Abstract
The DNA-dependent protein kinase (DNA-PK) is a pivotal component of the DNA repair machinery that governs the response to DNA damage, serving to maintain genome integrity. However, the DNA-PK kinase component was initially isolated with transcriptional complexes, and recent findings have illuminated the impact of DNA-PK-mediated transcriptional regulation on tumor progression and therapeutic response. DNA-PK expression has also been correlated with poor outcome in selected tumor types, further underscoring the importance of understanding its role in disease. Herein, the molecular and cellular consequence of DNA-PK will be considered, with an eye toward discerning the rationale for therapeutic targeting of DNA-PK.
Over two decades ago, the DNA-dependent protein kinase (DNA-PK) was discovered and characterized as part of SP1 transcription complexes [1] and as a regulatory component of transcriptionally poised RNA polymerase II [2], but has been extensively studied with regard to function in DNA double strand break repair. Recently, a plethora of studies unveiled critical functions of DNA-PK outside the DNA repair process, redefining the paradigm of DNA-PK activation and signaling. While much remains to be understood about the cellular consequence of DNA-PK, studies to date have dramatic implications for the role of this kinase in tumor progression and therapeutic response.
One mainstay of cancer treatment is induction of irreparable DNA damage. DNA-PK, a serine/threonine protein kinase complex composed of a heterodimer of Ku proteins (Ku70/Ku80) and the catalytic subunit DNA-PKcs, is a critical component of the response to damage; as such, it is not surprising that DNA-PK correlates with decreased response to DNA damaging agents and therapeutic resistance in multiple cancers [3–6]. Further, utilization of aptamer-shRNA chimeras identified DNA-PK as an ideal radiosensitization target in prostate cancer [7]. Less expected, however, is that DNA-PK is correlated with poor prognosis independent of damage induction in numerous tumor types. DNA-PK is elevated in esophageal cancer tissues compared to adjacent normal mucosae [8], and high tumor/normal expression ratio of DNA-PKcs in non-small cell lung cancer is associated with a 2.13-fold increased risk of death [9]. Additionally, DNA-PKcs expression and activity is higher in B-cell chronic lymphocytic leukemias that are positive for mutations known to predict for short survival and chemoresistance [10]. The overexpression of DNA-PK in cancer relative to normal tissue associated with poor prognosis has been suggested to result from deregulation of transcription factors controlling gene expression [11], though other mechanisms including gene amplification may play a role. These findings, combined with evidence that DNA-PKcs is a potentially actionable therapeutic target [12, 13], have prompted development of DNA-PK targeting strategies. Given the multitude of pathways that drive cancer cell survival and progression, understanding mechanisms of DNA-PK regulation and associated cellular consequences are critical to designing effective therapeutic regimens to suppress DNA-PK. Herein, the pleiotropic roles of DNA-PK in human tumorigenesis and progression will be discussed.
DNA-PK IN DNA REPAIR
DNA-PKcs is a member of the phosphatidylinositol 3-kinase-related (PIKK) family of protein kinases and is abundantly expressed in almost all mammalian cells [14]. Of the six members of the PIKK family of kinases, three play prominent roles in the response to damage: DNA-PKcs and ataxia telangiectasia mutated (ATM) function primarily in double strand break repair; while ataxia telangiectasia and Rad3 related (ATR) is activated by single strand breaks, all three kinases share substrate homology (phosphorylation of S/TQ motifs) [15]. Not surprisingly, these important kinases have been implicated in human diseases, as germline mutations in ATM and ATR result in ataxia-telangiectasia and Seckel syndrome, respectively [16, 17], and somatic mutations in ATM are frequently found in several tumor types [18]. It is now also appreciated that germline mutations targeting DNA-PKcs lead to severe combined immunodeficiency (SCID) [19], and selected somatic mutations in DNA-PKcs have been speculated to interfere with double strand break repair and promote genomic instability [20]. Identification of mutated DNA-PKcs, combined with the inverse correlations seen between DNA-PKcs levels and overall survival in human cancers, necessitates precise understanding of how DNA-PKcs functions both within and outside of the damage response to effectively design therapeutic strategies.
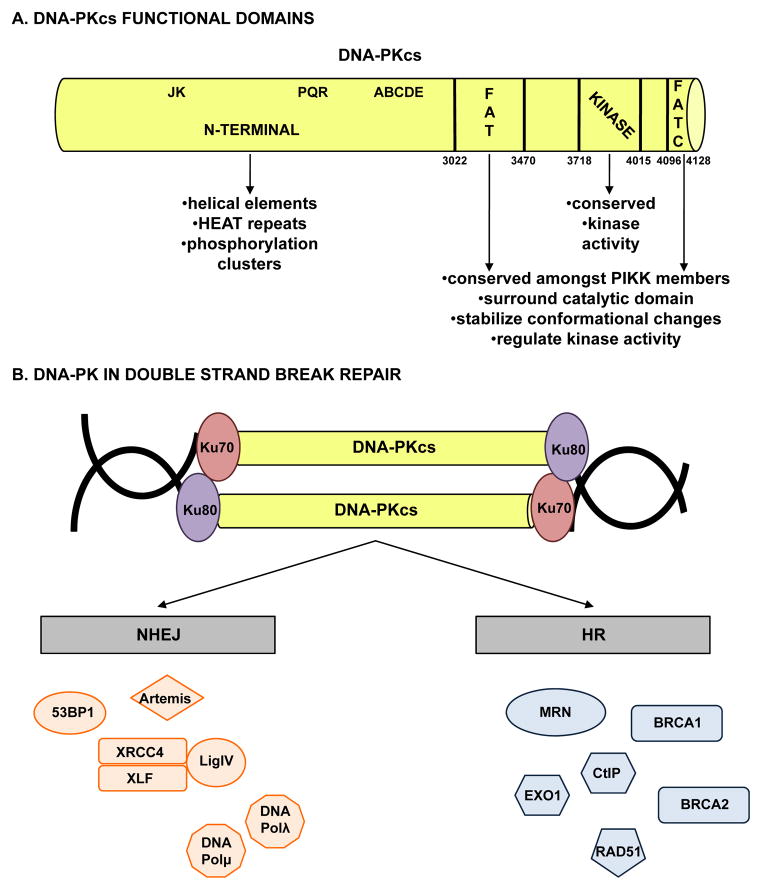
Ku70 and Ku80 (also called Ku86) are encoded by the XRCC6 and XRCC5 genes in humans, respectively, and have a strong affinity for free ends of DNA [21]. DNA-PKcs, a 469 kDa protein encoded by the PRKDC gene in humans, is composed of several distinct functional domains (Figure 1A); in addition to a highly conserved catalytic kinase domain, DNA-PKcs contains a large N-terminal domain containing mostly helical elements and HEAT (Huntingtin, Elongation Factor 3, PP2A, and TOR1) repeats along with the JK, PQR, and ABCDE phosphorylation clusters (groups of phosphorylation sites with important regulatory functions), and two additional regions conserved amongst PIKK family members: the FAT (named due to homology in this region between FAT, ATM, and TRRAP) domain, and a short C-terminal region termed the FATC (FAT at the extreme C terminus) domain [22]. The FAT and FATC domains surround the catalytic domain and serve to stabilize conformational changes to the catalytic core and regulate kinase activity [23]. As the kinase activity is crucial for roles both within and beyond the damage response, determination of the structure of DNA-PKcs has been critical in understanding regulation and function of the protein, most particularly as related to human malignancy.
Figure 1.
(A) DNA-PKcs is composed of multiple functional domains and (B) influences DNA double strand break repair through both the non-homologous end joining and homologous recombination pathways.
Role in NHEJ
The most well characterized function of DNA-PK is to mediate direct ligation of the broken ends of DNA double strand breaks generated both endogenously (e.g. via production of reactive oxygen species (ROS) during metabolism or via developmental VDJ recombination in the immune system) and exogenously (e.g. via radiomimetic drugs, ionizing radiation). Primarily, DNA-PK regulates repair of double strand breaks through the non-homologous end joining (NHEJ) DNA double strand break repair pathway (previously reviewed in depth [24]), a concept supported by the SCID phenotype of DNA-PKcs knockout mice resulting from lack of VDJ recombination during development [25]. Briefly, NHEJ is initiated through the recognition and binding of broken DNA ends by the ring shaped Ku70/Ku80 heterodimer, followed by recruitment of DNA-PKcs through its interaction with Ku and DNA (Figure 1B, left). Recruitment of DNA-PKcs facilitates movement of the Ku heterodimer into the DNA duplex, allowing DNA-PKcs to serve as a tether for the broken DNA ends and prevent exonucleolytic degradation [26]. Binding to DNA promotes activation of DNA-PKcs kinase activity, though the exact mechanism underlying this event remains poorly understood. Once activated, DNA-PKcs phosphorylates and alters the activity of proteins that mediate NHEJ, including Ku70, Ku80, Artemis, the X-ray cross complementing protein 4 (XRCC4), the XRCC4-like factor (XLF), and DNA ligase IV (LigIV); however, none of these phosphorylation events are independently required for efficient NHEJ repair [27]. Activated DNA-PKcs also phosphorylates Ser139 on histone variant H2AX (γH2AX) either directly or indirectly through Akt/GSK3b signaling [28]. γH2AX, a well known marker of DNA double strand breaks [29], aids in recruitment of repair factors to double strand breaks [30] and coordination of signaling cascades required for efficient repair [31].
Perhaps the most important substrate of DNA-PK is the kinase subunit itself, as autophosphorylation is critical for regulation of end processing, enzyme inactivation, and complex dissociation [32, 33]. The most well characterized autophosphorylation sites occur at Ser2056 (PQR cluster) and Thr2609 (ABCDE cluster) [27, 34]. Studies suggest that phosphorylation of the ABCDE cluster promotes access to DNA ends, while phosphorylation of the PQR cluster blocks end access [35]; additionally, phosphorylation of both clusters impacts dissociation of DNA-PKcs from DNA and subsequent loss of kinase activity, though additional yet to be determined phosphorylation sites are likely also involved [36, 37]. While these post-translational modifications are well understood, more than 40 phosphorylation sites have been identified in vitro [38], and the impact of each on DNA-PKcs function appears to be complex, leaving open the question as to which are critical for tumor-associated activities. Similarly, the order of recruitment and function of both processing and ligation factors involved in NHEJ after binding of Ku remain poorly defined, and the process is likely more fluid and dynamic than strictly regimented. Still, it is apparent that DNA-PKcs activation is required for repair, as pharmacologic inhibition of DNA-PK kinase activity results in inefficient repair and hypersensitivity to double strand break-inducing agents [12, 13]. Thus, DNA-PKcs is required for direct ligation of broken DNA ends, rendering it a critical factor in NHEJ.
Role in HR
In addition to its role in NHEJ, DNA-PK has also been implicated in the second major DNA double strand break repair pathway, homologous recombination (HR) (Figure 1B, right). Evidence for a definitive role for DNA-PK in alternative end-joining, a double strand break repair pathway thought to function when NHEJ and HR fail, remains inconclusive. While NHEJ is error-prone (since this class of repair does not utilize sequence homology and brings together broken ends of DNA, regardless of position or sequence) and occurs during all phases of cell cycle, HR involves Rad51-mediated strand invasion into a homologous strand of DNA that is employed as a template, resulting in accurate repair that occurs primarily during the G2/M phases of cell cycle when undamaged sister chromatids are available [39]. Outside of the obvious cell cycle constraints, the mechanisms underlying selection of HR or NHEJ for double strand break repair remain incompletely defined. One factor identified to influence this choice is DNA end resection, as blunt, minimally processed DNA ends are repaired by NHEJ, while 3′ end resection is required for loading of Rad51 in order for HR to occur [40]. End resection is controlled by interplay between 53BP1 and BRCA1, with 53BP1 supporting NHEJ by suppressing end resection while BRCA1 promotes CtBP-interacting protein (CtIP) and EXO1-mediated end resection through recruitment of the MRE11-RAD50-NBS1 (MRN) complex [41]. MRE11 is particularly important in the initial response to damage, as the endonuclease and exonuclease activities of MRE11 sequentially mediate resection of the DNA end [42] and MRE11 promotes pro-survival Akt-S473 phosphorylation in response to double strand breaks [43]. Factors influencing competition between 53BP1 and BRCA1 at double strand breaks include spatial dynamics and regulation by cell cycle specific factors, though other mechanisms yet to be determined likely are also involved [44]. Chromatin complexity likely also plays a role in selection of repair pathway, as HR repairs heterochromatin-associated double strand breaks during the G2 phase of cell cycle [45].
It has been postulated that NHEJ serves as the default pathway of double strand break repair, with HR occurring only when NHEJ fails [46]. This supposition is supported in part by the observation that Ku70, Ku80, and DNA-PKcs are abundant in mammalian cells [27] while HR factors are expressed to a lesser degree and in a cell cycle specific manner [47]. Moreover, Ku is recruited within minutes to sites of damage and NHEJ can be completed in approximately 30 minutes, while Rad51 foci form approximately 2 hours post damage induction and HR takes at least 7 hours to complete [48]. While NHEJ has been demonstrated to be the dominant mechanism of double strand break repair, a functional HR pathway is critical for viability [48], suggesting a mechanism by which HR can be activated even if NHEJ machinery is targeted to damage first. Initial attempts to address the question as to whether there is a direct role for DNA-PKcs in promoting HR failed to definitively determine whether it is the kinase activity of DNA-PKcs or simply the failure of NHEJ that promotes HR [27, 49]. Additional studies identified the JK cluster (Thr946 and Ser1004) and Thr3950 as DNA-PKcs phosphorylation sites that specifically promote HR while inhibiting NHEJ, thus providing a mechanism to protect certain DNA ends from repair by NHEJ [50]. While evidence suggests that phosphorylation of Thr3950 is an autophosphorylation event [35], the impact of different protein kinases on these regulatory sites remains under investigation [27]. In sum, these findings clearly define a function for DNA-PKcs outside of its classically defined role in NHEJ. Consonantly, it is increasingly appreciated that DNA-PKcs functions in a variety of capacities outside of NHEJ [51]. These novel functions of DNA-PKcs and mechanisms of regulation illustrate the versatility of the kinase and identify DNA-PK as a potential therapeutic target for a variety of human malignancies.
REGULATION OF DNA-PK ACTIVITY
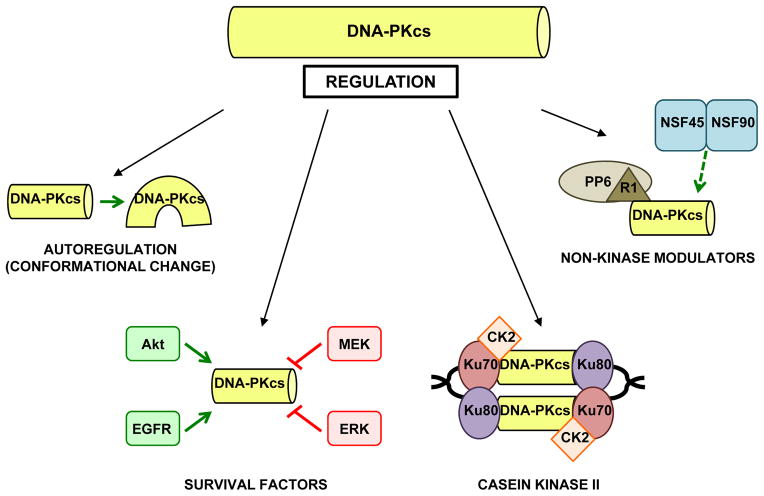
Given the critical role of DNA-PK in both NHEJ and HR, recent emphasis has been placed on identifying the mechanisms that control DNA-PKcs kinase activity. In addition to autoregulation, both kinase and non-kinase factors have been implicated as key modulators of DNA-PKcs activity in human cells.
Autoregulation
While it is well accepted that Ku-mediated binding of DNA-PKcs to broken DNA ends results in DNA-PKcs activation, the precise mechanism underlying the activation event remains unknown. Several recent reports provide insight into the mechanism and identify Ku-independent factors that modulate DNA-PKcs activity (Figure 2). Contrary to the paradigm that binding to DNA is a requirement for DNA-PKcs activation, it was demonstrated that DNA-PKcs autophosphorylation can be modulated by N-terminal conformational changes in the protein (such as that likely caused by DNA binding) [52]. This supposition was further supported by findings which revealed that the N-terminus (but not the C-terminus) is required for binding to the Ku-DNA complex and robust DNA-PKcs activity [53]. These collective studies support the concept that the N-terminus of DNA-PKcs is crucial for both targeting to Ku-bound DNA, and for conformational change-induced enzymatic activity.
Figure 2.
DNA-PKcs is activated through multiple mechanisms.
Cancer-associated survival factors
Consistent with a pro-tumorigenic role for DNA-PK, a number of canonical pro-survival signaling pathways have been shown to regulate DNA-PKcs kinase activity. The serine/threonine kinase Akt has been linked to NHEJ [54], and a recent report revealed that Akt1 forms a complex with DNA-PKcs as an initiating step in repair after IR. Akt1 promotes DNA-PKcs autophosphorylation and was critical for recruitment of the Akt1/DNA-PKcs complex to broken DNA ends [55]. Additionally, epidermal growth factor receptor (EGFR) can bind DNA-PKcs and enhance DNA-PKcs activity in response to damage [56]. As Akt can be activated by EGFR ligands [57], coordinated regulation between EGFR and Akt may be important for full DNA-PKcs activation in response to damage, though this putative pathway requires more rigorous depth of investigation. Further, as DNA-PKcs activity results in Akt-S473 phosphorylation in response to damage in a cell line specific context [58], there appears to be a positive feedback loop involving DNA-PKcs and Akt that may play a role in the response to DNA damage. Despite these intriguing findings, DNA-PKcs activation does not appear to be a general response to pro-survival signaling, as inhibition of MEK or knockdown of ERK1 or ERK2 results in enhanced etoposide induced activation of DNA-PKcs and increased double strand break repair, while constitutively active ERK reduced DNA-PKcs activation and subsequent double strand break repair [59]. Thus, DNA-PKcs activation is selectively regulated by factors involved in multiple pro-survival signaling pathways, which may serve as proximal effectors of the damage response and alter tumor cell behavior.
Casein kinase II
Kinases linked to enhanced cell cycle progression may also alter DNA-PKcs activation, including casein kinase II (CK2), a constitutively active serine/threonine kinase known to play a role in cancer-associated processes. CK2 was shown to interact with DNA-PKcs, and this interaction increases after DNA damage. Conversely, depletion of CK2 inhibits both DNA damage-induced DNA-PKcs S2056 autophosphorylation and double strand break repair in DNA-PKcs proficient cells [60], suggesting that CK2 plays a pivotal role in DNA-PKcs activation. The mechanism was recently identified, wherein CK2 was shown to co-localize with γH2AX at sites of DNA damage and likely stabilize the interaction between DNA-PKcs and Ku80 at broken DNA ends, resulting in enhanced DNA-PKcs activation in response to double strand breaks [61]. Notably, CK2 depletion results in induction of autophagy, decreased phosphorylation of S6 and Akt kinases, and increased phosphorylation and activity of ERK1/2 [62]; since Akt positively contributes to DNA-PKcs activation while MEK-ERK has the opposite effect, the data suggest that a coordinated mechanism of regulation between CK2 and the Akt and MEK-ERK signaling pathways may influence DNA-PKcs activation.
Non-kinase modulators
Non-kinase factors with roles in malignancy have also been identified as modulators of DNA-PKcs. Protein phosphatase 6 (PP6) is recruited to sites of damage and contributes to DNA-PKcs activation through direct interaction [63]. PP6 also influences the response to damage by dephosphorylating Ser139 of γH2AX, resulting in dissolution of IR induced γH2AX foci and release of cells from damage response associated checkpoints [64]. However, it was only recently determined that the regulatory subunit SAPSR1 (R1) of PP6 facilitates a bidentate mechanism of binding to DNA-PK that is necessary for the interaction with PP6 [65]. Thus, R1 influences DNA-PKcs activation by bridging PP6 and DNA-PK in response to damage.
Finally, the RNA-binding protein nuclear factor 90 (NF90) was recently shown to modulate DNA-PKcs activity. NF90 exists in a heterodimeric complex with NF45 and has been implicated in regulation of gene expression [66]. Surprisingly, immunodepletion or RNA digestion of NF90/NF45 reduced DNA end joining to a similar extent as DNA-PKcs immunodepletion, and NF90/NF45 depletion caused increased sensitivity to IR and elevated levels of γH2AX foci [67]. Though this does not directly implicate NF90/NF45 in DNA-PKcs activation, these findings do suggest that the NF90/NF45 complex may regulate DNA-PKcs mediated DNA damage repair. Combined, all of these observations indicate that DNA-PKcs activation is not merely the result of binding to broken DNA ends; rather, DNA-PKcs activation appears to be a complex process influenced by a variety of pro-tumorigenic factors, suggesting that cancer cells may select for enhanced DNA-PKcs activity. Clearly, additional studies are required for a complete understanding of the mechanisms governing DNA-PKcs activation and consequent downstream cellular effects of DNA-PKcs.
DNA-PK: CELLULAR CONSEQUENCES
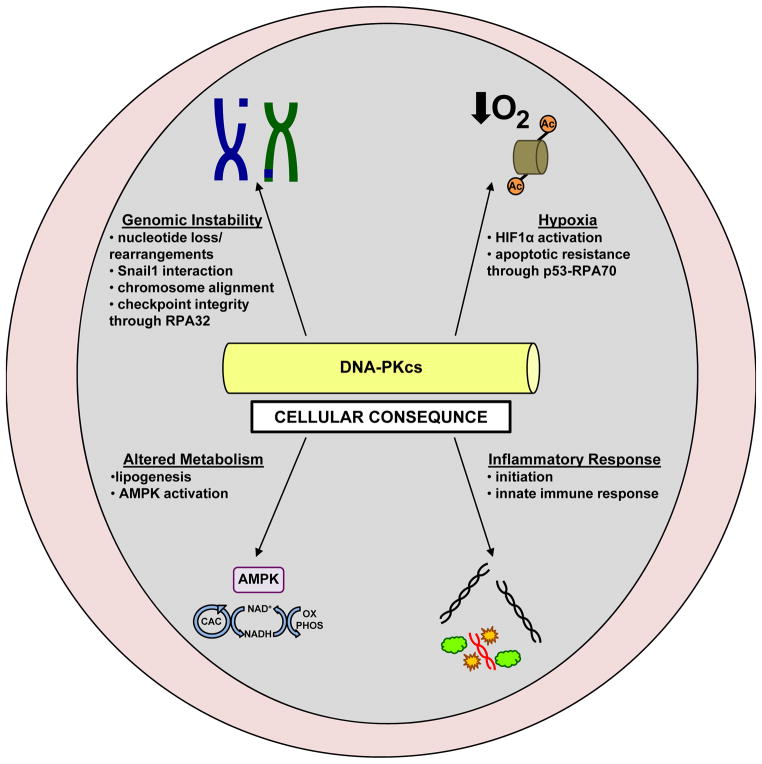
As the mechanisms regulating DNA-PKcs activation continue to be characterized, it is apparent that in addition to DNA double strand break repair, DNA-PKcs influences a variety of critical cellular processes associated with malignancy (Figure 3). Specifically, genomic instability, hypoxia, metabolism, and inflammatory response are all impacted by DNA-PKcs activity, and likely promote DNA-PKcs-associated tumor phenotypes.
Figure 3.
DNAP-Kcs modulates tumor-associated cellular pathways
Genomic stability
A process tightly associated with the response to DNA damage is maintenance of genomic stability. Primarily, the cellular response to damage is checkpoint activation, which serves to maintain genomic stability and protect against gross genomic rearrangements [68]. However, deregulation of cell cycle checkpoints in the presence of incomplete or inaccurate repair results in inappropriate cell proliferation and survival, and failure to efficiently repair damage promotes errors in DNA replication and chromosome segregation [69]. As NHEJ consists of indiscriminately ligating together broken DNA ends, this repair process can result in loss of nucleotides from the DNA strand. Further, as sequence homology is not required, direct ligation of double strand DNA breaks can promote deleterious rearrangements and chromosomal translocations at sites where multiple double strand breaks exist in close proximity, resulting in genomic instability. At a basic level, DNA-PKcs influences maintenance of genomic stability through its indispensible role in the error prone NHEJ pathway. However, recent reports indicate that DNA-PKcs may directly impact genomic stability through additional means.
One mechanism by which DNA-PK impacts genomic stability is through interaction with factors known to promote cancer progression. Snail1 is an E-box binding transcription factor known to promote epithelial-mesenchymal transition (EMT) during development and progression of cancer [70]. A recent report implicated interaction between Snail1 and DNA-PKcs as a driver of genomic instability and aggressive tumor characteristics [71]. DNA-PKcs was shown to interact with and phosphorylate Snail1 in response to damage, an event that stabilized Snail1 and reciprocally inhibited DNA-PKcs activity. Further, Snail1 overexpression increased chromosomal aberrations in both the absence and presence of damage and increased survival after damage, effects that were not observed in cell lines lacking DNA-PKcs. Combined, these findings suggest that DNA-PKcs plays a crucial role in Snail1-mediated genomic instability and resistance to genotoxic agents, possibly through direct interaction and reciprocal regulation between the two proteins.
DNA-PKcs also functions in chromosomal integrity during transition through mitosis. Exemplifying this, DNA-PKcs was found to be activated and closely associated with the spindle apparatus at centrosomes and kinetochores during progression through mitosis [72]. DNA-PKcs inhibition resulted in dysfunctional chromosome alignment, suggesting that DNA-PKcs activation is a critical regulator of microtubule dynamics during normal cell cycle progression through mitosis. Interestingly, a contrasting role for DNA-PKcs in regulation of mitotic entry during replication stress was identified. Interruption of replication machinery through a variety of mechanisms results in accumulation of single strand DNA bound by replication protein A (RPA), which triggers ATR activation and phosphorylation of the RPA32 subunit of RPA, resulting in checkpoint activation and cell cycle arrest [73]. Mutation in RPA32 phosphorylation sites targeted by DNA-PKcs resulted in decreased ATR signaling and increased proportion of cells in mitosis due to a G2/M arrest defect. Further, cells with mutant RPA32 or DNA-PKcs demonstrated inability to repair damage and inappropriate entry into mitosis in response to stress, revealing a critical role for DNA-PKcs in regulating mitotic entry in the presence of damage [74]. Thus, DNA-PKcs differentially regulates entry into and progression through mitosis dependent on the presence or absence of damage, defining a role for DNA-PKcs in maintaining chromosomal integrity and preventing genomic instability. In sum, while DNA-PKcs guards against chromosomal aberrations both in the presence and absence of damage, this complex may contribute to genomic instability through interaction with Snail1, underscoring the multi-faceted nature of DNA-PKcs activity. Perhaps these contrasting roles also reflect alterations in DNA-PKcs function in diseases including cancer, as the protective role may be lost and damaging role gained as a function of tumor progression.
Hypoxia
While mechanisms of activation in response to damage and maintenance of genomic stability are often considered parallel pathways, it has become evident that DNA-PK functions outside of the classical DNA damage response mechanisms. Hypoxia is a frequent physiological occurrence and hypoxic response plays a critical role in human cell biology and disease, particularly in cancer where solid tumors consume high amounts of oxygen despite inadequate vascularization. One critical regulator of the cellular response to hypoxia is hypoxia-inducible factor-1 alpha (HIF-1α), a transcription factor linked to numerous cancers that activates a number of oxygen-related genes required for adaptation to hypoxic conditions after induction by low oxygenation [75]. DNA-PKcs was found to be activated by hypoxia, independent of DNA double strand breaks, but dependent on histone acetylation, which is increased in hypoxic cells. DNA-PKcs positively regulated HIF-1α, resulting in modulation of oxygen-sensing and response pathways [76]. Moreover, hypoxia-induced γH2AX foci formation was delayed by depletion of HIF-1α or HIF-2α, though the factor directly responsible for phosphorylating H2AX was not investigated [77]. Combined, these studies present the novel concept that coordinated regulation between DNA-PKcs and the HIF family of transcription factors may influence cellular response to oxygen depletion.
DNA-PK activation by hypoxia also impacts cellular responses independently of HIF. Another factor recently implicated in hypoxic response is RPA70, a subunit of RPA (discussed above), which has been shown to increase DNA repair in cancer cells [78]. RPA70 is normally bound to the tumor suppressor p53, but this interaction is disrupted by hypoxia. A recent report demonstrated that DNA-PK phosphorylates p53 on Ser15, a crucial event in dissociating RPA70 from p53 [79]. RPA70 gene silencing resulted in increased apoptosis in resistant hypoxic cancer cells, revealing that DNA-PK phosphorylation of p53 results in p53-RPA70 dissociation and RPA70 mediated resistance to apoptosis under hypoxic conditions. In sum, these studies demonstrate that DNA-PK activation by histone acetylation during hypoxia influences cellular survival through both HIF-dependent and HIF-independent mechanisms.
Metabolism
A highly regulated and coordinated metabolic network of enzyme-catalyzed reactions exists in all living organisms to efficiently manage constant energy needs. Given the increasingly appreciated dynamic role of DNA-PK in cellular processes, it is not surprising that DNA-PK influences such a critical pathway. While DNA-PK has been implicated in transcriptional regulation of genes involved in lipogenesis [80], a recent report defines a general role for DNA-PK in metabolic regulation. AMP-activated protein kinase (AMPK) is an energy sensor that acts as a metabolic master switch influencing numerous processes [81]. DNA-PKcs interacts with the regulatory λ1 subunit of AMPK, and glucose-deprivation stimulated phosphorylation of AMPK is decreased in DNA-PKcs deficient cells [82]. Further, DNA-PKcs inhibition or depletion resulted in reduced AMPK phosphorylation and activation in response to glucose deprivation, suggesting that DNA-PKcs positively regulates AMPK phosphorylation and activation in glucose-deprived conditions. As AMPK is a major factor in cellular response to fluctuating energy levels, this finding reveals a significant role for DNA-PKcs in metabolic regulation, particularly in the high-energy need tumor environment.
Innate Immunity/Inflammatory Response
The innate immune system is a first line of defense against pathogens, protecting a host from infection in a non-specific way. The initial step in the innate immune response is detection of pathogen specific antigens (particularly viral nucleic acids), and recognition of single and double-stranded RNA substrates leads to transcriptional activation of anti-viral genes mediated by nuclear factor-kappa B (NF-κB) and interferon regulatory factor (IRF) families of transcription factors [83]. While DNA-PK has previously been demonstrated to be critical for NF-κB mediated expression of VCAM-1 in response to treatment with the cytokine tumor necrosis factor (TNF) [84], a novel role for DNA-PK in activation of innate immunity and inflammation response independent of NF-κB was revealed in studies wherein DNA from various pathogens was found to be bound by DNA-PK, resulting in IRF-3 mediated transcription of multiple cytokine and chemokine genes independent of DNA-PKcs kinase activity [85]. DNA-PKcs depletion produced diminished cytokine responses to DNA and DNA viruses but not to RNA or RNA virus infection, supporting the concept that DNA-PKcs recognition of foreign DNA is crucial. Further investigation revealed that stimulator of interferon genes (STING) interacts with the DNA-PK complex but is dissociated after introduction of DNA, suggesting that STING is released and able to activate IRF-3 mediated transcription after binding of DNA-PKcs to DNA. These unexpected findings reveal a critical role for DNA-PK in binding foreign DNA and activating the innate immune system and subsequent inflammatory response, and point to a role in DNA-PK potentially impacting tumor-associated inflammation.
Thus, DNA-PK modulates numerous important cellular processes, as the protein both guards against and promotes genomic stability in a context dependent fashion, regulates hypoxic response through HIF-dependent and independent mechanisms, impacts metabolism through interaction with AMPK, and activates the innate immune system in response to foreign DNA. Strikingly, however, a major function of DNA-PK in human disease is the ability to regulate gene expression.
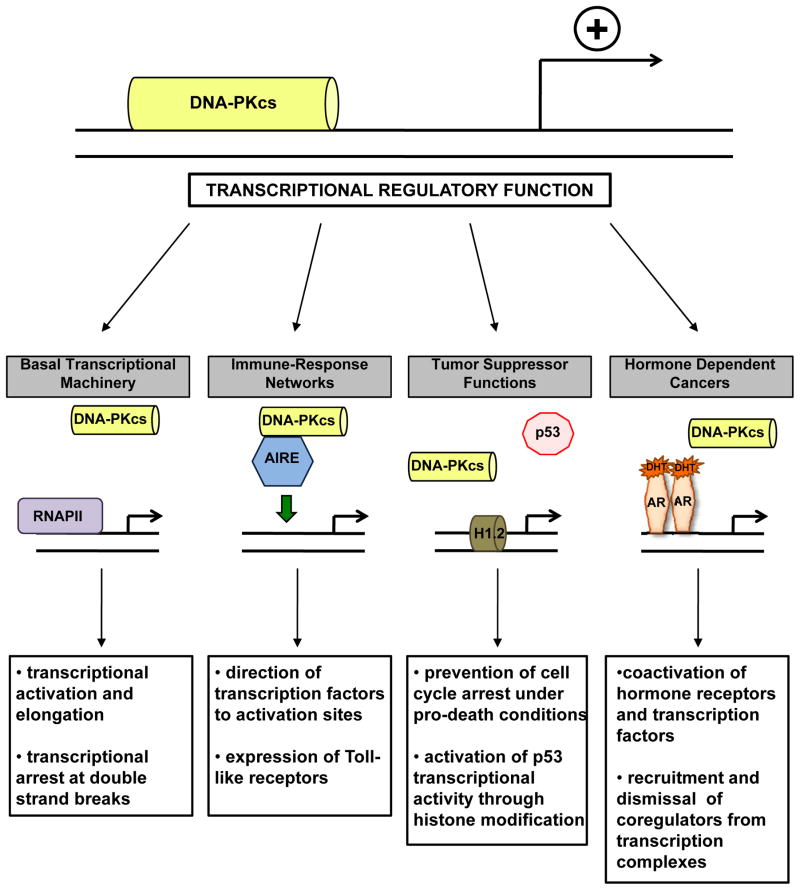
DNA-PK AS A TRANSCRIPTIONAL MODULATOR
Prior to characterization of the damage response role, DNA-PK was originally identified in complex with and shown to phosphorylate the transcription factor SP1 [1], along with being identified as a regulatory component of transcriptionally poised RNA polymerase II [2]. Further, it has been reported that activation of transcription results in induction of double strand breaks and recruitment of damage response machinery (including DNA-PK) [86], suggesting interaction between transcriptional networks and DNA-PK. Given the ability of Ku and DNA-PKcs to bind DNA double strand ends with high affinity, the context of binding (i.e. end binding versus transcription specific binding) should be considered and studies based on binding to double strand DNA interpreted with some degree of caution. Still, it is now clear that DNA-PK directly impacts transcriptional regulation, interacting with both the transcriptional machinery and transcription factors that alter cellular outcomes (Figure 4). Given that transcription is perhaps the most fundamental mechanism underlying cellular behavior and fate, the role of DNA-PK in regulating this process appears to have critical implications for cancer cell behavior and disease progression.
Figure 4.
DNA-PKcs regulates transcription through interaction with multiple factors in a variety of contexts.
Basal transcriptional machinery
RNA polymerase II (RNAPII), in complex with a set of general transcription factors and regulatory proteins, is recruited to promoter regions of DNA where it catalyzes transcription of RNA. It is generally appreciated that processively elongating RNAPII contains a hyperphosphorylated carboxy-terminal domain (CTD) while paused RNAPII possesses a hypophosphorylated CTD [87]. Previous observations have reported that DNA-PKcs phosphorylates the RNAPII CTD and that DNA-PKcs and Ku are components of the RNAPII holoenzyme [2, 88]. While the mechanism and functional consequence of DNA-PK communication with RNAPII has yet to be fully elucidated, two major lines of investigation found an interaction between DNA-PK and RNAPII that implicates DNA-PK as a transcriptional regulator. First, DNA-PK functionally interacts with RNAPII during human immunodeficiency virus (HIV) transcription. A recent study demonstrated that DNA-PK modulates HIV gene expression and interacts with the RNAPII complex at the HIV long terminal repeat (LTR), identical sequences used by viruses to insert genetic material into host genomes [89]. DNA-PK is recruited to and forms a complex with RNAPII both at the promoter and downstream regions of the HIV LTR, resulting in phosphorylation of specific residues on the RNAPII CTD. Knockdown of DNA-PKcs resulted in decreased HIV gene expression and replication, suggesting that DNA-PKcs plays an important role in both processes. Further, analysis of interplay between DNA-PK and Tat, a protein crucial for efficient HIV transcriptional elongation previously reported to interact with DNA-PK, revealed that DNA-PKcs mediated phosphorylation of Tat is required for full HIV LTR promoter transactivation. Thus, DNA-PKcs regulates transcription and replication of HIV directly through phosphorylation of the CTD of RNAPII and indirectly through phosphorylation of Tat, a factor critical for efficient transcriptional elongation. These findings provide rationale for treatment of HIV through DNA-PK inhibition, a concept that should be more thoroughly explored in future studies.
A more recent study revealed a role for DNA-PK in modulating RNAPII transcriptional activity at sites of broken DNA within coding regions of genes. Previous reports demonstrated that double strand breaks transiently repress RNAPI and RNAPII transcriptional machinery, and that breaks upstream of the promoter of an RNAPII reporter gene inhibited transcription elongation dependent on ATM signaling [90]. However, the impact of a single double strand break encountered during transcriptional elongation on RNAPII was not addressed. Introduction of a single double strand break by a site-specific meganuclease resulted in inhibition of transcription elongation followed by reinitiation [91]. Surprisingly, transcription inhibition was dependent on DNA-PKcs and not ATM, as RNAPII remained associated with the promoter and body of the genes being transcribed after inhibition of DNA-PKcs (but not ATM) activity. DNA-PKcs-dependent RNAPII removal was shown to require the proteasome, as treatment with the proteasome inhibitor MG132 or knockdown of a core subunit of the 19S proteasome complex (PSMD4) rescued transcriptional arrest. Combined, these results suggest that DNA-PKcs is required for eviction of RNAPII from DNA during elongation upon encountering a DNA lesion through a process that likely involves proteasome-mediated destabilization of the RNAPII complex. This defines an interaction between DNA-PKcs and RNAPII in a more general context, and demonstrates that DNA-PKcs may directly modulate RNAPII-mediated transcription in a variety of environments, including tumor-driven transcriptional networks.
Immune-related transcriptional networks
One important transcription factor regulated by DNA-PK is autoimmune regulator (AIRE), a transcriptional activator that mediates central tolerance in the thymus by activating expression of tissue-restricted antigen (TRA) genes [92]. Recruitment of AIRE to its transcriptional targets is dependent on DNA-PKcs, as assembly and activity of AIRE on DNA was abrogated in cells lacking DNA-PKcs [93]. Tethering of AIRE to DNA restored transcriptional activity in the absence of DNA-PKcs, suggesting that DNA-PKcs functions largely to direct AIRE to sites of transcriptional activation. An additional study utilizing AIRE-deficient cells stably transfected with AIRE demonstrated that DNA-PKcs depletion resulted in diminished transcriptional activity at promoters and decreased expression of Toll-like receptor (TLR) 1, TLR3, and TLR8, receptors known to play roles in the innate immune system [94]. As DNA-PKcs and AIRE were also demonstrated to directly interact, these findings suggest that DNA-PKcs regulates AIRE-mediated expression of TLRs. Combined, these studies reveal that DNA-PKcs is a critical modulator of the innate immune system through regulation of AIRE-mediated transcriptional networks.
Tumor suppressors
DNA-PK has also been implicated directly in cancer-associated transcriptional regulation, exemplified by influence on the p53 tumor suppressor. In general, p53-mediated activation of cell cycle arrest is dependent on expression and activation of the cyclin-dependent kinase inhibitor CDKN1A (p21Cip1) while apoptosis is mediated by expression and activation of pro-apoptotic genes including Bax and p53 upregulated modulator of apoptosis (PUMA). Analysis of the mechanism underlying p53-mediated expression of p21Cip1 revealed recruitment of DNA-PKcs and interaction with p53 at the p21Cip1 promoter after treatments that promoted apoptosis but not after treatments that promoted cell cycle arrest [95]. Inhibition of DNA-PKcs kinase activity blocked DNA-PKcs binding to p53 on the p21Cip1 promoter, resulting in increased p21Cip1 transcription and reduction in cell death after pro-death treatments. In sum, these findings demonstrated that DNA-PKcs negatively regulates p21Cip1 expression through modulation of p53 at the p21Cip1 promoter, resulting in induction of apoptosis after pro-death treatments. Interestingly, DNA-PKcs recruitment to the Bax or PUMA promoters after pro-death treatments were negligible relative to recruitment to the p21Cip1 promoter, revealing that DNA-PKcs regulation of p53 mediated induction of apoptosis is specific to certain genes. This specific recruitment of DNA-PKcs to negatively regulated p53 target gene promoters suggests that DNA-PKcs regulation of p53 targets occurs only when transcription is repressed, though the co-regulation between DNA-PKcs and p53 is likely complex and warrants further investigation. Further, negative regulation of p21Cip1 suggests that DNA-PKcs may coordinate expression of cell cycle machinery to damage response in conditions that promote cell cycle arrest and repair, though the DNA-PKcs-p53 connection is likely not a factor in G1 cell cycle arrest as DNA-PKcs deficient cells do not exhibit a defective G1 arrest [96].
Complementing these findings, it has been suggested that DNA-PK regulation of p53 transcriptional activity may also be dependent on histone modifications at target genes. Linker histone H1 regulates chromatin competency through binding to linker DNA [97]. One isoform, linker histone H1.2, inhibits p53-dependent transcription through alterations in chromatin remodeling, and detailed investigation revealed that p300-mediated p53 acetylation in combination with DNA-PK phosphorylation of histone H1.2 at T146 relieved H1.2 binding to p53, resulting in activation of p53 transcriptional activity [98]. Combined, these studies demonstrate that DNA-PK modulates p53-mediated transcription through both direct interaction with p53 at target genes and phosphorylation of histones that relieve repressive interactions between p53 and chromatin. Thus, while the mechanisms are likely complex and context dependent, DNA-PK plays a role in regulation of p53-mediated transcription in response to damage.
Hormone receptors and hormone dependent cancers
The class I nuclear hormone receptor superfamily is a group of related transcription factors, many of which are impacted by DNA-PK. Reports from over 15 years ago first demonstrated interaction between DNA-PK and hormone receptors, as DNA-PK was shown to phosphorylate both the rat glucocorticoid receptor and the chicken progesterone receptor, the latter of which impacted transcriptional activation of the receptor [99, 100]. However, only recently have new findings clearly defined a role for DNA-PK in transcriptional regulation of hormone receptors in human cells and demonstrated consequence for human malignancy.
The androgen receptor (AR) is a hormone receptor whose activity drives growth, survival, and progression of prostatic adenocarcinoma (PCa) at all stages of disease. A recent study demonstrated that AR is activated in response to genotoxic insult, driving expression of multiple genes critical for damage repair including DNA-PKcs [101]. AR activation was also shown to positively regulate DNA-PKcs activity, possibly through modulation of Ku70 levels. DNA-PKcs depletion or inhibition resulted in inability to repair damage and decreased overall survival in response to genotoxic insult, revealing that DNA-PKcs expression and activity is critical for efficient AR-mediated repair after damage. Further, DNA-PKcs was determined to be a coactivator of AR transcriptional activity, as DNA-PKcs depletion resulted in a significant decrease in expression of multiple AR target genes. Combined, these findings suggest that DNA-PKcs enhances AR transcriptional activation and that AR driven DNA-PKcs expression and activity is required for AR-mediated DNA repair, resulting in a positive feedback circuit that drives survival after damage.
The concept that DNA-PKcs modulates transcriptional activation in PCa is supported by findings that identified a role for DNA-PKcs in regulation of the ETS family of transcription factors, which are aberrantly expressed in a number of cancers. Notably, many prostate cancers exhibit gene rearrangements fusing AR target genes to ETS transcription factors, which result in elevated levels of ETS proteins driven by AR activity [102]. ETS factors induce a transcriptional program enriched for invasion-associated genes, and overexpression of ETS gene fusion proteins may contribute to cancer progression [103, 104]. ERG, the most frequently overexpressed ETS protein, was shown to interact with DNA-PKcs, and ERG-mediated transcription and consequential cell invasion, intravasation, and metastasis required DNA-PKcs and poly(ADP-ribose) polymerase 1 (PARP1) [105]. Functional association between DNA-PKcs and PARP1 is supported by studies reporting direct interaction between DNA-PKcs and PARP1 [106] and cooperation between DNA-PKcs and PARP1 to inhibit ribosomal RNA (rRNA) synthesis after DNA damage [107]. Thus, in addition to modulating AR transcriptional activity, DNA-PKcs cooperates with PARP1 to regulate ERG-mediated transcription, clearly defining DNA-PKcs as a regulator of transcription in PCa. Further, PARP1 has also been shown to be critical for AR transcriptional function and maintenance of AR-dependent cancer phenotypes, including tumor progression [108]. Combined, these findings suggest a model of coordinated regulation between AR, DNA-PKcs, and PARP1 that drives transcriptional networks in PCa.
Estrogen exerts its effects through estrogen receptor alpha and beta (ERα and ERβ), which differentially impact proliferation. ERα controls transcriptional programs driving cell cycle progression and proliferation and has been implicated in development and progression of disorders including breast cancer (BCa) and cardiovascular disease. Within the last several years, it was demonstrated that DNA-PK regulates ERα transcriptional activity. Initial findings demonstrated that in response to estrogen stimulation, DNA-PK interacts with and phosphorylates ERα at Ser118, resulting in stabilization of the ERα protein. Depletion of DNA-PKcs resulted in decreased ERα transcriptional activation, suggesting that ERα-mediated transcription is at least partially dependent on DNA-PKcs [109]. Additionally, a second report discovered two ERα binding sites in a region upstream of the DNA-PKcs transcriptional initiation site [110]. Estrogen-mediated ERα activation resulted in enhanced DNA-PKcs promoter activity and increased levels of DNA-PKcs while ERα inhibition or knockdown reduced DNA-PKcs expression, demonstrating that activated ERα transcriptionally regulates DNA-PKcs levels. Combined, these studies reveal that DNA-PKcs modulates ERα transactivation and define a circuit of regulation between ERα and DNA-PKcs, similar to that seen with AR and DNA-PKcs.
Most recently, cell-free biotin-estrogen response element (ERE) pulldown assays further characterized the mechanism underlying DNA-PKcs influence on ERα transcriptional activity, revealing that DNA-PKcs is responsible for recruitment of critical cofactors to regulatory regions of ERα target genes [111]. Addition of ATP to initially stable ERα-coregulator complexes resulted in immediate dismissal of five coregulators, including the corepressors RIP40 and the CtBPs, and phosphorylation of multiple coregulators, including transcriptionally activating sites on SRC-3, ERα, and MED1. DNA-PKcs depletion or inhibition prevented both dismissal of RIP140 and phosphorylation of the transcriptionally activating sites on SRC-3, MED1, and ERα, and similar complex formation was seen at a naturally occurring ERE containing enhancer. Thus, these findings build on previous studies reporting interaction between DNA-PKcs and ERα and provide mechanistic insight into how DNA-PKcs regulates ERα-mediated transcription, suggesting that DNA-PKcs phosphorylates SRC-3 and MED-1 for activation and RIP140 for dismissal from coregulator complexes.
TARGETING DNA-PK IN CANCER
Identification of defined roles for DNA-PK in tumor associated processes including genomic stability, hypoxia, metabolism, inflammatory response, and transcription supports targeting DNA-PK as a therapeutic intervention in human malignancy. The therapeutic efficacy of targeting DNA-PK has classically been gauged through potential to prevent repair of damage, given the importance of DNA-PK activity in NHEJ. Consistent with this concept, synthetic lethal interactions have been identified between DNA-PKcs and multiple damage response factors including ATM, MSH3, and numerous components of HR-mediated repair [112–114], suggesting that DNA-PK inhibitors may show efficacy as targeted agents for treatment of damage repair-defective human cancers. While many of the original DNA-PK inhibitors suffered from lack of specificity and poor pharmacokinetics, more recently developed inhibitors, including NU7026 and NU7441, demonstrate improved specificity and pharmaceutical properties and have been used preclinically to explore the impact of DNA-PK inhibition in multiple tumor types [115]. However, recent evidence implicating DNA-PK in pathways outside of NHEJ may help define novel strategies of therapeutic intervention, a concept supported by the reality that all of the agents targeting DNA-PK currently in clinical trial reflect the dynamic ability of DNA-PK to impinge on multiple pathways. CC-122 is a DNA-PK inhibitor in phase I clinical trial (NCT01421524) for solid tumors, Non-Hodgkin’s lymphoma, and multiple myeloma. CC-122 is described as a pleiotropic pathway modifier, echoing the ability of DNA-PK to modulate numerous pathways outside of double strand break repair. The two additional DNA-PK targeting molecules currently in clinical trial target both DNA-PK and the PI3K/Akt/mTOR signaling pathway. CC-115 is a dual inhibitor of DNA-PK and mTOR, and is currently in phase I trial (NCT01353625) for advanced solid tumors and hematologic malignancies while ZSTK474 is a PI3K inhibitor that also inhibits DNA-PK currently in phase I trials (NCT01280487 and NCT01682473) for advanced solid malignancies. As the only DNA-PK-targeting compounds currently in clinical trial impinge on pathways outside of DNA-PK-mediated damage repair, it will be important to consider novel DNA-PK regulated pathways when designing therapeutic strategies. For example, as a positive feedback circuit involving AR and DNA-PK drives PCa progression and survival, next generation means of targeting AR activity in combination with a DNA-PK inhibitor may represent a novel node of therapeutic intervention in advanced PCa. Though directly targeting cancer tissue is always important to consider with double strand break-inducing agents to avoid reduction of the therapeutic ratio, such combination strategies could afford new opportunities for precision therapies since DNA-PK seems to impact multiple tumor types.
FUTURE DIRECTIONS
Findings discussed herein illustrate that DNA-PK has dual functions in both DNA repair and gene regulation that promote tumor cell survival and progression, both in the presence and absence of genomic damage. These discoveries shed light on the potential mechanisms underlying DNA-PK-associated tumor phenotypes and poor outcomes associated with high DNA-PK expression. Key questions remain which will facilitate further understanding of DNA-PK-mediated tumor progression, and illuminate the overall consequence of targeting this kinase in human malignancy. First, precisely how is DNA-PKcs activated, and is the mechanism of activation altered in the context of cancer? While autophosphorylation of multiple sites denotes activation, the factors and events preceding the phosphorylation event are incompletely defined. Recent findings suggest that factors other than the Ku70/Ku80 heterodimer and DNA ends may assist in activation, and the factors influencing DNA-PKcs activation may be context dependent. Discovery and understanding of how and when different factors impact DNA-PKcs activation will provide insight into how DNA-PKcs impacts disease progression and identify new therapeutic targets. Second, what dictates the specificity of DNA-PKcs-mediated transcriptional regulation in cancer cells? DNA-PKcs has been defined as a transcriptional modulator in multiple contexts, but the global impact of DNA-PKcs on transcription has yet to be considered. Discovery of gene expression networks impacted by DNA-PKcs and understanding of specific cofactors involved will identify how DNA-PKcs damage-independent activation drives tumor survival. While recent studies have contributed some understanding to this issue, global discernment of the transcriptional function of DNA-PKcs in addition to the damage response function will provide disease-specific insight into progression. Third, which functions of DNA-PKcs should be considered when designing therapeutic strategies? Though the roles of DNA-PKcs in damage repair and genomic stability are currently integrated into therapeutic tactics, the transcriptional roles of DNA-PKcs have not been well considered when designing preclinical studies and clinical trials. Further, while the DNA-PK inhibitors entering clinical trial for cancer are known to inhibit the kinase activity of the protein, little is known regarding kinase-independent functions of DNA-PKcs that may impact therapeutic efficacy in targeting malignancy. Finally, does the function of DNA-PKcs change during cancer progression? One hallmark in progression of cancer (and other diseases) is alteration of signaling and regulatory pathways. While DNA-PKcs is generally considered a tumor suppressor under normal growth conditions, it acts more as a tumor promoter in cancerous cells, aiding in resistance to genotoxic therapeutics. Further, the effects of DNA-PKcs mutations recently identified in numerous cancer types should be determined in the contexts of transcriptional regulation and cancer progression. It will be critical to ascertain if modulators and/or substrates of DNA-PKcs change as cancers progress and mutate, particularly when designing therapeutic strategies aimed at specific stages of disease.
The continual discovery of novel means of regulation and roles of DNA-PK suggests that despite recent advances, thorough understanding of the cellular responsibilities of DNA-PK has yet to be realized. Additional studies regarding context dependent mechanisms of activation, global impact on transcription, kinase-independent functions, and alteration of function and regulation during cancer development and progression are needed to further characterize the function of DNA-PK, as each new advance may define a novel therapeutic strategy in any number of cancer types. While DNA-PK is certainly a critical component of the cellular response to DNA damage, it is clear that DNA-PK exerts pleiotropic cellular roles that yield major impact on human malignancies.
STATEMENT OF SIGNIFICANCE.
While classically considered a component of damage response, recent findings illuminate damage-independent functions of DNA-PK that impact multiple tumor-associated pathways and provide rationale for development of novel therapeutic strategies.
Acknowledgments
The authors thank members of the K. Knudsen laboratory for input and commentary.
GRANT SUPPORT
K.E. Knudsen is supported by grants from NIH (R01 CA159945 and R01 CA099996) and a Prostate Cancer Foundation Mazzone Challenge Award
Footnotes
Financial Disclosure: KEK receives research support from Celgene
References
- 1.Jackson SP, MacDonald JJ, Lees-Miller S, Tjian R. GC box binding induces phosphorylation of Sp1 by a DNA-dependent protein kinase. Cell. 1990;63:155–65. doi: 10.1016/0092-8674(90)90296-q. [DOI] [PubMed] [Google Scholar]
- 2.Dvir A, Peterson SR, Knuth MW, Lu H, Dynan WS. Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proc Natl Acad Sci U S A. 1992;89:11920–4. doi: 10.1073/pnas.89.24.11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shintani S, Mihara M, Li C, Nakahara Y, Hino S, Nakashiro K, et al. Up-regulation of DNA-dependent protein kinase correlates with radiation resistance in oral squamous cell carcinoma. Cancer Sci. 2003;94:894–900. doi: 10.1111/j.1349-7006.2003.tb01372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Um JH, Kwon JK, Kang CD, Kim MJ, Ju DS, Bae JH, et al. Relationship between antiapoptotic molecules and metastatic potency and the involvement of DNA-dependent protein kinase in the chemosensitization of metastatic human cancer cells by epidermal growth factor receptor blockade. J Pharmacol Exp Ther. 2004;311:1062–70. doi: 10.1124/jpet.104.070938. [DOI] [PubMed] [Google Scholar]
- 5.Beskow C, Skikuniene J, Holgersson A, Nilsson B, Lewensohn R, Kanter L, et al. Radioresistant cervical cancer shows upregulation of the NHEJ proteins DNA-PKcs, Ku70 and Ku86. Br J Cancer. 2009;101:816–21. doi: 10.1038/sj.bjc.6605201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouchaert P, Guerif S, Debiais C, Irani J, Fromont G. DNA-PKcs expression predicts response to radiotherapy in prostate cancer. Int J Radiat Oncol Biol Phys. 2012;84:1179–85. doi: 10.1016/j.ijrobp.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 7.Ni X, Zhang Y, Ribas J, Chowdhury WH, Castanares M, Zhang Z, et al. Prostate-targeted radiosensitization via aptamer-shRNA chimeras in human tumor xenografts. J Clin Invest. 2011;121:2383–90. doi: 10.1172/JCI45109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tonotsuka N, Hosoi Y, Miyazaki S, Miyata G, Sugawara K, Mori T, et al. Heterogeneous expression of DNA-dependent protein kinase in esophageal cancer and normal epithelium. Int J Mol Med. 2006;18:441–7. [PubMed] [Google Scholar]
- 9.Xing J, Wu X, Vaporciyan AA, Spitz MR, Gu J. Prognostic significance of ataxia-telangiectasia mutated, DNA-dependent protein kinase catalytic subunit, and Ku heterodimeric regulatory complex 86-kD subunit expression in patients with nonsmall cell lung cancer. Cancer. 2008;112:2756–64. doi: 10.1002/cncr.23533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willmore E, Elliott SL, Mainou-Fowler T, Summerfield GP, Jackson GH, O’Neill F, et al. DNA-dependent protein kinase is a therapeutic target and an indicator of poor prognosis in B-cell chronic lymphocytic leukemia. Clin Cancer Res. 2008;14:3984–92. doi: 10.1158/1078-0432.CCR-07-5158. [DOI] [PubMed] [Google Scholar]
- 11.Hosoi Y, Watanabe T, Nakagawa K, Matsumoto Y, Enomoto A, Morita A, et al. Up-regulation of DNA-dependent protein kinase activity and Sp1 in colorectal cancer. Int J Oncol. 2004;25:461–8. [PubMed] [Google Scholar]
- 12.Kurimasa A, Kumano S, Boubnov NV, Story MD, Tung CS, Peterson SR, et al. Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol Cell Biol. 1999;19:3877–84. doi: 10.1128/mcb.19.5.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Y, Thomas HD, Batey MA, Cowell IG, Richardson CJ, Griffin RJ, et al. Preclinical evaluation of a potent novel DNA-dependent protein kinase inhibitor NU7441. Cancer Res. 2006;66:5354–62. doi: 10.1158/0008-5472.CAN-05-4275. [DOI] [PubMed] [Google Scholar]
- 14.Hartley KO, Gell D, Smith GC, Zhang H, Divecha N, Connelly MA, et al. DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell. 1995;82:849–56. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- 15.Kim ST, Lim DS, Canman CE, Kastan MB. Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem. 1999;274:37538–43. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- 16.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–53. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 17.O’Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- 18.Lempiainen H, Halazonetis TD. Emerging common themes in regulation of PIKKs and PI3Ks. EMBO J. 2009;28:3067–73. doi: 10.1038/emboj.2009.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Burg M, Ijspeert H, Verkaik NS, Turul T, Wiegant WW, Morotomi-Yano K, et al. A DNA-PKcs mutation in a radiosensitive T-B- SCID patient inhibits Artemis activation and nonhomologous end-joining. J Clin Invest. 2009;119:91–8. doi: 10.1172/JCI37141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu FM, Zhang S, Chen BP. Role of DNA-dependent protein kinase catalytic subunit in cancer development and treatment. Transl Cancer Res. 2012;1:22–34. doi: 10.3978/j.issn.2218-676X.2012.04.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blier PR, Griffith AJ, Craft J, Hardin JA. Binding of Ku protein to DNA. Measurement of affinity for ends and demonstration of binding to nicks. J Biol Chem. 1993;268:7594–601. [PubMed] [Google Scholar]
- 22.Dobbs TA, Tainer JA, Lees-Miller SP. A structural model for regulation of NHEJ by DNA-PKcs autophosphorylation. DNA Repair (Amst) 2010;9:1307–14. doi: 10.1016/j.dnarep.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rivera-Calzada A, Maman JD, Spagnolo L, Pearl LH, Llorca O. Three-dimensional structure and regulation of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) Structure. 2005;13:243–55. doi: 10.1016/j.str.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 24.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jhappan C, Morse HC, 3rd, Fleischmann RD, Gottesman MM, Merlino G. DNA-PKcs: a T-cell tumour suppressor encoded at the mouse scid locus. Nat Genet. 1997;17:483–6. doi: 10.1038/ng1297-483. [DOI] [PubMed] [Google Scholar]
- 26.Yoo S, Dynan WS. Geometry of a complex formed by double strand break repair proteins at a single DNA end: recruitment of DNA-PKcs induces inward translocation of Ku protein. Nucleic Acids Res. 1999;27:4679–86. doi: 10.1093/nar/27.24.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neal JA, Meek K. Choosing the right path: does DNA-PK help make the decision? Mutat Res. 2011;711:73–86. doi: 10.1016/j.mrfmmm.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.An J, Huang YC, Xu QZ, Zhou LJ, Shang ZF, Huang B, et al. DNA-PKcs plays a dominant role in the regulation of H2AX phosphorylation in response to DNA damage and cell cycle progression. BMC Mol Biol. 2010;11:18. doi: 10.1186/1471-2199-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sak A, Stuschke M. Use of gammaH2AX and other biomarkers of double-strand breaks during radiotherapy. Semin Radiat Oncol. 2010;20:223–31. doi: 10.1016/j.semradonc.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 30.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10:886–95. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 31.Lukas C, Melander F, Stucki M, Falck J, Bekker-Jensen S, Goldberg M, et al. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 2004;23:2674–83. doi: 10.1038/sj.emboj.7600269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan DW, Chen BP, Prithivirajsingh S, Kurimasa A, Story MD, Qin J, et al. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 2002;16:2333–8. doi: 10.1101/gad.1015202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Douglas P, Sapkota GP, Morrice N, Yu Y, Goodarzi AA, Merkle D, et al. Identification of in vitro and in vivo phosphorylation sites in the catalytic subunit of the DNA-dependent protein kinase. Biochem J. 2002;368:243–51. doi: 10.1042/BJ20020973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meek K, Dang V, Lees-Miller SP. DNA-PK: the means to justify the ends? Adv Immunol. 2008;99:33–58. doi: 10.1016/S0065-2776(08)00602-0. [DOI] [PubMed] [Google Scholar]
- 35.Meek K, Douglas P, Cui X, Ding Q, Lees-Miller SP. trans Autophosphorylation at DNA-dependent protein kinase’s two major autophosphorylation site clusters facilitates end processing but not end joining. Mol Cell Biol. 2007;27:3881–90. doi: 10.1128/MCB.02366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Douglas P, Cui X, Block WD, Yu Y, Gupta S, Ding Q, et al. The DNA-dependent protein kinase catalytic subunit is phosphorylated in vivo on threonine 3950, a highly conserved amino acid in the protein kinase domain. Mol Cell Biol. 2007;27:1581–91. doi: 10.1128/MCB.01962-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merkle D, Douglas P, Moorhead GB, Leonenko Z, Yu Y, Cramb D, et al. The DNA-dependent protein kinase interacts with DNA to form a protein-DNA complex that is disrupted by phosphorylation. Biochemistry. 2002;41:12706–14. doi: 10.1021/bi0263558. [DOI] [PubMed] [Google Scholar]
- 38.Wang C, Lees-Miller SP. Detection and repair of ionizing radiation-induced DNA double strand breaks: new developments in nonhomologous end joining. Int J Radiat Oncol Biol Phys. 2013;86:440–9. doi: 10.1016/j.ijrobp.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–57. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 40.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–71. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 41.Escribano-Diaz C, Orthwein A, Fradet-Turcotte A, Xing M, Young JT, Tkac J, et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell. 2013;49:872–83. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 42.Shibata A, Moiani D, Arvai AS, Perry J, Harding SM, Genois MM, et al. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol Cell. 2014;53:7–18. doi: 10.1016/j.molcel.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fraser M, Harding SM, Zhao H, Coackley C, Durocher D, Bristow RG. MRE11 promotes AKT phosphorylation in direct response to DNA double-strand breaks. Cell Cycle. 2011;10:2218–32. doi: 10.4161/cc.10.13.16305. [DOI] [PubMed] [Google Scholar]
- 44.Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 45.Beucher A, Birraux J, Tchouandong L, Barton O, Shibata A, Conrad S, et al. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 2009;28:3413–27. doi: 10.1038/emboj.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delacote F, Han M, Stamato TD, Jasin M, Lopez BS. An xrcc4 defect or Wortmannin stimulates homologous recombination specifically induced by double-strand breaks in mammalian cells. Nucleic Acids Res. 2002;30:3454–63. doi: 10.1093/nar/gkf452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen F, Nastasi A, Shen Z, Brenneman M, Crissman H, Chen DJ. Cell cycle-dependent protein expression of mammalian homologs of yeast DNA double-strand break repair genes Rad51 and Rad52. Mutat Res. 1997;384:205–11. doi: 10.1016/s0921-8777(97)00020-7. [DOI] [PubMed] [Google Scholar]
- 48.Mao Z, Bozzella M, Seluanov A, Gorbunova V. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst) 2008;7:1765–71. doi: 10.1016/j.dnarep.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Convery E, Shin EK, Ding Q, Wang W, Douglas P, Davis LS, et al. Inhibition of homologous recombination by variants of the catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs) Proc Natl Acad Sci U S A. 2005;102:1345–50. doi: 10.1073/pnas.0406466102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neal JA, Dang V, Douglas P, Wold MS, Lees-Miller SP, Meek K. Inhibition of homologous recombination by DNA-dependent protein kinase requires kinase activity, is titratable, and is modulated by autophosphorylation. Mol Cell Biol. 2011;31:1719–33. doi: 10.1128/MCB.01298-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kong X, Shen Y, Jiang N, Fei X, Mi J. Emerging roles of DNA-PK besides DNA repair. Cell Signal. 2011;23:1273–80. doi: 10.1016/j.cellsig.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 52.Meek K, Lees-Miller SP, Modesti M. N-terminal constraint activates the catalytic subunit of the DNA-dependent protein kinase in the absence of DNA or Ku. Nucleic Acids Res. 2012;40:2964–73. doi: 10.1093/nar/gkr1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davis AJ, Lee KJ, Chen DJ. The N-terminal region of the DNA-dependent protein kinase catalytic subunit is required for its DNA double-stranded break-mediated activation. J Biol Chem. 2013;288:7037–46. doi: 10.1074/jbc.M112.434498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Toulany M, Kehlbach R, Florczak U, Sak A, Wang S, Chen J, et al. Targeting of AKT1 enhances radiation toxicity of human tumor cells by inhibiting DNA-PKcs-dependent DNA double-strand break repair. Mol Cancer Ther. 2008;7:1772–81. doi: 10.1158/1535-7163.MCT-07-2200. [DOI] [PubMed] [Google Scholar]
- 55.Toulany M, Lee KJ, Fattah KR, Lin YF, Fehrenbacher B, Schaller M, et al. Akt promotes post-irradiation survival of human tumor cells through initiation, progression, and termination of DNA-PKcs-dependent DNA double-strand break repair. Mol Cancer Res. 2012;10:945–57. doi: 10.1158/1541-7786.MCR-11-0592. [DOI] [PubMed] [Google Scholar]
- 56.Liccardi G, Hartley JA, Hochhauser D. EGFR nuclear translocation modulates DNA repair following cisplatin and ionizing radiation treatment. Cancer Res. 2011;71:1103–14. doi: 10.1158/0008-5472.CAN-10-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mukohara T, Kudoh S, Matsuura K, Yamauchi S, Kimura T, Yoshimura N, et al. Activated Akt expression has significant correlation with EGFR and TGF-alpha expressions in stage I NSCLC. Anticancer Res. 2004;24:11–7. [PubMed] [Google Scholar]
- 58.Toulany M, Schickfluss TA, Fattah KR, Lee KJ, Chen BP, Fehrenbacher B, et al. Function of erbB receptors and DNA-PKcs on phosphorylation of cytoplasmic and nuclear Akt at S473 induced by erbB1 ligand and ionizing radiation. Radiother Oncol. 2011;101:140–6. doi: 10.1016/j.radonc.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 59.Wei F, Yan J, Tang D, Lin X, He L, Xie Y, et al. Inhibition of ERK activation enhances the repair of double-stranded breaks via non-homologous end joining by increasing DNA-PKcs activation. Biochim Biophys Acta. 2013;1833:90–100. doi: 10.1016/j.bbamcr.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 60.Olsen BB, Issinger OG, Guerra B. Regulation of DNA-dependent protein kinase by protein kinase CK2 in human glioblastoma cells. Oncogene. 2010;29:6016–26. doi: 10.1038/onc.2010.337. [DOI] [PubMed] [Google Scholar]
- 61.BB, Wang SY, Svenstrup TH, Chen BP, Guerra B. Protein kinase CK2 localizes to sites of DNA double-strand break regulating the cellular response to DNA damage. BMC Mol Biol. 2012;13:7. doi: 10.1186/1471-2199-13-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olsen BB, Svenstrup TH, Guerra B. Downregulation of protein kinase CK2 induces autophagic cell death through modulation of the mTOR and MAPK signaling pathways in human glioblastoma cells. Int J Oncol. 2012;41:1967–76. doi: 10.3892/ijo.2012.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mi J, Dziegielewski J, Bolesta E, Brautigan DL, Larner JM. Activation of DNA-PK by ionizing radiation is mediated by protein phosphatase 6. PLoS One. 2009;4:e4395. doi: 10.1371/journal.pone.0004395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Douglas P, Zhong J, Ye R, Moorhead GB, Xu X, Lees-Miller SP. Protein phosphatase 6 interacts with the DNA-dependent protein kinase catalytic subunit and dephosphorylates gamma-H2AX. Mol Cell Biol. 2010;30:1368–81. doi: 10.1128/MCB.00741-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hosing AS, Valerie NC, Dziegielewski J, Brautigan DL, Larner JM. PP6 regulatory subunit R1 is bidentate anchor for targeting protein phosphatase-6 to DNA-dependent protein kinase. J Biol Chem. 2012;287:9230–9. doi: 10.1074/jbc.M111.333708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reichman TW, Muniz LC, Mathews MB. The RNA binding protein nuclear factor 90 functions as both a positive and negative regulator of gene expression in mammalian cells. Mol Cell Biol. 2002;22:343–56. doi: 10.1128/MCB.22.1.343-356.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shamanna RA, Hoque M, Lewis-Antes A, Azzam EI, Lagunoff D, Pe’ery T, et al. The NF90/NF45 complex participates in DNA break repair via nonhomologous end joining. Mol Cell Biol. 2011;31:4832–43. doi: 10.1128/MCB.05849-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–13. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 69.Huang L, Snyder AR, Morgan WF. Radiation-induced genomic instability and its implications for radiation carcinogenesis. Oncogene. 2003;22:5848–54. doi: 10.1038/sj.onc.1206697. [DOI] [PubMed] [Google Scholar]
- 70.Kajita M, McClinic KN, Wade PA. Aberrant expression of the transcription factors snail and slug alters the response to genotoxic stress. Mol Cell Biol. 2004;24:7559–66. doi: 10.1128/MCB.24.17.7559-7566.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pyun BJ, Seo HR, Lee HJ, Jin YB, Kim EJ, Kim NH, et al. Mutual regulation between DNA-PKcs and Snail1 leads to increased genomic instability and aggressive tumor characteristics. Cell Death Dis. 2013;4:e517. doi: 10.1038/cddis.2013.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee KJ, Lin YF, Chou HY, Yajima H, Fattah KR, Lee SC, et al. Involvement of DNA-dependent protein kinase in normal cell cycle progression through mitosis. J Biol Chem. 2011;286:12796–802. doi: 10.1074/jbc.M110.212969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Block WD, Yu Y, Lees-Miller SP. Phosphatidyl inositol 3-kinase-like serine/threonine protein kinases (PIKKs) are required for DNA damage-induced phosphorylation of the 32 kDa subunit of replication protein A at threonine 21. Nucleic Acids Res. 2004;32:997–1005. doi: 10.1093/nar/gkh265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu S, Opiyo SO, Manthey K, Glanzer JG, Ashley AK, Amerin C, et al. Distinct roles for DNA-PK, ATM and ATR in RPA phosphorylation and checkpoint activation in response to replication stress. Nucleic Acids Res. 2012;40:10780–94. doi: 10.1093/nar/gks849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wenger RH, Stiehl DP, Camenisch G. Integration of oxygen signaling at the consensus HRE. Sci STKE. 2005;2005:re12. doi: 10.1126/stke.3062005re12. [DOI] [PubMed] [Google Scholar]
- 76.Bouquet F, Ousset M, Biard D, Fallone F, Dauvillier S, Frit P, et al. A DNA-dependent stress response involving DNA-PK occurs in hypoxic cells and contributes to cellular adaptation to hypoxia. J Cell Sci. 2011;124:1943–51. doi: 10.1242/jcs.078030. [DOI] [PubMed] [Google Scholar]
- 77.Wrann S, Kaufmann MR, Wirthner R, Stiehl DP, Wenger RH. HIF mediated and DNA damage independent histone H2AX phosphorylation in chronic hypoxia. Biol Chem. 2013;394:519–28. doi: 10.1515/hsz-2012-0311. [DOI] [PubMed] [Google Scholar]
- 78.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 79.Madan E, Gogna R, Pati U. p53 Ser15 phosphorylation disrupts the p53-RPA70 complex and induces RPA70-mediated DNA repair in hypoxia. Biochem J. 2012;443:811–20. doi: 10.1042/BJ20111627. [DOI] [PubMed] [Google Scholar]
- 80.Wong RH, Chang I, Hudak CS, Hyun S, Kwan HY, Sul HS. A role of DNA-PK for the metabolic gene regulation in response to insulin. Cell. 2009;136:1056–72. doi: 10.1016/j.cell.2008.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Warden SM, Richardson C, O’Donnell J, Jr, Stapleton D, Kemp BE, Witters LA. Post-translational modifications of the beta-1 subunit of AMP-activated protein kinase affect enzyme activity and cellular localization. Biochem J. 2001;354:275–83. doi: 10.1042/0264-6021:3540275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Amatya PN, Kim HB, Park SJ, Youn CK, Hyun JW, Chang IY, et al. A role of DNA-dependent protein kinase for the activation of AMP-activated protein kinase in response to glucose deprivation. Biochim Biophys Acta. 2012;1823:2099–108. doi: 10.1016/j.bbamcr.2012.08.022. [DOI] [PubMed] [Google Scholar]
- 83.Pichlmair A, Reis e Sousa C. Innate recognition of viruses. Immunity. 2007;27:370–83. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 84.Ju J, Naura AS, Errami Y, Zerfaoui M, Kim H, Kim JG, et al. Phosphorylation of p50 NF-kappaB at a single serine residue by DNA-dependent protein kinase is critical for VCAM-1 expression upon TNF treatment. J Biol Chem. 2010;285:41152–60. doi: 10.1074/jbc.M110.158352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ferguson BJ, Mansur DS, Peters NE, Ren H, Smith GL. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. Elife. 2012;1:e00047. doi: 10.7554/eLife.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 87.O’Brien T, Hardin S, Greenleaf A, Lis JT. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature. 1994;370:75–7. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- 88.Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, et al. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature. 1996;381:86–9. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 89.Tyagi S, Ochem A, Tyagi M. DNA-dependent protein kinase interacts functionally with the RNA polymerase II complex recruited at the human immunodeficiency virus (HIV) long terminal repeat and plays an important role in HIV gene expression. J Gen Virol. 2011;92:1710–20. doi: 10.1099/vir.0.029587-0. [DOI] [PubMed] [Google Scholar]
- 90.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–81. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pankotai T, Bonhomme C, Chen D, Soutoglou E. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat Struct Mol Biol. 2012;19:276–82. doi: 10.1038/nsmb.2224. [DOI] [PubMed] [Google Scholar]
- 92.Anderson MS, Su MA. Aire and T cell development. Curr Opin Immunol. 2011;23:198–206. doi: 10.1016/j.coi.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zumer K, Low AK, Jiang H, Saksela K, Peterlin BM. Unmodified histone H3K4 and DNA-dependent protein kinase recruit autoimmune regulator to target genes. Mol Cell Biol. 2012;32:1354–62. doi: 10.1128/MCB.06359-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wu J, Zhu W, Fu H, Zhang Y, Sun J, Yang W, et al. DNA-PKcs interacts with Aire and regulates the expression of toll-like receptors in RAW264 7 cells. Scand J Immunol. 2012;75:479–88. doi: 10.1111/j.1365-3083.2012.02682.x. [DOI] [PubMed] [Google Scholar]
- 95.Hill R, Madureira PA, Waisman DM, Lee PW. DNA-PKCS binding to p53 on the p21WAF1/CIP1 promoter blocks transcription resulting in cell death. Oncotarget. 2011;2:1094–108. doi: 10.18632/oncotarget.378. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 96.Burma S, Kurimasa A, Xie G, Taya Y, Araki R, Abe M, et al. DNA-dependent protein kinase-independent activation of p53 in response to DNA damage. J Biol Chem. 1999;274:17139–43. doi: 10.1074/jbc.274.24.17139. [DOI] [PubMed] [Google Scholar]
- 97.Bustin M, Catez F, Lim JH. The dynamics of histone H1 function in chromatin. Mol Cell. 2005;17:617–20. doi: 10.1016/j.molcel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 98.Kim K, Jeong KW, Kim H, Choi J, Lu W, Stallcup MR, et al. Functional interplay between p53 acetylation and H1. 2 phosphorylation in p53-regulated transcription. Oncogene. 2012;31:4290–301. doi: 10.1038/onc.2011.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Weigel NL, Carter TH, Schrader WT, O’Malley BW. Chicken progesterone receptor is phosphorylated by a DNA-dependent protein kinase during in vitro transcription assays. Mol Endocrinol. 1992;6:8–14. doi: 10.1210/mend.6.1.1738374. [DOI] [PubMed] [Google Scholar]
- 100.Giffin W, Kwast-Welfeld J, Rodda DJ, Prefontaine GG, Traykova-Andonova M, Zhang Y, et al. Sequence-specific DNA binding and transcription factor phosphorylation by Ku Autoantigen/DNA-dependent protein kinase. Phosphorylation of Ser-527 of the rat glucocorticoid receptor. J Biol Chem. 1997;272:5647–58. doi: 10.1074/jbc.272.9.5647. [DOI] [PubMed] [Google Scholar]
- 101.Goodwin JF, Schiewer MJ, Dean JL, Schrecengost RS, de Leeuw R, Han S, et al. A hormone-DNA repair circuit governs the response to genotoxic insult. Cancer Discov. 2013;3:1254–71. doi: 10.1158/2159-8290.CD-13-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 103.Helgeson BE, Tomlins SA, Shah N, Laxman B, Cao Q, Prensner JR, et al. Characterization of TMPRSS2:ETV5 and SLC45A3:ETV5 gene fusions in prostate cancer. Cancer Res. 2008;68:73–80. doi: 10.1158/0008-5472.CAN-07-5352. [DOI] [PubMed] [Google Scholar]
- 104.Zong Y, Xin L, Goldstein AS, Lawson DA, Teitell MA, Witte ON. ETS family transcription factors collaborate with alternative signaling pathways to induce carcinoma from adult murine prostate cells. Proc Natl Acad Sci U S A. 2009;106:12465–70. doi: 10.1073/pnas.0905931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Brenner JC, Ateeq B, Li Y, Yocum AK, Cao Q, Asangani IA, et al. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 2011;19:664–78. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Spagnolo L, Barbeau J, Curtin NJ, Morris EP, Pearl LH. Visualization of a DNA-PK/PARP1 complex. Nucleic Acids Res. 2012;40:4168–77. doi: 10.1093/nar/gkr1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Calkins AS, Iglehart JD, Lazaro JB. DNA damage-induced inhibition of rRNA synthesis by DNA-PK and PARP-1. Nucleic Acids Res. 2013;41:7378–86. doi: 10.1093/nar/gkt502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schiewer MJ, Goodwin JF, Han S, Brenner JC, Augello MA, Dean JL, et al. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov. 2012;2:1134–49. doi: 10.1158/2159-8290.CD-12-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Medunjanin S, Weinert S, Schmeisser A, Mayer D, Braun-Dullaeus RC. Interaction of the double-strand break repair kinase DNA-PK and estrogen receptor-alpha. Mol Biol Cell. 2010;21:1620–8. doi: 10.1091/mbc.E09-08-0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Medunjanin S, Weinert S, Poitz D, Schmeisser A, Strasser RH, Braun-Dullaeus RC. Transcriptional activation of DNA-dependent protein kinase catalytic subunit gene expression by oestrogen receptor-alpha. EMBO Rep. 2010;11:208–13. doi: 10.1038/embor.2009.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Foulds CE, Feng Q, Ding C, Bailey S, Hunsaker TL, Malovannaya A, et al. Proteomic analysis of coregulators bound to ERalpha on DNA and nucleosomes reveals coregulator dynamics. Mol Cell. 2013;51:185–99. doi: 10.1016/j.molcel.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gurley KE, Kemp CJ. Synthetic lethality between mutation in Atm and DNA-PK(cs) during murine embryogenesis. Curr Biol. 2001;11:191–4. doi: 10.1016/s0960-9822(01)00048-3. [DOI] [PubMed] [Google Scholar]
- 113.Riabinska A, Daheim M, Herter-Sprie GS, Winkler J, Fritz C, Hallek M, et al. Therapeutic targeting of a robust non-oncogene addiction to PRKDC in ATM-defective tumors. Sci Transl Med. 2013;5:189ra78. doi: 10.1126/scitranslmed.3005814. [DOI] [PubMed] [Google Scholar]
- 114.Dietlein F, Thelen L, Jokic M, Jachimowicz RD, Ivan L, Knittel G, et al. A Functional Cancer Genomics Screen Identifies a Druggable Synthetic Lethal Interaction between MSH3 and PRKDC. Cancer Discov. 2014;4:592–605. doi: 10.1158/2159-8290.CD-13-0907. [DOI] [PubMed] [Google Scholar]
- 115.Davidson D, Amrein L, Panasci L, Aloyz R. Small Molecules, Inhibitors of DNA-PK, Targeting DNA Repair, and Beyond. Front Pharmacol. 2013;4:5. doi: 10.3389/fphar.2013.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]