Abstract
Flavoparmelia caperata (L.) Hale is medicinally very important and possesses antifungal and antibacterial activities. F. caperata is the only species found in India. Inter simple sequence repeat (ISSR) and Directed amplification of minisatellite DNA (DAMD) methods were used to analyze the genetic variability within F. caperata from the Western Himalayan region of India. Eleven ISSR and 10 DAMD primers produced 139 and 117 polymorphic bands, and detected 91.44 and 82.34 % polymorphisms, respectively. Cumulative band data generated for ISSR and DAMD markers resulted in 86.86 % polymorphism across all the accessions of F. caperata. The average Polymorphic information content (PIC) value obtained with ISSR, DAMD, and cumulative band data were 0.28, 0.27, and 0.27, respectively. The clustering of the F. caperata accessions in the UPGMA dendrogram showed that these accessions are intermingled with each other in different subclusters irrespective of their geographical affiliations. The pattern of genetic variations within F. caperata accessions could be due to free exchange of spores that might have taken place among these accessions in the wild. ISSR and DAMD markers efficiently and reliably resulted in discrete banding patterns and polymorphic profiles. These markers despite targeting different regions of genome, revealed almost similar levels of polymorphism across all the accessions. The wide range of genetic distance and high level of polymorphism detected by ISSR and DAMD reflected a high genetic variability among the different accessions of F. caperata.
Keywords: DAMD, Flavoparmelia caperata, Genetic variations, ISSR
Introduction
The genus Flavoparmelia belongs to a medium to large foliose lichenized fungi within the family Parmeliaceae. The genus is characterized by a distinctive pale or yellow-green upper cortex when dry (Nash et al. 2002). There are about 38 species under the genus Flavoparmelia in the world, which predominantly grow on bark of trees and rocks (Crespo et al. 2010), and Flavoparmelia caperata (L.) Hale is the only species found in India. The species is distributed in the states of Andhra Pradesh, Assam, Himachal Pradesh, Jammu and Kashmir, Manipur, Meghalaya, Nagaland, Sikkim, Tamil Nadu, Uttarakhand, and West Bengal in India. F. caperata is medicinally very important and reported to possess antifungal and antibacterial activities (Pennington 1963; Cobanoglu et al. 2010). Secondary metabolites like usnic acid, atranorin, protocetraric acid, and caperatic acid have also been reported in F. caperata, which shows anti-inflammatory activities (Nash et al. 2002). Orange-brown to yellow dye is obtained from the thallus of F. caperata, which is used to dye animal skins, fabrics, crafts, and hair (Uphof 1959).
A large number of methods varying from morphological to molecular have been used to analyze the individuals, populations, and species. Most species in lichens do not show any unambiguous infraspecific morphological attributes (Printzen 2002). Phenotypic variability of the thallus has been used very often to discriminate among lichenized fungi; it can, however, be somewhat variable within a species and can clearly be influenced by the habitat and environment (Grube and Hawksworth 2007). Molecular markers could, therefore, be helpful to explore the mode and effectiveness of dispersal, gene exchange, and intra as well as infraspecific studies in lichens (Printzen 2002). DNA fingerprinting methods are known to generate markers that are simple, rapid, and unaffected by the environment, and have been successfully employed to determine genetic variability and relationships at species and population level in many lichenized fungi (Murtagh et al. 1999; Honegger et al. 2004; Bayraktar et al. 2008; Yuzbasioglu et al. 2011; Fontaine et al. 2013). In the present study, we used inter simple sequence repeats (ISSR) (Provost and Wilkinson 1999) and directed amplification of minisatellite DNA (DAMD) (Heath et al. 1993) methods to estimate the genetic variability in F. caperata. ISSR and DAMD profiles are generated from a single primer PCR reaction. As they are distributed throughout the genome, therefore, the chances of amplifying large numbers of polymorphic bands are reasonably high (Hamada and Kakunaga 1982; Tautz and Renz 1984; Heath et al. 1993; Zietkiewicz et al. 1994; Zhou et al. 1997). The main advantages of the ISSR and DAMD methods are that they are quick, inexpensive, highly polymorphic, and randomly distributed throughout the genomes. As both these markers are PCR-based, therefore, they only require small amounts of template DNA. Furthermore, they do not require any sequence information for primer construction. As such, there is no information available on application of ISSR and DAMD markers in F. caperata; therefore, the objective of the present study was to estimate the genetic variability within F. caperata in the Western Himalayan region of India using these markers.
Materials and methods
Plant materials and DNA isolation
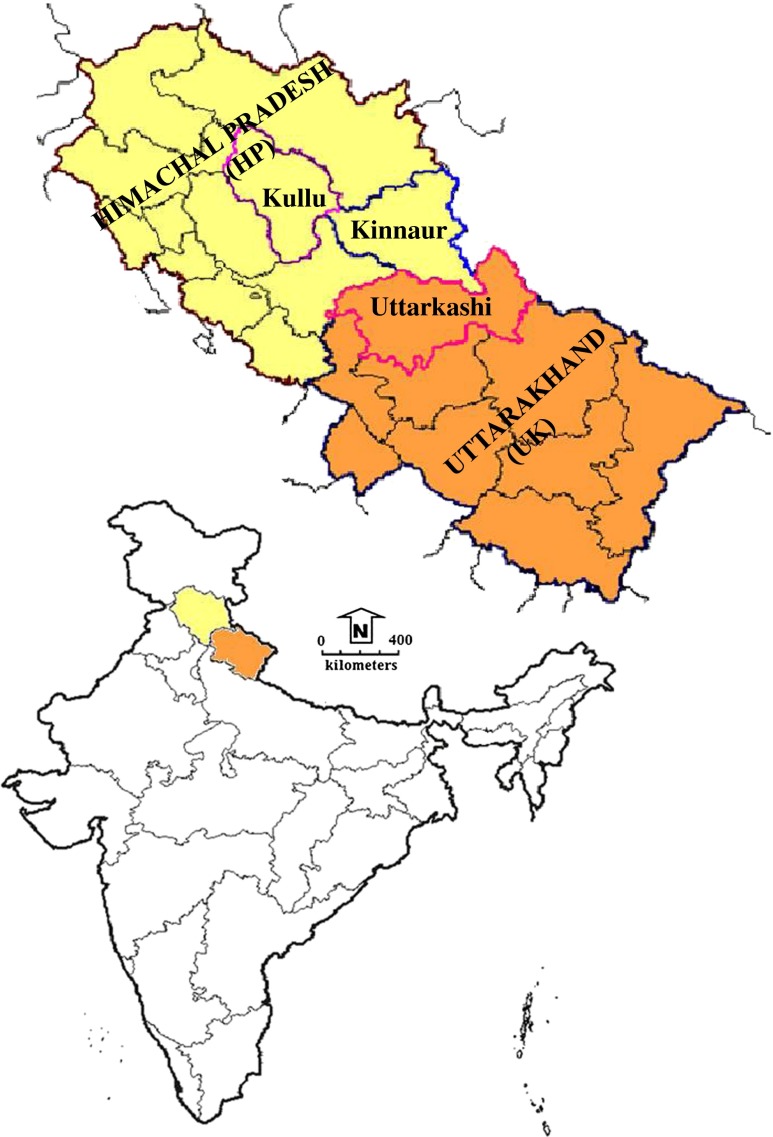
Thirty-nine accessions of F. caperata considered in the present study were collected in the wild from the Western Himalayan region particularly Himachal Pradesh (HP) and Uttarakhand (UK) states of India. Everniastrum cirrhatum (Ec), a closely related taxon, was considered as the out group. Each individual was collected, segregated, and kept in a separate packet in the field itself. The sampling sites and geographic coordinates of the accessions are provided in Fig. 1 and Table 1. Voucher specimens for each accession have also been prepared for all the collected materials and deposited in the herbarium of the CSIR-National Botanical Research Institute (LWG), Lucknow. Total genomic DNA was isolated using the DNeasy Plant Mini Kit (Qiagen, Hilden Germany), according to the instructions of the manufacturer. Isolated genomic DNA was checked for its quality and quantity by gel electrophoresis on 0.8 % agarose gel, staining with ethidium bromide, and comparison was made with a set of known DNA concentration standard, and also by UV spectroscopy using a Nanodrop ND-1000 Spectrophotometer (NanoDrop Technologies Inc. USA).
Fig. 1.
Map showing the collection sites of Flavoparmelia caperata in the Western Himalayan region (HP and UK) of India
Table 1.
Geographical details of Flavoparmelia caperata accessions used in the present study
| S. no. | Sample code | Voucher no. | Locality | Latitude (°′N) | Longitude (°′E) | Altitude (m) | Substratum |
|---|---|---|---|---|---|---|---|
| 1 | Fc01 | 12-017421 | UK; Uttarkashi (GWLS) | 31° 06′ 50.66″ | 78° 20′ 25.58″ | 2,592 | Rock |
| 2 | Fc02 | 12-018567 | UK; Uttarkashi (GWLS) | 31° 06′ 55.09″ | 78° 20′ 38.03″ | 2,582 | Bark |
| 3 | Fc03 | 12-018568 | UK; Uttarkashi (GWLS) | 31° 06′ 59.35″ | 78° 20′ 45.02″ | 2,586 | Rock |
| 4 | Fc04 | 12-016606 | UK; Uttarkashi (GWLS) | 31° 04′ 25.51″ | 78° 06′ 51.44″ | 1,536 | Bark |
| 5 | Fc05 | 12-018963 | UK; Uttarkashi (GWLS) | 31° 04′ 46.31″ | 78° 07′ 20.56″ | 1,582 | Rock |
| 6 | Fc06 | 12-018736 | UK; Uttarkashi (GWLS) | 31° 04′ 35.49″ | 78° 08′ 07.46″ | 1,870 | Bark |
| 7 | Fc07 | 12-018964 | UK; Uttarkashi (GWLS) | 31° 04′ 44.93″ | 78° 11′ 04.26″ | 1,915 | Bark |
| 8 | Fc08 | 12-018980 | UK; Uttarkashi (GWLS) | 31° 00′ 54.21″ | 78° 02′ 11.06″ | 1,205 | Bark |
| 9 | Fc09 | 12-016605 | UK; Uttarkashi (GWLS) | 31° 04′ 44.97″ | 78° 11′ 04.91″ | 1,908 | Bark |
| 10 | Fc10 | 12-018863 | UK; Uttarkashi (GWLS) | 31° 07′ 25.67″ | 78° 04′ 41.71″ | 1,560 | Bark |
| 11 | Fc11 | 12-018906 | UK; Uttarkashi (GWLS) | 31° 07′ 26.71″ | 78° 04′ 41.67″ | 1,564 | Bark |
| 12 | Fc12 | 12-016618 | UK; Uttarkashi (GWLS) | 31° 04′ 44.91″ | 78° 11′ 19.39″ | 1,714 | Rock |
| 13 | Fc13 | 12-018790 | UK; Uttarkashi (GWLS) | 31° 06′ 22.75″ | 78° 04′ 46.02″ | 1,512 | Bark |
| 14 | Fc14 | 12-018909 | UK; Uttarkashi (GWLS) | 31° 04′ 36.53″ | 78° 10′ 50.21″ | 1,912 | Bark |
| 15 | Fc15 | 12-018876 | UK; Uttarkashi (GWLS) | 30° 55′ 08.82″ | 78° 03′ 18.17″ | 2,009 | Bark |
| 16 | Fc16 | 12-018775 | UK; Uttarkashi (GWLS) | 31° 01′ 56.70″ | 78° 07′ 44.70″ | 2,340 | Bark |
| 17 | Fc17 | 12-018891 | UK; Uttarkashi (GWLS) | 31° 00′ 55.33″ | 78° 01′ 55.76″ | 1,260 | Bark |
| 18 | Fc18 | 12-018988 | UK; Uttarkashi (GWLS) | 31° 00′ 39.66″ | 78° 01′ 43.56″ | 1,252 | Rock |
| 19 | Fc19 | 12-018809 | UK; Uttarkashi (GWLS) | 31° 04′ 44.93″ | 78° 11′ 03.61″ | 1,915 | Bark |
| 20 | Fc20 | 12-018871 | UK; Uttarkashi (GWLS) | 30° 52′ 43.01″ | 78° 04′ 27.61″ | 1,468 | Bark |
| 21 | Fc21 | 12-016615 | UK; Uttarkashi (GWLS) | 31° 04′ 27.64″ | 78° 06′ 18.88″ | 1,344 | Rock |
| 22 | Fc22 | 12-018800 | UK; Uttarkashi (GWLS) | 31° 04′ 44.98″ | 78° 11′ 05.49″ | 1,906 | Bark |
| 23 | Fc23 | 13-019821 | UK; Uttarkashi (GWLS) | 31° 06′ 41.34″ | 78° 20′ 16.84″ | 2,664 | Rock |
| 24 | Fc24 | 13-019822 | UK; Uttarkashi (GWLS) | 31° 06′ 41.56″ | 78° 19′ 05.99″ | 2,507 | Bark |
| 25 | Fc25 | 13-019483 | UK; Uttarkashi (GWLS) | 31° 07′ 15.37″ | 78° 21′ 20.23″ | 2,723 | Bark |
| 26 | Fc26 | 13-019821 | UK; Uttarkashi (GWLS) | 31°06′ 41.38″ | 78° 19′ 40.00″ | 2,476 | Rock |
| 27 | Fc27 | 13-019825 | UK; Uttarkashi (GWLS) | 31° 04′ 51.47″ | 78° 14′ 53.01″ | 2,075 | Bark |
| 28 | Fc28 | 13-019803 | UK; Uttarkashi (GWLS) | 31° 06′ 36.63″ | 78° 14′ 46.43″ | 2,587 | Bark |
| 29 | Fc29 | 13-019486 | UK; Uttarkashi (GWLS) | 31° 04′ 43.76″ | 78° 15′ 04.53″ | 2,104 | Bark |
| 30 | Fc30 | 13-023658 | HP; Kinaur (Chitkul) | 31° 21′ 01.15″ | 78° 26′ 03.99″ | 3,420 | Bark |
| 31 | Fc31 | 13-019779 | HP; Kullu (Kot) | 31° 30′ 55.40″ | 77° 25′ 19.80″ | 2,311 | Bark |
| 32 | Fc32 | 13-019793 | HP; Kullu (Jaloritop) | 31° 32′ 36.86″ | 77° 22′ 46.96″ | 2,951 | Bark |
| 33 | Fc33 | 13-023364 | HP; Kullu (Kothi) | 32° 19′ 19.11″ | 77° 11′ 38.47″ | 2,555 | Bark |
| 34 | Fc34 | 13-023259 | HP; Kullu (Marhi) | 32° 20′ 47.11″ | 77° 13′ 14.07″ | 3,270 | Bark |
| 35 | Fc35 | 13-023266 | HP; Kullu (Gulaba) | 32° 19′ 57.17″ | 77° 11′ 48.35″ | 2,620 | Bark |
| 36 | Fc36 | 13-019791 | HP; Kullu (Gulaba) | 32° 19′ 53.89″ | 77° 11′ 50.40″ | 2,653 | Bark |
| 37 | Fc37 | 13-019787 | HP; Kullu (Kasol) | 32° 00′ 39.19″ | 77° 19′ 05.15″ | 1,580 | Rock |
| 38 | Fc38 | 13-023355 | HP; Kullu (Kasol) | 32° 00′ 53.29″ | 77° 19′ 37.06″ | 1,670 | Bark |
| 39 | Fc39 | 13-019765 | HP; Kullu (Manikaran) | 32° 01′ 31.32″ | 77° 21′ 17.99″ | 1,770 | Bark |
| 40 | Ec | 13-019803 | UK; Uttarkashi (GWLS) | 31° 06′ 36.63″ | 78° 14′ 46.43″ | 2,587 | Bark |
GWLS Govind Wild Life Sanctuary, HP Himachal Pradesh, UK Uttarakhand, Ec Everniastrum cirrhatum out group taxon used in the present study
ISSR analysis
In ISSR analysis, two accessions of F. caperata were selected at random to carry out preliminary experiments to determine optimal amplification reaction conditions (annealing temperature and cycling time); reaction components like quantity of template, primers, Deoxynucleotide triphosphates (dNTPs), and Mg+2 ions; and primer screening reactions for ISSR. One hundred ISSR primers (University of British Columbia, Canada) were initially screened with two template DNAs. Screening of the primers revealed that 24 primers resulted in inconsistent profiles, 39 primers gave fuzzy profiles, 26 primers did not result in any amplification, and only 11 primers out of the 100 primers screened, resulted in discrete profiles consisting of well-separated fragments (Table 2). PCR amplification of 30 ng DNA was performed in 20-μl reaction containing 1.6 μl (10 mM) dNTP mix (2.5 mM each dNTP), 1.6 μl (25 mM) MgCl2, 2 μl (Buffer F) 10× assay buffer supplied along with the enzyme, 0.33 μl (15 mM) primer, and 0.33 μl (3 U/μl) Taq DNA polymerase (Merck Specialities Pvt. Ltd. India.) using PTC 200 thermo cycler (MJ Research, Inc., USA). After initial denaturation at 94 °C for 4 min, each cycle consisted of 1 min denaturation at 94 ° C, 1 min of annealing at 54 °C, 2 min extension at 72 °C along with 7 min extension at 72 °C at the end of 35 cycles.
Table 2.
ISSR and DAMD primers, total number of bands, polymorphic bands, and polymorphic information content (PIC)
| Primer name | Sequence (5′—3′) | Loci amplified | Polymorphic loci | Percentage polymorphism | PIC | Approx band size (bp) |
|---|---|---|---|---|---|---|
| ISSR | ||||||
| UBC 807 | (AG)8T | 17 | 15 | 88.24 | 0.20 | 280–1,200 |
| UBC 808 | (AG)8C | 12 | 11 | 91.67 | 0.34 | 300–1,500 |
| UBC 809 | (AG)8G | 13 | 12 | 92.31 | 0.28 | 200–2,000 |
| UBC 810 | (GA)8T | 15 | 15 | 100 | 0.27 | 290–1,200 |
| UBC 811 | (GA)8C | 14 | 13 | 92.86 | 0.26 | 250–2,500 |
| UBC 826 | (AC)8C | 13 | 11 | 84.62 | 0.23 | 360–2,000 |
| UBC 835 | (AG)8YC | 17 | 17 | 100 | 0.30 | 280–2,500 |
| UBC 836 | (AG)8YA | 14 | 14 | 100 | 0.36 | 280–1,200 |
| UBC 840 | (GA)8YT | 12 | 10 | 83.33 | 0.27 | 260–1,700 |
| UBC847 | (CA)8RC | 11 | 9 | 81.82 | 0.40 | 290–1,600 |
| UBC 861 | (ACC)6 | 14 | 12 | 85.71 | 0.21 | 300–2,000 |
| Total | 11 | 152 | 139 | 91.44 | 0.28 | 200–2,500 |
| DAMD | ||||||
| 14C2 | GGCAGGATTGAAGC | 12 | 9 | 75 | 0.25 | 200–1,500 |
| M-13 | GAGGGTGGCGGTTCT | 14 | 12 | 85.71 | 0.19 | 300–2,500 |
| 33.6 | AGGGCTGGAGG | 9 | 7 | 77.78 | 0.32 | 200–1,500 |
| HVA | AGGATGGAAAGGAGGC | 12 | 9 | 75 | 0.29 | 200–2,200 |
| 6.2H+ | AGGAGGAGGGGAAGG | 11 | 8 | 72.73 | 0.38 | 120–2,000 |
| OGRB01 | AGGGCTGGAGGAGGGC | 23 | 18 | 78.26 | 0.12 | 200–1,800 |
| FVII ex8 | ATGCACACACACAGG | 14 | 14 | 100 | 0.34 | 280–2,000 |
| FVII ex8c | CCTGTGTGTGTGCAT | 16 | 15 | 93.75 | 0.28 | 200–1,800 |
| HBV3 | GGTGAAGCACAGGTG | 16 | 15 | 93.75 | 0.28 | 200–2,200 |
| HVR | CCTCCTCCCTCCT | 14 | 10 | 71.43 | 0.20 | 200–1,500 |
| Total | 10 | 141 | 117 | 82.34 | 0.27 | 120–2,500 |
| ISSR + DAMD | 21 | 293 | 256 | 86.86 | 0.27 | 120–2,500 |
Y = (C, T); R = (A, G)
DAMD analysis
The DAMD primers were custom synthesized from Sigma Aldrich Chemicals Pvt. Ltd. India, and are listed in Table 2. DNA amplification was carried out according to Zhou et al. (1997). The reaction (20 μl) contained 1.6 μl (10 mM) dNTP mix (2.5 mM each dNTP), 0.4 μl (25 mM) MgCl2, 2 μl (Buffer A) 10× assay buffer with (15 mM) MgCl2 supplied along with the enzyme, 0.8 μl (20 uM) primer and 0.40 μl (3 U/μl) Taq DNA polymerase (Merck Specialities Pvt. Ltd. India), and approximately 40 ng genomic DNA. Optimal DNA amplification was obtained through 40 cycles (92 °C for 1 min, 55 °C for 2 min, and 72 °C for 2 min) in a thermal cycler (PTC 200, MJ Research, Inc., USA).
Agarose gel electrophoresis
The amplified PCR products were resolved on 1.5 % agarose gel using 0.5X TBE buffer (1X TBE buffer: Tris-borate 89 mM; 2 mM EDTA, pH 8.3) at constant voltage of 5 V/cm. After electrophoresis, the gel was stained in ethidium bromide and then visualized and archived using Gel Documentation System (UV Tech, UK). The gel profiles were photographed and stored as digital images in gel documentation system.
Data analysis
The binary data were scored as presence (1) or absence (0) of a band, and only distinct and well-separated bands were considered in the present analysis. The polymorphic information content (PIC) was calculated according to Botstein et al. (1980). Pair wise distances were computed using Jaccard’s coefficient in the Free Tree program (ver. 0.9.1.5) (Pavlicek et al. 1999). Jaccard’s coefficient of similarity (Jsim) and dissimilarity (Jdist) was calculated by using the formula Jdis = 1-Jsim. Unweighted pair group method with arithmetic mean (UPGMA) tree was generated after allowing a 1,000 replicate bootstrap test using the same program. All the trees were viewed annotated and printed using Tree View (ver.1.6.5) (Page 2001). The Principal coordinate analysis (PCA) and The Mantel Z-statistic were used to test the correlation between two datasets of the molecular markers used in the present study using the NTSYS-pc software version 2.02e (Rohlf 1998).
Results and discussion
ISSR analysis
In the present study, 100 ISSR primers were screened to examine their amplification efficiency and 11 ISSR primers were chosen for further DNA amplification of F. caperata samples. The selected ISSR primers amplified a total of 152 DNA bands, of which 139 bands were polymorphic, with 91.44 % polymorphism throughout the F. caperata accessions. Primers 807 and 835 revealed the highest number of bands (17), while primer 847 produced the lowest number of bands (11). The primers 810, 835, and 836 gave 100 % polymorphic bands, whereas primer 847 had fewer polymorphic bands (81.82 %). The PIC were calculated for each primer, and primer 847 revealed maximum PIC value (0.40), while corresponding minimum PIC value (0.20) was obtained with primer 807. The mean PIC value obtained for 11 ISSR primers was 0.28 (Table 2). This level of intraspecific polymorphism is significant and suggests that populations of F. caperata that grow in the Western Himalayan region have significant genetic variability.
DAMD analysis
Initially, 20 DAMD primers were screened with two template DNAs, out of which 10 DAMD primers produced clear and reproducible banding patterns. The 10 selected DAMD primers amplified a total 141 DNA bands, of which 117 bands were polymorphic, with 82.34 % polymorphism across the F. caperata accessions. The highest number of amplified bands (23) was obtained with primer OGRB-01, whereas the primer 33.6 resulted in the lowest number (9) of amplified bands. The primer FvII ex8 gave 100 % polymorphic bands, whereas primer HVR revealed 71.34 % polymorphism. Primer 6.2H+ produced the highest PIC value (0.38) while primer OGRB-01 resulted in minimum PIC value (0.12). The mean PIC value recorded for 10 DAMD primers was 0.27 (Table 2).
Cumulative analysis
A total of 21 primers (11 ISSR and 10 DAMD) resulted in to 293 bands, of which 256 bands were polymorphic, with 86.86 % polymorphism across the F. caperata accessions. The mean PIC value recorded for 21 cumulative primers was 0.27. The approx band size range for 21 cumulative primers was 120–2,500 bp. The cumulative (ISSR + DAMD) genetic distance showed a distance range from 0.15 to 0.66 with an average value of 0.42 among all the accessions of F. caperata. The maximum intraspecific average genetic distance was (0.66) between Fc08, Fc26, and Fc31 accessions, while the corresponding least genetic distance was (0.15) between Fc32 and Fc34 accessions, respectively (Data not shown).
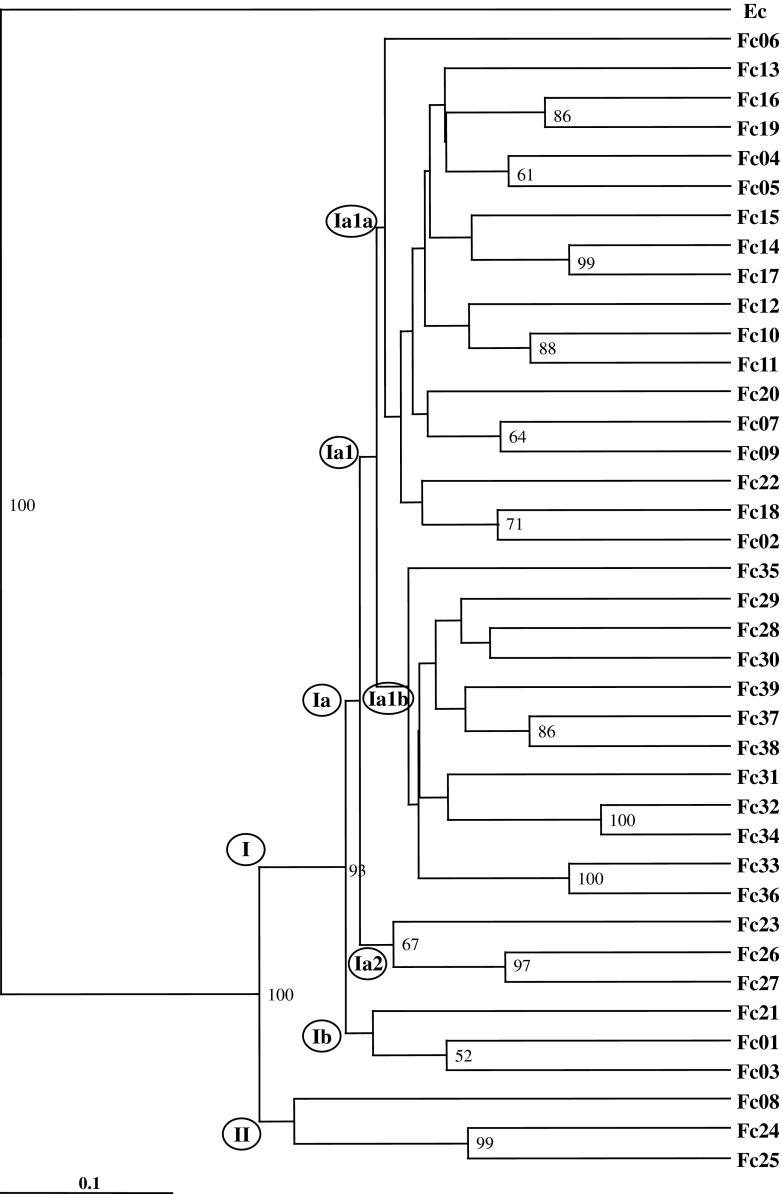
The genetic distance data were used to generate the UPGMA dendrogram, and UPGMA dendrogram showed two major clusters, cluster I and cluster II. The out group taxon (E. cirrhatum) was separated out clearly from the rest of the F. caperata accessions. Cluster I was divided into six subclusters (Ia, Ib, Ia1, Ia2, Ia1a, Ia1b) with low genetic differences between them. Subcluster Ia was further divisible into four subclusters (Ia1, Ia2, Ia1a, and Ia1b), and Ia1a grouped all the accessions from Uttarakhand (UK), whereas subcluster Ia1b clustered together all the accessions from Himachal Pradesh (HP) in two distinct groups except accession Fc35, which did not show any affinity with other accessions from HP. The accessions Fc01, Fc02, and Fc21 from UK grouped together in subcluster Ia2, and did not show any affinity with other accessions of the UK state. Cluster II consisted of three accessions (Fc08, Fc24, Fc25) from Naitwar, Osla, and Harki Doon (UK), respectively (Fig. 2). The clustering of the F. caperata accessions in the UPGMA dendrogram revealed that the groupings are, however, in congruence with the geographical affinities of the accessions but with weak bootstrap support. In the present investigation, accessions did not show any altitudinal specific preferences in UPGMA dendrogram and PCA plot.
Fig. 2.
UPGMA dendrogram generated for the cumulative data of ISSR and DAMD methods. Numbers at the node are bootstrap values. Fc1 to Fc39 denotes the accession numbers of Flavoparmelia caperata and Ec (Everniastrum cirrhatum) is the out group taxon
Mantel’s test (Mantel 1967) was performed for three data sets (ISSR, DAMD, and cumulative), which revealed a significant correlation between cumulative versus DAMD and showed the maximum (0.91) correlation coefficient (r), followed by cumulative versus ISSR (0.90), and ISSR versus DAMD (0.65), respectively. These values revealed that DAMD and cumulative data have good correlation and are best fit to each other (Table 3). Two-dimensional plot generated from principal coordinate analysis (PCA) of ISSR and DAMD data showed two major clusters. Cluster I grouped together all the accessions from Uttarakhand (UK), except Fc23 to Fc29, and some of these accessions (Fc23, Fc26, Fc27, Fc28, and Fc29) are intermingled with Cluster II, which is primarily a Himachal Pradesh (HP) cluster. Accession numbers Fc8, Fc24, and Fc25 from UK did not show any relationship with accessions of HP and UK (Fig. 3). Two-dimensional plot generated from principal coordinate analysis (PCA) of ISSR and DAMD data also supported the clustering patterns in UPGMA dendrogram.
Table 3.
Mantel correlation between the genetic distances obtained from ISSR and DAMD and cumulative data among Flavoparmelia caperata accessions used in the present study
| Marker pairs | Correlation coefficient (r) | p value |
|---|---|---|
| ISSR vs DAMD | 0.6465 | 0.0769 |
| ISSR vs cumulativea | 0.9042 | 0.0769 |
| DAMD vs cumulativea | 0.9102 | 0.0769 |
aCombined ISSR and DAMD band data
Fig. 3.
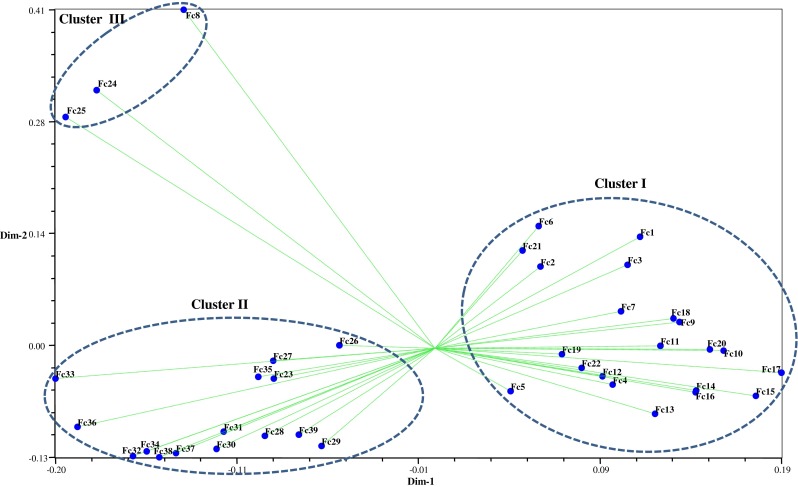
Principal coordinates analysis representing relationships among Flavoparmelia caperata accessions
ISSR and DAMD data were pooled together and analyzed to generate a cumulative UPGMA dendrogram, (Fig. 2), based on Jaccard’s similarity coefficient. ISSR and DAMD methods revealed information for F. caperata genomes in the regions rich in microsatellite and minisatellite repetitive DNA sequences, respectively, ensuring wider coverage of the genomes and providing a better reflection of the relationships and affinities of the F. caperata accessions used in this study. All the accessions of the F. caperata separated into two major clusters and five subclusters. The clustering of the F. caperata accessions in the UPGMA dendrogram revealed that these accessions are intermingled with each other in different subclusters irrespective of their geographical affiliations. This pattern of distribution of genetic variation among F. caperata accessions might have been due to free exchange of spores that have taken place among these accessions in the wild. ISSR and DAMD markers efficiently and reliably resulted in discrete banding patterns and polymorphic profiles. These markers, despite targeting different regions of genome, revealed almost similar levels of polymorphism across all the accessions. The wide range of genetic distance and high level of polymorphism detected by ISSR and DAMD reflected a high genetic variability among the different accessions of F. caperata. Similar level of polymorphism was also revealed with RAPD and ISSR markers in Lobathalia radiosa (Yuzbasioglu et al. 2011) and Diplosphaera chodatii based on internal transcribed spacer (ITS) of ribosomal DNA (rDNA) and actin gene sequences (Fontaine et al. 2013). The present study establishes that the ISSR and DAMD are the suitable markers to study the genetic variation in F. caperata. Genetic variability forms the basis of the evolutionary potential of a species to respond to environmental changes. Loss of genetic variation reduces the ability of a species to adopt in response to environmental changes (Lande 1988; Frankham 1996, 1999). Therefore, maintenance of genetic variation within a species is very important for long term survival (Lande 1988; Shah et al. 2008). The present study significantly revealed high genetic variability within F. caperata in the Western Himalayan region of India, and appears to be a maiden attempt in this direction.
Acknowledgments
The authors are thankful to the Director of the CSIR-National Botanical Research Institute, Lucknow for faculties and encouragements. The financial support (No. BT/PR1457/39/204/2011) received from the Department of Biotechnology, New Delhi is gratefully acknowledged.
References
- Bayraktar H, Dolar FS, Maden S. Use of RAPD and ISSR markers in detection of genetic variation and population structure among Fusarium oxysporum f. sp. ciceris isolates on chickpea. Turk J Phytopathol. 2008;156:146–154. doi: 10.1111/j.1439-0434.2007.01319.x. [DOI] [Google Scholar]
- Botstein D, White RL, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphism. Am J Hum Genet. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- Cobanoglu G, Sesal C, Gokmen B, Cakar S. Evaluation of the antimicrobial properties of some lichens. SW J Hort Biol Environ. 2010;1:153–158. [Google Scholar]
- Crespo A, Kauff F, Divakar PK, et al. Phylogenetic generic classification of parmelioid lichens (Parmeliaceae, Ascomycota) based on molecular, morphological and chemical evidence. Taxon. 2010;59:1735–1753. [Google Scholar]
- Fontaine KM, Stocker-Worgotter ET, Booth T, Piercey-Normore MD. Genetic diversity of the lichen-forming alga, Diplosphaera chodatii, in North America and Europe. Lichenologist. 2013;45(6):799–813. doi: 10.1017/S0024282913000510. [DOI] [Google Scholar]
- Frankham R. Relationship of genetic variation to population size in wildlife. Conserv Biol. 1996;10:1500–1508. doi: 10.1046/j.1523-1739.1996.10061500.x. [DOI] [Google Scholar]
- Frankham R. Quantitative genetics in conservation biology. Genet Res. 1999;74:237–244. doi: 10.1017/S001667239900405X. [DOI] [PubMed] [Google Scholar]
- Grube M, Hawksworth DL. Trouble with lichen: the re-evaluation and re-interpretation of thallus form and fruit body types in the molecular era. Mycol Res. 2007;111:1116–1132. doi: 10.1016/j.mycres.2007.04.008. [DOI] [PubMed] [Google Scholar]
- Hamada H, Kakunaga T. Potential Z-DNA forming sequences are highly dispersed in the human genome. Nature. 1982;298:396–398. doi: 10.1038/298396a0. [DOI] [PubMed] [Google Scholar]
- Heath DD, Iwama GK, Devlin RH. PCR primed with the VNTR core sequences yields species specific patterns and hypervariable probes. Nucleic Acids Res. 1993;21:5782–5785. doi: 10.1093/nar/21.24.5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honegger R, Zipper U, Scherrer, Dyer PS. Genetic diversity in Xanthoria parietina (L.) Th. Fr. (Lichen-forming ascomycete) from worldwide locations. Lichenologist. 2004;36(6):381–390. doi: 10.1017/S002428290401477X. [DOI] [Google Scholar]
- Lande R. Genetics and demography in biological. Conserv Sci. 1988;241:1455–1460. doi: 10.1126/science.3420403. [DOI] [PubMed] [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- Murtagh GJ, Dyer PS, McClure PC, Crittenden PD. Use of randomly amplified polymorphic DNA markers as a tool to study variation in lichen-forming fungi. Lichenologist. 1999;31(3):257–267. doi: 10.1017/S0024282999000365. [DOI] [Google Scholar]
- Nash TH, III, Ryan BD, Gries C, Bungartz F. Lichen flora of the greater sonoran desert region. Tempe: Lichens Unlimited, Arizona State University; 2002. [Google Scholar]
- Page RDM (2001) TreeView (Win32), Ver. 1.6.5. Available from: http://taxonomy.zoology.gla.ac.uk/rod/treeview.html
- Pavlicek A, Hrda S, Flegr J. Free Tree—freeware program for construction of phylogenetic trees on the basis of distance data and bootstrapping/ jackknife analysis of the tree robustness. Application in the RAPD analysis of the genus Frenkelia. Folia Biol (Praha) 1999;45:97–99. [PubMed] [Google Scholar]
- Pennington CW. The Tarahumar of Mexico: their environment and material culture. Salt Lake City: Univ of Utah Press; 1963. [Google Scholar]
- Printzen C. Fungal specific primers for PCR-amplification of mitochondrial LSU in lichens. Mol Ecol Notes. 2002;2:130–132. doi: 10.1046/j.1471-8286.2002.00179.x. [DOI] [Google Scholar]
- Provost A, Wilkinson MJ. A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genet. 1999;98:107–112. doi: 10.1007/s001220051046. [DOI] [Google Scholar]
- Rohlf FJ. NTSYS-pc: numerical taxonomy and multivariate analysis system, version 2.02e. Setauket: Applied Biostatistics Inc., Exeter Software; 1998. [Google Scholar]
- Shah A, Li DZ, Gao LM, Li HT, Möller M. Genetic diversity within and among populations of the endangered species Taxus fauna (Taxaceae) from Pakistan and implications for its conservation. Biochem Syst Ecol. 2008;36:183–193. doi: 10.1016/j.bse.2007.09.012. [DOI] [Google Scholar]
- Tautz D, Renz M. Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res. 1984;12:4127–4138. doi: 10.1093/nar/12.10.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uphof JCT. Dictionary of economic plants. New York: Hafner; 1959. [Google Scholar]
- Yuzbasioglu E, Halici MG, Karabacak M, Aksoy A. RAPD and ISSR markers indicate high genetic variation within Lobathallia radiosa in Turkey. Mycol Prog. 2011;10:219–228. doi: 10.1007/s11557-010-0691-4. [DOI] [Google Scholar]
- Zhou Z, Bebeli PJ, Somers DJ, Gustafson JP. Direct amplification of minisatellite-region DNA with VNTR core sequences in the genus Oryza. Theor Appl Genet. 1997;95:942–949. doi: 10.1007/s001220050645. [DOI] [Google Scholar]
- Zietkiewicz E, Rafalski A, Labuda D. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics. 1994;20:176–183. doi: 10.1006/geno.1994.1151. [DOI] [PubMed] [Google Scholar]