Abstract
Methylation of lysine 27 on histone 3 (H3K27me), a modification associated with gene repression, plays a critical role in regulating the expression of genes that determine the balance between cell differentiation and proliferation. Alteration of the level of this histone modification has emerged as a recurrent theme in many types of cancer, demonstrating that either excess or lack of H3K27 methylation can have oncogenic effects. Cancer genome sequencing has revealed the genetic basis of H3K27me deregulation, including mutations of the components of the H3K27 methyltransferase complex PRC2 and accessory proteins, and deletions and inactivating mutations of the H3K27 demethylase UTX in a wide variety of neoplasms. More recently, mutations of lysine 27 on histone 3 itself were shown to prevent H3K27me in pediatric glioblastomas. Aberrant expression or mutations in proteins that recognize H3K27me3 also occur in cancer and may result in misinterpretation of this mark. Additionally, due to the crosstalk between different epigenetic modifications, alterations of chromatin modifiers controlling H3K36me, or even mutations of this residue, can ultimately regulate H3K27me levels and distribution across the genome. The significance of mutations altering H3K27me is underscored by the fact that many tumors harboring such lesions often have a poor clinical outcome. New therapeutic approaches targeting aberrant H3K27 methylation include small molecules that block the action of mutant EZH2 in germinal center-derived lymphoma. Understanding the biological consequences and gene expression pathways affected by aberrant H3K27 methylation may also lead to other new therapeutic strategies.
Background
The phenotype and fate of a given cell depends on a precise control of gene expression that determines the set of genes that are expressed at a specific moment. Local chromatin configuration at gene promoters and enhancers determines DNA accessibility to transcription machinery and factors involved in chromatin looping that brings promoters and enhancers into close proximity. Epigenetic modifications, namely DNA methylation at CpG sites, and covalent modifications of the N-terminal tails of core histones, are critical regulators of chromatin structure and ultimately gene expression. Over the past decade, aberrant epigenetic regulation and alteration of histone modifications have emerged as a recurrent theme in malignancy.
The amount and distribution of a specific histone modification can be pathologically altered by aberrant expression or function of the enzymes that place the modification (“writers”), dysfunction of the enzymes that remove the mark (“erasers”), or by mutations of the histone that prevent the residue from being modified. Histone modifications are intricately coordinated, and alterations of a histone mark can affect the levels and distribution of other modifications. In addition, alterations in factors that recognize the modification (“readers”) can result in an aberrant functional outcome of the mark. In this review, we will focus on mechanisms that lead to altered methylation of lysine 27 on histone H3 (H3K27me), a common feature in many types of cancer.
The presence of trimethylation of H3K27 (H3K27me3) at promoter regions is associated with gene repression. This modification is generated by the Polycomb repressive complex 2 (PRC2), composed of the SET domain-containing histone methyltransferase (HMT) EZH2 (enhancer of zeste homolog 2) or its functional homologue EZH1, and core accessory proteins (EED, SUZ12, and RbAp48) (Fig. 1A). The H3K27me3 mark is recognized by the PRC1 complex, which represses transcription by several mechanisms, including ubiquitination of histone H2A on lysine K119 and chromatin compaction (1). Despite its repressive function, H3K27me3 is found together with the activation-associated H3K4me3 mark at the promoters of “bivalent genes”. These genes, characteristic of embryonic stem cells (ESC) (2), are poised for either activation or repression. Upon differentiation, either H3K27me3 or H3K4me3 is lost, leading to gene activation or repression, respectively. Accordingly, EZH2 and the PRC2 complex are essential for normal differentiation of ESCs (3).
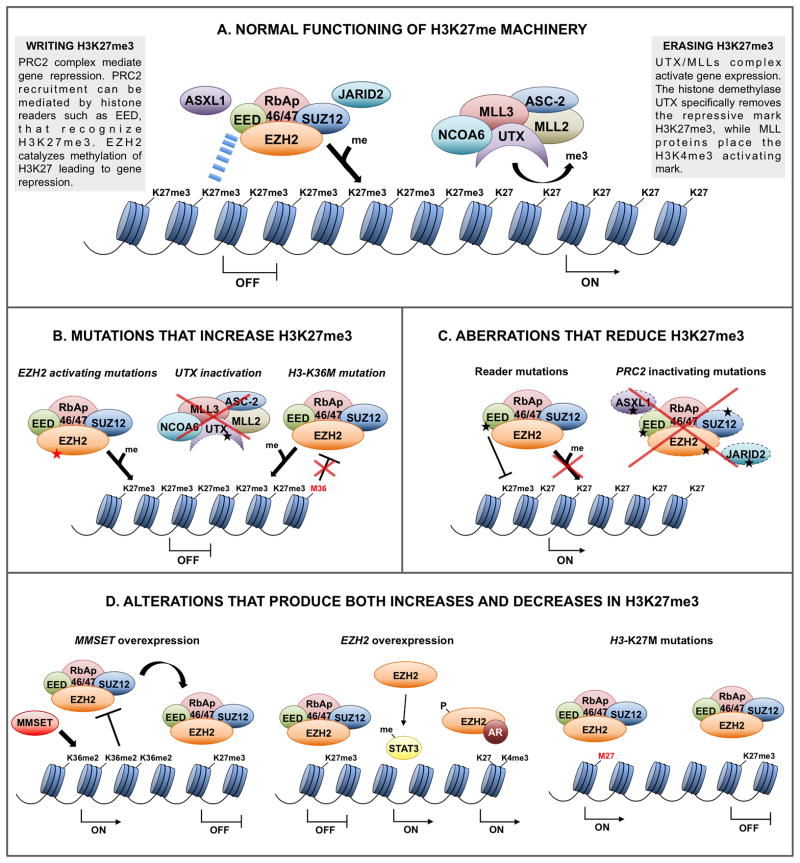
Figure 1.
A. H3K27me3 writing is carried out by the PRC2 complex accompanied by accessory proteins such as JARID2 and ASXL1. One mechanism of PRC2 recruitment is the recognition of H3K27me3 by the PRC2-component EED, which leads to self-propagation of this mark. EZH2 catalyzes H3K27 methylation leading to H3K27me3 and gene repression. H3K27me3 erasing is done by the histone demethylase UTX, which specifically removes the repressive mark H3K27me3 leading to gene activation. B. Alterations that produce increases in the levels of H3K27me3 include: activating mutations of EZH2, inactivating mutations of the histone demethylase UTX that lead to failure in the removal of this mark, and K36M mutations in histone H3. H3-K36M mutant provokes a decreased in global levels of H3K36me. The decrease of this mark will likely result in a lack of inhibition of PRC2, increased levels of H3K27me3 and gene repression. C. Aberrations that lead to reduced levels of H3K27me3 include inactivating mutations of the PRC2 components and accessory proteins, and mutations in readers of H3K27me3. For example, mutations in EED inhibit the recognition of H3K27me3 by this factor, leading to a failure in the recruitment of PRC2 complex and the propagation of H3K27me3. D. Some aberrations lead to both increases and decreases of H3K27me3 at different loci. MMSET induces higher global levels of H3K36me2, which impede PRC2 binding. PRC2 is displaced and further represses loci that are able to maintain H3K27me. Overexpression of EZH2 increases PRC2 activity and H3K27me3 at specific loci. High levels of EZH2 also methylate and activate STAT3 to promote gene activation. Phosphorylated EZH2 associates with androgen receptor (AR) and activates gene expression. H3.3-K27M mutations sequester the PRC2 complex provoking a global decrease of H3K27me3, although some loci show increased H3K27me3 and gene inactivation due to PRC2 redistribution.
Removal of di and tri-methyl groups from H3K27 is performed by the histone demethylases UTX/KMD6A and JMJD3/KDM6B, which contain a JmjC (Jumonji) catalytic domain (4, 5) (Fig. 1A). UTX is encoded on the X chromosome but escapes X inactivation in females (6). This protein is part of a transcriptional activator complex including the MLL2/MLL3 H3K4 methyltransferases, suggesting a concerted mechanism in which repressive H3K27 methyl marks are removed and H3K4 is methylated to activate transcription (5). UTX (7) and JMJD3 (8) are also required for ESC differentiation, underscoring that the regulated resolution of the bivalent state is critical for normal development. Given the critical role of the H3K27me in temporal and spatial control of gene expression, it is not surprising that mutations of the machinery that creates, removes and interprets this chromatin modification play a role in malignancy (1).
Clinical-Translational Advances
Alterations in H3K27me “writers”
EZH2 overexpression
Overexpression of EZH2 is found in a wide variety of cancers (9). In solid tumors, high levels of this factor are associated with aggressive biology, metastasis and poor clinical outcome (9–11). Accordingly, experimental overexpression of EZH2 promotes proliferation, anchorage-independent growth, migration, invasion and metastasis (9), with oncogenic activity dependent on the SET domain, and presumably, on its ability to mark histones for gene repression. Genome-wide analyses demonstrated that of in cancer-associated EZH2 overexpression leads to increased H3K27me3 and repression of genes involved in the differentiation of cancer SCs, such as p16 and p19, and tumor suppressor genes such as RUNX3 and CDH1 (9, 12) (Fig. 1D). In contrast, overexpression of EZH2 was more recently associated with lower global levels of H3K27me3 in castration-resistant PCa (13). In this system, phosphorylated EZH2 did not associate with the PRC2 complex but bound the androgen receptor; and was present at actively transcribed genes (Fig. 1D). This activity required the SET domain of EZH2 (13), suggesting that methylation of non-histone targets may also be relevant for the oncogenic action of EZH2. As another example of this alternative action, in glioblastoma, phosphorylated EZH2 methylates STAT3, increasing activation of this oncogenic transcription factor (14) (Fig. 1D).
EZH2 mutations
In 2010, cancer genome sequencing identified heterozygous somatic missense mutations of EZH2 in 7% of follicular lymphoma and up to 22% of germinal center B-cell-like diffuse large B-cell lymphoma (GCB-DLBCL) (15). These mutations are found within the SET domain of EZH2 at tyrosine 641 (Y641N, F, S, or H), alanine 677 (A677G) and alanine 687 (A687V) (15–17). Such mutations result in a change-of-function of EZH2 and, unlike EZH2 overexpression, lead to a marked genome-wide increase in the abundance of H3K27me3 (18). While wild-type EZH2 efficiently monomethylates H3K27, it has weaker activity for the subsequent reactions that lead to di- and tri-methylation. By contrast, mutations in Y641 block the ability of EZH2 to modify unmethylated substrates while increasing its efficiency in converting mono- or dimethylated histones, created by wild-type EZH2, to the tri-methylated state (19, 20) (Fig. 1B). EZH2 A687V works similarly, whereas EZH2 A677G has enhanced catalytic activity converting all un-, mono- and dimethylated histones to H3K27me3 (16, 17). During B-cell development, EZH2 is expressed in GC cells where it establishes a specific chromatin landscape, comprised of repressed and bivalent loci, silencing of cell cycle checkpoints and differentiation factors and allowing for the expansion of B-cells. With B-cell maturation, EZH2 activity is overcome, leading to gene activation and allowing cells to exit the GC compartment, differentiate into plasma cells and cease division. In GC B-cells, the Y641 mutation enhances H3K27 trimethylation and results in an exaggerated silencing of EZH2 targets, blocking differentiation and promoting proliferation and tumor formation (18).
In contrast to lymphoma, in T-cell acute lymphoblastic leukemia (T-ALL) and myeloid malignancies, a range missense, nonsense and frameshift mutations of EZH2 occur (21–23). These lesions can be homozygous, are found throughout the gene and generally are predicted to disable HMT activity. Accordingly, cell lines harboring inactivating EZH2 mutations show a global decrease in H3K27me3 (21) (Fig.1C). The biological consequences of these mutations were elucidated in T-ALL, where the PRC2 complex normally competes with NOTCH1, a major driver of this disease, at specific target genes. Thus, loss of EZH2 favors NOTCH1-driven oncogenic gene activation in T-ALL (23). In myeloid neoplasms, EZH2 mutations are associated with poor outcome, and are more common in patients with myelodysplastic syndromes (MDS) and myeloproliferative neoplasms (MPN) and rare in de novo acute myeloid leukemia (AML) (21, 24–26). While targets in myeloid human malignancies have yet to be identified, deletion of Ezh2 in mice produces a drastic reduction in H3K27me and de-repression of potential oncogenes such as Hmga2, Pbx3, Lmo1 and Myc target genes, leading to an MDS/MPN-like phenotype (27).
The identification of EZH2 mutations with opposing outcomes on H3K27me highlights the importance of the balance of this histone mark for cell homeostasis, and suggests that EZH2 target genes may differ among tissues. Moreover, while EZH2 inhibition may be therapeutic in lymphoma, it could be deleterious in other cell-types, emphasizing the need for comprehensive preclinical mechanistic studies before embarking on such epigenetic therapies.
Mutations in Polycomb-group associated proteins
Alterations in other members of the PRC2 complex and associated proteins have also been identified. Missense and inactivating mutations of SUZ12, EED and the PCR2-associated factor JARID2 were found in MPN and MDS, and mimic EZH2 mutations, leading to reduced PRC2 activity (28–30). Recurrent somatic mutations and deletions of ASXL1, encoding another PRC2-associated factor (31), occur in patients with MDS, MPN, and AML, and predict poor survival (24, 25, 32). Most of these mutations are found within the last exon of the gene, yielding a truncated protein. Accordingly, truncated forms of ASXL1 inhibit myeloid differentiation and induce MDS-like disease in mice (33). ChIP-seq studies suggest that in the absence of ASXL1, PRC2 neither binds chromatin nor methylates H3K27, resulting in a global loss of H3K27me3. As a result, many genes are aberrantly activated, including the HOXA cluster, contributing to myeloid transformation (31) (Fig. 1C).
Alterations in H3K27me erasers
While aberrations in JMJD3 are rare (34, 35), somatic inactivating mutations and deletions of UTX are more common in hematological malignancies and solid tumors (36–38). UTX lesions tend to be homozygous in females, and to be accompanied by genomic loss of its homologue UTY in males, suggesting a tumor suppressor role (39). Supporting this idea, loss of UTX enhances proliferation of cancer cells in many contexts (39–41). However, this may be a tissue-specific effect, as UTX overexpression in breast cancer promotes proliferation and invasion (42). UTX expression rises in parallel with MLL2/KMT2D in breast cancer (42), whereas inactivating mutations of UTX and MLL2 in bladder cancer are mutually exclusive (35). These findings suggest that the balance of H3K27/H3K4 methylation promoted by the UTX/MLL2 complex is critical for cell homeostasis, and thus, overrepresentation or disruption of this complex can lead to oncogenesis. Aberrations in this complex may explain the presence of cancer-specific bivalent genes, which show decreased expression and are associated with drug resistance and tumor progression in ovarian cancer (43). While in Drosophila UTX mutants have increased global levels of H3K27me3, UTX aberrations in human cancers lead to only local alterations of this mark (39, 44, 45) (Fig. 1B). Retinoblastoma (RB)-binding proteins and HOX genes are regulated by UTX in fibroblasts and ESCs, and their aberrant repression in the absence of UTX alters cell cycle and differentiation regulation (7, 41). In breast cancer, pro-invasive and cell cycle genes are upregulated upon UTX overexpression, correlating with H3K27me3 loss (42). These studies suggest that the subset of genes showing altered H3K27me3 upon UTX deregulation is cell-type specific, and that similarly to EZH2, UTX may act as an oncogene or tumor suppressor depending on the cellular context. Thus, additional effort is needed to understand how alterations of UTX deregulate the epigenome in specific cancers.
Histone H3 mutations
Remarkably, recurrent disease-driving mutations are even found in the genes encoding histones. A prime example is H3F3A, the gene encoding the histone variant H3.3, in which heterozygous mutations (K27M and G34R/V) are found in diffuse intrinsic pontine glioma and in up to 60% of cases of pediatric glioblastoma multiforme (46, 47), where the presence of the K27M-H3.3 predicts poor survival (47–50). Cell lines and human specimens harboring the K27M-H3.3 mutation display reduced levels of H3K27me2/3, and DNA hypomethylation at many loci (48, 51, 52). Similarly, enforced expression of H3K27M reduces genome-wide levels of H3K27me3. This can be explained by the ability of the H3K27M mutant to inhibit the PRC2 complex by interacting with the SET domain of EZH2, as well as SUZ12, competing with normal histone binding and turnover. However, mutant cells also display enhanced EZH2 recruitment, increased H3K27me3 and repression of specific loci. Hence, aberrant gene repression as well as activation may occur through redistribution of the PRC2 complex (Fig. 1D). Accordingly, glioblastoma samples harboring histone mutations show a distinct gene expression and DNA methylation profile (47, 50, 53).
Histone mutations may also affect H3K27me3 in an indirect way. In a high fraction of cases of chondroblastoma and giant cell bone tumors, a mutation of lysine 36 histone 3 to methionine (K36M) is found (54). There is crosstalk between the H3K36 and H3K27 methyl marks, as the PRC2 complex is unable to methylate H3K27 in the presence of H3K36me2/me3 (55, 56). Thus, the global reduction of H3K36 methylation that K36M mutants provoke (51) may result in a lack of PRC2-inhibition and expansion of the H3K27me3 mark, leading to aberrant repression of many loci (Fig. 1B). Experiments are needed to determine the nature of genes deregulated in such tumors.
Alterations in H3K27me readers
The functional outcome of methylated H3K27 depends on the recruitment of effector molecules that recognize this modification through WD40 and chromo domains. Aberrations in these “readers” may therefore result in lack of H3K27me interpretation. The PRC2 component EED recognizes H3K27me3, promoting the self-propagation of this modification. Mutations in the WD40 motifs of EED are found in myeloid malignancies (30, 57, 58), and interrupt the interaction between EED and H3K27me3 leading to reduced PRC2 activity (57) (Fig. 1C). The chromo domain protein CDYL also participates in the propagation of H3K27me3, bringing EZH2 to sites harboring this mark (59). Loss of heterozygosity of the CDYL locus is frequent in cervical cancer and correlates with poor prognosis. Furthermore, decreased CDYL expression contributes to transformation by promoting de-repression of the proto-oncogene TrkC (60). Missense and nonsense mutations in the chromo domain of CDYL and its homolog CDYL2 (35), may also affect H3K27me3 recognition. CBX proteins are components of the PRC1 complex that recognize H3K27me3 and target PRC1 to specific loci. CBX7 is critical for the formation of H3K9me3, which is recognized by HP-1 (heterochromatin protein 1) leading to gene silencing. CBX7 is a tumor suppressor that is downregulated in many types of cancer, leading to de-repression of cell cycle genes such as CCNE1 (61). Paradoxically, overexpression of CBX7 is also pathogenic as it enhances self-renewal and induces leukemia, an effect that requires binding to H3K27me3 (62). Misinterpretation of H3K27me3 by these and other readers may therefore result in effects as drastic as those produced by aberrations in writers and erasers. However, more efforts are needed to fully characterize the mechanisms of action of these mutations.
Alterations of other chromatin regulators
Due to the balance between H3K27 and H3K36 methylation, aberrations in chromatin modifiers regulating H3K36me2/3 may indirectly affect H3K27me. MMSET/NSD2 is a H3K36-specific HMT overexpressed in multiple myeloma (MM) cases harboring the translocation t(4;14), which is associated with poor prognosis (63). MMSET drives cancer cell proliferation, clonogenicity and an invasive phenotype (64, 65). MMSET overexpression provokes a genome-wide increase in H3K36me2 and a concomitant reduction in H3K27me3. In these cells, EZH2 is unable to bind and methylate sites that show increased H3K36me2, and is relocated to loci that maintain H3K27me, leading to increased H3K27me3 and further repression of those genes (66) (Fig. 1D). MMSET-overexpressing cells show higher sensitivity to small molecules targeting EZH2 (EZH2i), indicating that H3K27me3-mediated repression is relevant to the molecular pathogenesis of this form of malignancy. Recently, a recurrent mutation (E1099K) in MMSET was identified in pediatric B-ALL and mantle cell lymphoma (67–69). This mutation lies within the catalytic SET domain of MMSET and mimics the effects of MMSET overexpression, promoting cell growth and the chromatin switch between H3K36me2 and H3K27me3.
NSD1, a close homologue of MMSET, is fused to the NUP98 locus in rare cases of AML to generate the NUP98-NSD1 fusion protein. Expression of NUP98-NSD1 in mice stimulates expression of Hoxa9 and Meis1 proto-oncogenes and induces AML. These effects depend on the HMT activity of NSD1, which provokes local increases in H3K36me3, and concomitant loss of EZH2 and H3K27me3 enrichment across these genes (70).
Several studies suggest that the chromatin remodeling complex SWI/SNF antagonizes polycomb proteins in the control of gene expression. Loss of the SWI/SNF component SNF5 occurs in aggressive cancers including malignant rhabdoid tumors. SNF5 loss produces loci-specific increases in H3K27me3, due to a lack of polycomb protein displacement by SWI/SNF and increased EZH2 levels (71).
Targeting deregulated H3K27me
Due to the reversible nature of epigenetic modifications, chromatin regulators such as EZH2 have emerged as potential therapeutic targets. The first agent identified to target EZH2 was DZNep (Deazaneplanocin A), which inhibits SAH-hydrolase, a required cofactor for several HMTs. DZNep leads to degradation of EZH2 and associated proteins, reactivation of PRC2 target genes and antitumor activity (9). Nevertheless, its short plasma half-life and lack of specificity led to the development of highly selective molecules (GlaxoSmithKline, Epizyme) that directly target EZH2 and compete with the cofactor SAM binding at the active site (18, 72–74). Some of these compounds have high selectivity for mutant-EZH2 lymphoma cells, leading to de-repressed expression of PRC2 target genes, decreased tumor growth, and apoptosis (18, 72). Moreover, GCB but not activated B-cell-type DLBCL cells respond to these inhibitors irrespectively of EZH2 status (18). Interestingly, high global levels of H3K27me3 may not always dictate sensitivity to EZH2i. MMSET overexpression decreases global levels of H3K27me3 but sensitizes MM cells to EZH2i. Local increases of this mark at specific loci seem to guide their sensitivity, presumably due to the functional outcome of those repressed genes. Rhabdoid tumors lacking the SNF5 SWI/SNF protein, which promotes local H3K27me3-increases, display enhanced sensitivity to EZH2i (75). In the case of loss of UTX, it remains to be elucidated whether localized increases in H3K27me3 also sensitize these cells to EZH2i. A more detailed understanding these altered epigenomes is critical to ascertain which cancer subtypes could benefit from these drugs.
T-ALL and myeloid malignancies show decreased H3K27me due to inactivation of different PRC2 components. Finding a therapeutic strategy for such tumors may require the identification of biologically relevant transcription factors that normally compete with the PRC2 complex (such as NOTCH1 in T-ALL), and/or genes whose upregulation upon H3K27me loss contributes to transformation. Another therapeutic approach to be explored is the use of Jumonji H3K27 inhibitor molecules (76), which would impede the removal of the aberrantly low H3K27me levels and perhaps restore repression. Developing an epigenetic targeting strategy for tumors bearing K27M-H3.3 mutations also seems complex, as these aberrations lead to both global decreases and local increases in H3K27me2/3. Careful study of the pathways and genes altered in these cells will aid in the identification of putative therapeutic approaches.
Taken together, alteration of H3K27me has emerged as a hallmark of transformation in many types of cancer, and a platform for development of new therapies to overcome the aberrant function of the writers or erasers of the modification. One of the most recent approaches to epigenetic therapy has been the development of molecules that block the “readers” of the H4K20 methyl mark (77). Likewise, compounds targeting H3K27me “readers” may be developed to treat cancers showing a gain of function of these factors. Lastly, elucidation of the pathways deregulated by alterations of H3K27me in specific tumor types may lead to new understanding of cancer biology and development of targeted therapy.
Acknowledgments
This work was supported in part by a Leukemia & Lymphoma Society Specialized Center of Research award (7006-13; to J.D. Licht), the Multiple Myeloma Research Foundation Epigenetic Program (J.D. Licht), and an NCI Physical Sciences Oncology Center grant (U54CA143869; to J.D. Licht).
Footnotes
Disclosure of Potential Conflicts of Interest
J.D. Licht reports receiving a commercial research grant from Epizyme and is a consultant/advisory board member for Abbvie, Bristol-Myers Squibb, Celgene, and GlaxoSmithKline. No potential conflicts of interest were disclosed by the other author.
References
- 1.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–9. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 3.Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–79. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci U S A. 2007;104:18439–44. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–4. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 6.Greenfield A, Carrel L, Pennisi D, Philippe C, Quaderi N, Siggers P, et al. The UTX gene escapes X inactivation in mice and humans. Hum Mol Genet. 1998;7:737–42. doi: 10.1093/hmg/7.4.737. [DOI] [PubMed] [Google Scholar]
- 7.Morales Torres C, Laugesen A, Helin K. Utx is required for proper induction of ectoderm and mesoderm during differentiation of embryonic stem cells. PLoS One. 2013;8:e60020. doi: 10.1371/journal.pone.0060020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burgold T, Spreafico F, De Santa F, Totaro MG, Prosperini E, Natoli G, et al. The histone H3 lysine 27-specific demethylase Jmjd3 is required for neural commitment. PLoS One. 2008;3:e3034. doi: 10.1371/journal.pone.0003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deb G, Singh AK, Gupta S. EZH2: Not EZHY (easy) to deal. Mol Cancer Res. 2014;12:639–53. doi: 10.1158/1541-7786.MCR-13-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100:11606–11. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–9. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 12.Kim J, Yu J. Interrogating genomic and epigenomic data to understand prostate cancer. Biochim Biophys Acta. 2012;1825:186–96. doi: 10.1016/j.bbcan.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465–9. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23:839–52. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–5. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCabe MT, Graves AP, Ganji G, Diaz E, Halsey WS, Jiang Y, et al. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27) Proc Natl Acad Sci U S A. 2012;109:2989–94. doi: 10.1073/pnas.1116418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Majer CR, Jin L, Scott MP, Knutson SK, Kuntz KW, Keilhack H, et al. A687V EZH2 is a gain-of-function mutation found in lymphoma patients. FEBS Lett. 2012;586:3448–51. doi: 10.1016/j.febslet.2012.07.066. [DOI] [PubMed] [Google Scholar]
- 18.Beguelin W, Popovic R, Teater M, Jiang Y, Bunting KL, Rosen M, et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell. 2013;23:677–92. doi: 10.1016/j.ccr.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, Richon VM, et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci U S A. 2010;107:20980–5. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yap DB, Chu J, Berg T, Schapira M, Cheng SW, Moradian A, et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117:2451–9. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–6. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 22.Nikoloski G, Langemeijer SM, Kuiper RP, Knops R, Massop M, Tonnissen ER, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010;42:665–7. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 23.Ntziachristos P, Tsirigos A, Van Vlierberghe P, Nedjic J, Trimarchi T, Flaherty MS, et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med. 2012;18:298–301. doi: 10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abdel-Wahab O, Pardanani A, Patel J, Wadleigh M, Lasho T, Heguy A, et al. Concomitant analysis of EZH2 and ASXL1 mutations in myelofibrosis, chronic myelomonocytic leukemia and blast-phase myeloproliferative neoplasms. Leukemia. 2011;25:1200–2. doi: 10.1038/leu.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bejar R, Stevenson K, Abdel-Wahab O, Galili N, Nilsson B, Garcia-Manero G, et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364:2496–506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel JP, Gonen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–89. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muto T, Sashida G, Oshima M, Wendt GR, Mochizuki-Kashio M, Nagata Y, et al. Concurrent loss of Ezh2 and Tet2 cooperates in the pathogenesis of myelodysplastic disorders. J Exp Med. 2013;210:2627–39. doi: 10.1084/jem.20131144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brecqueville M, Cervera N, Adelaide J, Rey J, Carbuccia N, Chaffanet M, et al. Mutations and deletions of the SUZ12 polycomb gene in myeloproliferative neoplasms. Blood Cancer J. 2011;1:e33. doi: 10.1038/bcj.2011.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brecqueville M, Rey J, Bertucci F, Coppin E, Finetti P, Carbuccia N, et al. Mutation analysis of ASXL1, CBL, DNMT3A, IDH1, IDH2, JAK2, MPL, NF1, SF3B1, SUZ12, and TET2 in myeloproliferative neoplasms. Genes Chromosomes Cancer. 2012;51:743–55. doi: 10.1002/gcc.21960. [DOI] [PubMed] [Google Scholar]
- 30.Score J, Hidalgo-Curtis C, Jones AV, Winkelmann N, Skinner A, Ward D, et al. Inactivation of polycomb repressive complex 2 components in myeloproliferative and myelodysplastic/myeloproliferative neoplasms. Blood. 2012;119:1208–13. doi: 10.1182/blood-2011-07-367243. [DOI] [PubMed] [Google Scholar]
- 31.Abdel-Wahab O, Adli M, LaFave LM, Gao J, Hricik T, Shih AH, et al. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell. 2012;22:180–93. doi: 10.1016/j.ccr.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boultwood J, Perry J, Pellagatti A, Fernandez-Mercado M, Fernandez-Santamaria C, Calasanz MJ, et al. Frequent mutation of the polycomb-associated gene ASXL1 in the myelodysplastic syndromes and in acute myeloid leukemia. Leukemia. 2010;24:1062–5. doi: 10.1038/leu.2010.20. [DOI] [PubMed] [Google Scholar]
- 33.Inoue D, Kitaura J, Togami K, Nishimura K, Enomoto Y, Uchida T, et al. Myelodysplastic syndromes are induced by histone methylation-altering ASXL1 mutations. J Clin Invest. 2013;123:4627–40. doi: 10.1172/JCI70739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cbioportal.org. cBioPortal for Cancer Genomics. 2014 [cited; Available from: http://www.cbioportal.org/public-portal/
- 36.Van der Meulen J, Speleman F, Van Vlierberghe P. The H3K27me3 demethylase UTX in normal development and disease. Epigenetics. 2014;9:658–668. doi: 10.4161/epi.28298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43:875–8. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ross JS, Wang K, Al-Rohil RN, Nazeer T, Sheehan CE, Otto GA, et al. Advanced urothelial carcinoma: next-generation sequencing reveals diverse genomic alterations and targets of therapy. Mod Pathol. 2014;27:271–80. doi: 10.1038/modpathol.2013.135. [DOI] [PubMed] [Google Scholar]
- 39.van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–3. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ho AS, Kannan K, Roy DM, Morris LG, Ganly I, Katabi N, et al. The mutational landscape of adenoid cystic carcinoma. Nat Genet. 2013;45:791–8. doi: 10.1038/ng.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang JK, Tsai MC, Poulin G, Adler AS, Chen S, Liu H, et al. The histone demethylase UTX enables RB-dependent cell fate control. Genes Dev. 2010;24:327–32. doi: 10.1101/gad.1882610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim JH, Sharma A, Dhar SS, Lee SH, Gu B, Chan CH, et al. UTX and MLL4 Coordinately regulate transcriptional programs for cell proliferation and invasiveness in breast cancer cells. Cancer Res. 2014;74:1705–17. doi: 10.1158/0008-5472.CAN-13-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chapman-Rothe N, Curry E, Zeller C, Liber D, Stronach E, Gabra H, et al. Chromatin H3K27me3/H3K4me3 histone marks define gene sets in high-grade serous ovarian cancer that distinguish malignant, tumour-sustaining and chemo-resistant ovarian tumour cells. Oncogene. 2013;32:4586–92. doi: 10.1038/onc.2012.477. [DOI] [PubMed] [Google Scholar]
- 44.Herz HM, Madden LD, Chen Z, Bolduc C, Buff E, Gupta R, et al. The H3K27me3 demethylase dUTX is a suppressor of Notch- and Rb-dependent tumors in Drosophila. Mol Cell Biol. 2010;30:2485–97. doi: 10.1128/MCB.01633-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jankowska AM, Makishima H, Tiu RV, Szpurka H, Huang Y, Traina F, et al. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, and DNMT3A. Blood. 2011;118:3932–41. doi: 10.1182/blood-2010-10-311019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44:251–3. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, et al. Driver mutations in histone H3. 3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–31. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 48.Chan KM, Fang D, Gan H, Hashizume R, Yu C, Schroeder M, et al. The histone H3. 3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 2013;27:985–90. doi: 10.1101/gad.217778.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khuong-Quang DA, Buczkowicz P, Rakopoulos P, Liu XY, Fontebasso AM, Bouffet E, et al. K27M mutation in histone H3. 3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol. 2012;124:439–47. doi: 10.1007/s00401-012-0998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22:425–37. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 51.Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–61. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bender S, Tang Y, Lindroth AM, Hovestadt V, Jones DT, Kool M, et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013;24:660–72. doi: 10.1016/j.ccr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 53.Bjerke L, Mackay A, Nandhabalan M, Burford A, Jury A, Popov S, et al. Histone H3.3 mutations drive pediatric glioblastoma through upregulation of MYCN. Cancer Discov. 2013 doi: 10.1158/2159-8290.CD-12-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Behjati S, Tarpey PS, Presneau N, Scheipl S, Pillay N, Van Loo P, et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet. 2013;45:1479–82. doi: 10.1038/ng.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng Y, Sweet SM, Popovic R, Martinez-Garcia E, Tipton JD, Thomas PM, et al. Total kinetic analysis reveals how combinatorial methylation patterns are established on lysines 27 and 36 of histone H3. Proc Natl Acad Sci U S A. 2012;109:13549–54. doi: 10.1073/pnas.1205707109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem. 2011;286:7983–9. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ueda T, Sanada M, Matsui H, Yamasaki N, Honda ZI, Shih LY, et al. EED mutants impair polycomb repressive complex 2 in myelodysplastic syndrome and related neoplasms. Leukemia. 2012;26:2557–60. doi: 10.1038/leu.2012.146. [DOI] [PubMed] [Google Scholar]
- 58.Khan SN, Jankowska AM, Mahfouz R, Dunbar AJ, Sugimoto Y, Hosono N, et al. Multiple mechanisms deregulate EZH2 and histone H3 lysine 27 epigenetic changes in myeloid malignancies. Leukemia. 2013;27:1301–9. doi: 10.1038/leu.2013.80. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y, Yang X, Gui B, Xie G, Zhang D, Shang Y, et al. Corepressor protein CDYL functions as a molecular bridge between polycomb repressor complex 2 and repressive chromatin mark trimethylated histone lysine 27. J Biol Chem. 2011;286:42414–25. doi: 10.1074/jbc.M111.271064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mulligan P, Westbrook TF, Ottinger M, Pavlova N, Chang B, Macia E, et al. CDYL bridges REST and histone methyltransferases for gene repression and suppression of cellular transformation. Mol Cell. 2008;32:718–26. doi: 10.1016/j.molcel.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Forzati F, Federico A, Pallante P, Abbate A, Esposito F, Malapelle U, et al. CBX7 is a tumor suppressor in mice and humans. J Clin Invest. 2012;122:612–23. doi: 10.1172/JCI58620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Klauke K, Radulovic V, Broekhuis M, Weersing E, Zwart E, Olthof S, et al. Polycomb Cbx family members mediate the balance between haematopoietic stem cell self-renewal and differentiation. Nat Cell Biol. 2013;15:353–62. doi: 10.1038/ncb2701. [DOI] [PubMed] [Google Scholar]
- 63.Keats JJ, Maxwell CA, Taylor BJ, Hendzel MJ, Chesi M, Bergsagel PL, et al. Overexpression of transcripts originating from the MMSET locus characterizes all t(4;14)(p16;q32) -positive multiple myeloma patients. Blood. 2005;105:4060–9. doi: 10.1182/blood-2004-09-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martinez-Garcia E, Popovic R, Min DJ, Sweet SM, Thomas PM, Zamdborg L, et al. The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood. 2011;117:211–20. doi: 10.1182/blood-2010-07-298349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ezponda T, Popovic R, Shah MY, Martinez-Garcia E, Zheng Y, Min DJ, et al. The histone methyltransferase MMSET/WHSC1 activates TWIST1 to promote an epithelial-mesenchymal transition and invasive properties of prostate cancer. Oncogene. 2013;32:2882–90. doi: 10.1038/onc.2012.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Popovic R, Martinez E, Zhang Q, Ezponda T, Jiang Y, Shah MY, et al. MMSET dysregulates gene expression in myeloma through global and focal changes in H3K36 and H3K27 methylation. ASH Annual Meeting Abstracts. 2012;120:523. [Google Scholar]
- 67.Oyer JA, Huang X, Zheng Y, Shim J, Ezponda T, Carpenter Z, et al. Point mutation E1099K in MMSET/NSD2 enhances its methyltranferase activity and leads to altered global chromatin methylation in lymphoid malignancies. Leukemia. 2014;28:198–201. doi: 10.1038/leu.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jaffe JD, Wang Y, Chan HM, Zhang J, Huether R, Kryukov GV, et al. Global chromatin profiling reveals NSD2 mutations in pediatric acute lymphoblastic leukemia. Nat Genet. 2013;45:1386–91. doi: 10.1038/ng.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bea S, Valdes-Mas R, Navarro A, Salaverria I, Martin-Garcia D, Jares P, et al. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc Natl Acad Sci U S A. 2013;110:18250–5. doi: 10.1073/pnas.1314608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol. 2007;9:804–12. doi: 10.1038/ncb1608. [DOI] [PubMed] [Google Scholar]
- 71.Wilson BG, Wang X, Shen X, McKenna ES, Lemieux ME, Cho YJ, et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell. 2010;18:316–28. doi: 10.1016/j.ccr.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–12. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 73.Kim W, Bird GH, Neff T, Guo G, Kerenyi MA, Walensky LD, et al. Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nat Chem Biol. 2013;9:643–50. doi: 10.1038/nchembio.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Knutson SK, Kawano S, Minoshima Y, Warholic NM, Huang KC, Xiao Y, et al. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol Cancer Ther. 2014;13:842–54. doi: 10.1158/1535-7163.MCT-13-0773. [DOI] [PubMed] [Google Scholar]
- 75.Knutson SK, Warholic NM, Wigle TJ, Klaus CR, Allain CJ, Raimondi A, et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc Natl Acad Sci U S A. 2013;110:7922–7. doi: 10.1073/pnas.1303800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 2012;488:404–8. doi: 10.1038/nature11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.James LI, Barsyte-Lovejoy D, Zhong N, Krichevsky L, Korboukh VK, Herold JM, et al. Discovery of a chemical probe for the L3MBTL3 methyllysine reader domain. Nat Chem Biol. 2013;9:184–91. doi: 10.1038/nchembio.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]