Summary
The tumor suppressor protein BRCA1 promotes homologous recombination (HR), a high fidelity mechanism to repair DNA double-strand breaks (DSBs) that arise during normal replication and in response to DNA damaging agents. Recent genetic experiments indicate that BRCA1 also performs an HR-independent function during the repair of DNA interstrand crosslinks (ICLs). Here, we show that BRCA1 is required to unload the CMG helicase complex from chromatin after replication forks collide with an ICL. Eviction of the stalled helicase allows leading strands to be extended toward the ICL, followed by endonucleolytic processing of the crosslink, lesion bypass, and DSB repair. Our results identify BRCA1-dependent helicase unloading as a critical, early event in ICL repair.
Introduction
Mutations in BRCA1 predispose individuals to hereditary breast and ovarian cancers (Narod and Foulkes, 2004). Growing evidence also indicates that BRCA1 loss plays an important role in the development of sporadic cancers (Chalasani and Livingston, 2013; Leeneer et al., 2011). In the absence of BRCA1, cells develop multiple chromosomal abnormalities, implicating genome maintenance in tumor suppression (Zhang, 2013). Consistent with this, BRCA1 has been linked to various aspects of the DNA damage response (Wu et al., 2010) including error-free repair of DNA double-strand breaks (DSBs) (Bekker-Jensen and Mailand, 2010).
BRCA1 forms a heterodimeric complex with BARD1 (BRCA1-associated RING domain protein 1), which is required for BRCA1 stability and function (Choudhury et al., 2004; Westermark et al., 2003). BRCA1 activity is also modulated by numerous protein interactions that form distinct BRCA1-containing complexes (Silver and Livingston, 2012; Wang, 2012). In response to DSBs, BRCA1 regulates repair pathway choice, promoting template-directed repair by homologous recombination (HR) over non-homologous end joining (NHEJ), an error-prone pathway (Kass and Jasin, 2010). BRCA1 is thought to support resection of DSB ends, leading to the generation of a 3’ single-stranded DNA (ssDNA) tail that is bound by the RAD51 recombinase. BRCA1 also associates with BRCA2 (via PALB2/FANCN) (Zhang et al., 2009), which stimulates RAD51 loading onto ssDNA (Jensen et al., 2010; Liu et al., 2010).
BRCA1-deficient cells are sensitive to various DNA damaging agents, including DNA interstrand cross-links (ICLs) (Bhattacharyya et al., 2000). ICLs covalently link the two strands of the double helix, thereby blocking cellular processes that require strand separation, such as DNA replication and transcription. Cellular resistance to ICLs is dependent on both the BRCA and Fanconi anemia (FANC) proteins, which act together in a common DNA repair pathway (Kim and D'Andrea, 2012). ICL repair involves a DSB intermediate, which is formed after replication forks collide with an ICL (Raschle et al., 2008)(McHugh et al., 2001). As such, ICL sensitivity in BRCA1-deficient cells has been attributed primarily to BRCA1’s HR functions.
Recent genetic data indicate that BRCA1 has an additional function during ICL repair that is distinct from its established role in HR. In 2010, Nussenzweig’s group showed that the HR defect in BRCA1-deficient cells is almost completely reversed by mutation of 53BP1 (Bunting et al., 2010), an NHEJ protein that modulates chromatin structure at DNA breaks. These results argued that the primary function of BRCA1 in DSB repair is to promote resection by antagonizing 53BP1. More recently, they discovered that loss of 53BP1 does not rescue the ICL sensitivity observed in BRCA1-deficient cells, even though RAD51 foci formation was largely restored (Bunting et al., 2012). These results argue that BRCA1 performs an additional function in ICL repair that is independent of DSB resection, RAD51 loading, and 53BP1. Notably, FANCD2 foci formation was impaired in BRCA1-deficient cells after exposure to DNA crosslinking agents (Bunting et al., 2012), suggesting that BRCA1’s HR-independent function might involve recruitment of FANCD2 to ICLs.
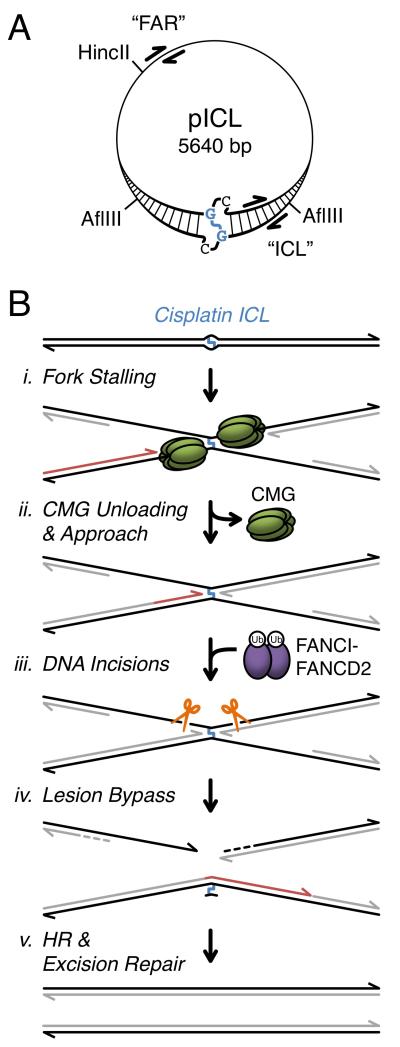
Using Xenopus egg extracts, we previously established a cell-free system that recapitulates replication-coupled repair of a single, site-specific cisplatin ICL on a plasmid (pICL, Figure 1A) (Raschle et al., 2008). Error-free removal of the crosslink regenerates a SapI restriction site, which is used to assay repair. Upon addition of pICL to egg extracts, replication initiates at a random location, and two replication forks rapidly converge on the ICL and stall (Figure 1B, i). The 3’ ends of the two stalled leading strands are initially located ~20-40 nucleotides from the crosslink (“−20 position”). After a ~15 minute delay, the leading strands are extended to within one nucleotide of the crosslink (“−1 position”). Extension of leading strands from −20 to −1 (“Approach”, Figure 1B, ii) occurs concurrently with unloading of the CMG replicative DNA helicase (Fu et al., 2011), which is comprised of Cdc45, MCM2-7, and GINS (Ilves et al., 2010). Based on this correlation, we proposed that leading strand stalling at −20 is due to steric hindrance by CMG and that Approach requires CMG unloading (Fu et al., 2011). Concurrent with Approach, the Fanconi anemia pathway is activated, leading to mono-ubiquitylation of the FANCI-FANCD2 complex. Ubiquitylated FANCI-FANCD2 promotes incisions by XPF-ERCC1 and possibly other endonucleases, creating a DSB in one sister chromatid (Figure 1B, iii) (Douwel et al., 2014; Knipscheer et al., 2009). The leading strand is then extended past the unhooked ICL by translesion DNA polymerases (Figure 1B, iv), creating an intact template for recombination-mediated repair of the DSB (Figure 1B, v) (Long et al., 2011). Finally, the unhooked adduct is probably removed by excision repair (Muniandy et al., 2010), although this event does not occur in egg extracts.
Figure 1. ICL repair in Xenopus egg extract.
(A) pICL schematic. ICL and cross-linked nucleotides are shown in blue. ChIP primer pairs are shown for “ICL” (25-132 bp from ICL) and “FAR” (2523-2622 bp from ICL) loci. (B) Model of ICL repair (Fu et al., 2011; Knipscheer et al., 2009; Long et al., 2011; Raschle et al., 2008). Parental DNA strands are black, and nascent strands are gray, or red for emphasis. CMG (replicative helicase complex comprised of Cdc45, MCM7, and Sld5), Ub (ubiquitin). See Figure S7 for revised model.
Here, we show that ubiquitin signaling targets BRCA1 to ICL-stalled forks where BRCA1 promotes unloading of the CMG helicase, allowing Approach and FANCI-FANCD2-dependent endonucleases to excise the crosslink and facilitate repair. Our results identify CMG unloading as a critical, early event in ICL repair and identify a new function for BRCA1 in the DNA damage response.
Results
Ubiquitin signaling is required for chromatin unloading of the replicative helicase
Ubiquitin signaling plays an integral role in targeting repair factors to sites of damaged chromatin (Pinder et al., 2013). To investigate the role of ubiquitin signaling in ICL repair, we employed ubiquitin vinyl-sulfone (UbVS), a highly specific, irreversible inhibitor of deubiquitylating enzymes (Borodovsky et al., 2001). Incubation of Xenopus egg extract with UbVS blocks ubiquitin turnover, leading to the depletion of free ubiquitin (Dimova et al., 2012). Extracts were incubated with buffer, UbVS, or UbVS and excess free ubiquitin prior to addition of pICL. Although DNA synthesis was not significantly inhibited by the addition of UbVS (Figure 2A), ICL repair was abolished (Figure 2B). Only a limited amount of repair was rescued by the addition of free ubiquitin, suggesting that turnover of ubiquitylated substrates is important for repair, even in the presence of excess ubiquitin (Nijman et al., 2005; Oestergaard et al., 2007). Consistent with this idea, addition of free ubiquitin reversed the FANCD2 ubiquitylation defect caused by UbVS but did not restore the FANCD2 de-ubiquitylation that is normally observed late in the reaction (Figure S1A and B).
Figure 2. Ubiquitin signaling is required for replicative helicase unloading.
pICL was replicated in egg extract supplemented with buffer (+Buffer), 14 μM UbVS (+UbVS), or 14 μM UbVS and 50 μM ubiquitin (+UbVS+Ub). Samples were analyzed by agarose gel electrophoresis to determine the efficiency of (A) replication and (B) ICL repair (described in Methods). (C-E) Protein recruitment to the ICL was analyzed by ChIP with the indicated antibodies. (F) Schematic of leading strand intermediates from the rightward moving fork as it bypasses the ICL. (G) Nascent strand products were analyzed by denaturing polyacrylamide gel electrophoresis. (H-J) ICL recruitment was analyzed by ChIP with the indicated antibodies. Note that the MCM7 ChIP signal starts high because MCM2-7 has already been loaded onto DNA at the 0 minute time point as a result of licensing in HSS extract (see Methods). All data shown was analyzed from a single experiment. See Figure S1 for primary gel data, ChIP recovery at the FAR locus, quantification of nascent strand products, and experimental replicates.
DSBs trigger a histone modification cascade that includes histone ubiquitylation and subsequent recruitment of various repair factors to the site of damage, including Rap80, BRCA1, and FANCD2 (Wang et al., 2004; Yan and Jetten, 2008). To determine whether a similar response is activated during ICL repair in egg extracts, we used chromatin immunoprecipitation (ChIP) to analyze protein recruitment to pICL. As shown in Figures 2C-E, Rap80, BRCA1, and FANCD2 were each recruited to ICLs, but not when UbVS was present. Recruitment was rescued by the addition of free ubiquitin, indicating that recruitment defects were due to ubiquitin depletion.
To investigate how UbVS affects ICL repair, nascent strand products (Figure 2F) were analyzed by denaturing polyacrylamide gel electrophoresis. UbVS treatment had no effect on the arrival of leading strands at the ICL (Figure 2G, compare lanes 7, 13, and 19; Figure S1K-Q for experimental replicates), consistent with replication proceeding normally (Figure 2A). In contrast, UbVS completely blocked the Approach of leading strands to the −1 position, as well as formation of all downstream nascent strand products (Figure 2G, compare lanes 7-11 with 13-17). Addition of free ubiquitin with UbVS restored Approach, Insertion, and Extension, albeit with delayed kinetics (Figure 2G, lanes 19-23).
We showed previously that Approach correlates with dissociation of the CMG helicase (Fu et al., 2011). These results suggested that failure of the Approach step after ubiquitin depletion might be caused by persistence of CMG at the ICL. To test this idea, several helicase components were analyzed by ChIP. Strikingly, unloading of Cdc45, MCM7, and Sld5 was severely delayed in UbVS-treated reactions (Figure 2H-J). As for Approach, CMG unloading was partially restored by the addition of free ubiquitin. Together, these results demonstrate that ubiquitin signaling is required to remove the CMG helicase from replication forks after collision with an ICL, and they support our previous hypothesis (Fu et al., 2011) that helicase removal is an essential, early, and active process associated with ICL repair.
BRCA1 functions at stalled forks prior to DSB formation
We wanted to know which ubiquitin-dependent pathway promotes CMG unloading. We showed previously that failure to ubiquitylate FANCD2 blocks incisions, which occur downstream of Approach and CMG unloading (Fu et al., 2011; Knipscheer et al., 2009). Defective FANCD2 ubiquitylation therefore cannot account for the effect of UbVS. Notably, recent evidence indicates that BRCA1 has an HR-independent role in ICL repair (Bunting et al., 2012) and that it contributes to fork stability (Silver and Livingston, 2012). Given that ubiquitin signaling is required for BRCA1 recruitment (Figure 2D), we postulated that BRCA1 might function as an effector of ubiquitin signaling in ICL repair.
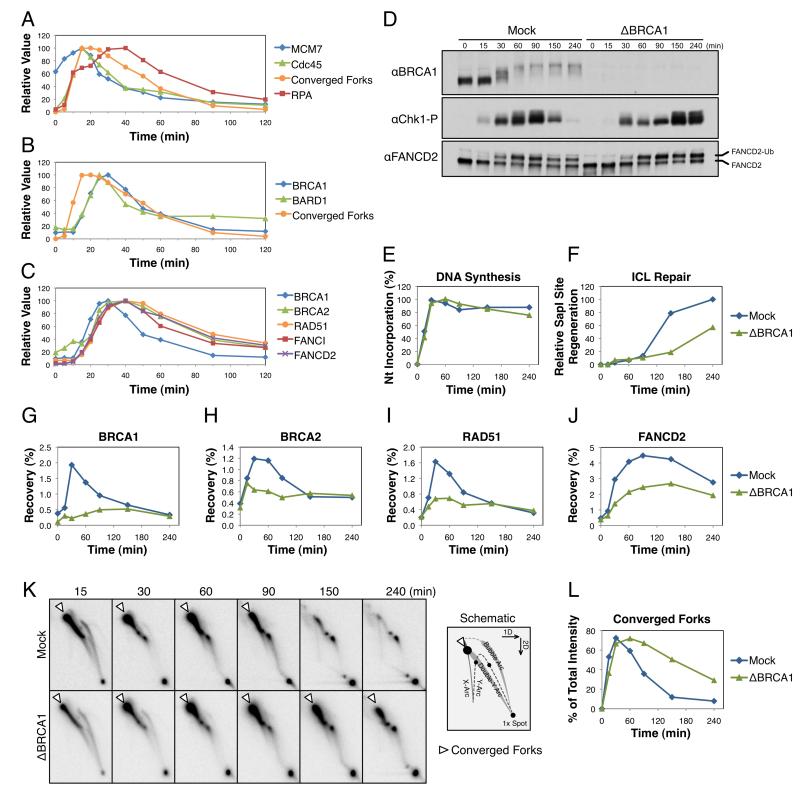
To investigate how BRCA1 contributes to ICL repair, we first used ChIP to address when BRCA1 is recruited to ICLs relative to other events of repair. Fifteen minutes after replication of pICL was initiated, MCM7 and Cdc45 accumulated at the crosslink, coincident with fork convergence (Figure 3A) (Fu et al., 2011). The ssDNA-binding protein RPA initially accumulated at the ICL with converging forks, but after a short delay, its abundance increased further (Figure 3A, red trace), likely due to lagging strand resection (Raschle et al., 2008). BRCA1 and its binding partner BARD1 were recruited to the crosslink ~7-10 minutes after fork convergence and well before the disappearance of converged fork structures, which are lost as a result of dual incisions (Figure 3B). Notably, BRCA2, RAD51, FANCI, and FANCD2 were all recruited ~5 minutes after BRCA1 (Figure 3C), consistent with BRCA1’s established role in the recruitment of these proteins to sites of DNA damage (Bhattacharyya et al., 2000; Garcia-Higuera et al., 2001; Greenberg et al., 2006; Smogorzewska et al., 2007). Collectively, the data are consistent with BRCA1 having an early role at stalled forks prior to DSB formation (Bunting et al., 2012).
Figure 3. BRCA1 has an early role at ICL-stalled forks.
(A-C) pICL was replicated in egg extract and ICL recruitment of various proteins was analyzed by ChIP. Samples were also analyzed for accumulation of ICL-stalled forks (Converged Forks) by agarose gel electrophoresis. Relative recovery shown with data normalized to peak accumulation. All data shown was analyzed from a single experiment with Converged Forks duplicated in A and B, and BRCA1-ChIP duplicated in B and C. pICL was replicated in mock-depleted (Mock) or BRCA1-depleted (ΔBRCA1) extract. Samples from the same reaction were analyzed by: Western blot with the indicated antibodies (D), agarose gel electrophoresis to determine the efficiency of replication (E) and ICL repair (F), ChIP with the indicated antibodies (G-J), and two-dimensional agarose gel electrophoresis (2DGE) (K) to analyze accumulation of converged forks (open arrowhead, see schematic and Figure 1B, i), which is quantified in (L). See Figure S2 for primary gel data, replicates of ICL repair data, and ChIP recovery at the FAR locus.
To determine whether BRCA1 is required for cell-free ICL repair, pICL was replicated in mock-depleted or BRCA1-depleted egg extract (Figure 3D). Although replication of pICL occurred with similar kinetics in both reactions (Figure 3E), ICL repair was delayed by at least one hour in BRCA1-depleted extracts (Figure 3F). A small amount of BRCA1 was still recruited to the crosslink at late times in BRCA1-depleted reactions (Figure 3G, green trace). As such, the delayed appearance of repair products (Figure 3F, green trace) may be due to residual BRCA1 not removed by depletion.
Consistent with immunofluorescence localization studies in mammalian cells (Bhattacharyya et al., 2000; Garcia-Higuera et al., 2001; Greenberg et al., 2006), recruitment of BRCA2, RAD51, and FANCD2 to ICLs was reduced in the absence of BRCA1 (Figure 3H-J). These defects were not due to co-depletion of BRCA2, RAD51, or FANCD2 from egg extract (Figure S2A). Although FANCD2 recruitment was impaired by BRCA1 depletion, FANCD2 ubiquitylation occurred normally (Figure 3D), consistent with previous reports (Bunting et al., 2012; Vandenberg et al., 2003). Loss of BRCA1 led to a severe incision defect (Figure 3K and L), indicating that FANCD2 ubiquitylation is not sufficient for DNA incisions without its localization to ICLs. High-level Chk1 phosphorylation was also delayed (Figure 3D, compare 60 minute time points), consistent with a defect in DSB formation. Together, these results indicate that BRCA1 functions at ICL-stalled replication forks, where it recruits BRCA2, RAD51, and FANCD2.
BRCA1 is not required for resection at ICL-stalled forks
Given that BRCA1 has been implicated in resection of DSBs (Bouwman et al., 2010; Bunting et al., 2010; Schlegel et al., 2006; Yun and Hiom, 2009), we examined the BRCA1-dependence of this process in our cell-free system. Depletion of BRCA1 from extract led to a slight increase in the recruitment of RPA to ICLs (Figure 4A). However, when the amount of single-stranded DNA on pICL was analyzed directly by quantitative PCR, similar levels of ssDNA were detected in mock-depleted and BRCA1-depleted reactions (Figure 4B-D). These results argue that loss of BRCA1 does not compromise resection of ICL-stalled forks. Instead, defective RAD51 binding in the absence of BRCA1 (see Figure 3G) may elevate the amount of RPA present on chromatin.
Figure 4. Analysis of strand resection during ICL repair.
Mock-depleted and BRCA1-depleted samples from Figure 3D-L were used to analyze: (A) RPA recruitment by ChIP, and (B) the presence of ssDNA by quantitative PCR (described in Methods). The table indicates the distance in base pairs from the 3’ end of the Test primer to the ICL (“0”). The same information is also graphically indicated in (C). (D) Stalled fork schematic showing the 118 ssDNA Test primer (wavy orange line), which is extended (dashed orange line) after annealing to ssDNA. Amplification of the extended Test primer with Left and Right primers (red arrows) produces a 146 base pair product that is analyzed by quantitative PCR.
BRCA1 promotes CMG unloading
We then analyzed the formation of nascent strand products in mock- and BRCA1-depleted reactions. As seen for UbVS-treated reactions, depletion of BRCA1 severely compromised the Approach of leading strands to the −1 position (Figure 5A; Figure S3G-J for experimental replicates). BRCA1 depletion also inhibited CMG unloading (Figure 5B-D; Figure S3N-P for experimental replicates), as seen in UbVS-treated reactions. Notably, CMG unloading and Approach are not dependent on RAD51 (Long et al., 2011) or FANCD2 (Knipscheer et al., 2009), indicating that defective helicase removal in the absence of BRCA1 is not an indirect consequence of defective RAD51 or FANCD2 recruitment. Moreover, recruitment of BRCA1 to the ICL occurred shortly after the arrival of forks at −20 and just before the Approach to −1 (Figure S3A), consistent with BRCA1 playing a direct role in promoting Approach. Importantly, BRCA1 depletion had no significant effect on helicase unloading during replication of undamaged plasmids (Figure S3B-D). Together, these results indicate that BRCA1 is required to unload the replicative helicase from ICL-stalled forks but not from forks undergoing termination.
Figure 5. BRCA1 depletion inhibits leading strand Approach and helicase unloading.
Mock-depleted or BRCA1-depleted samples from Figure 3 were analyzed by denaturing polyacrylamide gel electrophoresis (A) and by ChIP with the indicated antibodies (B-D). See Figure S3 for quantification of nascent strand products, ChIP recovery at the FAR locus, and experimental replicates.
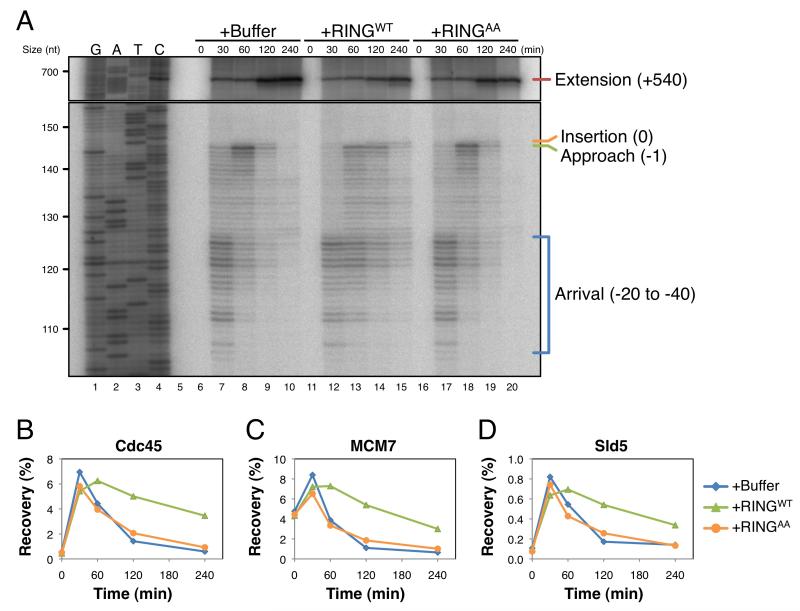
When BRCA1-depleted extracts were supplemented with recombinant BRCA1-BARD1 heterodimer (Joukov et al., 2006), helicase eviction was not restored (data not shown), suggesting that the activity of the complex is dependent on additional binding factors or specific modifications (Silver and Livingston, 2012; Wang, 2012). Therefore, to further investigate whether BRCA1 is required for CMG unloading, BRCA1 activity was inhibited with a fragment of BARD1 (Westermark et al., 2003). BRCA1 and BARD1 interact through their respective RING domains, with two α-helices from each domain combining to form a four-helix bundle (Brzovic et al., 2001). In cells, expression of a RING peptide was shown to inhibit BRCA1 function, leading to defects in HR and hyper-sensitivity to DNA cross-linking agents (Westermark et al., 2003). As reported previously (Joukov et al., 2001), BRCA1 antibodies quantitatively immunodepleted BARD1 from egg extract and vice versa (Figure S3E, lanes 4 and 6), demonstrating that BRCA1 and BARD1 are present as a stable 1:1 complex. Importantly, BARD1 RING peptide (RINGWT) recovered BRCA1 but not BARD1 (Figure S3F, lane 5), arguing that the peptide disrupted the BRCA1-BARD1 complex. Insertion of a single alanine residue into each α-helix of the RING domain disrupted its binding to BRCA1 (Figure S3F, lane 7), and this mutant peptide (RINGAA) served as a negative control for BRCA1 inhibition.
When the RINGWT peptide was added to egg extracts, it only slightly delayed Approach and CMG unloading (data not shown). To improve the efficacy of peptide inhibition, we partially depleted BRCA1 prior to peptide addition. Depletion of BRCA1 to ~25% of endogenous levels (Figure S4A) by itself had little or no effect on any aspect of ICL repair measured (Figure 6A-D, Figure S4B-K, and data not shown). However, when the partially depleted extract was supplemented with RINGWT peptide, Approach and helicase unloading were both impaired (Figure 6A-D; Figure S4B-K for experimental replicates). Importantly, the RINGAA peptide caused no inhibition. Together, the data indicate that the integrity of the BRCA1-BARD1 complex is required to promote unloading of the CMG helicase complex and subsequent leading strand Approach.
Figure 6. The BRCA1-BARD1 complex is required to promote leading strand Approach and helicase unloading.
pICL was replicated in extracts that were partially depleted of BRCA1 (Figure S4A), then supplemented with buffer (+Buffer), wild-type RING peptide (+RINGWT), or RING peptide containing two alanine insertions (+RINGAA). Samples from the same reaction were analyzed by denaturing polyacrylamide gel electrophoresis (A) and by ChIP with the indicated antibodies (B-C). See Figure S4 for quantification of nascent strand products, ChIP recovery at the FAR locus, and experimental replicates.
During DSB repair, BRCA1 is recruited to DNA through a phospho-specific interaction with Abraxas (Wang et al., 2007). This interaction is mediated through BRCA1’s tandem BRCT domains, which bind to a phospho-SXXF motif at the C-terminus of Abraxas. To investigate the role that this interaction plays during ICL repair, egg extracts were supplemented with peptides containing the phospho-SXXF motif from Abraxas (pSPTF), or a non-phosphorylated control S4Q). BRCA1 recruitment was only partially inhibited by the SPTF peptide, consistent with the non-phosphorylated peptide having reduced affinity for BRCA1 (Wang et al., 2007). Notably, compared to SPTF, pSPTF had a greater inhibitory effect on Approach (Figure S4L, compare lanes 14 and 19) and CMG unloading (Figure S4R-S, compare 80 minute time points). Together, these results argue that BRCA1’s helicase unloading activity is dependent on BRCT-mediated recruitment to chromatin.
CMG unloading and leading strand Approach support DNA incisions
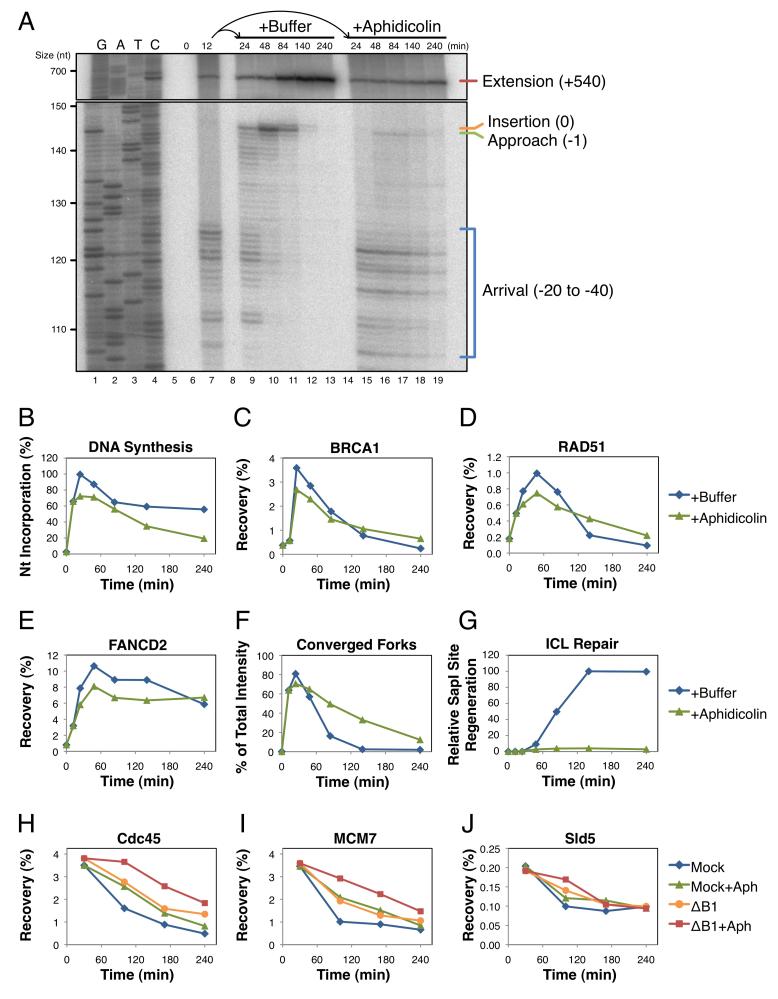
To determine the role that Approach plays in ICL repair, we sought to block this event by a direct and independent means that does not involve perturbation of BRCA1 or the ubiquitin system. To this end, pICL was replicated for 12 minutes to allow the majority of forks to arrive at the −20 position. Reactions were then split and supplemented with buffer or the DNA polymerase inhibitor aphidicolin (Errico et al., 2007). Aphidicolin-treated samples exhibited little or no Approach (Figure 7A), as well as a ~25% decrease in total nucleotide incorporation due to degradation of some forks that had not yet stalled at the crosslink (Figure 7B). ChIP showed that BRCA1, RAD51, and FANCD2 were still recruited to the ICL in aphidicolin-treated samples (Figure 7C-E), although total recovery was also decreased by ~25%. In contrast, DNA incisions were inhibited, as measured by persistence of the converged fork structure (Figure 7F and Figure S5I), and this mirrored what we observed in BRCA1-depleted reactions (Figure 3L). Together, these results indicate that Approach and, by extension, CMG unloading are required for incisions and downstream repair events (Figure 7G). In addition, they show that BRCA1 helps recruit RAD51 and FANCD2 independently of Approach.
Figure 7. BRCA1 and DNA polymerase independently promote helicase unloading.
pICL was replicated in egg extract for 12 minutes. The reaction was then split and supplemented with buffer (+Buffer) or 50 μM aphidicolin (+Aphidicolin) to block polymerase activity. Samples from the same reaction were analyzed by: (A) denaturing polyacrylamide gel electrophoresis, (B) agarose gel electrophoresis to determine the efficiency of replication, (C-E) ChIP with the indicated antibodies, (F) 2DGE to visualize the accumulation of converged forks, and (G) agarose gel electrophoresis to determine the efficiency of ICL repair. See Figure S5 for primary gel data, quantification of nascent strand products, and ChIP recovery at the FAR locus. (H-J) pICL was replicated in mock-depleted (Mock) or BRCA1-depleted (ΔB1) extract for 30 minutes. Each reaction was then split and supplemented with buffer or aphidicolin (+Aph) to block polymerase extension. Protein recruitment to the ICL was analyzed by ChIP with the indicated antibodies. See Figure S6 for primary experimental data.
Both BRCA1 and polymerase extension contribute to helicase unloading
Interestingly, blocking the Approach step with aphidicolin delayed CMG unloading (Figure S5J-L). These results suggested that the DNA polymerase also contributes to helicase removal. To investigate the relationship between BRCA1-dependent and polymerase-dependent helicase unloading, pICL was replicated in mock- or BRCA1-depleted extract until forks had stalled at the ICL. Each reaction was then split and supplemented with buffer or aphidicolin, as in Figure 7A. Analysis of the helicase complex by ChIP showed that BRCA1 depletion and aphidicolin treatment caused additive inhibition of CMG unloading (Figure 7H-J). These results indicate that BRCA1 and DNA polymerase can promote CMG unloading through independent mechanisms.
Discussion
The CMG helicase is a highly processive molecular motor that binds tightly to DNA. Little is known about how CMG is dismantled from chromatin, both when replication forks meet during termination and in response to certain forms of replication stress. We previously found that when forks encounter an ICL, the Approach of leading strands from the −20 to the −1 position correlates with dissociation of CMG from the site of damage (Fu et al., 2011). Here, we show that inhibiting CMG dissociation in various ways also blocks Approach and ICL repair. These data establish CMG unloading as a critical, early step in ICL repair.
We further present multiple lines of evidence that CMG unloading requires the BRCA1-BARD1 complex. BRCA1 immunodepletion and disruption of the BRCA1-BARD1 complex with a dominant negative peptide both inhibit CMG unloading. In addition, preventing BRCA1 inhibits CMG unloading. Finally, the timing of BRCA1 binding to ICLs and CMG unloading are highly correlated. Together, our results identify helicase eviction from stalled forks as a novel function for the BRCA pathway in DNA repair. Importantly, BRCA1 is not required for helicase unloading during replication termination (Figure S3B-D). Perhaps there is a difference in the arrangement of CMG helicases during ICL repair and termination that allows BRCA1-BARD1 to discriminate between the two situations.
Our data uncover other interesting mechanistic features of ICL repair. When we added aphidicolin immediately after replication forks stalled at the ICL, Approach failed and ICL repair was inhibited (Figure 7A). Interestingly, aphidicolin also caused a delay in CMG unloading in both mock-depleted and BRCA1-depleted reactions (Figure 7H-J). One interpretation of this observation is that the polymerase contributes to helicase eviction by exerting mechanical force on the stalled CMG complex (positioned immediately in front of the polymerase). Alternatively, aphidicolin treatment may stabilize the interaction between DNA polymerase and DNA (Cheng and Kuchta, 1993), thereby indirectly preventing another BRCA1-independent mechanism of helicase unloading. In either case, both BRCA1- and DNA polymerase-linked mechanisms likely cooperate to achieve efficient helicase eviction. One attractive model is that the E3 ligase activity of BRCA1-BARD1 (Hashizume et al., 2001) ubiquitylates one or more CMG components, destabilizing the complex and/or helping facilitate its displacement by DNA polymerase.
We recently showed that FANCI-FANCD2 promotes ICL incisions by recruiting the XPF-ERCC1 nuclease to sites of damage (Douwel et al., 2014; Knipscheer et al., 2009). Interestingly, in the presence of aphidicolin, FANCD2 was still localized to ICLs but incisions were severely inhibited. Therefore, the recruitment of FANCD2 to ICLs is not sufficient for incision when Approach is blocked (Figure 7A). We speculate that extension of the leading strand to the −1 position after CMG unloading creates a DNA structure that is recognized by FANCI-FANCD2-dependent nucleases. Alternatively, the delay in CMG unloading seen in the presence of aphidicolin might account for the incision defect. Thus, the presence of CMG at ICLs might shield the structure from endonucleolytic processing.
A future challenge is to determine how the various genome maintenance functions of BRCA1 contribute to tumor suppression. Interestingly, the crosslink sensitivity of BRCA1-deficient cells is less severe than those carrying mutations in other ICL repair factors (Bridge et al., 2005; Niedzwiedz et al., 2004; Ohashi et al., 2005; Qing et al., 2011). This could be explained by the fact that a polymerase-linked mechanism can promote helicase unloading in the absence of BRCA1. Moreover, the Fanconi anemia pathway is still activated in the absence of BRCA1, as evidenced by normal FANCD2 ubiquitylation in response to ICLs (Bunting et al., 2012). As such, repair might still proceed, albeit with reduced efficiency and higher propensity for error.
In conclusion, our results show that the BRCA and Fanconi anemia pathways execute an ordered series of fork processing events (helicase eviction, DNA incisions, and homologous recombination) that promote error-free removal of ICLs from DNA (Figure S7).
Experimental Procedures
Xenopus egg extracts and DNA replication
Preparation of Xenopus egg extracts was performed as described previously (Lebofsky et al., 2009). For DNA replication, plasmids were first incubated in a high-speed supernatant (HSS) of egg cytoplasm (final concentration 7.5 ng DNA/μL extract) for 20 minutes at 21°C, leading to the formation of pre-replication complexes (pre-RCs). Next, two volumes of nucleoplasmic egg extract (NPE) was added to one volume of HSS, initiating Cdk2-dependent replication at pre-RCs. For all figures, the 0 minute time point corresponds to NPE addition. For DNA labeling, reactions were supplemented with [α-32P]dATP, which is incorporated into nascent strands during replication. For UbVS reactions, NPE was supplemented with 14 μM ubiquitin vinyl sulfone alone, or with 50 μM ubiquitin (both from Boston Biochem, Cambridge, MA, USA) prior to mixing with HSS. Reactions were stopped with 10 volumes Stop Solution A (0.5% SDS, 25 mM EDTA, 50 mM Tris-HCl pH 7.5) and replication intermediates were purified as described (Raschle et al., 2008). Replication and repair intermediates were separated by 0.8% native agarose gels and visualized using a phosphorimager to determine replication efficiency (Lebofsky et al., 2009). All experiments were performed at least twice, and a representative result is shown.
ICL repair assay
Repair efficiency was calculated essentially as described (Raschle et al., 2008). pICL contains a single, site-specific cisplatin ICL that interrupts a SapI recognition site (see (Enoiu et al., 2012) for description and preparation). ICL repair is assayed by SapI cleavage, which requires error-free removal of the crosslink. To quantify the formation of SapI-cleavable products, DNA samples were digested with either HincII alone, or HincII and SapI, then separated by a native agarose gel and visualized using a phosphorimager. SapI cleavage of HincII-linearized molecules produces two fragments that are 2.3 and 3.3 kb in size. Fragments of similar size are also generated when ICL-stalled fork arms are broken or cleaved. Since these intermediates do not represent ICL repair products, they were quantified in the HincII-digested samples and subtracted from the HincII/SapI-generated fragments. This yields the amount of fragments produced exclusively by SapI cleavage. To determine the efficiency of repair as a percentage of the total DNA replicated, radioactivity in each sample is normalized to correct for variation introduced during sample preparation. To this end, a small amount of an unrelated, undamaged plasmid (pQuant) was included in the reaction (0.375 ng DNA/μL final concentration in HSS) to serve as an internal standard for quantification. The percentage of SapI-cleavable products is then calculated by comparing the normalized value of SapI fragments to the radioactivity present in the known amount of pQuant (which is 1/20th the amount of pICL added to the reaction). ICL repair data are shown with peak values set to 100% and background SapI fragments from contaminating uncrosslinked plasmid subtracted out.
ChIP and quantitative real-time PCR
ChIP was performed essentially as described (Long et al., 2011). Reaction samples were cross-linked in egg lysis buffer (ELB: 10 mM HEPES-KOH pH 7.7, 2.5 mM MgCl2, 50 mM KCl, 250 mM sucrose, and 1 mM DTT) containing 1% formaldehyde for 10 minutes at 21°C. Cross-linking was stopped by adding glycine to a final concentration of 125 mM followed by passage through a Micro Bio-Spin 6 Chromatography column (Bio-Rad, Hercules, CA, USA) to remove excess formaldehyde. The flowthrough was diluted to 500 μL with sonication buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 2 mM EDTA, 0.5% NP-40, 5 μg/mL Aprotinin+Leupeptin, and 2 mM PMSF) and subjected to sonication, yielding DNA fragments ~300-500 bp in size. Following immunoprecipitation with the indicated antibodies, formaldehyde cross-links were reversed and DNA was purified for analysis by quantitative real-time PCR with the following primer pairs: “ICL” (5’-AGCCAGATTTTTCCTCCTCTC-3’ and 5’-CATGCATTGGTTCTGCACTT-3’) and “FAR” (5’-AACGCCAATAGGGACTTTCC-3’ and 5’-GGGCGTACTTGGCATATGAT-3’). Antibodies for ChIP were purified using Protein A Sepharose beads (GE Healthcare, Piscataway, NJ, USA).
2D gel electrophoresis
Purified pICL intermediates were digested with HincII and then analyzed by native/native 2DGE. The first-dimension gel consisted of 0.4% agarose run in 1xTBE buffer at 0.75 V/cm for 26 hours at 21°C. The desired lane was then cast across the top of the second-dimension gel, which consisted of 1% agarose with 0.3 μg/mL ethidium bromide, and run in 1xTBE containing 0.3 μg/mL ethidium bromide at 4.5 V/cm for 14 hours at 4°C. DNA from the resulting gel was transferred to a 0.45 μm positively charged nylon transfer membrane (GE Healthcare, Piscataway, NJ, USA), cross-linked with a 120 mJ/cm2 UV exposure, and visualized using a phosphorimager.
Single-stranded DNA analysis
ssDNA was detected by quantitative PCR as described (Holstein and Lydall, 2012). Native DNA samples were first incubated at low temperature, allowing ssDNA to anneal with a “Test” primer that contains a unique sequence at its 5’ end. A single round of primer extension then creates a novel DNA product whose amount is proportional to the original amount of ssDNA. The novel product is then amplified using “Left” and “Right” primers at high temperatures to prevent annealing of the Test primer. ssDNA quantity is calculated using a ssDNA standard curve. Primer locations denote the distance from the 3’ end of the Test primer to the ICL. The “118”, “400”, “1454”, and “2925” Test primers anneal to the lagging strand template, while the “Anti-204” Test primer anneals to the leading strand template, serving as a control for dsDNA. See Figure 4D for schematic.
118: Test 5’-TGACTGCGCACCAGGCATAGTCAGGAGAGGA-3’, Left 5’-TTCCATAGAAAAGCCTTGACTTGAGGT-3’, Right 5’-TGACTGCGCACCAGGCATAG-3’. 400: Test 5’-TGACTGCGCACAGCGTACGAGTGAGAGACAC-3’, Left 5’-CCCTGGCTCACAAATACCACTGAG-3’, Right 5’-TGACTGCGCACAGCGTACGA-3’. 1454: Test 5’-TGACTGCGCACCAGGCATAGTTGATGAAGGA-3’, Left 5’-GCTCCATGGCTTCCAAGGTGT-3’, Right 5’-TGACTGCGCACCAGGCATAG-3’. 2925: Test 5’-TGACTGCGCACCAGGCATAGCGATGACTAAT-3’, Left 5’-TGCCAAGTACGCCCCCTATTG-3’, Right 5’-TGACTGCGCACCAGGCATAG-3’. Anti-204: Test 5’-TGACTGCGCACCAGGCATGACTTGAGGTTAG-3’, Left 5’-TGACTGCGCACCAGGCATGA-3’, Right 5’-TCAGGAGAGGAGGAAAAATCTGG-3’.
Nascent strand analysis
Nascent strand analysis was performed as described (Raschle et al., 2008). Briefly, pICL was replicated in the presence of [α-32P]dATP and purified pICL intermediates were digested with AflIII, followed by addition of 0.5 volumes Stop Solution B (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, and 0.05% xylene cyanol). Radiolabeled nascent strands were then separated by a 7% denaturing polyacrylamide gel, transferred to filter paper, dried, and visualized using a phosphorimager. Sequencing ladders were generated with primer S (5’-CATGTTTTACTAGCCAGATTTTTCCTCCTCTCCTG-3’) using the Cycle Sequencing kit (USB, Cleveland, OH, USA).
Antibodies and immunodepletion
The following rabbit polyclonal antibodies were described previously: BRCA1 (raised against X.l.BRCA1 residues 1001-1192) (Joukov et al., 2001), BARD1 (Joukov et al., 2001), RAD51 (Long et al., 2011), FANCI (Knipscheer et al., 2009), FANCD2 (Raschle et al., 2008), RPA (Walter and Newport, 2000), PCNA (Kochaniak et al., 2009), Cdc45 (Walter and Newport, 2000), MCM7 (Fang and Newport, 1993), Sld5 (Kubota et al., 2003) (provided by H. Takisawa, Osaka University), and Polε (Waga et al., 2001) and Polδ (Fukui et al., 2004) (both provided by S. Waga, Osaka University). Chk1-P (S345) was purchased from Cell Signaling Technology, Danvers, MA, USA. BRCA2 antibodies were raised in rabbits against X.l.BRCA2 residues 1842-2080. Rap80 antibodies were raised in rabbits against a 223 residue X.l.Rap80 protein fragment (see GenBank Accession CX130807). The cDNA encoding this fragment was generated from a Xenopus laevis mRNA library by PCR with primers: Left, 5’-CCGGAATTCGTACAGGAAATAGATGATCAATGCTCA-3’ and Right, 5’-CATAGTTTAGCGGCCGCTGGCTCCAGATCCGTTCCTGCACC-3’. The PCR fragment was digested with EcoRI and NotI, then cloned into the corresponding sites of a pET29a vector. To deplete BRCA1, Xenopus egg extracts were incubated with antibodies pre-bound to Protein A Sepharose beads (50 μg total IgGs from serum per μL of beads) at a 4:1 ratio of extract to beads for 40 minutes at 4°C for three rounds.
BRCA1 inhibitory peptides
X.l.BARD1 residues 2-195 corresponding to H.s.BARD1 residues 2-202 containing the RING domain were cloned into pGEX-6P-1 to create an N-terminal GST fusion construct. The RINGAA alanine insertions (after amino acids 35 and 100) were introduced by Site-Directed Mutagenesis (Agilent Technologies, Santa Clara, CA, USA). Recombinant RING-GST fusions were expressed using BL21 cells induced by 0.5 mM IPTG and purified using Glutathione Sepharose 4B (GE Healthcare, Piscataway, NJ, USA). Where indicated, extract was supplemented with 38 μM RINGWT or RINGAA peptide. The following peptides (X.l.Abraxas residues 398-408) were synthesized by Tufts University Core Facility: VEVSRSKpSPTF (pSPTF) and VEVSRSKSPTF (SPTF). Where indicated, extract was supplemented with 3 mM pSPTF or SPTF peptide. Due to nonspecific inhibitory effects, SPTF peptides were added to extract after replication forks had converged on the ICL.
BRCA1-BARD1 purification
The X.l.BRCA1-BARD1 heterodimeric complex was purified from insect cells as in (Joukov et al., 2006). Full-length FLAG-BRCA1 and HA-BARD1 were each cloned into pFastBac and used to generate the corresponding recombinant baculoviruses (Bac-to-Bac Baculovirus Expression System, Life Technologies, Carlsbad, CA, USA). Sf9 cells were co-infected with both viruses and the heterodimer was purified from cell lysates by sequential affinity chromatography using anti-FLAG M2 agarose and anti-HA agarose (both from Sigma-Aldrich, St. Louis, MO, USA). The eluted heterodimer was then dialyzed (20 mM HEPES pH 7.6, 100 mM KCl, 2.5 mM MgCl2, 250 mM sucrose, 1 mM DTT) and frozen at −80°C.
Supplementary Material
Highlights.
Replicative helicase unloading is an essential, early event in ICL repair.
Ubiquitin signaling is required for helicase unloading after fork collision.
BRCA1 promotes unloading of the replicative helicase complex.
Replicative DNA polymerases contribute to helicase unloading.
Acknowledgments
We thank A. D’Andrea, D. Livingston, and R. Scully for critical reading of the manuscript. We also thank D. Livingston for the generous gift of BRCA1 and BRCA2 antibodies. V.J. generated the X.l.BARD1 peptide construct and antibodies for X.l.BRCA1, BRCA2, and Rap80; M.B. made the initial observation of aphidicolin’s effect on MCM7 unloading; D.T.L. and J.C.W. designed and analyzed experiments; D.T.L. performed experiments; D.T.L. and J.C.W. prepared the manuscript. This work was supported by: American Cancer Society postdoctoral fellowship PF-10-146-01-DMC and NIH award K99GM102325 to D.T.L., Department of Defense Breast Cancer Research Program Award W81XWH-04-1-0524 to V.J., Human Frontiers Science Program long-term fellowship LT000773/2010-L to M.B., and NIH grants HL098316 and GM80676 to J.C.W. J.C.W. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bekker-Jensen S, Mailand N. Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair. 2010;9:1219–1228. doi: 10.1016/j.dnarep.2010.09.010. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. The Breast Cancer Susceptibility Gene BRCA1 Is Required for Subnuclear Assembly of Rad51 and Survival following Treatment with the DNA Cross-linking Agent Cisplatin. Journal of Biological Chemistry. 2000;275:23899–23903. doi: 10.1074/jbc.C000276200. [DOI] [PubMed] [Google Scholar]
- Borodovsky A, Kessler BM, Casagrande R, Overkleeft HS, Wilkinson KD, Ploegh HL. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 2001;20:5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nature Structural & Molecular Biology. 2010;17:688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge WL, Vandenberg CJ, Franklin RJ, Hiom K. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nature Genetics. 2005;37:953–957. doi: 10.1038/ng1627. [DOI] [PubMed] [Google Scholar]
- Brzovic PS, Rajagopal P, Hoyt DW, King M, Klevit RE. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nature Structural Biology. 2001;8:833–837. doi: 10.1038/nsb1001-833. [DOI] [PubMed] [Google Scholar]
- Bunting SF, Callén E, Kozak ML, Kim JM, Wong N, López-Contreras AJ, Ludwig T, Baer R, Faryabi RB, Malhowski A, et al. BRCA1 Functions Independently of Homologous Recombination in DNA Interstrand Crosslink Repair. Molecular Cell. 2012;46:125–135. doi: 10.1016/j.molcel.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callén E, Wong N, Chen H, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 Inhibits Homologous Recombination in Brca1-Deficient Cells by Blocking Resection of DNA Breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalasani P, Livingston DM. Differential Chemotherapeutic Sensitivity for Breast Tumors With “BRCAness”: A Review. The Oncologist. 2013;18:909–916. doi: 10.1634/theoncologist.2013-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Kuchta RD. DNA Polymerase epsilon: Aphidicolin Inhibition and the Relationship between Polymerase and Exonuclease Activity. Biochemistry. 1993;32:8568–8574. doi: 10.1021/bi00084a025. [DOI] [PubMed] [Google Scholar]
- Choudhury AD, Xu H, Baer R. Ubiquitination and Proteasomal Degradation of the BRCA1 Tumor Suppressor Is Regulated during Cell Cycle Progression. Journal of Biological Chemistry. 2004;279:33909–33918. doi: 10.1074/jbc.M403646200. [DOI] [PubMed] [Google Scholar]
- Dimova NV, Hathaway NA, Lee B-H, Kirkpatrick DS, Berkowitz ML, Gygi SP, Finley D, King RW. APC/C-mediated multiple monoubiquitylation provides an alternative degradation signal for cyclin B1. Nature Cell Biology. 2012;14:168–176. doi: 10.1038/ncb2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douwel DK, Boonen RACM, Long DT, Szypowska AA, Raschle M, Walter JC, Knipscheer P. XPF-ERCC1 acts in unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.03.015. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoiu M, Ho TV, Long DT, Walter JC, Schärer OD. Construction of Plasmids Containing Site-Specific DNA Interstrand Cross-Links for Biochemical and Cell Biological Studies. Methods in Molecular Biology. 2012;920:203–219. doi: 10.1007/978-1-61779-998-3_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errico A, Costanzo V, Hunt T. Tipin is required for stalled replication forks to resume DNA replication after removal of aphidicolin in Xenopus egg extracts. Proceedings of the National Academy of Sciences. 2007;104:14929–14934. doi: 10.1073/pnas.0706347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F, Newport JW. Distinct roles of cdk2 and cdc2 in RP-A phosphorylation during the cell cycle. J Cell Sci. 1993;106:983–994. doi: 10.1242/jcs.106.3.983. [DOI] [PubMed] [Google Scholar]
- Fu Yu V., Yardimci H, Long David T., Guainazzi A, Bermudez Vladimir P., Hurwitz J, van Oijen A, Schärer Orlando D., Walter Johannes C. Selective Bypass of a Lagging Strand Roadblock by the Eukaryotic Replicative DNA Helicase. Cell. 2011;146:931–941. doi: 10.1016/j.cell.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui T, Yamauchi K, Muroya T, Akiyama M, Maki H, Sugino A, Waga S. Distinct roles of DNA polymerases delta and epsilon at the replication fork in Xenopus egg extracts. Genes to Cells. 2004;9:179–191. doi: 10.1111/j.1356-9597.2004.00716.x. [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD. Interaction of the Fanconi Anemia Proteins and BRCA1 in a Common Pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- Greenberg RA, Sobhian B, Pathania S, Cantor SB, Nakatani Y, Livingston DM. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes & Development. 2006;20:34–46. doi: 10.1101/gad.1381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashizume R, Fukuda M, Maeda I, Nishikawa H, Oyake D, Yabuki Y, Ogata H, Ohta T. The RING Heterodimer BRCA1-BARD1 Is a Ubiquitin Ligase Inactivated by a Breast Cancer-derived Mutation. Journal of Biological Chemistry. 2001;276:14537–14540. doi: 10.1074/jbc.C000881200. [DOI] [PubMed] [Google Scholar]
- Holstein E-M, Lydall D. Quantitative Amplification of Single-Stranded DNA. Methods in Molecular Biology. 2012;920:323–339. doi: 10.1007/978-1-61779-998-3_23. [DOI] [PubMed] [Google Scholar]
- Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 Helicase by Association with Cdc45 and GINS Proteins. Molecular Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joukov V, Chen J, Fox EA, Green JBA, Livingston DM. Functional communication between endogenous BRCA1 and its partner, BARD1, during Xenopus laevis development. Proceedings of the National Academy of Sciences. 2001;98:12078–12083. doi: 10.1073/pnas.211427098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joukov V, Groen AC, Prokhorova T, Gerson R, White E, Rodriguez A, Walter JC, Livingston DM. The BRCA1/BARD1 Heterodimer Modulates Ran-Dependent Mitotic Spindle Assembly. Cell. 2006;127:539–552. doi: 10.1016/j.cell.2006.08.053. [DOI] [PubMed] [Google Scholar]
- Kass EM, Jasin M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Letters. 2010;584:3703–3708. doi: 10.1016/j.febslet.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, D'Andrea AD. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes & Development. 2012;26:1393–1408. doi: 10.1101/gad.195248.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC. The Fanconi Anemia Pathway Promotes Replication-Dependent DNA Interstrand Cross-Link Repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochaniak AB, Habuchi S, Loparo JJ, Chang DJ, Cimprich KA, Walter JC, van Oijen AM. Proliferating Cell Nuclear Antigen Uses Two Distinct Modes to Move along DNA. Journal of Biological Chemistry. 2009;284:17700–17710. doi: 10.1074/jbc.M109.008706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y, Takase Y, Komori Y, Hashimoto Y, Arata T, Kamimura Y, Araki H, Takisawa H. A novel ring-like complex of Xenopus proteins essential for the initiation of DNA replication. Genes & Development. 2003;17:1141–1152. doi: 10.1101/gad.1070003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebofsky R, Takahashi T, Walter JC. DNA Replication in Nucleus-Free Xenopus Egg Extracts. Methods in Molecular Biology. 2009;521:229–252. doi: 10.1007/978-1-60327-815-7_13. [DOI] [PubMed] [Google Scholar]
- Leeneer K, Coene I, Crombez B, Simkens J, Broecke R, Bols A, Stragier B, Vanhoutte I, Paepe A, Poppe B, et al. Prevalence of BRCA1/2 mutations in sporadic breast/ovarian cancer patients and identification of a novel de novo BRCA1 mutation in a patient diagnosed with late onset breast and ovarian cancer: implications for genetic testing. Breast Cancer Research and Treatment. 2011;132:87–95. doi: 10.1007/s10549-011-1544-9. [DOI] [PubMed] [Google Scholar]
- Liu J, Doty T, Gibson B, Heyer W-D. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nature Structural & Molecular Biology. 2010;17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long DT, Raschle M, Joukov V, Walter JC. Mechanism of RAD51-Dependent DNA Interstrand Cross-Link Repair. Science. 2011;333:84–87. doi: 10.1126/science.1204258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh PJ, Spanswick VJ, Hartley JA. Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. The Lancet Oncology. 2001;2:483–490. doi: 10.1016/S1470-2045(01)00454-5. [DOI] [PubMed] [Google Scholar]
- Muniandy PA, Liu J, Majumdar A, Liu S.-t., Seidman MM. DNA interstrand crosslink repair in mammalian cells: step by step. Critical Reviews in Biochemistry and Molecular Biology. 2010;45:23–49. doi: 10.3109/10409230903501819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nature Reviews Cancer. 2004;4:665–676. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- Niedzwiedz W, Mosedale G, Johnson M, Ong CY, Pace P, Patel KJ. The Fanconi Anaemia Gene FANCC Promotes Homologous Recombination and Error-Prone DNA Repair. Molecular Cell. 2004;15:607–620. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Nijman SMB, Huang TT, Dirac AMG, Brummelkamp TR, Kerkhoven RM, D'Andrea AD, Bernards R. The Deubiquitinating Enzyme USP1 Regulates the Fanconi Anemia Pathway. Molecular Cell. 2005;17:331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Oestergaard VH, Langevin F, Kuiken HJ, Pace P, Niedzwiedz W, Simpson LJ, Ohzeki M, Takata M, Sale JE, Patel KJ. Deubiquitination of FANCD2 Is Required for DNA Crosslink Repair. Molecular Cell. 2007;28:798–809. doi: 10.1016/j.molcel.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi A, Zdzienicka MZ, Chen J, Couch FJ. Fanconi Anemia Complementation Group D2 (FANCD2) Functions Independently of BRCA2- and RAD51-associated Homologous Recombination in Response to DNA Damage. Journal of Biological Chemistry. 2005;280:14877–14883. doi: 10.1074/jbc.M414669200. [DOI] [PubMed] [Google Scholar]
- Pinder JB, Attwood KM, Dellaire G. Reading, writing, and repair: the role of ubiquitin and the ubiquitin-like proteins in DNA damage signaling and repair. Frontiers in Genetics. 2013;4 doi: 10.3389/fgene.2013.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing Y, Yamazoe M, Hirota K, Dejsuphong D, Sakai W, Yamamoto KN, Bishop DK, Wu X, Takeda S. The Epistatic Relationship between BRCA2 and the Other RAD51 Mediators in Homologous Recombination. PLoS Genetics. 2011;7:e1002148. doi: 10.1371/journal.pgen.1002148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith J, Ellenberger T, Scharer O, Walter J. Mechanism of Replication-Coupled DNA Interstrand Crosslink Repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel BP, Jodelka FM, Nunez R. BRCA1 Promotes Induction of ssDNA by Ionizing Radiation. Cancer Research. 2006;66:5181–5189. doi: 10.1158/0008-5472.CAN-05-3209. [DOI] [PubMed] [Google Scholar]
- Silver DP, Livingston DM. Mechanisms of BRCA1 Tumor Suppression. Cancer Discovery. 2012;2:679–684. doi: 10.1158/2159-8290.CD-12-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D'Andrea AD, et al. Identification of the Fanconi anemia (FANC) I protein, a monoubiquitinated FANCD2 paralog required for crosslink repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg CJ, Gergely F, Ong CY, Pace P, Mallery DL, Hiom K, Patel KJ. BRCA1-Independent Ubiquitination of FANCD2. Molecular Cell. 2003;12:247–254. doi: 10.1016/s1097-2765(03)00281-8. [DOI] [PubMed] [Google Scholar]
- Waga S, Masuda T, Takisawa H, Sugino A. DNA polymerase is required for coordinated and efficient chromosomal DNA replication in Xenopus egg extracts. Proceedings of the National Academy of Sciences. 2001;98:4978–4983. doi: 10.1073/pnas.081088798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Newport J. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- Wang B. BRCA1 tumor suppressor network: focusing on its tail. Cell & Bioscience. 2012;2:6. doi: 10.1186/2045-3701-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Matsuoka S, Ballif B, Zhang D, Smogorzewska A, Giyi S, Elledge SJ. Abraxas and Rap80 form a novel BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Andreassen PR, D'Andrea AD. Functional Interaction of Monoubiquitinated FANCD2 and BRCA2/FANCD1 in Chromatin. Molecular and Cellular Biology. 2004;24:5850–5862. doi: 10.1128/MCB.24.13.5850-5862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermark UK, Reyngold M, Olshen AB, Baer R, Jasin M, Moynahan ME. BARD1 Participates with BRCA1 in Homology-Directed Repair of Chromosome Breaks. Molecular and Cellular Biology. 2003;23:7926–7936. doi: 10.1128/MCB.23.21.7926-7936.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Lu L-Y, Yu X. The role of BRCA1 in DNA damage response. Protein & Cell. 2010;1:117–123. doi: 10.1007/s13238-010-0010-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Jetten AM. RAP80 and RNF8, key players in the recruitment of repair proteins to DNA damage sites. Cancer Letters. 2008;271:179–190. doi: 10.1016/j.canlet.2008.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Fan Q, Ren K, Andreassen PR. PALB2 Functionally Connects the Breast Cancer Susceptibility Proteins BRCA1 and BRCA2. Molecular Cancer Research. 2009;7:1110–1118. doi: 10.1158/1541-7786.MCR-09-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. The role of BRCA1 in homologous recombination repair in response to replication stress: significance in tumorigenesis and cancer therapy. Cell & Bioscience. 2013;3:11. doi: 10.1186/2045-3701-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.