Abstract
RNA interference (RNAi) is a powerful tool for target identification and can lead to novel therapies for pharmacologically intractable targets such as KRAS. RNAi therapy must combine potent siRNA payloads with reliable in vivo delivery for efficient target inhibition. We employed a functional “Sensor” assay to establish a library of potent siRNAs against RAS pathway genes and show they efficiently suppress their targets at low dose. This reduces off-target effects and enables combination gene knockdown. We administered Sensor siRNAs in vitro and in vivo and validated the delivery of KRAS siRNA alone and siRNA targeting the complete RAF effector node (A/B/C-RAF) as promising strategies to treat KRAS-mutant colorectal cancer. We further demonstrate that improved therapeutic efficacy is achieved by formulating siRNA payloads that combine both single-gene siRNA and node-targeted siRNAs (KRAS+PIK3C-A/B). The customizable nature of Sensor siRNA payloads offers a universal platform for combination target identification and development of RNAi therapeutics.
Keywords: RNAi, KRAS, nanoparticle, colorectal cancer, RNAi off-target effects
Introduction
Mutations in the Ras family of small GTPases, particularly KRAS, occur in 30% of all human cancers and are often associated with resistance to chemo- and targeted- therapy. Somatic mutations lock Ras in the GTP-bound state, leading to constitutive activation of its downstream effector pathways. Despite intense efforts, pharmacological inhibition of KRAS itself (1) and inhibition of individual effector kinases downstream of KRAS such as RAF (2, 3) and MEK (4-6) have so far been unsuccessful in treating KRAS-mutant tumors. Combinations of MEK and PI3K inhibitors are currently being evaluated in clinical trials, though toxicity in normal tissue could limit their therapeutic window (7). Thus, effective and targeted treatment against KRAS-driven cancer is an urgent and unmet clinical need.
RNA interference (RNAi) provides an alternative therapeutic approach to small molecule and antibody-based therapeutics for inhibiting gene function. RNAi can, in principle, be applied to reversibly silence any target gene [reviewed in (8, 9)], thereby increasing the druggable landscape from 10% to virtually 100% of the genome (10, 11). Since all siRNAs bear structural similarity and are likely to have comparable pharmacokinetic profiles, their use as therapeutics would facilitate drug formulation, pre-clinical testing and development of combination therapies. While delivery is still a major challenge, more reliable lipid and polymeric nanoparticles are in development to deliver siRNA payloads to target tissues [reviewed in (12)]. Regardless, treatment efficacy would benefit from developing optimized siRNA payloads that potently and specifically silence well-validated target genes at low dose.
Beyond its potential as a therapeutic modality, RNAi is a useful tool for identifying and validating new drug targets. This has proven particularly powerful in cancer research, where shRNA or siRNA screens have been employed to identify genes that are selectively required for the proliferation and survival of cancer cells. However, identification and current in silico prediction of effective shRNAs and siRNAs remains imprecise, resulting in low success rates. Consequently, screening of high-order sh/siRNA combinations is not possible without first establishing a collection of functionally validated RNAi triggers. Additionally, off-target effects, which can be due to sequence-dependent and -independent gene deregulation [reviewed (13)], must be minimized for meaningful interpretation of phenotypic outcomes.
To overcome these limitations, we employed a previously described “Sensor” assay (14) to generate a functionally validated library of RNAi sequences against RAS pathway genes. We show that Sensor siRNAs efficiently ablate their gene targets at low nanomolar concentrations in vitro, which decreased off-target effects and enabled the use of high-order siRNA combinations to co-deplete multiple genes. By applying Sensor siRNAs in vitro and in vivo, we identify single-gene and combination-gene payloads that inhibit the growth of KRAS-mutant colorectal cancer. Thus, the use of Sensor siRNAs for target discovery and development of customized siRNA payloads can lead to nanoparticle-based treatments for KRAS-mutant cancer and provide a blueprint for similar strategies to target other key nodes in cancer maintenance.
Results
A functionally validated RNAi library targeting KRAS pathway genes
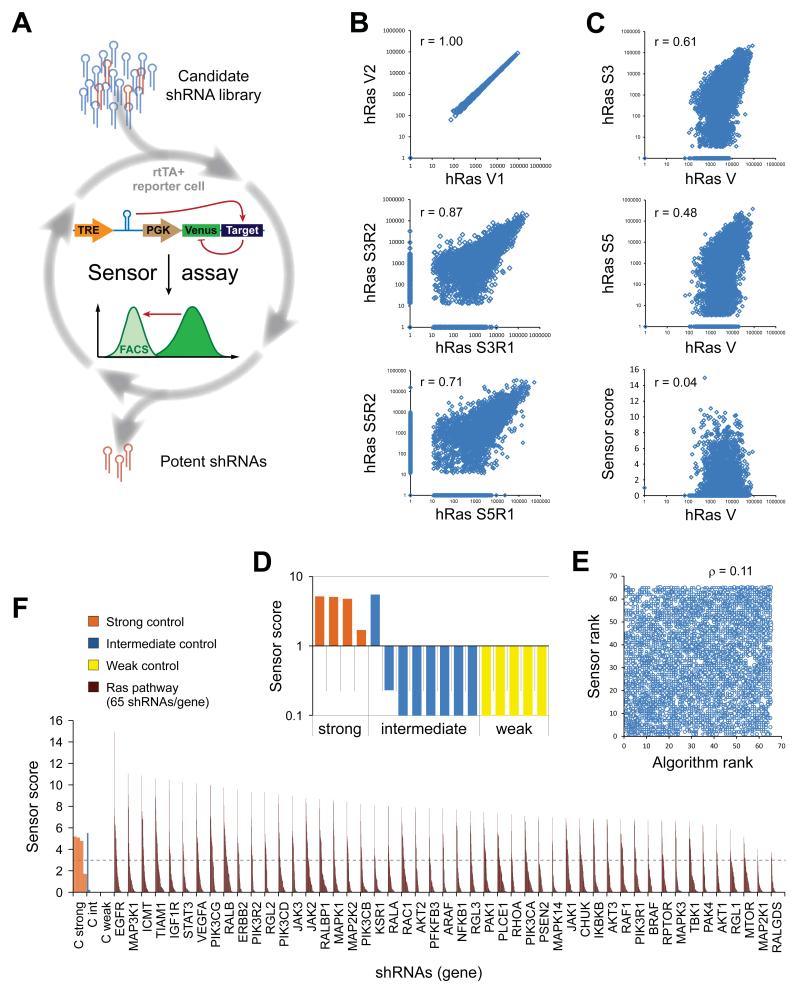
To identify potent shRNAs targeting the Ras network, we applied the Sensor assay to evaluate candidate shRNA sequences targeting 75 human genes and their mouse orthologs (Fig. 1A). These genes encode many classes of proteins, including kinases, GTPases and transcription factors. The Sensor assay interrogates large numbers of shRNAs under conditions of single genomic integration (“single-copy”) for their ability to repress a cognate target sequence placed downstream of a fluorescent reporter expressed in cis (14). While we have previously established this assay and shown its potential to identify very potent shRNA sequences through full gene tiling, here we apply it to evaluate the efficiency of pre-selected candidate sequences targeting a much larger set of genes to establish a functionally validated RNAi library. To assemble the initial candidates, 65 shRNAs per gene were selected using a combination of bioinformatics predictions (15) and “Sensor rules” requiring shRNA-specific features (14). This resulted in a total of ~10,000 shRNAs (split into sets targeting mouse and human genes), including 17 control shRNAs added at 1× representation to monitor the functional behavior and stoichiometric fluctuations of the assay. Deep sequencing after two-step cloning of the shRNA-Sensor libraries (14) revealed that >99.8% of all designed human (4958/4967) and mouse (4828/4837) Ras set vectors were successfully constructed.
Figure 1. Functionally validated Ras pathway RNAi libraries for potent and specific gene silencing.
(A) The Sensor assay enables the generation of functionally validated shRNA libraries. The potency of candidate shRNAs was biologically probed in a pooled assay by quantifying knockdown of a Venus reporter cDNA fused to the shRNA’s cognate target site. (B-F) Human Ras set (hRas) Sensor assay results. Comparable results were obtained for the mouse Ras set (mRas, Table S1 and S2). (B) Correlations in read numbers of technical vector pool replicates (V1, V2), and biological duplicates (R1, R2) at different selection stages of the Sensor assay (S3, Sort 3; S5, Sort 5; r, Pearson correlation coefficient). (C) Correlations in read numbers between the initial vector library (mean of technical duplicates) and the endpoint populations after the indicated sorts (geometric mean of biological replicates), and the final Sensor scores. (D) Sensor scores of control shRNAs of strong (orange), intermediate (blue) or weak (yellow) potency. (E) Rank correlation between input (Algorithm rank) and output library (Sensor rank), showing that the Sensor rank is not predicted by the informatics tool (ρ, Spearman rank correlation coefficient). (F) Sensor shRNA scores for a selection of direct “Ras effector” genes (for full list see Table S2). The dotted line indicates a threshold for potent shRNAs, based on known controls. 65 shRNAs/gene; controls are represented multiple times for better visibility.
To evaluate shRNA efficiency through enrichment of potent shRNAs and depletion of non-functional constructs, five iterative rounds of fluorescence-activated cell sorting (FACS) were run (Sort 1-5) with gates set to progressively select for only the most potent, functional shRNAs (14). Compared to our previous libraries generated by complete tiling of transcripts, the preselected Ras pathway libraries showed a less drastic reduction in shRNA complexity during early selection cycles (Fig. S1A and S1B), indicating that the bioinformatics shRNA pre-selection process enriched the pools for potent shRNAs. Importantly, shRNA representation in independent biological replicates correlated throughout the sorting procedure (Fig. 1B and Table S1), while the correlation to the initial population was progressively lost, showing that the assay can reproducibly identify potent shRNAs regardless of their initial representation in the library (Fig. 1C and Table S1).
The recovery of all positive control shRNAs indicates that the assay robustly enriched for potent single-copy shRNAs, while depleting weak and intermediate ones (Fig. 1D and Table S1). Strikingly, there was nearly no correlation between the initial algorithmic rank and the rank obtained through the biological Sensor assay (Fig. 1E), showing that prediction tools alone are still insufficient to select the most potent shRNAs (see also Figure S1C and S1D). Conversely, the Sensor assay identified potent shRNAs for nearly every gene, with top ranked shRNAs showing similar scores across all genes (Fig. 1F and Table S2). More than 91% of the top 5 human (344/375) and >88% of the top 5 mouse (333/375) shRNAs scored >3 (Fig. S1C and S1D), a threshold score defined by positive control shRNAs. This indicates that the pre-selection process of 65 shRNAs per gene provided sufficient coverage to identify several very potent shRNAs per gene.
Sensor siRNAs are potent and on-target
Synthetic shRNAs and siRNAs enter the endogenous microRNA pathway at different stages, but ultimately use the same conserved machinery to down-regulate their target genes. We thus hypothesized that potent shRNA sequences could be directly converted into potent siRNA triggers. Based on top-scoring Sensor shRNA sequences, we generated corresponding 22-mer Sensor siRNAs targeting human KRAS, KSR1, ARAF, BRAF, RAF1/CRAF, MAP2K1, MAP2K2, MAPK1, MAPK3, PIK3CA, PIK3CB, PIK3CD, AKT1, AKT2 and AKT3. We then measured the knockdown efficiency of three top Sensor shRNAs and two corresponding siRNAs targeting endogenous KRAS and found that Sensor siRNAs retain >80% mRNA knockdown when transfected at concentrations as low as 0.5 nM (Fig. 2A, 2B, and S2A). We extended this validation to ARAF, BRAF and RAF1 siRNAs in a human osteosarcoma cell line, U2OS, which does not have mutations in the RAS/MAPK pathway and is not sensitive to knockdown of these genes. Transfection of 4 out of 6 of these Sensor siRNAs resulted in >70% mRNA and >80% protein knockdown at 0.1 nM, and all 6 siRNAs conferred >80% protein knockdown at 0.5 nM and 2 nM (Fig. 2C and S2A). Lastly, we tested at least 2 of the top scoring siRNAs for select genes in U2OS cells. Among 46 Sensor siRNAs tested, ~90% (41) knocked down their target mRNA at >70% when transfected at 2 nM (Fig. 2D). The Sensor assay thus serves as a powerful and reliable strategy to identify potent siRNA sequences to generate functionally validated sh/siRNA libraries.
Figure 2. Sensor siRNA potency and KRAS siRNA sensitivity in CRC lines.
(A) SW1116 cells expressing an shRNA-resistant HA-KRASG12V cDNA were infected with one of three KRAS shRNAs or a negative control (sh.Ren.713), and knockdown of endogenous KRAS protein was measured. (B-C) U2OS cells were transfected with varying concentrations of Sensor siRNAs against KRAS (B) or RAF kinases (C) for 72h and knockdown of endogenous protein was measured (See Figure S2A for quantification). (D) Top-ranked siRNAs against selected Ras pathway genes were transfected at 2nM into U2OS cells, and mRNA levels were measured by RT-qPCR 72h post-transfection. (E) Colorectal cancer cell lines with the indicated KRAS and BRAF mutational status were transfected with siKRAS_234 or siKRAS_355 at 5nM, and cell viability was correlated between the two siRNAs. Four of the 5 KRAS mutant lines were sensitive to KRAS depletion whereas KRAS WT lines were resistant. (F) SW1116 cells carrying a doxycycline-inducible siRNA-resistant HA-KRASG12V construct were treated ± doxycycline (100ng/ml) 60 hours prior to siRNA transfection. Cell lysates were collected 48h post-siRNA transfection for western blot analysis. (G) SW1116 cells carrying doxycycline-inducible HA-KRASG12V or HA-KRASWT constructs were treated ± doxycycline (100ng/ml) 60 hours prior to siRNA transfection. Cell viability (normalized to uninduced cells) was assessed 5 days post-siRNA transfection.
To biologically validate KRAS Sensor siRNAs for specificity, we transfected two siRNAs, siKRAS_234 and siKRAS_355, into a panel of 9 human colorectal cancer (CRC) cell lines with or without mutant KRAS alleles. We observed a strong correlation between siKRAS-induced cell death and KRAS mutational status and a strong correlation between the behaviors of the two siRNAs (Fig. 2E). The anti-proliferative effect of the siRNAs in KRAS mutant cells was attributable to apoptosis (Fig. S2B), though as previously reported, heterogeneity amongst cell lines was observed (16). To verify that the cell death observed in KRAS mutant cells was an on-target effect, we generated SW1116 cells (KRAS-mutant and KRAS-dependent) stably expressing an siRNA-resistant, tetracycline-inducible KRASG12V cDNA (Table S2). As expected, both KRAS siRNAs effectively knocked down endogenous KRAS, which led to a decrease in phospho-MEK and phospho-ERK (Fig. 2F) and decreased cell viability (Fig. 2G). Doxycycline-induced expression of siRNA-resistant KRASG12V, but not siRNA-resistant KRASWT cDNA, was sufficient to restore phospho-MEK and phospho-ERK (Fig. 2F) and rescue cell viability (Fig. 2G). Together, these results indicate that siKRAS_234 and siKRAS_355 are potent at low dose and their cytotoxic effect on KRAS mutant cells is due to KRAS depletion.
Low dose shRNA and siRNA minimizes off-target effects
Off-target effects remain a major concern with RNAi experiments and can occur through several mechanisms, including (i) sequence-dependent cross-hybridization to unintended mRNA targets (17, 18); (ii) sequence-independent saturation of the RNAi machinery (19); and (iii) other as yet undefined mechanisms. As competition of RNAi triggers for incorporation and retention in RISC is concentration dependent (20) and since bulk transfected siRNAs can outcompete endogenous miRNAs for RISC loading (19), we set out to specifically investigate dose-dependent RNAi off-target effects by testing shRNAs and siRNAs in cells where their cognate target genes are absent. In such a system, any changes in gene or miRNA expression should result from off-target effects.
We analyzed shRNA-based off-target effects by transducing Trp53−/− MEFs (21) at single- and high-copy with six well-characterized, potent and weak Trp53 shRNAs (14). Small RNA sequencing showed that high-copy shRNA transduction resulted on average in 20-70 fold more shRNA-derived mature small RNAs than single-copy transduction (Fig. 3A), suggesting that each high-copy transduced cell contained at least 20-70 viral integrations. Interestingly, endogenous microRNA levels were unchanged at both conditions (Fig. S3A and S3B), raising the possibility that common gene perturbations might be due to unintended competing endogenous RNA (ceRNA)-like sponge effects of vector-based mRNA transcripts (22) or increased engagement of the transcription and translation machinery. Gene expression analysis revealed several hundred sequence-based off-target effects at high-copy (550 up and 301 down, p-value <0.05), whereas only 7 altered genes were found under single-copy conditions (Fig. S3C). Gene perturbations commonly found across all 6 shRNAs numbered in the thousands at high-copy transduction (1821 up and 2910 down, p-value <0.05), but again were absent at single-copy (Fig. 3B and S3D). Interestingly, potent and weak shRNAs perturbed gene expression to a similar degree. We also observed that potent Sensor shRNAs lead to preferential retention of the designed guide strands (Fig. S3E) (14), which might reduce passenger strand mediated off-target effects, and thus designed all Sensor siRNAs as 22-mers – rather than conventional 21-mers – to guard the same sequence properties for both strands.
Figure 3. Low dose shRNA and siRNA minimizes off-target effects.
(A) Trp53−/− MEFs were infected at single- or high-copy with one of 6 Trp53 shRNAs. RNA deep-sequencing showed that high-copy shRNA transduction resulted in increased levels of mature Trp53 shRNAs in all cases. Endogenous microRNA expression was unperturbed (see also S3A, S3B, S3E and Table S3 for details; r, Pearson correlation coefficient). (B) Microarray analysis was performed on Trp53−/− MEFs not expressing any shRNA (WT and empty vector) or transduced at single- or high-copy with one of 6 Trp53 shRNAs. Shown are the top 1500 genes that are sequence-independently up- and down-regulated across all 6 shRNAs. Significant perturbations were observed only at high-copy transduction (p<0.05). For more details see Figure S3C and S3D. (C) Gene expression changes were analyzed in Kras−/− mouse embryonic fibroblasts transfected with Sensor siKRAS_234 or siKRAS_355 at various concentrations after 72h. Unsupervised clustering of significantly downregulated genes (>2-fold) showed a similar gene expression pattern between control cells and cells transfected with low concentrations (0.2 and 2 nM) of siRNAs. More profound gene perturbation occurred in cells transfected with high concentrations (20 and 50 nM) of siRNAs (see also Figure S3F-H).
To test dose-dependent off-target effects of such Sensor-derived siRNAs, we transfected siKRAS_234 and siKRAS_355, which target both human and mouse KRAS, into Kras−/− mouse embryonic fibroblasts (23) at 0.2, 2, 20, and 50 nM and measured global gene expression changes. Higher concentrations (20 and 50nM) of siRNAs caused significant gene expression changes (>2-fold down-regulated genes: 12-16 for siKRAS_234 and 63-93 for siKRAS_355), whereas lower concentrations (0.2 and 2 nM) perturbed only a limited numbers of genes (>2-fold down-regulated genes: 0-1 for siKRAS_234 and 3-6 for siKRAS_355; Fig. 3C and Fig. S3F-G). Additionally, we found that at 20 and 50 nM, siKRAS_355 down-regulated more genes than siKRAS_234 and that the differentially down-regulated genes were largely non-overlapping (Fig. S3H), indicating that these effects are sequence-dependent. Together, our data show that while both shRNAs and siRNAs can induce aberrant gene perturbations at high concentrations, lowering their dosage does not eliminate but critically reduces both sequence-specific and general off-target effects.
Sensor siRNAs can be used in high-order combinations to identify therapeutic siRNA cocktails
We next tested whether the high potency of Sensor siRNAs allows for low-dose transfection of multiple siRNA species without affecting the efficiency of individual siRNAs. We performed high-order combination siRNA delivery into U2OS cells, such that individual siRNAs were transfected at 2 nM each in a pool of 4 different siRNAs. The majority of Sensor siRNAs exhibited no decrease in potency when transfected in combination compared to when transfected alone (Fig. S4A). At the protein level, transfection of a complex pool of 7 potent Sensor siRNAs at 2 nM each, targeting ARAF, BRAF, RAF1/CRAF, MEK1, MEK2, ERK1 and ERK2 resulted in strikingly efficient depletion of all 7 proteins simultaneously (Fig. 4A). Notably, the potency of each siRNA was not compromised as siRNA complexity increased. By contrast, for siRNAs that are less potent, their knockdown efficiency was only preserved when pooled with equally weak siRNAs, but further declined when pooled with potent siRNAs (Fig. S4B). Apparently, only siRNAs of equally high potency can be used in high-order combinations to consistently achieve robust multi-gene knockdown, further emphasizing the value of the Sensor assay in identifying such equally potent sequences across various genes.
Figure 4. Sensor siRNAs can be used in high-order combinations.
(A) U2OS cells were transfected with Sensor siRNAs targeting MAPK pathway genes. Each siRNA was transfected at 2nM ± a non-targeting siNEG for a total of 14nM siRNA. Target protein knockdown was assessed by western blot 72h post-transfection. (B) Viability of CRC cells was measured 5 days post-siRNA transfection or treatment with PLX4032 (1□M). (C) Apoptosis of CRC cells was assessed by measuring caspase-3/7 activity 72h post-siRNA transfection. Apoptosis was adjusted by viability and normalized to a siNEG-transfected control. (D) CRC cells were treated for 72h with 5nM of the indicated siRNA(s) ± siNEG for a total of 30nM siRNA. Where indicated, cells were treated with AZD6244 (1□M), BEZ235 (1□M) or PLX4032 (10□M for SW48, Caco2, SW620 and SW1116; 1□M for RKO and LS411N) for 24h. Lysates were analyzed for pMEK, pERK, pAKT and pS6 levels with the ViBE bioanalyzer (ND, not determined).
Next, we set out to use Sensor siRNAs to identify siRNA combinations that could selectively inhibit the proliferation of KRAS-mutant cell lines. As shown above (Fig. 2E), siRNAs targeting KRAS itself are one viable option. To identify alternative targets, we assessed the functional dependency of different RAF and PI3K isoforms in CRC cells by depleting single and multiple isoforms in cell lines carrying wild-type KRAS and BRAF alleles (Caco-2 and SW48), mutant KRAS alleles (SW1116 and SW620) and mutant BRAF alleles (RKO and LS411N). Cell viability was measured to identify lethal siRNA combinations and downstream signaling was monitored to validate on-target knockdown.
We first investigated how KRAS mutational status affects sensitivity to knockdown of various RAF isoforms. Caco-2 and SW48 cells, two KRAS-wildtype cell lines, were relatively resistant to individual and combinations of RAF isoform knockdown, as measured by cell viability. As expected, the two BRAF-mutant cell lines, RKO and LS411N, were uniquely sensitive to BRAF depletion (Fig. 4B), indicative of their addiction to the BRAF oncogene. The KRAS-mutant cell lines, SW1116 and SW620, showed partial sensitivity to the depletion of either BRAF or CRAF (Fig. 4B). This finding is consistent with recent reports that CRAF plays a critical role in RAS-driven lung carcinoma and melanoma (24-26). Combined depletion of BRAF and CRAF in these cells resulted in additive toxicity, though depletion of all three RAF isoforms led to the strongest reduction in cell viability that was comparable to that of KRAS depletion.
To investigate the mechanism by which co-depletion of multiple RAF isoforms in KRAS-mutant cells phenocopies KRAS knockdown, we analyzed apoptosis, ERK phosphorylation and the accumulation of the pro-apoptotic protein BIM in these cells. Progressive depletion of RAF isoforms in the KRAS-mutant SW620 cells led to progressive reduction in phospho-ERK (pERK) levels, such that B/CRAF and A/B/CRAF co-depletion reduced pERK to levels comparable to those seen upon KRAS knockdown (Fig. S4C). This correlated with higher accumulation of BIM protein (Fig. S4C). Furthermore, co-depletion of multiple RAFs led to a stronger apoptotic response in the KRAS-mutant cells (Figure 4C), indicating that depletion of multiple RAF isoforms is necessary to effectively inhibit ERK activation and induce apoptosis in the KRAS-mutant context. In the KRAS-wildtype Caco-2 cells, individual and combined RAF depletion also down-regulated pERK, but this did not strongly induce BIM (Figure S4C) or lead to a significant apoptotic response (Fig 4C). Lastly, in the BRAF-mutant LS411N cells, pERK was primarily sensitive to BRAF depletion (Figure S4C and Fig. 4C). These results emphasize the importance of inhibiting complete effector nodes, including all related gene isoforms, presumably due to functional redundancy that can lead to compensation or feedback reactivation of the pathway. In comparison, the small molecule BRAF inhibitor PLX4720 did not inhibit the proliferation of KRAS-mutant cells (Figure 4B), as previously reported (2, 3, 26). Thus RAF inhibition by siRNA and small molecules may exert distinct biological effects, likely owing to the unique mechanism of RAF activation (27, 28).
We next investigated how depletion of various PI3K isoforms in combination with RAF depletion would modulate cell viability. Although the KRAS-wildtype Caco-2 and SW48 cells were resistant to individual and combinations of RAF isoform knockdown (Fig. 4B), the PI3K node on the other hand, in particular the PIK3CA gene, is critical for their viability (Fig. 4D). Across all cell lines tested, ablation of all three PI3K isoforms decreased cell viability to some extent, and the combined knockdown of all RAF and PI3K isoforms most severely decreased viability (Fig. 4D). However, the fact that this siRNA cocktail impaired viability in all cell lines tested, indicates a lack of selectivity for KRAS-mutant cells. Hence, our experiments support the notion that targeting complete effector nodes, particularly the RAF kinases, can inhibit proliferation in KRAS-mutant CRC cells and confer a selective therapeutic benefit in KRAS-mutant colorectal cancer.
Sensor siRNAs can be effectively delivered in vivo
To test the therapeutic efficacy of Sensor siRNAs in vivo, we used previously characterized cyclodextrin-polymer nanoparticles (29, 30) to deliver Sensor siRNAs to tumors in mice. These particles are transferrin-tagged for endocytic uptake in tumor cells and size-selected to extravasate from leaky tumor vessels. Preclinical studies indicate that they are safe in animals (31, 32) and, indeed, these particles have been used in human clinical trials for the treatment of solid tumors (30). To carefully monitor nanoparticle-payload delivery, we developed a fluorescent reporter system to non-invasively and longitudinally monitor nanoparticle transduction using stable expression of DsRed and EGFP in tumors. Successful transduction of cells with nanoparticles carrying siDsRed resulted in loss of DsRed expression as monitored in real-time (Fig. S5A). This system thus reports on the terminal step of nanoparticle transduction, which is target knockdown within tumor cells. EGFP fluorescence can be concomitantly tracked to monitor tumor size and treatment efficacy.
These cyclodextrin nanoparticles assemble at a fixed polymer to payload ratio of 3:1. Given that high dose RNAi leads to significant changes in gene expression (Fig. 3 and S3), we designed a non-targeting, non-RISC binding “filler” siRNA, siANC_22, (Fig. S5B) that facilitates particle assembly through charge neutralization, but does not compete with processing of Sensor siRNAs when transfected at up to 40-fold higher concentrations (Fig. S5C). To monitor nanoparticle delivery, nanoparticles containing siANC_22 and siDsRed, hereafter denoted NP(ANC_22+DsRed), were intravenously administered to mice bearing DsRed;EGFP-positive subcutaneous xenografts at varying frequencies. Untreated mice displayed minor fluctuations in DsRed fluorescence, while mice treated 2 or 3 times a week displayed 2 or 3 distinct reductions in DsRed fluorescence per week, respectively (Fig. S5D). Moreover, untreated tumor sections displayed homogenous DsRed and EGFP staining, while treated tumors displayed heterogeneous DsRed staining, with particularly low DsRed levels proximal to blood vessels (Fig. S5E). Together, these results indicate that the nanoparticles were capable of successfully delivering a siRNA payload.
To test the processing efficiency of a combination siRNA payload targeting two genes, we treated mice with nanoparticles packaging siKRAS and siDsRed, NP(KRAS+DsRed), and harvested the tumors 2 days later. Tumors were enzymatically digested into single cell suspensions and FACS sorted for transduced (EGFPhigh;DsRedlow) and untransduced (EGFPhigh;DsRedhigh) populations (Fig. S5F). Western blot analysis showed that both KRAS and DsRed were suppressed in transduced cells compared to untransduced cells, confirming successful combination-siRNA payload delivery.
Nanoparticle-mediated delivery of siKRAS to KRAS-mutant tumors impairs tumor growth
To test the therapeutic efficacy of siKRAS_234 delivery to tumors, we treated mice bearing DsRed;EGFP-positive SW620 xenografts (KRASG12V) with nanoparticles packaging differing doses of siKRAS_234 (Fig. 5A). Mice were treated twice a week beginning when tumors reached 100 mm3, and tumor growth and nanoparticle uptake was monitored daily. Over three weeks, siKRAS 4 mg/kg treatment significantly slowed tumor growth compared to control siANC_22 treatment (Fig. 5B). Lower doses of 2 mg/kg and 1 mg/kg siKRAS also elicited anti-tumor effects, though with greater variability (Fig. S5G). We suspect the amount of siRNA released into each cell to be limiting, which emphasizes the importance of using very potent siRNAs that achieve target knockdown at low concentration.
Figure 5. SW620 xenografts treated with varying doses of siKRAS.
(A) Three doses of NP(siKRAS+siDsRed) were prepared by varying the ratios of siKRAS_234 and siANC_22. Nanoparticles were administered via tail vein injection on days 1, 3, 8, 10 and 15, beginning when tumors reached 100mm3. (B) NP treatment of siKRAS at 4mg/kg was able to significantly slow tumor growth compared to control siANC_22 treated tumors. (C) At day 19, whole tumor lysate was analyzed by western blot for KRAS expression and markers of MAPK signaling and cell cycle progression. (D) Tumor volume and relative tumor viability were measured daily with optical imaging of EGFP fluorescence. Raw EGFP images of 2 representative tumors from each treatment group are shown (left), where NP-siRNAs were injected on day 1 and day 3. Viability of transduced cells (right) is monitored by concomitant tracking of DsRed and EGFP. Concurrent drops in DsRed and EGFP signals (synchronous arrowheads) illustrate siRNA-induced lethality, while DsRed drops that do not induce EGFP drops (opposing arrowheads) indicated no siRNA effect on viability. (E) Tumors were analyzed by immunohistochemistry for pERK and pS6 levels. (F) Immunohistochemistry staining and quantification of Ki67 and cleaved caspase-3 (CC3) positive cells in tumors (***p<0.001; ****p<0.0001).
In most siKRAS-treated tumors, resected tumor tissue (Fig. 5C) and FACS-isolated transduced tumor cells (Fig. S5H) showed decreased levels of KRAS protein. Accordingly, concomitant decreases in pERK and pCRAF and increased levels of p21 were also observed (Fig. 5C). To monitor the effects of KRAS knockdown during tumorigenesis, EGFP fluorescence was measured daily to track tumor cell viability. siKRAS-treated tumors displayed a dose-dependent decrease in EGFP signal during the first week of treatment, indicating tumor regression (Fig. 5D, left). Synchronous dips in DsRed and EGFP fluorescence further indicated that siRNA delivery and cell death occur concordantly, thus validating our real-time reporter approach (Fig. 5D, right).
Tumor regression was not maintained after the first week of treatment. However the growth-suppressive effects of siKRAS treatment persisted throughout the treatment course. Immunohistochemistry on tumor sections harvested at the terminal timepoint revealed decreased pERK staining in 4 mg/kg siKRAS-treated tumors, particularly in regions proximal to blood vessels (Fig. 5E). Ki67-positivity was also significantly decreased in 4 mg/kg and 2 mg/kg siKRAS-treated tumors, and caspase-3 (CC3) positivity was increased in 4 mg/kg siKRAS-treated tumors compared to siANC-treated tumors (Fig. 5F). These data collectively indicate that loss of KRAS protein via siKRAS_234 delivery attenuates, but does not ablate, SW620 tumor growth through inhibition of the MAPK pathway, decreased proliferation and increased apoptosis.
Nanoparticle-mediated delivery of combination payloads allows for multi-gene knockdown in vivo
As we were able to concomitantly knock down up to 7 genes in vitro using high-order combinations of Sensor siRNAs, we evaluated the possibility of using Sensor siRNAs to concurrently knock down multiple genes in vivo. Our in vitro data indicated that combined knockdown of the RAF effector node (A/B/C-RAF) mimics KRAS knockdown in SW1116 and SW620 CRC cell lines (Fig. 4B). We thus packaged 2 mg/kg of each individual siRNA in various combinations into nanoparticles and administered these formulations three times a week to animals carrying SW1116 (KRASG12A) xenografts (Fig. 6A).
Figure 6. SW1116 xenografts treated with siKRAS or combination siA/B/C-RAF.
(A) Four NP treatments delivering 2mg/kg of each siRNA species were prepared as depicted. Nanoparticles were administered via tail vein injection on days 1, 3, 5, 8, 10, 12 and 15, beginning when tumors reached 100mm3. (B) siKRAS and siA/B/C-RAF treatment slowed tumor growth to a similar extent, compared to siANC_22 and siB/C-RAF treated tumors. The inset (bottom) shows growth of all individual tumors analyzed. (C) At day 17, whole tumor lysate was analyzed by western blot for KRAS and RAF isoform expression. (D) Tumor volume and relative viability were measured daily with optical imaging in the EGFP channel, and DsRed fluorescence tracked nanoparticle delivery, as described in Figure 5C. NP-siRNAs were injected on days 8, 10 and 12. (E) Tumors were analyzed by immunohistochemistry for Ki67 and cleaved caspase-3 positive cells (**p<0.01; ***p<0.001; ****p<0.0001).
Over the 2.5-week treatment course, siKRAS and siA/B/C-RAF treatment equally inhibited tumor growth (Fig. 6B), whereas siB/C-RAF treatment had no therapeutic efficacy, as predicted by in vitro experiments (Fig. 4B). Resected tumor tissue (Fig. 6C) and FACS-sorted, transduced tumor cells (Fig. S5I) showed decreased levels of all intended target genes, albeit with some variability at this low dose. It is also possible that the low vascularity of this xenograft model prevented penetration of the nanoparticles to the entire tumor. Accordingly, siKRAS and siA/B/C-RAF treatment only inhibited tumor growth in this xenograft model and did not induce regression, as indicated by relatively stable levels of EGFP in tumor tissue (Fig. 6D). Nevertheless, a significant decrease in Ki67-positivity and corresponding increase in CC3-positivity was observed in siKRAS and siA/B/C-RAF-treated tumors compared to siANC or siB/C-RAF-treated tumors (Fig. 6E). These data demonstrate that four genes can be concurrently knocked down in tumor tissue, allowing for inhibition of complete effector nodes and the potential to rationally design complex combination therapies.
Concomitant knockdown of KRAS and PI3K improves KRAS-mutant tumor growth inhibition
To improve the therapeutic efficacy of single-agent siKRAS treatment in SW620 tumors (Fig. 5), we formulated a combination payload targeting both KRAS and the PI3K node. In mouse lung, PI3K activation is dependent on oncogenic KRAS and is required for tumor maintenance (33, 34). However in other tissues, PI3K can be activated through RAS-independent mechanisms such as direct binding to receptor tyrosine kinases. To more thoroughly eliminate PI3K activity, we sought to target the PI3K node directly in combination with KRAS. We administered nanoparticles targeting KRAS alone, PIK3C-A/B, KRAS+PIK3C-A/B or ANC_22 three times a week for two weeks at 2 mg/kg per individual siRNA (Fig. 7A). As observed previously (Fig. 5B,D), siKRAS alone induced an initial tumor regression that was not sustained over time (Fig. 7B). Co-targeting of PIK3C-A/B together with KRAS induced a similar level of tumor inhibition during the early stages of treatment but importantly, provided significantly better inhibition of tumor growth during later stages of treatment, even surpassing the efficacy of the high dose (4 mg/kg) siKRAS treatment (Fig. 5B).
Figure 7. SW620 xenografts treated with combination siKRAS+siPIK3CA/B.
(A) Four NP treatments delivering 2mg/kg of each siRNA species were prepared as depicted. Nanoparticles were administered via tail vein injection on days 1, 3, 5, 8, 10, and 12, beginning when tumors reached 70-100mm3. (B) siKRAS and siPIK3CA/B treatment induced mild tumor regression and slowed the rate of tumor growth during the first week of treatment compared to siANC_22-treated tumors. siKRAS+PIK3CA/B treatment potentiated the growth inhibition throughout the course of treatment. The inset (bottom) shows growth of all individual tumors analyzed. (C) Whole tumor lysate was collected on day 15 and analyzed by western blot for KRAS, p110□ and p110□ expression. (D) Tumor volume and relative viability were measured daily with optical imaging in the DsRed channel, and EGFP fluorescence tracked nanoparticle delivery, as described in Figure 5C. NP-siRNAs were injected on days 1, 3 and 5. (E) Tumors were analyzed by immunohistochemistry for Ki67 and cleaved caspase-3 positive cells (*p<0.05; **p<0.01; ***p<0.001; ****p<0.0001).
Resected tumor tissue showed at least partial knockdown of all intended target genes (Fig. 7C), and for all experimental treatment arms, concomitant dips in both the red and green channels indicate loss of cell viability concurrent with target knockdown (Fig. 7D). As shown previously, loss of cell viability in siKRAS-treated tumors resulted from decreased proliferation and increased apoptosis (Fig. 7E), whereas siPIK3C-A/B-treated tumors primarily underwent increased apoptosis. Accordingly, the combination therapy only slightly further decreased proliferation and increased apoptosis but not in an additive manner. This indicates that the sustained growth inhibition was likely a consequence of perturbing other KRAS and PI3K-regulated cellular functions. Taken together, these data show that the therapeutic efficacy of anti-KRAS RNAi therapies can be increased through the delivery of combination payloads. In the absence of small molecules against most putative cancer targets, the ability to validate such combination targets in vivo with siRNA also has the potential to accelerate both RNAi therapy and small molecule development.
Discussion
In this study we used the Sensor assay (14) to build two focused, functionally validated RNAi libraries targeting 75 human Ras pathway genes and their mouse orthologs (Table S2). Inhibiting the RAS signaling pathway has been a challenging but important area in translational research. Our library of KRAS pathway genes was curated to contain both direct KRAS effectors (RAF, PI3K, RAL) as well as indirect and non-canonical effectors (apoptosis, metabolism and stress pathway genes), and we believe this library will be a useful tool for the broader research community. Sensor siRNAs against RAS pathway genes efficiently depleted their targets at nanomolar concentrations in vitro (Fig. 2 and 3) and accordingly reduced both sequence-dependent and -independent off-target effects. Furthermore, low dose activity facilitates higher-order combinatorial gene knockdown by eliminating interference between siRNA species.
Using Sensor siRNAs targeting KRAS, we demonstrate KRAS addiction in a panel of KRAS-mutant colorectal cancer (CRC) cell lines and provide in vivo proof-of-principle evidence that direct targeting of KRAS by RNAi is a viable strategy to inhibit KRAS-mutant cancer (Fig. 2 and 5). These results are corroborated by a parallel study from Xue et al, where nanoparticle-mediated delivery of KRAS siRNA and other small RNAs was shown to inhibit the growth of KRAS-mutant lung cancers (35). We further show that targeting the complete RAF effector node by co-depleting all three RAF kinases, but not any single RAF kinase or dual combination, can phenocopy the therapeutic efficacy of siKRAS in KRAS-mutant CRC cells in vitro and in vivo (Fig. 4 and 6). Functional redundancy between RAF isoforms likely allows for compensatory kinase activity when fewer than all three genes are targeted, thereby incompletely shutting off RAF signaling. As most major RAS effectors are indeed comprised of multi-isoform families (MEK1/2, ERK1/2, PIK3CA/B/D, RALA/B, etc.), it may be important to consider entire effector nodes as targets for drug development. While such combination targeting can most easily be achieved using RNAi, pan-specific small molecules may achieve similar ends.
The ability to multiplex Sensor siRNAs for combination target knockdown is particularly useful in the context of KRAS-driven cancers, where numerous effector pathways are activated. We show that KRAS knockdown alone conferred considerable growth inhibition in vivo. However we further show that targeting KRAS in combination with the PI3K effector node potentiated this effect (Fig. 7). Thus the delivery of a combination payload targeting both the driver oncogene and an effector node offers the potential to more thoroughly disable mitogenic and survival signaling pathways. Many KRAS effectors, including RAF, PI3K, RAL and RAC, have RAS-independent means of activation (36), which can be circumvented through direct targeting of effector nodes in addition to KRAS. Furthermore, concurrent targeting of effector nodes may also be effective in disabling feedback-mediated pathway reactivation or in delaying acquired resistance. Our system provides a robust and rapid means for exploration of such combinatorial space, allowing for the rational discovery and validation of optimal target combinations for Ras-driven cancers.
Materials and Methods
Sensor library design and construction
The shRNA-Sensor libraries were constructed as previously described (14), with a few improvements to enhance pooled cloning performance. A recipient vector for the first pooled cloning step, pTNL (TREtight-NeoR-5′miR30-BamHI-MfeI-EcoRI), was cloned by replacing the XhoI/ClaI fragment in pSENSOR with a linker containing a BamHI-MfeI-EcoRI multiple cloning site (CTCGAGGGATCCCAATTGGAATTCATCGAT). To generate the Ras pathway shRNA-Sensor libraries, we designed ~5,000 185-mer oligonucleotides per library, each containing a 101-mer miR30-shRNA fragment, an EcoRI/BamHI/MluI cloning site, the cognate 50 nt Sensor cassette and an 18 nt primer binding site (CCTAGGATCGACGCGGAC). For the 75 human Ras pathway genes and their 75 mouse orthologs (Table S2), we generated 65 candidate shRNAs each by taking the common transcript of all gene variants and designing the Top300 DSIR predictions (15). We then further selected and ranked these shRNAs using Sensor rules (14) that demand features recurrently found in potent shRNAs and excluded sequences containing restriction sites of endonucleases used for cloning. The 65 top ranked candidates for each gene (Table S2) were synthesized on custom 27k oligonucleotide arrays (Agilent Technologies) that also contained 17 control shRNAs-Sensor oligonucleotides included at 1× representation. The final libraries were constructed according to the previously described two-step cloning procedure, with an additional MfeI digestion of the ligation product from cloning step one and BamHI digestion of the ligation product from cloning step two. Briefly, in step one, oligonucleotides were PCR amplified with the Sens5′Xho (TACAATACTCGAGAAGGTATATTGCTGTTGACAGTGAGCG) and Sens3′Mfe (ATTCATCACAATTGTCCGCGTCGATCCTAGG) primers, XhoI/MfeI digested and ligated into XhoI/EcoRI cut pTNL backbone vector. The ligation product was then digested with MfeI to reduce background noise from concatemers. In step two, the missing 3′miR30-PGK-Venus fragment was cloned into the EcoRI/MluI site separating each shRNA and its cognate target Sensor, followed by BamHI digestion of the ligation product to further reduce background noise. During each cloning step, a representation of at least 1000 fold the complexity of the library was maintained.
Cell lines and reagents
The human osteosarcoma cell line U2OS and human colorectal cancer (CRC) cell lines SW620, SW403, SW1116, LoVo, LS123, Caco-2, SW48, RKO, and LS411N were obtained from Dr. Thomas Reid (NCI) and grown in McCoy’s 5A media (Lonza) supplemented with 10% fetal bovine serum (FBS). Trp53−/− MEFs (21) were grown in DMEM supplemented with 10% FBS. KRAS−/− MEFs were obtained from Dr. Mariano Barbacid (CNIO). All cell lines were cultured at 37°C in a humidified 5% CO2 incubator. No cell line authentication was performed by the authors.
Sensor siRNA transfection and validation
Cells were transfected with Sensor siRNAs at the indicated concentrations by using Lipofectamine RNAiMAX (Invitrogen) following the manufacture’s instruction. AllStars Negative Control siRNA and AllStars Human Cell Death Control siRNA (Qiagen) were transfected as controls. To validate the Sensor siRNAs and to test knockdown efficiency, U2OS cells were transfected with Sensor siRNAs at the indicated concentrations. Cells were collected 72 hours post-transfection for total cell lysate preparation or total RNA isolation using RNeasy Mini Kit (Qiagen) followed by cDNA conversion using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). To measure mRNA level, quantitative real-time PCR was performed using SYBR Green method on a 7900HT Fast Real-Time PCR System platform (Applied Biosystems). GAPDH mRNA was measured as an endogenous control. Each quantitative RT-PCR assay was performed in triplicate. To measure protein level, total cell lysates were subjected to immunoblotting as previously described (37).
Cell viability and apoptosis assay
Cell viability was measured 5 days post-transfection by performing CellTiter-Glo Luminescent Cell Viability Assay (Promega), while apoptosis was measured 3 days post-transfection by performing Caspase-Glo 3/7 Assay (Promega) following manufacturer’s instructions. Cell viability and apoptosis rate were normalized against AllStars Negative Control siRNA-transfected cells.
Off-target effects and gene expression analysis
Trp53−/− MEFs were transduced with LMP (38) expressing Trp53 shRNAs at single copy (11-21% infection efficiency) and high copy (>98% infection efficiency), selected on puromycin and grown in absence of the selection agent before harvest. Uninfected Trp53−/− MEFs and Trp53−/− MEFs infected with an empty vector control served as “no shRNA” reference. Total RNA was extracted from at least 5×106 cells per sample using TRIzol (Invitrogen), followed by acidic phenol:chloroform:IAA (125:24:1, pH 4.5, Ambion) purification and isopropanol precipitation. All samples were DNase I (Roche) treated and again purified by acid phenol:chloroform:IAA extraction and isopropanol precipitation. Gene expression analysis was performed on MoGene-1_0-st-v1 microarrays (Affymetrix). To test siRNA off-target effect, RAS-less MEFs stably expressing exogenous NRAS were transfected with siRNA KRAS_234 and KRAS_355 at 0.2, 2, 20, and 50 nM in triplicates for 72h. Total RNA was extracted and subjected to microarray hybridization on Mouse Gene ST 2.0 microarrays (Affymetrix). Total RNA samples extracted from cells transfected with lipid only were used as gene expression baseline control. Gene expression analysis was performed using Partek Genomics Suite 6.0. The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession numbers GSE59952 and GSE59823.
Xenografts and tumor imaging
SW620 and SW1116 cells stably expressing DsRed and EGFP were made by lentiviral infection. Cells were FACS sorted for double positive cells and pooled populations were propagated for engraftment. All mouse experiments were done in compliance with UCSF and Institutional Animal Care and Use Committee (IACUC) policies. 2-3×106 cells in PBS/Matrigel (1:1) were subcutaneously injected bilaterally into each rear flank of nude mice. Upon engraftment, tumor volume was measured daily with calipers (volume=0.5233×W×L×H). Additionally, tumors were optically imaged for DsRed and EGFP fluorescence daily using a G1 Xenogen IVIS 100 System. Optical data was analyzed using Living Image software v. 4.2 (PerkinElmer).
Supplementary Material
Statement of Significance.
To advance RNAi therapy for KRAS-mutant cancer, we developed a validated siRNA library against RAS pathway genes that enables combination gene silencing. Using an in vivo model for real-time siRNA delivery tracking, we show that siRNA-mediated inhibition of KRAS or RAF/PI3K combinations can impair KRAS-mutant colorectal cancer in xenograft models.
Acknowledgements
We would like to thank Beicong Ma and Colin Merrifield for excellent technical assistance, Mark Davis, Jeremy Heidel and Thomas Schluep for providing nanoparticles and helpful discussion, and Mariano Barbacid for cell line reagents. S.W.L. is an investigator of the Howard Hughes Medical Institute. C.F. and S.W.L. are founders of Mirimus Inc., and C.F. is an employee of Mirimus Inc., a company focused on producing RNAi transgenic mice that has licensed some of the shRNA technology used here. This work was supported by funding from the Susan G. Komen Foundation (KG111338 to T.L.Y.), from Daiichi Sankyo (to T.L.Y. and F.M.), from the NCI Intramural Research Program (1ZIABC011437 to J.L.) and from the Howard Hughes Medical Institute (to S.W.L).
Funding: This work was supported by funding from the Susan G. Komen Foundation (KG111338 to T.L.Y.), from Daiichi Sankyo (to T.L.Y. and F.M.), from the NCI Intramural Research Program (1ZIABC011437 to J.L.) and from the Howard Hughes Medical Institute (to S.W.L).
#.
References
- 1.Gysin S, Salt M, Young A, McCormick F. Therapeutic strategies for targeting ras proteins. Genes Cancer. 2011;2:359–72. doi: 10.1177/1947601911412376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–5. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 3.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–30. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rinehart J, Adjei AA, Lorusso PM, Waterhouse D, Hecht JR, Natale RB, et al. Multicenter phase II study of the oral MEK inhibitor, CI-1040, in patients with advanced non-small-cell lung, breast, colon, and pancreatic cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2004;22:4456–62. doi: 10.1200/JCO.2004.01.185. [DOI] [PubMed] [Google Scholar]
- 5.Haura EB, Ricart AD, Larson TG, Stella PJ, Bazhenova L, Miller VA, et al. A phase II study of PD-0325901, an oral MEK inhibitor, in previously treated patients with advanced non-small cell lung cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2010;16:2450–7. doi: 10.1158/1078-0432.CCR-09-1920. [DOI] [PubMed] [Google Scholar]
- 6.Hainsworth JD, Cebotaru CL, Kanarev V, Ciuleanu TE, Damyanov D, Stella P, et al. A phase II, open-label, randomized study to assess the efficacy and safety of AZD6244 (ARRY-142886) versus pemetrexed in patients with non-small cell lung cancer who have failed one or two prior chemotherapeutic regimens. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2010;5:1630–6. doi: 10.1097/JTO.0b013e3181e8b3a3. [DOI] [PubMed] [Google Scholar]
- 7.Shimizu T, Tolcher AW, Papadopoulos KP, Beeram M, Rasco DW, Smith LS, et al. The clinical effect of the dual-targeting strategy involving PI3K/AKT/mTOR and RAS/MEK/ERK pathways in patients with advanced cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18:2316–25. doi: 10.1158/1078-0432.CCR-11-2381. [DOI] [PubMed] [Google Scholar]
- 8.Hannon GJ. RNA interference. Nature. 2002;418:244–51. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–30. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 11.Patel MN, Halling-Brown MD, Tym JE, Workman P, Al-Lazikani B. Objective assessment of cancer genes for drug discovery. Nat Rev Drug Discov. 2013;12:35–50. doi: 10.1038/nrd3913. [DOI] [PubMed] [Google Scholar]
- 12.Zhou J, Shum KT, Burnett JC, Rossi JJ. Nanoparticle-Based Delivery of RNAi Therapeutics: Progress and Challenges. Pharmaceuticals (Basel) 2013;6:85–107. doi: 10.3390/ph6010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fellmann C, Lowe SW. Stable RNA interference rules for silencing. Nat Cell Biol. 2014;16:10–8. doi: 10.1038/ncb2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fellmann C, Zuber J, McJunkin K, Chang K, Malone CD, Dickins RA, et al. Functional Identification of Optimized RNAi Triggers Using a Massively Parallel Sensor Assay. Mol Cell. 2011;41:733–46. doi: 10.1016/j.molcel.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vert JP, Foveau N, Lajaunie C, Vandenbrouck Y. An accurate and interpretable model for siRNA efficacy prediction. BMC Bioinformatics. 2006;7:520. doi: 10.1186/1471-2105-7-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–7. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 18.Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, et al. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- 19.Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol. 2009;27:549–55. doi: 10.1038/nbt.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nat Struct Mol Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- 21.Livingstone LR, White A, Sprouse J, Livanos E, Jacks T, Tlsty TD. Altered cell cycle arrest and gene amplification potential accompany loss of wild-type p53. Cell. 1992;70:923–35. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- 22.Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–57. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drosten M, Dhawahir A, Sum EY, Urosevic J, Lechuga CG, Esteban LM, et al. Genetic analysis of Ras signalling pathways in cell proliferation, migration and survival. Embo J. 2010;29:1091–104. doi: 10.1038/emboj.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blasco RB, Francoz S, Santamaria D, Canamero M, Dubus P, Charron J, et al. c-Raf, but not B-Raf, is essential for development of K-Ras oncogene-driven non-small cell lung carcinoma. Cancer Cell. 2011;19:652–63. doi: 10.1016/j.ccr.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karreth FA, Frese KK, DeNicola GM, Baccarini M, Tuveson DA. C-Raf is required for the initiation of lung cancer by K-Ras(G12D) Cancer Discov. 2011;1:128–36. doi: 10.1158/2159-8290.CD-10-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–21. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freeman AK, Ritt DA, Morrison DK. Effects of Raf dimerization and its inhibition on normal and disease-associated Raf signaling. Molecular Cell. 2013;49:751–8. doi: 10.1016/j.molcel.2012.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holderfield M, Merritt H, Chan J, Wallroth M, Tandeske L, Zhai H, et al. RAF Inhibitors Activate the MAPK Pathway by Relieving Inhibitory Autophosphorylation. Cancer Cell. 2013;23:594–602. doi: 10.1016/j.ccr.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 29.Davis ME. The first targeted delivery of siRNA in humans via a self-assembling, cyclodextrin polymer-based nanoparticle: from concept to clinic. Mol Pharm. 2009;6:659–68. doi: 10.1021/mp900015y. [DOI] [PubMed] [Google Scholar]
- 30.Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–70. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu-Lieskovan S, Heidel JD, Bartlett DW, Davis ME, Triche TJ. Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastatic Ewing’s sarcoma. Cancer Res. 2005;65:8984–92. doi: 10.1158/0008-5472.CAN-05-0565. [DOI] [PubMed] [Google Scholar]
- 32.Heidel JD, Yu Z, Liu JY, Rele SM, Liang Y, Zeidan RK, et al. Administration in non-human primates of escalating intravenous doses of targeted nanoparticles containing ribonucleotide reductase subunit M2 siRNA. Proc Natl Acad Sci U S A. 2007;104:5715–21. doi: 10.1073/pnas.0701458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castellano E, Sheridan C, Thin MZ, Nye E, Spencer-Dene B, Diefenbacher ME, et al. Requirement for interaction of PI3-kinase p110alpha with RAS in lung tumor maintenance. Cancer Cell. 2013;24:617–30. doi: 10.1016/j.ccr.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gupta S, Ramjaun AR, Haiko P, Wang Y, Warne PH, Nicke B, et al. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell. 2007;129:957–68. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 35.Xue W, Dahlman JE, Tammela T, Khan OF, Sood S, Dave A, et al. Small RNA combination therapy for lung cancer. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1412686111. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–31. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weng MT, Lee JH, Wei SC, Li Q, Shahamatdar S, Hsu D, et al. Evolutionarily conserved protein ERH controls CENP-E mRNA splicing and is required for the survival of KRAS mutant cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E3659–67. doi: 10.1073/pnas.1207673110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nature Genetics. 2005;37:1289–95. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.