renewal and ESC maintenance (Figure S6). Although the phenotype of Snai1 depletion in ESCs is relatively minor, consistent and dramatic effects were noted following Snai1 depletion in combination with RA differentiation using multiple siRNAs targeting distinct regions of the Snai1 mRNA, which allowed us to identify Snai1 as an RNAi hit that positively regulates the Nanog-GFP promoter (Figures 1 and S1), further highlighting the power of our unique RNAi screen platform.
In summary, our data demonstrate that cooperative binding of Nanog and Snai1 through their novel partnership to downstream targets in pre-iPSCs is a critical event during the final transition to naive pluripotency leading to transcriptional activation of pluripotency-associated miRNAs and genes (dubbed “iPSC genes”) and simultaneous repression of inhibitory miRNAs and “pre-iPSC genes” (Figure 6A–C). Interestingly, an independent study observed positive roles of Snai1 in early stage of MEF reprogramming by directly repressing let-7 family miRNAs (George Daley, Children’s Hospital Boston, personal communication). Future studies are needed to understand how the novel Nanog-Snai1 partnership endows the typical transcriptional repressor Snai1 with an activator function in promoting the Nanog-driven final stage of reprogramming, and whether such a novel connection of Snai1 (but not Snai2) with the pluripotency program may relate to the differential function of Snai1/2 in development and cancer. Of note, deletion of Snai1 results in embryonic lethality due to gastrulation defects (Carver et al., 2001), and conditional knockout of Snai1 at the epiblast stage also results in embryonic lethality (Lomeli et al., 2009). By contrast, Snai2 germline knockout mice are viable with impaired postnatal defects (Parent et al., 2010; Perez-Losada et al., 2002). In addition, ectopic expression of Snai1 was found to positively regulate Nanog expression during EMT in non-small-cell lung cancer (Liu et al., 2014).
EXPERIMENTAL PROCEDURES
RNAi Library and Screening Strategy
The mouse siGENOME library from Thermo Scientific, covering siRNA targets for 16,872 genes with pools of 4 sequences per target gene was used to screen for genes that either positively or negatively regulate Nanog-GFP fluorescence. For the RNAi screen, 384-well Aurora tissue culture treated screening microplates (Brooks Automation) were coated with 0.1% gelatin for 15 minutes. Gelatin was aspirated using a 24-channel manifold (Drummond Scientific). An average of 750 mouse ESCs in a volume 40 μL of mouse ES media with LIF were added with a MultiDrop Combi liquid dispenser (Thermo Scientific) to 10 μL siRNA/transfection mixture for reverse-transfection for a final volume of 50 μL containing 1:667 transfection reagent and 25 nM siRNA. For the next 2 days of culture, media was changed to mouse ES media without LIF and containing 10 nM retinoic acid. Three days after siRNA transfection, cells were fixed in 2% formaldehyde in PBS for 15 minutes, stained with Hoechst 33342 5 μg/mL for 15 minutes at RT and washed with PBS twice prior to confocal fluorescence imaging. For additional details see Extended Experimental Procedures.
Reprogramming Assays Using Mouse Embryonic Fibroblasts (MEFs) and Neural stem (NS) Cells Derived Reprogramming Intermediates
MEF- and NS-derived pre-iPSCs were employed for reprogramming tests in the presence or absence of Snai1/2 ectopic expression or depletion using a medium switch from serum/LIF to 2i/LIF as described (Costa et al., 2013).
Chromatin Immunoprecipitation Coupled with Quantitative Real-Time PCR (ChIP-qPCR)
ChIP was performed with anti-Nanog or anti-FLAG in pre-iPSCs followed by qPCR on the defined loci. See Supplemental Table S3 for all the primers used in this study.
Immunoprecipitation and Western Blot Analysis
Whole cell extracts and nuclear extracts were prepared as previously reported (Costa et al., 2013). For coIP in HEK293T cells, we transiently co-transfected cells with plasmids expressing 3xFlag-tagged Snai1 or Snai2 and V5-his tagged Nanog. Two days after transfection, nuclear extracts were prepared and incubated with EZview™ Red anti-Flag M2 Affinity Gel or anti-V5 Agarose Affinity gel antibody overnight. Co-immunoprecipitated 3xFlag-Snai1, 3xFlag-Snai2 or V5his-Nanog were identified by Western blot using anti-Flag M2 and anti-V5 HRP (Invitrogen, cat#1030648), respectively. Gapdh antibody (Proteintech Group, cat #10494-1-AP) was also used for protein sample loading control. Additional antibodies used in western blot and coIP/IP analyses are: anti-Nanog (Bethyl Labotatories, cat #A300-397A), anti-Myc (Cell Signaling, cat#2276).
Analysis of miRNA Target Predictions
Microarray datasets for genes differentially expressed in pre-iPSCs and iPSCs were downloaded from GEO (Accession number: GSE14012) (Sridharan et al., 2009). The set of mouse genes that are potentially regulated by the miR-290-295 cluster (miR-290-3p, miR-292-3p, miR-293, miR-204 and miR-295) and miR-let-7 family (miR-let-7a, miR-let-7b, miR-let-7c1, miR-let-7c2, miR-let-7d, miR-let-7f, miR-let-7g, and miR-let-7i) were download from microRNA.org (http://www.microrna.org/microrna/home.do). Note that the same binding sites for miR-let-7g (Figure S7E) are also predicted for other mature miRNAs from the same family, such as let-7a–f, and I. Genes differentially expressed and predicted to be targets of microRNAs are provided in Table S4. We arbitrarily selected genes with expression value (log2) difference greater than 2 between iPS and pre-iPS cells.
Analysis of E-box Enrichment
Nanog ChIP-Seq datasets for analysis of E-box enrichment were downloaded from GEO with accession number of GSE11431 (Chen et al., 2008) and GSE11724 (Marson et al., 2008). Reads were uniquely aligned to the mouse (mm9) genome by Bowtie and Nanog peaks were determined in MACS software. Peak sequences were retrieved by using getfasta in Bedtools software (version 2.18.1). The frequency of the canonical E-Box motif “CANNTG” in Nanog peak sequences was calculated using in-house Python (version 2.7.6) scripts with Biopython modules.
Supplementary Material
Figure S1. Additional Unpooled siRNA and shRNA Validation of Candidates with Immunofluorescence and Flow Cytometry. Related to Figure 1.
(A) Results from the RNAi primary screen on median cell fluorescence. A selected subset of the outlier and control genes is labeled.
(B) Representative fluorescent images from a Non-Targeting siRNA-treated well. For each well treated with a given siRNA, nuclei-stained cells are identified and levels of GFP are measured.
(C) Levels of GFP for each siRNA-treated well and controls. Dots denote effect averaged across technical triplicate. Short black horizontal lines are average of 4 unpooled siRNA effects. Territory outside of long horizontal lines in grey denotes FDR <0.05 using the empty controls. Single star before gene name indicates 2 of 4 distinct siRNAs are significant with FDR<0.05, with two stars if 3–4 of distinct siRNAs are significant. Further details are in the Supplementary Information.
(D) Relative knockdown efficiencies of shRNAs in pLKO.1 vector. Values are normalized to shLuciferase (shLuci). Error bars indicate average ± SD.
(E) Venn diagram for genes assayed (left) and changed >2-fold following 2 days 2 μM RA or EB differentiation or considered RNAi screen candidates (right). Genes in shaded central region on left are assayed in all datasets.
(F) Contingency table for genes assayed in all datasets and changed following RA or EB differentiation or considered RNAi screen candidates. “+” and “−” indicate inclusion (overlapping) and exclusion (non-overlapping), respectively, in the pairwise comparisons.
Figure S2. Loss-of-Function Studies of Snai1 and Snai2 in Nanog-Driven Pre-iPSC Reprogramming. Related to Figure 2.
(A) Schematic overview of Nanog-dependent NS pre-iPSC reprogramming.
(B) Relative gene expression during the pre-iPSC reprogramming process. The time points of gene expression analysis are indicated. Relative gene expression in NS, rOKM and rOKM+Nanog (N) cells was measured by qRT-PCR. Data are normalized to Gapdh.
(C) Schematic overview of pre-iPSC reprogramming.
(D) Representative images of Nanog±shSnai2 reprogramming of Oct4-GFP reporter pre-iPSCs. shRNA is marked by mCherry. Data from one shRNA are shown.
(E) Images of AP stained colonies generated as indicated in D.
(F) Quantitation of AP+ colonies at day 10 after reprogramming with indicated shRNAs. Data are represented as average ± SD of 2 independent shRNAs against each Snai1 and Snai2.
(G) Schematic overview of pre-iPSC reprogramming in MEFs harboring the Nanog-GFP reporter transgene.
(H) Representative images of shSnai2 reprogramming of MEF-derived pre-iPSCs. shRNA vector is marked by mCherry and final iPSCs are positive for Nanog-GFP reporter activity.
(I) Quantitation of Nanog-GFP+ colonies at day 10 after reprogramming with indicated shRNAs. Data are represented as average ± SD of 2 independent shRNAs against each Snai1 and Snai2.
(J) Cell cycle analyses by flow cytometry upon Snai1/2 depletion in pre-iPSCs (rOKM) with or without ectopic Nanog expression.
Figure S3. Gain-of-Function Studies of Snai1 and Snai2 in Nanog-Driven Pre-iPSC Reprogramming. Related to Figure 3.
(A) Combinations of empty vector (EV), Nanog, Snai1, and Snai2 were used to generate stable pre-iPSC clones. Western blots confirm transgenic Nanog, 3xFlag-Snai1 or 3xFlag-Snai2 protein expression in pre-iPSCs cultured in serum+LIF.
(B) Representative images of iPSCs generated from overexpression of Snai1 and Snai2 in the presence or absence or ectopic Nanog for both Oct4-GFP (left) and AP staining (right) at day 10 of 2i/LIF treatment.
(C) Cell cycle analyses by flow cytometry upon Snai1/2 ectopic expression in pre-iPSCs (rOKM) with or without ectopic Nanog expression.
(D) Fluorescence and bright-field images of transgene-free Nanog and Nanog+Snai1 iPSCs cultured under serum+LIF or 2i/LIF for 30 days.
(E) Immunofluorescence analysis against H3K27me3 in female transgene-free iPSCs under serum/LIF or 2i/LIF culture. Nuclei were stained with DAPI.
(F) Alkaline phosphatase (AP) and immunofluorescence staining for SSEA-1, Nanog, Oct4, and Sox2 protein levels in transgene-free Nanog and Nanog+Snai1 iPSCs.
(G) Similar expression of pluripotency and lineage specific marker genes in transgene-free Nanog and Nanog+Snai1 iPSCs. Error bars indicate average ± SD. Note that the similar Snai1 levels between Nanog+EV and Nanog+Snai1 samples indicate the complete removal of the ectopic Snai1 in Nanog+Snai1 iPSCs.
(H) Similar gene expression pattern of transgene-free Nanog and Nanog+Snai1 iPSCs during differentiation induced by LIF-withdrawal, retinoic acid (RA) treatment, or embryoid body (EB) formation. Gene expression analysis was measured by qRT-PCR, and data were normalized to Gapdh.
(I) Schematic overview of the reprogramming strategy in MEF-derived pre-iPSCs with Snai1/2 gain-of-function and Nanog ectopic expression.
(J) Quantification of Nanog-GFP+ colonies for noted treatments.
(K) CoIP validation of the Nanog-Snai1 interaction in 293T cells.
Figure S4. The Nanog and Snai1 Partnership in Transcriptional Regulation of Pluripotency-Associated Genes and miRNAs. Related to Figure 4.
(A) Relative expression levels of RNAi hit genes that are potential targets of miR-290–295 (left) and miR-Let-7 (right) in pre-iPSCs and iPSCs. Scatter plots of global gene expression arrays from published microarrays (Sridharan et al., 2009) showing highest (“pre-iPSC genes”, left panel) or lowest (“iPSC genes”, right panel) relative expression (log2>2) in pre-iPSCs compared to iPSCs. Predicted target genes for the mouse miR-290–295 cluster are shown as blue dots, and those predicted for the mouse miR-Let7 family are shown as red dots. Blue and red bars indicate percentages of total pre-iPSC genes and iPS genes that are predicted targets of miR-290–295 and miR-Let7, respectively. Specific genes are indicated with arrows.
(B) Relative expression of the “pre-iPS genes” (left panel) or “iPSC genes” (right panel) measured by quantitative RT-PCR in the NS-derived reprogramming intermediates transduced with the indicated shRNA and cultured in serum/LIF.
Figure S5. Snai2 Does Not Compete with Snai1 in Binding to the Nanog Sites at the Lin28 and miR-290–295 Loci. Related to Figure 5.
(A) Pie plots showing the percentage of Nanog peaks containing the consensus E-Box motif (CANNTG) in two published Nanog ChIP-Seq datasets. Number and frequency of individual E-Box motifs within the Nanog peaks are listed.
(B) Nanog ChIP and qPCR analyses on the miR-290–295 and Lin28 loci. Snai2 does not affect Nanog binding to the regulatory regions in both miR-290–295 and Lin28a genes in pre-iPSCs ectopically expressing the indicated factors. Peaks “1–7” and “Peak2” are Nanog positive binding peaks on miR290–295 and Lin28a regulatory regions, respectively. Peaks “a”, “g” and “Peak1” are negative control regions. Error bars indicate average ± SD.
Figure S6. Snai1 is Dispensable for ESC Maintenance. Related to Figure 6.
(A) Snai1 knockdown (left panel) and transgenic (right) efficiency by shRNA and ectopic expression in CCE ESCs. Knockdown efficiency represents the average of two independent shRNAs against Snai1. Error bars indicate average ± SD.
(B) Pluripotency and lineage-specific marker analysis in ESCs transduced with two independent shRNAs against Snai1 or shEV (left panel) and in transgenic stable lines with ectopic expression of Snai1 or EV (right panel). Error bars indicate average ± SD.
(C) Colony formation assay (CFA) demonstrating lack of effect of Snai1 depletion (left panel) or enforced expression (right panel) on self-renewal. Colonies stained for AP (Alkaline Phosphatase) were scored into three categories (uniformly undifferentiated, partially differentiated, and fully differentiated). Error bars indicate average ± SD (n=3).
(D) ESC proliferation is independent of Snai1. Growth curve of CCE ESCs transduced with a shRNA against Snai1 or shEV (left panel) or with enforced Snai1 or EV expression (right panel). Cell numbers were counted every another day. Error bars indicate average ± SD (n=3).
(E) Nanog and Rex1 expression are not affected by Snai1 depletion (top panel) or enforced expression (bottom panel). Two independent ESC lines were used to measure the GFP activity under the Nanog (NG4 ES line) or Rex1 (Rex1-dGFP ES line) promoter.
Table S1. RNAi Screen Studies in ESCs, Related to Figure 1.
Supplementary Table S3, Related to Figures 1–5.
List of qRT-PCR, ChIP-qPCR primer sequences as well as siRNA/shRNA sequences used in this study.
Supplementary Table S4, Related to Figure S4.
List of genes differentially expressed in pre-iPSCs and iPSCs that are predicted to be targets of miR-290-295 cluster and Let-7 family miRNAs.
Acknowledgments
We thank members of the Wang laboratory for critically reading the manuscript, Dr. Jose Silva (University of Cambridge, United Kingdom) for the pre-iPSCs, and Dr. Michael Detmar (ETH Zurich, Switzerland) for the miR-290-295 constructs. This research was funded by grants from the National Institutes of Health (NIH) to I.R.L. (5R01GM078465) and to J.W. (1R01-GM095942), the Empire State Stem Cell Fund through New York State Department of Health (NYSTEM) to I.R.L. (C024176), C.S. (C024410) and to J.W. (C026420, C028103, C028121). J.W. is also a recipient of Irma T. Hirschl and Weill-Caulier Trusts Career Scientist Award. J.A.G. thanks the NIH MSTP Grant GM007280 to the Medical Scientist Training Program and the DSCB NRSA Training Grant. D.-F.L. is a New York Stem Cell Foundation Stanley and Fiona Druckenmiller Fellow.
Footnotes
Supplemental Information includes Extended Experimental Procedures, Figures, and Tables.
AUTHOR CONTRIBUTIONS
J.A.G. and M.F. conceived, designed and conducted the studies and wrote the manuscript draft. Z.L., H.Z., D.-F.L., and D.P.F. conducted the siRNA screen. D.G., Z.S., F.F., and C.S. designed and performed experiments. A.W. and X.H. performed bioinformatics analyses. I.R.L. and J.W. conceived the project, designed the experiments, prepared and approved the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could a3ect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aoi T, Yae K, Nakagawa M, Ichisaka T, Okita K, Takahashi K, Chiba T, Yamanaka S. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008;321:699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- Buckley SM, Aranda-Orgilles B, Strikoudis A, Apostolou E, Loizou E, Moran-Crusio K, Farnsworth CL, Koller AA, Dasgupta R, Silva JC, et al. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell. 2012;11:783–798. doi: 10.1016/j.stem.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. Single-Cell Expression Analyses during Cellular Reprogramming Reveal an Early Stochastic and a Late Hierarchic Phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter Ava C, Davis-Dusenbery Brandi N, Koszka K, Ichida Justin K, Eggan K. Nanog-Independent Reprogramming to iPSCs with Canonical Factors. Stem Cell Reports. 2014;2:119–126. doi: 10.1016/j.stemcr.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver EA, Jiang R, Lan Y, Oram KF, Gridley T. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol Cell Biol. 2001;21:8184–8188. doi: 10.1128/MCB.21.23.8184-8188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Chen Y, Liersch R, Detmar M. The miR-290-295 cluster suppresses autophagic cell death of melanoma cells. Scientific reports. 2012;2:808. doi: 10.1038/srep00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia NY, Chan YS, Feng B, Lu X, Orlov YL, Moreau D, Kumar P, Yang L, Jiang J, Lau MS, et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010 doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- Costa Y, Ding J, Theunissen TW, Faiola F, Hore TA, Shliaha PV, Fidalgo M, Saunders A, Lawrence M, Dietmann S, et al. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature. 2013;495:370–374. doi: 10.1038/nature11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger AK, de Vries I, Kittler R, Junqueira M, Shevchenko A, Schulz H, et al. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell. 2009;4:403–415. doi: 10.1016/j.stem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Dos Santos RL, Tosti L, Radzisheuskaya A, Caballero IM, Kaji K, Hendrich B, Silva JC. MBD3/NuRD Facilitates Induction of Pluripotency in a Context-Dependent Manner. Cell Stem Cell. 2014 doi: 10.1016/j.stem.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidalgo M, Faiola F, Pereira CF, Ding J, Saunders A, Gingold J, Schaniel C, Lemischka IR, Silva JC, Wang J. Zfp281 mediates Nanog autorepression through recruitment of the NuRD complex and inhibits somatic cell reprogramming. Proc Natl Acad Sci U S A. 2012;109:6. doi: 10.1073/pnas.1208533109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailesellasse Sene K, Porter CJ, Palidwor G, Perez-Iratxeta C, Muro EM, Campbell PA, Rudnicki MA, Andrade-Navarro MA. Gene function in early mouse embryonic stem cell differentiation. BMC genomics. 2007;8:85. doi: 10.1186/1471-2164-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson J, Rafiee M, Reiland S, Polo J, Gehring J, Okawa S, Huber W, Hochedlinger K, Krijgsveld J. Highly Coordinated Proteome Dynamics during Reprogramming of Somatic Cells to Pluripotency. Cell reports. 2012;2:1579–1592. doi: 10.1016/j.celrep.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27:459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006;8:285–292. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- Kataoka H, Murayama T, Yokode M, Mori S, Sano H, Ozaki H, Yokota Y, Nishikawa S, Kita T. A novel snail-related transcription factor Smuc regulates basic helix-loop-helix transcription factor activities via specific E-box motifs. Nucleic Acids Res. 2000;28:626–633. doi: 10.1093/nar/28.2.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch H, McEwen K, Turp A, Encheva V, Carroll T, Grabole N, Mansfield W, Nashun B, Knezovich J, Smith A, et al. Naive pluripotency is associated with global DNA hypomethylation. Nature structural & molecular biology. 2013;20:311–316. doi: 10.1038/nsmb.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardo TR, Schultheisz HL, Loring JF, Laurent LC. The functions of microRNAs in pluripotency and reprogramming. Nat Cell Biol. 2012;14:1114–1121. doi: 10.1038/ncb2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Lin Y, Li XY, Willis AL, Liu C, Chen G, Weiss SJ. Snail1-dependent control of embryonic stem cell pluripotency and lineage commitment. Nat Commun. 2014;5:3070. doi: 10.1038/ncomms4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CW, Li CH, Peng YJ, Cheng YW, Chen HW, Liao PL, Kang JJ, Yeng MH. Snail regulates Nanog status during the epithelial-mesenchymal transition via the Smad1/Akt/GSK3beta signaling pathway in non-small-cell lung cancer. Oncotarget. 2014;5:3880–3894. doi: 10.18632/oncotarget.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Sun H, Qi J, Wang L, He S, Liu J, Feng C, Chen C, Li W, Guo Y, et al. Sequential introduction of reprogramming factors reveals a time-sensitive requirement for individual factors and a sequential EMT-MET mechanism for optimal reprogramming. Nat Cell Biol. 2013;15:829–838. doi: 10.1038/ncb2765. [DOI] [PubMed] [Google Scholar]
- Lomeli H, Starling C, Gridley T. Epiblast-specific Snai1 deletion results in embryonic lethality due to multiple vascular defects. BMC research notes. 2009;2:22. doi: 10.1186/1756-0500-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent AE, Newkirk KM, Kusewitt DF. Slug (Snai2) expression during skin and hair follicle development. J Invest Dermatol. 2010;130:1737–1739. doi: 10.1038/jid.2010.22. [DOI] [PubMed] [Google Scholar]
- Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol. 2004;24:306–319. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Losada J, Sanchez-Martin M, Rodriguez-Garcia A, Sanchez ML, Orfao A, Flores T, Sanchez-Garcia I. Zinc-finger transcription factor Slug contributes to the function of the stem cell factor c-kit signaling pathway. Blood. 2002;100:1274–1286. [PubMed] [Google Scholar]
- Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Hochedlinger K. When fibroblasts MET iPSCs. Cell Stem Cell. 2010;7:5–6. doi: 10.1016/j.stem.2010.05.018. [DOI] [PubMed] [Google Scholar]
- Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, Viukov S, Mansour AA, Caspi I, Krupalnik V, Zerbib M, et al. Deterministic direct reprogramming of somatic cells to pluripotency. Nature. 2013 doi: 10.1038/nature12587. [DOI] [PubMed] [Google Scholar]
- Reynolds N, Latos P, Hynes-Allen A, Loos R, Leaford D, O’Shaughnessy A, Mosaku O, Signolet J, Brennecke P, Kalkan T, et al. NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell Stem Cell. 2012;10:583–594. doi: 10.1016/j.stem.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Schaniel C, Ang YS, Ratnakumar K, Cormier C, James T, Bernstein E, Lemischka IR, Paddison PJ. Smarcc1/Baf155 couples self-renewal gene repression with changes in chromatin structure in mouse embryonic stem cells. Stem Cells. 2009;27:2979–2991. doi: 10.1002/stem.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaniel C, Lee DF, Lemischka IR. Exploration of self-renewal and pluripotency in ES cells using RNAi. Methods in enzymology. 2010;477:351–365. doi: 10.1016/S0076-6879(10)77018-X. [DOI] [PubMed] [Google Scholar]
- Schwarz BA, Bar-Nur O, Silva JC, Hochedlinger K. Nanog Is Dispensable for the Generation of Induced Pluripotent Stem Cells. Curr Biol. 2014 doi: 10.1016/j.cub.2013.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemens H, Jackstadt R, Hunten S, Kaller M, Menssen A, Gotz U, Hermeking H. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10:4256–4271. doi: 10.4161/cc.10.24.18552. [DOI] [PubMed] [Google Scholar]
- Silva J, Barrandon O, Nichols J, Kawaguchi J, Theunissen TW, Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS biology. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, Wray J, Yamanaka S, Chambers I, Smith A. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian V, Klattenhoff CA, Boyer LA. Screening for novel regulators of embryonic stem cell identity. Cell Stem Cell. 2009;4:377–378. doi: 10.1016/j.stem.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tanabe K, Nakamura M, Narita M, Takahashi K, Yamanaka S. Maturation, not initiation, is the major roadblock during reprogramming toward pluripotency from human fibroblasts. Proc Natl Acad Sci U S A. 2013;110:12172–12179. doi: 10.1073/pnas.1310291110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka-Matakatsu M, Uemura T, Oda H, Takeichi M, Hayashi S. Cadherin-mediated cell adhesion and cell motility in Drosophila trachea regulated by the transcription factor Escargot. Development. 1996;122:3697–3705. doi: 10.1242/dev.122.12.3697. [DOI] [PubMed] [Google Scholar]
- Theunissen TW, van Oosten AL, Castelo-Branco G, Hall J, Smith A, Silva JC. Nanog overcomes reprogramming barriers and induces pluripotency in minimal conditions. Curr Biol. 2011;21:65–71. doi: 10.1016/j.cub.2010.11.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- Wu Y, Zhou BP. Snail: More than EMT. Cell adhesion & migration. 2010;4:199–203. doi: 10.4161/cam.4.2.10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S-H, Kalkan T, Morrisroe C, Smith A, Sharrocks A. A genome-wide RNAi screen reveals MAP kinase phosphatases as key ERK pathway regulators during embryonic stem cell differentiation. PLoS genetics. 2012:8. doi: 10.1371/journal.pgen.1003112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Kang Y. Multilayer control of the EMT master regulators. Oncogene. 2013 doi: 10.1038/onc.2013.128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Additional Unpooled siRNA and shRNA Validation of Candidates with Immunofluorescence and Flow Cytometry. Related to Figure 1.
(A) Results from the RNAi primary screen on median cell fluorescence. A selected subset of the outlier and control genes is labeled.
(B) Representative fluorescent images from a Non-Targeting siRNA-treated well. For each well treated with a given siRNA, nuclei-stained cells are identified and levels of GFP are measured.
(C) Levels of GFP for each siRNA-treated well and controls. Dots denote effect averaged across technical triplicate. Short black horizontal lines are average of 4 unpooled siRNA effects. Territory outside of long horizontal lines in grey denotes FDR <0.05 using the empty controls. Single star before gene name indicates 2 of 4 distinct siRNAs are significant with FDR<0.05, with two stars if 3–4 of distinct siRNAs are significant. Further details are in the Supplementary Information.
(D) Relative knockdown efficiencies of shRNAs in pLKO.1 vector. Values are normalized to shLuciferase (shLuci). Error bars indicate average ± SD.
(E) Venn diagram for genes assayed (left) and changed >2-fold following 2 days 2 μM RA or EB differentiation or considered RNAi screen candidates (right). Genes in shaded central region on left are assayed in all datasets.
(F) Contingency table for genes assayed in all datasets and changed following RA or EB differentiation or considered RNAi screen candidates. “+” and “−” indicate inclusion (overlapping) and exclusion (non-overlapping), respectively, in the pairwise comparisons.
Figure S2. Loss-of-Function Studies of Snai1 and Snai2 in Nanog-Driven Pre-iPSC Reprogramming. Related to Figure 2.
(A) Schematic overview of Nanog-dependent NS pre-iPSC reprogramming.
(B) Relative gene expression during the pre-iPSC reprogramming process. The time points of gene expression analysis are indicated. Relative gene expression in NS, rOKM and rOKM+Nanog (N) cells was measured by qRT-PCR. Data are normalized to Gapdh.
(C) Schematic overview of pre-iPSC reprogramming.
(D) Representative images of Nanog±shSnai2 reprogramming of Oct4-GFP reporter pre-iPSCs. shRNA is marked by mCherry. Data from one shRNA are shown.
(E) Images of AP stained colonies generated as indicated in D.
(F) Quantitation of AP+ colonies at day 10 after reprogramming with indicated shRNAs. Data are represented as average ± SD of 2 independent shRNAs against each Snai1 and Snai2.
(G) Schematic overview of pre-iPSC reprogramming in MEFs harboring the Nanog-GFP reporter transgene.
(H) Representative images of shSnai2 reprogramming of MEF-derived pre-iPSCs. shRNA vector is marked by mCherry and final iPSCs are positive for Nanog-GFP reporter activity.
(I) Quantitation of Nanog-GFP+ colonies at day 10 after reprogramming with indicated shRNAs. Data are represented as average ± SD of 2 independent shRNAs against each Snai1 and Snai2.
(J) Cell cycle analyses by flow cytometry upon Snai1/2 depletion in pre-iPSCs (rOKM) with or without ectopic Nanog expression.
Figure S3. Gain-of-Function Studies of Snai1 and Snai2 in Nanog-Driven Pre-iPSC Reprogramming. Related to Figure 3.
(A) Combinations of empty vector (EV), Nanog, Snai1, and Snai2 were used to generate stable pre-iPSC clones. Western blots confirm transgenic Nanog, 3xFlag-Snai1 or 3xFlag-Snai2 protein expression in pre-iPSCs cultured in serum+LIF.
(B) Representative images of iPSCs generated from overexpression of Snai1 and Snai2 in the presence or absence or ectopic Nanog for both Oct4-GFP (left) and AP staining (right) at day 10 of 2i/LIF treatment.
(C) Cell cycle analyses by flow cytometry upon Snai1/2 ectopic expression in pre-iPSCs (rOKM) with or without ectopic Nanog expression.
(D) Fluorescence and bright-field images of transgene-free Nanog and Nanog+Snai1 iPSCs cultured under serum+LIF or 2i/LIF for 30 days.
(E) Immunofluorescence analysis against H3K27me3 in female transgene-free iPSCs under serum/LIF or 2i/LIF culture. Nuclei were stained with DAPI.
(F) Alkaline phosphatase (AP) and immunofluorescence staining for SSEA-1, Nanog, Oct4, and Sox2 protein levels in transgene-free Nanog and Nanog+Snai1 iPSCs.
(G) Similar expression of pluripotency and lineage specific marker genes in transgene-free Nanog and Nanog+Snai1 iPSCs. Error bars indicate average ± SD. Note that the similar Snai1 levels between Nanog+EV and Nanog+Snai1 samples indicate the complete removal of the ectopic Snai1 in Nanog+Snai1 iPSCs.
(H) Similar gene expression pattern of transgene-free Nanog and Nanog+Snai1 iPSCs during differentiation induced by LIF-withdrawal, retinoic acid (RA) treatment, or embryoid body (EB) formation. Gene expression analysis was measured by qRT-PCR, and data were normalized to Gapdh.
(I) Schematic overview of the reprogramming strategy in MEF-derived pre-iPSCs with Snai1/2 gain-of-function and Nanog ectopic expression.
(J) Quantification of Nanog-GFP+ colonies for noted treatments.
(K) CoIP validation of the Nanog-Snai1 interaction in 293T cells.
Figure S4. The Nanog and Snai1 Partnership in Transcriptional Regulation of Pluripotency-Associated Genes and miRNAs. Related to Figure 4.
(A) Relative expression levels of RNAi hit genes that are potential targets of miR-290–295 (left) and miR-Let-7 (right) in pre-iPSCs and iPSCs. Scatter plots of global gene expression arrays from published microarrays (Sridharan et al., 2009) showing highest (“pre-iPSC genes”, left panel) or lowest (“iPSC genes”, right panel) relative expression (log2>2) in pre-iPSCs compared to iPSCs. Predicted target genes for the mouse miR-290–295 cluster are shown as blue dots, and those predicted for the mouse miR-Let7 family are shown as red dots. Blue and red bars indicate percentages of total pre-iPSC genes and iPS genes that are predicted targets of miR-290–295 and miR-Let7, respectively. Specific genes are indicated with arrows.
(B) Relative expression of the “pre-iPS genes” (left panel) or “iPSC genes” (right panel) measured by quantitative RT-PCR in the NS-derived reprogramming intermediates transduced with the indicated shRNA and cultured in serum/LIF.
Figure S5. Snai2 Does Not Compete with Snai1 in Binding to the Nanog Sites at the Lin28 and miR-290–295 Loci. Related to Figure 5.
(A) Pie plots showing the percentage of Nanog peaks containing the consensus E-Box motif (CANNTG) in two published Nanog ChIP-Seq datasets. Number and frequency of individual E-Box motifs within the Nanog peaks are listed.
(B) Nanog ChIP and qPCR analyses on the miR-290–295 and Lin28 loci. Snai2 does not affect Nanog binding to the regulatory regions in both miR-290–295 and Lin28a genes in pre-iPSCs ectopically expressing the indicated factors. Peaks “1–7” and “Peak2” are Nanog positive binding peaks on miR290–295 and Lin28a regulatory regions, respectively. Peaks “a”, “g” and “Peak1” are negative control regions. Error bars indicate average ± SD.
Figure S6. Snai1 is Dispensable for ESC Maintenance. Related to Figure 6.
(A) Snai1 knockdown (left panel) and transgenic (right) efficiency by shRNA and ectopic expression in CCE ESCs. Knockdown efficiency represents the average of two independent shRNAs against Snai1. Error bars indicate average ± SD.
(B) Pluripotency and lineage-specific marker analysis in ESCs transduced with two independent shRNAs against Snai1 or shEV (left panel) and in transgenic stable lines with ectopic expression of Snai1 or EV (right panel). Error bars indicate average ± SD.
(C) Colony formation assay (CFA) demonstrating lack of effect of Snai1 depletion (left panel) or enforced expression (right panel) on self-renewal. Colonies stained for AP (Alkaline Phosphatase) were scored into three categories (uniformly undifferentiated, partially differentiated, and fully differentiated). Error bars indicate average ± SD (n=3).
(D) ESC proliferation is independent of Snai1. Growth curve of CCE ESCs transduced with a shRNA against Snai1 or shEV (left panel) or with enforced Snai1 or EV expression (right panel). Cell numbers were counted every another day. Error bars indicate average ± SD (n=3).
(E) Nanog and Rex1 expression are not affected by Snai1 depletion (top panel) or enforced expression (bottom panel). Two independent ESC lines were used to measure the GFP activity under the Nanog (NG4 ES line) or Rex1 (Rex1-dGFP ES line) promoter.
Table S1. RNAi Screen Studies in ESCs, Related to Figure 1.
Supplementary Table S3, Related to Figures 1–5.
List of qRT-PCR, ChIP-qPCR primer sequences as well as siRNA/shRNA sequences used in this study.
Supplementary Table S4, Related to Figure S4.
List of genes differentially expressed in pre-iPSCs and iPSCs that are predicted to be targets of miR-290-295 cluster and Let-7 family miRNAs.